Abstract
Allelic variation in FLOWERING LOCUS C (FLC), a central repressor of flowering, contributes to natural differences in flowering behavior among Arabidopsis accessions. The weak nature of the FLC allele in the Ler accession is due to low levels of FLC RNA resulting, through an unknown mechanism, from a transposable element inserted in an intron of FLC. Here we show that the transposable element renders FLC-Ler subject to repressive chromatin modifications mediated by short interfering RNAs generated from homologous transposable elements in the genome. Our studies have general implications for the role of transposable elements in eukaryotic gene expression and evolution.
Keywords: Transposon, siRNA, intron, FLC, HEN1, H3-K9 methylation
The transition from vegetative to reproductive growth in plants, that is, floral transition or flowering, is controlled by endogenous as well as environmental signals. Longer day length and prolonged cold temperature (known as vernalization) are two of the major environmental signals that induce flowering in Arabidopsis. While a complex network of genes controls flowering in Arabidopsis, a few genes appear to act as integrators of key flowering pathways (for review, see Boss et al. 2004). One such gene, FLOWERING LOCUS C (FLC), is a central repressor of flowering and is negatively regulated by vernalization and by genes in the so-called autonomous pathway, and positively regulated by a gene named FRIGIDA (FRI) (Michaels and Amasino 1999; Sheldon et al. 1999).
Arabidopsis accessions exhibit two major types of flowering behavior. While many are late flowering unless they have been vernalized, a few accessions flower early in the absence of vernalization. The natural variation in flowering behavior is largely conferred by allelic variation at two loci, FRI and FLC (for review, see Boss et al. 2004). Late-flowering accessions have dominant, functional alleles at both loci, whereas early-flowering accessions have recessive alleles at one or both loci. The two commonly used accessions, Landsberg erecta (Ler) and Columbia (Col), are early flowering and contain fri-null alleles (Johanson et al. 2000). The Col FLC allele, FLC-Col, is functional, and when combined with functional FRI (such as FRI-SF2) is expressed at high levels and causes extremely late flowering (Michaels and Amasino 1999). The Ler FLC allele, FLC-Ler, responds minimally to FRI (Lee et al. 1993a; Michaels and Amasino 1999), although this allele does not contain missense or nonsense mutations in the FLC coding region (Gazzani et al. 2003; Michaels et al. 2003). FLC-Ler has a 1224-bp nonautonomous Mutator-like transposable element (TE) inserted in the first intron (Gazzani et al. 2003; Michaels et al. 2003), and this insertion acts in cis to cause FLC-Ler to be expressed at low levels (Michaels et al. 2003). However, the underlying mechanism by which the intronic TE insertion affects the steady-state mRNA level of the host gene is unclear.
Small RNAs 21-25 nt long play regulatory roles in a variety of eukaryotic organisms including plants. One class of small RNAs, microRNAs (miRNAs), was first identified in Caenorhabditis elegans through genetic analyses (Lee et al. 1993b; Reinhart et al. 2000) and later found in both animals and plants (for review, see Bartel 2004). MiRNAs are generated from longer hairpin precursors by the ribonuclease III-like enzyme Dicer and are incorporated into a protein complex named RISC, in which they guide the sequence-specific regulation of target mRNAs through translation inhibition or transcript cleavage (for review, see Bartel 2004). Another class of small RNAs, siRNAs, was first identified in plants undergoing post-transcriptional gene silencing (PTGS) and subsequently found in metazoans and fungi undergoing RNA silencing (Hamilton and Baulcombe 1999; for review, see Hannon 2002). siRNAs are derived from long double-stranded precursor RNAs by Dicer, and are incorporated into RISC, in which the siRNA mediates the sequence-specific cleavage of the target RNA (for review, see Hannon 2002). SiRNAs corresponding to transposons and repeat sequences have been found in vivo (Hamilton et al. 2002; Reinhart and Bartel 2002; Xie et al. 2004). They lead to DNA methylation and/or histone methylation at the homologous chromatin (Hamilton et al. 2002; Volpe et al. 2002; Zilberman et al. 2003; Xie et al. 2004).
Here we show that the intronic TE in FLC in the Ler accession renders FLC-Ler subject to siRNA-mediated regulation. This results in reduced FLC-Ler expression and vernalization-independent early flowering of Ler. Our studies demonstrate that an siRNA-based mechanism contributes to the evolution of a fast-cycling accession from a late-flowering accession. In addition, our studies have profound implications in gene expression regulated by intronic TEs in general because eukaryotic genomes contain numerous TEs, many of which are found in introns of genes (Nekrutenko and Li 2001).
Results and Discussion
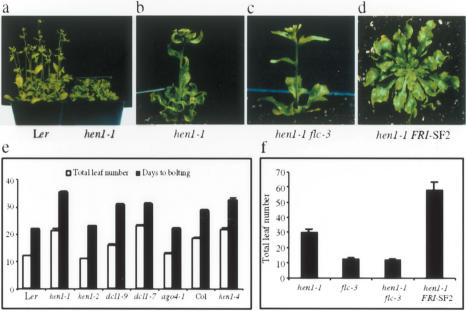
In previous studies, we isolated recessive mutations in a gene named HEN1 (Chen et al. 2002), which was later shown to act in microRNA (miRNA) biogenesis, in the accumulation of siRNAs in sense-transgene post-transcriptional gene silencing (S-PTGS), and in the accumulation of siRNAs from transposons in Arabidopsis (Park et al. 2002; Boutet et al. 2003; Xie et al. 2004). The two hen1 alleles in the Ler accession, hen1-1 and hen1-2, have different flowering time phenotypes. hen1-1 mutants exhibit delayed flowering, whereas plants with the weaker hen1-2 allele are normal in flowering time (Fig. 1a,e; Chen et al. 2002). In F2 populations of a hen1-1 × Ler cross, the late-flowering phenotype co-segregates with the hen1-1 mutation. In contrast to the large delay of flowering in Ler by hen1-1, a strong hen1 allele, hen1-4, causes only a slight delay in the floral transition in the Col accession (Fig. 1e). These findings implicate the role of small RNAs in the regulation of the floral transition in Arabidopsis, particularly in the Ler accession.
Figure 1.
Flowering time of various genotypes under long-day (LD) conditions. (a) One-month-old Ler and hen1-1 plants. (b) A 50-d hen1-1 plant. (c) A 30-d hen1-1 flc-3 plant. (d) A 50-d hen1-1 FRI-SF2 plant. (e,f) Flowering time (total leaf number and/or days to bolting) of the indicated genotypes. n = 90 (e) or 20-30 (f).
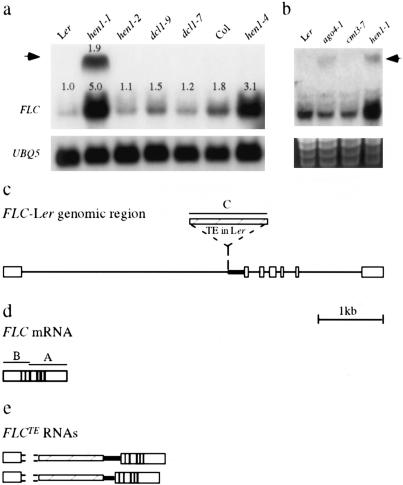
To incorporate HEN1 into the regulatory networks controlling flowering in Arabidopsis (Boss et al. 2004), we examined the flowering behavior of hen1-1 plants under various physiological conditions (Supplementary Fig. S1). These studies place HEN1 into the broad category of the so-called autonomous pathway (Boss et al. 2004), whose primary target is FLC (Michaels and Amasino 2001). We thus examined the expression of FLC at the RNA level. Indeed, hen1-1, but not hen1-2, exhibits elevated accumulation of FLC mRNA relative to Ler (Fig. 2a). This elevated FLC expression underlies the late-flowering behavior of hen1-1 plants, since flc-3, an flc-null allele (Michaels and Amasino 1999), suppresses the late-flowering phenotype of hen1-1 plants (Fig. 1b,c,f).
Figure 2.
Analysis of FLC expression. (a,b) FLC RNA filter hybridization with probe A (d). The numbers in a indicate the abundance of the RNAs relative to FLC mRNA in Ler. An image of a stained RNA gel in b indicates the amount of RNAs in each lane. (c) A diagram of FLC-Ler genomic region, with white boxes representing exons, lines representing introns, and the TE represented by the striped box. The thick, black line represents the intron 1 sequence downstream of the TE. (d) FLC mRNA. (e) Two FLCTE RNAs that differ in intron 1 sequences 3′ to the TE. The sequence at the junction of exon 1 and the TE was not determined, but RT-PCR experiments indicate the presence of exon 1 and the lack of intron 1 sequence 5′ to the TE. A-C indicate probes for RNA filter hybridization.
A larger FLC transcript of ∼2200 nt is detectable by filter hybridization in hen1-1, but not hen1-2, dcl1-9, or dcl1-7 plants in the Ler accession (arrow in Fig. 2a). This transcript is not detectable in hen1-4 in the Col accession (Fig. 2a). This RNA species can hybridize to probes corresponding to FLC exons 4-7 (probe A), exons 1-3 (probe B), and the TE (probe C) (Fig. 2a,c,d; Supplementary Fig. S2), but not to probes corresponding to sequences 5′ to the TE in intron 1, intron 6, or sequences upstream or downstream of the FLC mRNA (data not shown). Sequences of cloned RT-PCR products from the TE-containing RNAs indicate the presence of two major RNA species containing exon 1, the TE, intron 1 sequences 3′ to the TE, and exons 2-7 (Fig. 2e). One species contains the entire intron 1 sequence 3′ to the insertion, while the other species has part of the intron 1 sequence 3′ to the insertion. The second species is apparently generated by an alternative splicing event using a cryptic splice donor site in the TE and the normal intron 1 splice acceptor site. Therefore, these RNAs are aberrantly spliced products that retain the TE and partial intron 1 sequences and will be referred to as FLCTE RNAs. They would potentially generate a severely truncated, presumably nonfunctional protein. FLCTE RNAs can be detected, at much lower levels, in Ler plants by RT-PCR (Supplementary Fig. S3), indicating that they can be generated independently of the hen1-1 mutation. This suggests that the generation of FLCTE RNAs is intrinsic to the presence of the TE in FLC-Ler, which would be consistent with the well-documented ability of plant TEs to affect RNA processing (for review, see Weil and Wessler 1990).
Is the regulation of FLC RNA levels by HEN1 mediated by miRNAs or siRNAs? miRNA biogenesis in Arabidopsis requires DCL1, a Dicer homolog (Park et al. 2002; Reinhart et al. 2002). dcl1-7 and dcl1-9, two mutations in DCL1 (in the Ler accession), strongly affect the accumulation of miRNAs but have little effect on the accumulation of endogenous siRNAs (Xie et al. 2004) or siRNAs from transgenes undergoing S-PTGS (Finnegan et al. 2003). While dcl1-7 and dcl1-9, like hen1-1, cause delayed floral transition (Fig. 1e; Ray et al. 1996), FLC mRNA abundance is only slightly increased in these plants (Fig. 2a). Therefore, an miRNA pathway plays a small role in the regulation of FLC RNA abundance. The Col FLC allele (FLC-Col) does not contain the TE. There is only a small increase in FLC RNA abundance in hen1-4 relative to Col (0.7-fold increase), indicating that it is the intronic TE that renders FLC-Ler subject to regulation by HEN1. Given the role of RNA silencing and siRNAs in the regulation of transposable elements (for review, see Plasterk 2002) and the established role of HEN1 in siRNA metabolism (Boutet et al. 2003; Xie et al. 2004), we evaluated whether an siRNA-based mechanism regulates the TE-containing FLC-Ler gene.
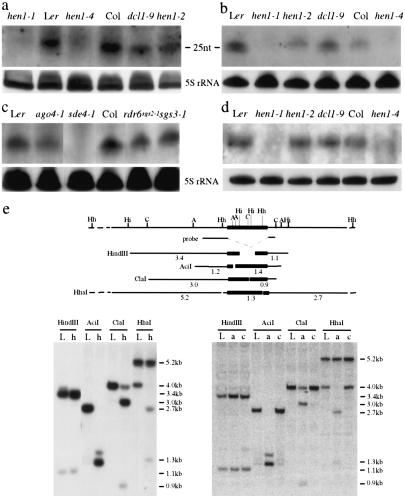
We first searched for siRNAs corresponding to FLC-Ler. Indeed, small RNAs corresponding to the TE (referred to as siRNATEs), but not other regions in FLC-Ler (data not shown), were detected with both antisense and sense riboprobes in Ler (Fig. 3a,b). The TE in FLC-Ler is present in several copies elsewhere in the Arabidopsis genome (Gazzani et al. 2003; Michaels et al. 2003). siRNATEs also accumulate in Col (Fig. 3a-c), which does not have the TE in the FLC gene. This suggests that siRNATEs are generated from homologous TEs elsewhere in the genome. siRNATEs are present in dcl1-9 and hen1-2 but are barely detectable in hen1-1 (Fig. 3a,b). The accumulation of siRNAs corresponding to another transposon, AtSN1 (Hamilton et al. 2002), is also much reduced in hen1-1 but not affected by dcl1-9 (Fig. 3d; Xie et al. 2004). ago4-1 and sde4-1 mutations are also known to affect the accumulation of siRNAs from AtSN1 (Hamilton et al. 2002; Zilberman et al. 2003). We found that siRNATEs are reduced in abundance in ago4-1 and are not detectable in sde4-1 plants (Fig. 3c). In addition, rdr6sgs2-1 and sgs3-1, mutations known to affect siRNA accumulation from transgenes undergoing S-PTGS (Dalmay et al. 2000; Mourrain et al. 2000; Boutet et al. 2003), do not abolish the accumulation of siRNATEs (Fig. 3c). Therefore, the accumulation of siRNATEs requires SDE4, HEN1, and to some extent AGO4.
Figure 3.
Transposon siRNA accumulation and DNA methylation in various genotypes. Sense (a,c) or antisense (b) probes both detected siRNATEs. (d) Detection of siRNAs corresponding to AtSN1 in various genotypes. The 5S rRNA served as an internal control. rdr6sgs2-1, sgs3-1, and hen1-4 are in the Col accession; sde4-1 is in the C24 accession; and all other genotypes are in the Ler accession. The size of the siRNATEs was estimated with a 10-nt RNA ladder (Ambion) and by probing the same blot to visualize miR173 of known size (22 nt). (e) Determination of DNA methylation status at FLC-Ler in Ler (L), hen1-1 (h), ago4-1 (a), and cmt3-7 (c). A diagram of FLC-Ler genomic region is shown on top with the positions of restriction sites as indicated: (Hh) HhaI; (Hi) HindIII; (C) ClaI; (A) AciI; (Hh) HhaI. The probe corresponds to sequences flanking the TE (black box). The fragments that can hybridize to the probe in various digests are diagramed. The size of the fragments is indicated by numbers (in kilobases) underneath the fragments.
The levels of FLC mRNA and FLCTE RNAs are low in Ler, hen1-2, dcl1-9, ago4-1 (in Ler) (Fig. 2a,b), genotypes that accumulate siRNATEs. Conversely, the levels of FLC mRNA and FLCTE RNAs are high in hen1-1, a genotype that does not accumulate siRNATEs (Fig. 3a,b). The large increase in FLCTE RNAs in a hen1 mutant in Ler, together with the fact that in hen1-4 in the Col background there is very little effect on the mRNA levels of the TE-free FLC-Col allele (Fig. 2a), support a model in which siRNATEs target FLC-Ler for silencing. Because siRNATEs are present in both Col and Ler, the siRNATEs can be generated from other copies of the TE in the genome and are likely to target FLC-Ler due to the presence of a TE in FLC-Ler.
Two hypotheses can best explain the concomitant increase in abundance of FLC mRNA and FLCTE RNAs in hen1-1. First, FLC is transcribed at a higher rate in hen1-1, resulting in a higher abundance of FLC pre-mRNA, which undergoes normal splicing to give rise to FLC mRNA and aberrant splicing to produce FLCTE RNAs. Second, the hen1-1 mutation leads to increased stability of the FLC pre-mRNA or both FLC mRNA and FLCTE RNAs. Notably, siRNATEs belong to the 24-25-nt size class that has been correlated with histone H3-K9 methylation and DNA methylation (Hamilton et al. 2002; Zilberman et al. 2003; Xie et al. 2004), hallmarks of transcriptionally repressive chromatin (Hall et al. 2002; Volpe et al. 2002), suggesting that siRNATEs may lead to transcriptional silencing of FLC-Ler. We first examined DNA methylation at FLC-Ler by Southern hybridization. A probe flanking the TE in the FLC-Ler genomic region was used to detect the TE in FLC and its surrounding DNA (Fig. 3e). While the methylation-insensitive enzyme HindIII results in identical patterns in Ler and hen1-1, enzymes sensitive to methylation at CG sites (AciI, ClaI, and HhaI) result in patterns of digestion consistent with the presence of DNA methylation in the TE in FLC-Ler but not in its surrounding genomic DNA and with the release of TE methylation in hen1-1 (Fig. 3e). Additional probes in FLC-Ler in combination with these and other restriction enzymes (HphI, BstBI, Fnu4HI, and MspI) did not detect DNA methylation in FLC-Ler outside the TE (including 1.5 kb of the FLC promoter). Consistent with previous findings that CMT3 is primarily responsible for CNG methylation (Bartee et al. 2001; Lindroth et al. 2001), cmt3-7 (in Ler) does not affect TE methylation at these sites (Fig. 3e). DNA methylation of the TE is also reduced or released in ago4-1 (Fig. 3e). Because ago4-1 has low levels of FLC mRNA (Fig. 2b) and flowers early (Fig. 1e), loss of DNA methylation at the TE cannot explain the elevated levels of FLC RNA or the lateness to flowering in hen1-1. However, it is possible that DNA methylation at other sites within the TE causes transcriptional silencing of FLC-Ler.
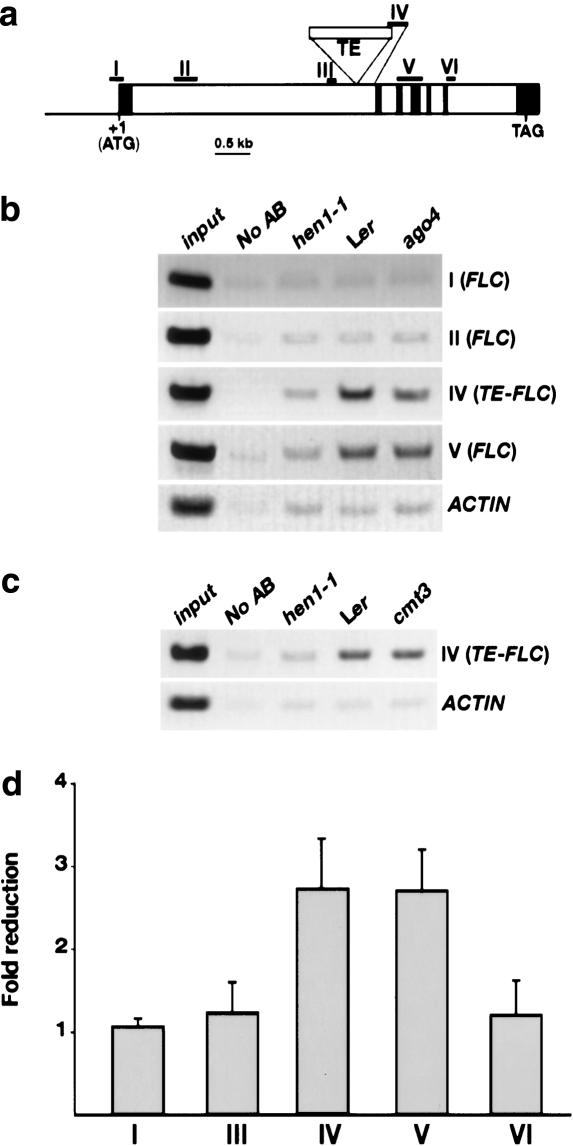
Next, we performed chromatin immunoprecipitation (ChIP) assays to determine the status of histone H3-K9 dimethylation at the FLC-Ler locus in Ler, hen1-1, ago4-1, and cmt3-7 plants. The level of H3-K9 dimethylation in a region of FLC-Ler chromatin including the TE (region IV) and a region downstream of the TE (region V) was elevated in Ler (Fig. 4). The level of H3-K9 dimethylation in this region was much reduced in the hen1-1 mutant (Fig. 4). However, there was no reproducible difference in the level of H3-K9 dimethylation between Ler and cmt3-7 (Fig. 4c) and only a slight difference between Ler and ago4-1 (Fig. 4b). The levels of histone H3-K9 methylation at the FLC-Ler locus in these four genotypes inversely correlate with the levels of FLC mRNA (Fig. 2a,b). Therefore, it is likely that the TE in FLC-Ler results in transcriptional silencing of FLC-Ler through histone H3-K9 methylation triggered by siRNATEs. Consistent with transcriptional rather than post-transcriptional gene silencing in Ler, the TE affects FLC-Ler in cis in an FLC-Ler/FLC-Col heterozygous plant (Michaels et al. 2003). Interestingly, the relatively high level of histone H3-K9 dimethylation at the FLC-Ler locus is confined to the TE and a region around the TE. At least two other regions of the first intron, which contains multiple regulatory elements (Sheldon et al. 2002; He et al. 2003), are not affected (Fig. 4). Thus, the presence of the TE appears to create an island of heterochromatin that attenuates, but does not prevent, gene expression. This probably permits expression of FLC-Ler to be modulated by other regulators: FLC-Ler can still respond to genes in the autonomous flowering pathway, vernalization, and, to some degree, FRI (Lee et al. 1993a; Rouse et al. 2002).
Figure 4.
Chromatin immunoprecipitation (ChIP) to detect histone H3-K9 dimethylation at the FLC-Ler locus. (a) Schematic structure of the genomic FLC-Ler region. I-VI represent the regions for which H3-K9 dimethylation states were examined by ChIP. The translation initiation point is +1. The filled boxes represent exons, and open boxes represent introns. (b) ChIP analyses of the H3-K9 dimethylation state of various genomic FLC regions. The input is Ler chromatin before immunoprecipitation. “No AB” refers to the control sample lacking antibody. ACTIN served as an internal control. (c) ChIP analyses of the H3-K9 dimethylation state of FLC chromatin in Ler, hen1-1, and cmt3-7. (d) The fold reduction of dimethylated H3-K9 of FLC chromatin in hen1-1 compared to Ler at the indicated regions. The fold reduction of hen1-1 compared to Ler was calculated as follows: FLC in Ler was first normalized to ACTIN; the normalized FLC in Ler was then divided by the normalized FLC in hen1-1 to obtain the fold reduction in the mutant. The error bars represent standard deviations of three independent immunoprecipitations.
While a release of transcriptional silencing of FLC-Ler is probably responsible for the elevated levels of FLC mRNA in hen1-1, it likely only partially accounts for the elevated levels of FLCTE RNAs in this genotype. If transcriptional silencing of FLC-Ler were the only silencing mechanism in Ler, it would be expected that the increase in abundance for both FLCTE RNAs and FLC mRNA in hen1-1 relative to Ler be comparable. However, the increase in FLCTE RNAs in hen1-1 relative to Ler is much greater than that of FLC mRNA (Fig. 2a; Supplementary Fig. S3), suggesting that an additional mechanism reduces the levels of FLCTE RNAs in Ler and this mechanism is released in hen1-1. Perhaps siRNATEs also guide the degradation of FLCTE RNAs. Intriguingly, FLCTE RNAs can be detected by filter hybridization in ago4-1 (Fig. 2a), although FLC mRNA does not accumulate to high levels in this genotype. This also supports the notion that FLCTE RNAs are regulated by an additional, perhaps PTGS, mechanism.
Consistent with the conclusion that siRNA-mediated silencing of FLC-Ler is the primary contributor to the weak nature of the FLC-Ler allele, relieving FLC-Ler from silencing converts the FLC-Ler allele into a functional allele such that it is highly responsive to FRI regulation: hen1-1 FRI-SF2 plants are extremely late flowering (Fig. 1d,f). Interestingly, modulation of FLC expression by intronic TE insertion appears to have occurred at least twice in evolution to accelerate flowering (Michaels et al. 2003). Two summer annual Arabidopsis accessions, Ler and Da (1)-12, harbor independent TE insertions in FLC intron 1 (Michaels et al. 2003). In fact, an H3mK9 island is also present in FLC-Da (1)-12 at the transposon (Supplementary Fig. S4). Therefore, an siRNA-based silencing mechanism has been used as a way to evolve new developmental traits.
Although TEs inserted in introns of genes are known to affect RNA processing, we demonstrate here that an intronic TE can render its host gene susceptible to siRNA-mediated transcriptional gene silencing and that this mechanism has been adopted to evolve new developmental traits. A recent study showed that transposons affect the expression of nearby genes at the genome scale in Arabidopsis (Lippman et al. 2004). These studies confirm McClintock's view that transposons can serve as regulators of gene expression by introducing heterochromatin into genes in which they insert (McClintock 1950).
Most eukaryotic genomes are littered with repeat sequences and transposable elements, and many TEs or remnants of TEs are in introns of genes or have been recruited into exons of genes during evolution (Nekrutenko and Li 2001). For example, the majority of all known genes in the human genome have Alu insertions in their introns and/or UTRs (Grover et al. 2004). Given the universality of siRNA-based regulation, we propose that a TE integrated in the transcribed region of a gene may confer siRNA-based regulation to its host gene if the TE itself or its homologs in the genome are actively repressed by siRNA-mediated gene silencing.
Materials and methods
RNA filter hybridization
Forty micrograms of total RNA from 20-d-old plants was resolved on denaturing formaldehyde gels, transferred to nylon membranes, and hybridized with randomly labeled probes corresponding to different portions of FLC genomic DNA or cDNA. The probes and primers used were as follows: Probe A was amplified by PCR from FLC cDNA using primers p4 (5′-CTGAACTATGGTTCACACTATGAGCTA-3′) and p5 (5′-AGATCTCGATGCAATTCTCACACGAA-3′). Probe B was amplified by PCR from FLC cDNA with primers p1 (5′-CGCTTAGTATCTCCGGCGACTTGAAC-3′) and p41 (5′-GTTCAAGTAGCTCATAGTGTGAACCA-3′). Probe C was amplified by PCR from Ler genomic DNA with primers p52 (5′-GGGTGAATTGCAAAAATAACCTATTTGGAA-3′) and p53 (5′-GGGAAATTTGGAAAACTAACCTACAAA-3′).
RNA filter hybridization was also carried out with RNAs isolated from 10-d-old plants of the genotypes used in the chromatin immunoprecipitation experiments. The relative levels of FLC RNAs among the genotypes are similar to those of 20-d-old plants.
siRNA detection
Small RNA enrichment, gel electrophoresis, blotting, and hybridization were carried out as described (Hamilton et al. 2002). The RNAs were isolated from 20-d-old plants. The TE DNA was amplified by PCR with p52 and p53, cloned into pGEMTE (Promega), and used as templates for in vitro transcription to generate sense and antisense probes. The pCR4-AtSN1 plasmid was a gift from Steve Jacobsen (University of California, Los Angeles).
Chromatin Immunoprecipitation (ChIP) Assays
The chromatin immunoprecipitation experiments were performed as described by Johnson et al. (2002) using 10-d-old seedlings. The anti-dimethyl-histone H3-K9 and ChIP assay kit were from Upstate Biotechnology. Each of the immunoprecipitations was replicated three times. The amounts of genomic DNA immunoprecipitated were determined by quantitative PCR. The primer pair HL1 (5′-CAAGTTGAAGGAGTCTGTGGTCT-3′) and HL2 (5′-GGATCTTGACCAGGCTGGAGAG-3′) was used to amplify the junction between the 3′-end of the Mutator-like transposon and FLC-Ler (region IV); the primer pair HL3 (5′-TTCATGGGCAGGATCATCAGTCAA-3′) and HL4 (5′-GCTTCAACATGAGTTCGGTCTGCA-3′) was used to amplify region V of FLC-Ler. The primer pair HD1 (5′-GAGTCGTGTCTGTTACACAGTTGT-3′) and HD2 (5′-GTTATGAAAGAAGAACAGAGTAAGAACGA-3′) was used to amplify the retrotransposon in FLC-Da. HD1 and HD3 (5′-AAATATAGGAAATTGGAACCTCACAGT-3′) were used to amplify the junction between the 3′-end of the retrotransposon and FLC-Da. Aliquots of the PCR reactions were resolved by electrophoresis in 1.6% agarose gels, and quantified with ImageQuant software (Molecular Dynamics, Inc).
Acknowledgments
We are indebted to Vicki Vance for advice on the project and David Baulcombe for sharing unpublished information. We thank Alan Herr, David Baulcombe, Daniel Zilberman, Xiaofeng Cao, Steve Jacobsen, Scott Michaels, and Hervé Vaucheret for genetic materials; Yulan Cheng, Hugo Dooner, Abram Gabriel, Randy Kerstetter, Junjie Li, and Ruth Steward for comments on the manuscript; and Xiaofeng Cao and Zhizhong Gong for technical advice. This work was supported by a grant from the National Institutes of Health to X.C. (GM61146) and by grants from the U.S. Department of Agriculture National Research Initiative Competitive Grants Program and the National Science Foundation to R.A.
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1217304.
References
- Bartee L., Malagnac, F., and Bender, J. 2001. Arabidopsis cmt3 chromo-methylase mutations block non-CG methylation and silencing of an endogenous gene. Genes & Dev. 15 1753-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116 281-297. [DOI] [PubMed] [Google Scholar]
- Boss P.K., Bastow, R.M., Mylne, J.S., and Dean, C. 2004. Multiple pathways in the decision to flower: Enabling, promoting, and resetting. Plant Cell 16 Suppl: S18-S31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutet S., Vazquez, F., Liu, J., Beclin, C., Fagard, M., Gratias, A., Morel, J.B., Crete, P., Chen, X., and Vaucheret, H. 2003. Arabidopsis HEN1. A genetic link between endogenous miRNA controlling development and siRNA controlling transgene silencing and virus resistance. Curr. Biol. 13 843-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Liu, J., Cheng, Y., and Jia, D. 2002. HEN1 functions pleiotropically in Arabidopsis development and acts in C function in the flower. Development 129 1085-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmay T., Hamilton, A., Rudd, S., Angell, S., and Baulcombe, D.C. 2000. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101 543-553. [DOI] [PubMed] [Google Scholar]
- Finnegan E.J., Margis, R., and Waterhouse, P.M. 2003. Posttranscriptional gene silencing is not compromised in the Arabidopsis carpel factory (dicer-like1) mutant, a homolog of Dicer-1 from Drosophila. Curr. Biol. 13 236-240. [DOI] [PubMed] [Google Scholar]
- Gazzani S., Gendall, A.R., Lister, C., and Dean, C. 2003. Analysis of the molecular basis of flowering time variation in Arabidopsis accessions. Plant Physiol. 132 1107-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover D., Mukerji, M., Bhatnagar, P., Kannan, K., and Brahmachari, S.K. 2004. Alu repeat analysis in the complete human genome: Trends and variations with respect to genomic composition. Bioinformatics 20 813-817. [DOI] [PubMed] [Google Scholar]
- Hall I.M., Shankaranarayana, G.D., Noma, K., Ayoub, N., Cohen, A., and Grewal, S.I. 2002. Establishment and maintenance of a heterochromatin domain. Science 297 2232-2237. [DOI] [PubMed] [Google Scholar]
- Hamilton A.J. and Baulcombe, D.C. 1999. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286 950-952. [DOI] [PubMed] [Google Scholar]
- Hamilton A., Voinnet, O., Chappell, L., and Baulcombe, D. 2002. Two classes of short interfering RNA in RNA silencing. EMBO J. 21 4671-4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon G.J. 2002. RNA interference. Nature 418 244-251. [DOI] [PubMed] [Google Scholar]
- He Y., Michaels, S., and Amasino, R. 2003. Regulation of flowering time by histone acetylation in Arabidopsis. Science 302 1751-1754. [DOI] [PubMed] [Google Scholar]
- Johanson U., West, J., Lister, C., Michaels, S., Amasino, R., and Dean, C. 2000. Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290 344-347. [DOI] [PubMed] [Google Scholar]
- Johnson L., Cao, X., and Jacobsen, S. 2002. Interplay between two epigenetic marks. DNA methylation and histone H3 lysine 9 methylation. Curr. Biol. 12 1360-1367. [DOI] [PubMed] [Google Scholar]
- Lee I., Bleecker, A., and Amasino, R.M. 1993a. Analysis of naturally occurring late flowering in Arabidopsis thaliana. Mol. Gen. Genet. 237 171-176. [DOI] [PubMed] [Google Scholar]
- Lee R.C., Feinbaum, R.L., and Ambros, V. 1993b. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75 843-854. [DOI] [PubMed] [Google Scholar]
- Lindroth A.M., Cao, X., Jackson, J.P., Zilberman, D., McCallum, C.M., Henikoff, S., and Jacobsen, S.E. 2001. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 292 2077-2080. [DOI] [PubMed] [Google Scholar]
- Lippman Z., Gendrel, A.V., Black, M., Vaughn, M.W., Dedhia, N., McCombie, W.R., Lavine, K., Mittal, V., May, B., Kasschau, K.D., et al. 2004. Role of transposable elements in heterochromatin and epigenetic control. Nature 430 471-476. [DOI] [PubMed] [Google Scholar]
- McClintock B. 1950. The origin and behavior of mutable loci in maize. Proc. Natl. Acad. Sci. 36 344-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S.D. and Amasino, R.M. 1999. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11 949-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2001. Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 13 935-941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S., He, Y., Scortecci, K.C., and Amasino, R. 2003. Attenuation of FLOWERING LOCUS C activity as a mechanism for the evolution of summer-annual flowering behavior in Arabidopsis. Proc. Natl. Acad. Sci. 100 10102-10107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourrain P., Beclin, C., Elmayan, T., Feuerbach, F., Godon, C., Morel, J.B., Jouette, D., Lacombe, A.M., Nikic, S., Picault, N., et al. 2000. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101 533-542. [DOI] [PubMed] [Google Scholar]
- Nekrutenko A. and Li, W.-H. 2001. Transposable elements are found in a large number of human protein-coding genes. Trends Genet. 17 619-621. [DOI] [PubMed] [Google Scholar]
- Park W., Li, J., Song, R., Messing, J., and Chen, X. 2002. CARPEL FAC TORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12 1484-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasterk R.H. 2002. RNA silencing: The genome's immune system. Science 296 1263-1265. [DOI] [PubMed] [Google Scholar]
- Ray A., Lang, J.D., Golden, T., and Ray, S. 1996. SHORT INTEGUMENT (SIN1), a gene required for ovule development in Arabidopsis, also controls flowering time. Development 122 2631-2638. [DOI] [PubMed] [Google Scholar]
- Reinhart B.J. and Bartel, D.P. 2002. Small RNAs correspond to centromere heterochromatic repeats. Science 297 1831. [DOI] [PubMed] [Google Scholar]
- Reinhart B.J., Slack, F.J., Basson, M., Pasquinelli, A.E., Bettinger, J.C., Rougvie, A.E., Horvitz, H.R., and Ruvkun, G. 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403 901-906. [DOI] [PubMed] [Google Scholar]
- Reinhart B.J., Weinstein, E.G., Rhoades, M.W., Bartel, B., and Bartel, D.P. 2002. MicroRNAs in plants. Genes & Dev. 16 1616-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse D.T., Sheldon, C.C., Bagnall, D.J., Peacock, W.J., and Dennis, E.S. 2002. FLC, a repressor of flowering, is regulated by genes in different inductive pathways. Plant J. 29 183-191. [DOI] [PubMed] [Google Scholar]
- Sheldon C.C., Burn, J.E., Perez, P.P., Metzger, J., Edwards, J.A., Peacock, W.J., and Dennis, E.S. 1999. The FLF MADS box gene: A repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11 445-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon C.C., Conn, A.B., Dennis, E.S., and Peacock, W.J. 2002. Different regulatory regions are required for the vernalization-induced repression of FLOWERING LOCUS C and for the epigenetic maintenance of repression. Plant Cell 14 2527-2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe T.A., Kidner, C., Hall, I.M., Teng, G., Grewal, S.I., and Martienssen, R.A. 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297 1833-1837. [DOI] [PubMed] [Google Scholar]
- Weil C.F. and Wessler, S.R. 1990. The effects of plant transposable element insertion on transcription initiation and RNA processing. Annu. Rev. Plant Physiol. Plant Mol. Biol. 41 527-552. [Google Scholar]
- Xie Z., Johansen, L.K., Gustafson, A.M., Kasschau, K.D., Lellis, A.D., Zilberman, D., Jacobsen, S.E., and Carrington, J.C. 2004. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2 e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D., Cao, X., and Jacobsen, S.E. 2003. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299 716-719. [DOI] [PubMed] [Google Scholar]