Abstract
Protein localization is crucial for cellular morphogenesis and intracellular signal transduction cascades. Here we describe an interaction between two membrane proteins expressed in different cells of the Bacillus subtilis sporangium, the mother cell protein SpoIIIAH and the forespore protein SpoIIQ. We used affinity chromatography, coimmunoprecipitation, and the yeast two-hybrid system to demonstrate that the extracellular domains of these proteins interact, tethering SpoIIIAH to the sporulation septum, and directing its assembly with SpoIIQ into helical arcs and foci around the forespore. We also demonstrate that this interaction can direct proteins made in the same cell to active division sites, as when SpoIIQ is made in the mother cell, it localizes to nascent septa in a SpoIIIAH-dependent manner. Both SpoIIIAH and SpoIIQ are necessary for activation of the second forespore-specific transcription factor (σG) after engulfment, and we propose that the SpoIIIAH-SpoIIQ complex contributes to a morphological checkpoint coupling σG activation to engulfment. In keeping with this hypothesis, SpoIIIAH localization depends on the first step of engulfment, septal thinning. The SpoIIQ-SpoIIIAH complex reaches from the mother cell cytoplasm to the forespore cytoplasm and is ideally positioned to govern the activity of engulfment-dependent transcription factors.
Keywords: Sporulation, protein localization, signal transduction, bacterial development
Dynamic protein localization is crucial for bacterial development, signal transduction cascades, and the cell cycle. In recent years, microbial cell biologists have discovered tubulin-like filaments involved in cell shape and chromosome segregation (Carballido-Lopez and Errington 2003a,b; Shih et al. 2003; Gerdes et al. 2004), oscillating proteins that govern division site selection (Hu and Lutkenhaus 1999; Raskin and de Boer 1999a,b), chromosome segregation, and cellular differentiation (Glaser et al. 1997; Lin et al. 1997; Autret and Errington 2003), as well as localized proteins required for pathogenesis, cell division, development, and signal transduction cascades (for review, see Lybarger and Maddock 2001; Shapiro et al. 2002; Errington et al. 2003; Ryan and Shapiro 2003). Although bacterial cells lack membrane-enclosed compartments, a growing body of evidence suggests that the activity of signal transduction proteins can be efficiently compartmentalized by being confined to a specific subcellular address. For example, in Caulobacter crescentus, cell division results in the production of morphologically distinct daughter cells: the motile swarmer cell and the sessile stalked cell, which is the only daughter capable of immediately initiating a new cell cycle (McAdams and Shapiro 2003; Quardokus and Brun 2003; Jacobs-Wagner 2004; Skerker and Laub 2004). These divergent cell cycles are controlled by two oscillating regulatory proteins that accumulate in opposite daughter cells: one represses chromosome replication and the cell cycle in the swarmer cell while the other promotes the cell cycle in the stalked cell (Holtzendorff et al. 2004). Therefore, it is clearly important to understand the spatial and temporal distribution of bacterial proteins as well as their biochemical properties.
Protein localization is also important in B. subtilis sporulation, during which the synthesis of an asymmetrically positioned septum produces a smaller forespore, which will become the spore, and a larger mother cell, which will lyse after contributing to spore development (for review, see Errington 2003). A variety of proteins involved in morphogenesis and intracellular signal transduction localize to the septum between these cells (Driks et al. 1994; Arigoni et al. 1995; Resnekov et al. 1996; Fawcett et al. 1998; Perez et al. 2000; Abanes-De Mello et al. 2002; van Ooij and Losick 2003; Rubio and Pogliano 2004) and in many cases disrupting localization has severe consequences for development (King et al. 1999).
In theory, localized membrane proteins could either be directly inserted into their target membrane domain, or be inserted elsewhere and diffuse to their correct location. It appears that both pathways operate during sporulation. In the forespore, even membrane proteins that are ultimately randomly distributed throughout the forespore membrane initially localize to the septum, suggesting, although not conclusively demonstrating, that membrane protein insertion is restricted to the forespore septum (Rubio and Pogliano 2004). In contrast, in the mother cell, it has been proposed that membrane protein insertion is nonlocalized (Rudner et al. 2002), since mother cell membrane proteins engineered to be expressed before division are initially randomly distributed, but then relocalize from this nonlocalized pool to the septum (Rudner et al. 2002; van Ooij and Losick 2003). Both direct and random insertion require the capture of membrane proteins at the correct location to prevent their rapid diffusion, which could be achieved if they interact with another molecule that is stationary and localized.
Bacillus subtilis sporulation generates a unique bacterial cell that has two separate membrane-enclosed compartments, a consequence of engulfment (for review, see Errington 2003). During engulfment, the membrane of the mother cell migrates around the forespore, until the leading edge meets and fuses to release the forespore into the mother cell cytoplasm (Fig. 1). After engulfment, the forespore is bounded by two membranes: its cytoplasmic membrane and a second membrane derived from the mother cell, the outer forespore membrane. The generation of a cell with two separate membrane compartments could have important consequences for the localization of membrane proteins targeted to the outer forespore membrane and is a morphological checkpoint essential for the activation of two late transcription factors: σG in the forespore and σK in the mother cell (for review, see Kroos and Yu 2000; Errington 2003).
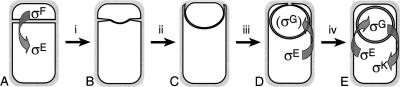
Figure 1.
Stages of sporulation and cell-specific gene expression. (A) Following synthesis of the sporulation septum, σF becomes active in the smaller cell, the forespore, followed by activation of σE in the larger mother cell. (B) During the first step of engulfment [(i) septal thinning] septal peptidoglycan (gray) is removed, starting at the septal midpoint. (C) Subsequently, the mother cell membrane migrates around the forespore (steps ii-iii), meets (D) then fuses (E) at the forespore pole to release the forespore into the mother cell cytoplasm (step iv), at which time σG and σK become active in the forespore and mother cell, respectively.
SpoIIQ is an attractive candidate for a protein that senses the completion of engulfment. This forespore-expressed membrane protein is conserved in all endospore-forming bacteria (Stragier 2002), required for synthesis and activation of σG, and under some conditions, for engulfment (Londoño-Vallejo et al. 1997; Sun et al. 2000). Previous work has shown that SpoIIQ localizes to the septum, tracks the engulfing mother cell membrane, assembles helical arcs and foci surrounding the forespore, and is degraded after engulfment (Rubio and Pogliano 2004). Interestingly, retention of SpoIIQ at the septum requires mother cell gene expression, suggesting that its extracellular domain interacts with that of a mother cell tether (Rubio and Pogliano 2004). This interaction between SpoIIQ and a mother cell tether provides a potential route for signaling between the mother cell and forespore that is necessary for σG activation (Partridge and Errington 1993).
A second conserved group of proteins required for σG activity is the proteins encoded by the spoIIIA operon (Kellner et al. 1996). This operon encodes eight proteins, one of which, SpoIIIAA, shows homology to the AAA family of ATPases, whereas the rest are predicted to be membrane proteins. To investigate the role of these proteins in σG activation and determine if they interact with SpoIIQ, we initiated studies of their subcellular distribution, obtaining functional fusions between the protein encoded by the last gene in the operon, SpoIIIAH and the Flag epitope. We here demonstrate that SpoIIIAH is localized to the septum by a direct interaction between its extracellular domain and that of SpoIIQ and that the two proteins colocalize in arcs and foci surrounding the forespore. Furthermore, SpoIIIAH localization depends on engulfment, suggesting that this dynamic complex plays a central role sensing engulfment.
Results
Localization of SpoIIIAH
The final gene in the spoIIIA operon is spoIIIAH, which encodes a protein predicted by HMMTOP (Tusnady and Simon 2001) and TM pred (Hofmann and Stoffel 1993) to have one membrane-spanning segment and a large extracellular domain. As the last gene in the spoIIIA operon, it was relatively simple to construct a Flag epitope fusion to the 3′-end of spoIIIAH and integrate a plasmid into the B. subtilis chromosome by a single recombination event. The resulting strain expressed SpoIIIAH-Flag (but not SpoIIIAH) from its native promoter without disrupting other genes in the spoIIIA operon. This strain produced wild-type levels of spores, indicating that SpoIIIAH-Flag was functional; fractionation data demonstrated that it was membrane-associated (data not shown). We localized SpoIIIAH-Flag using immunofluorescence microscopy and used the membrane stain FM 4-64 to correlate localization with engulfment. In sporangia that have initiated engulfment and have slightly curved septa, SpoIIIAH-Flag localized to the septum, often with discrete foci at the septum and faint mother cell fluorescence (Fig. 2A, arrow). During engulfment, SpoIIIAH-Flag tracked the engulfing mother cell membrane as it moved around the forespore (Fig. 2A, arrowhead). In engulfed sporangia, SpoIIIAH-Flag localized around the forespore in discrete foci (Fig. 2B, arrow). Thus, SpoIIIAH displays a dynamic localization pattern, first localizing to the septum, and then assembling foci as the engulfing membrane surrounds the forespore (Fig. 2H).
Figure 2.
Localization of the mother cell protein SpoIIIAH. Immunofluorescence microscopy was used to localize SpoIIIAH-Flag (green) with the membranes stained with FM 4-64 (red). Sporulation was initiated by resuspension and samples harvested after 2 h (t2; A,C,E,G) and 3 h (t3; B,D,F) of sporulation. (A,B) SpoIIIAH-Flag in wild type (KP856). Bar, 2 μm. (C,D) SpoIIIAH-Flag localization in the spoIIQ null (KP857). (E,F) SpoIIIAH-Flag localization in spoIIIAA::EZ::TN-(kan) (KP858). (G) SpoIIIAH-Flag expressed during growth from the xylose promoter (KP886). (H) Cartoon depicting localization of SpoIIIAH-Flag (green circles) in wild-type sporangia throughout sporulation. (I) Cartoon depicting SpoIIIAH-Flag localization without SpoIIQ.
To determine if SpoIIIAH synthesized during growth could subsequently localize to the sporulation septum, we constructed a strain in which the xylose promoter controlled expression of SpoIIIAH-Flag (Pxyl-spoIIIAH-Flag). This strain was grown in the presence of xylose, then washed and resuspended in sporulation medium without xylose to prevent further synthesis of SpoIIIAH-Flag. In cells without a sporulation septum, SpoIIIAH-Flag was present throughout the membranes (Fig. 2G, arrow), but once the sporulation septum was synthesized, the majority of SpoIIIAH-Flag was localized to the septum (Fig. 2G, arrowhead). SpoIIIAH-Flag remained localized after engulfment, forming disorganized foci in ∼50% of sporangia (Fig. 2G, double arrowhead). Thus, SpoIIIAH can reach the sporulation septum by diffusion from the cytoplasmic membrane.
SpoIIIAH localization depends on SpoIIQ, but not SpoIIIAA-AF or SpoIIIJ
SpoIIIAH is required to activate the second forespore transcription factor σG after engulfment, as are SpoIIIJ, SpoIIQ, and the remaining SpoIIIA proteins. We were interested in determining whether localization of SpoIIIAH-Flag would be affected by the absence of any of these proteins, or by the absence of σG itself. Localization of SpoIIIAH-Flag was not affected in strains with mutations in the gene encoding σG (spoIIIG) (data not shown) or in spoIIIJ (Supplementary Fig. 1); therefore, σG-dependent proteins are not required for the stages of SpoIIIAH localization described here. In contrast, SpoIIIAH-Flag appeared randomly distributed throughout the mother cell membrane in the absence of SpoIIQ (Fig. 2C,D, arrows); with a slight enrichment at the engulfing membrane (Fig. 2D, arrowhead), likely due to the presence of a double layer of mother cell membrane formed as the leading edge migrates around the forespore (Fig. 2I). SpoIIIAH localized identically in the spoIIQ null and in the absence of forespore-specific gene expression (Supplementary Fig. 1), suggesting that SpoIIQ is the primary forespore-specific localization determinant of SpoIIIAH.
To determine if SpoIIIAH localized in the absence of the first six SpoIIIA proteins (SpoIIIAA-AF), we introduced spoIIIAH-Flag into strains containing an EZ::TN(kan) insertion in the first gene in the operon, spoIIIAA (Materials and Methods), and with in-frame deletions in each spoIIIA gene. Immunoblot analysis indicated that SpoIIIAH-Flag reached similar levels in the spoIIIAA::EZ::TN(kan) strain as in wild type (data not shown), supporting the existence of a second spoIIIAH promoter. Immunofluorescence microscopy showed that many early sporangia had normal SpoIIIAH localization (Fig. 2E, arrowhead), while others showed little septal localization (Fig. 2E, arrow). Later, most spoIIIAA:: EZ::TN(kan) sporangia, as well as strains with in-frame deletions in spoIIIAA, spoIIIAB, spoIIIAC-AD, spoIIIAE, spoIIIAF, and spoIIIAG showed SpoIIIAH-Flag foci apparently identical to wild type (Fig. 2F, arrow). Thus, the spoIIIAA::EZ::TN(kan) insertion delays the initial localization of SpoIIIAH to the septum, but ultimately SpoIIIAH assembles normally.
Evidence for a direct interaction between SpoIIQ and SpoIIIAH
SpoIIQ and SpoIIIAH each track the engulfing mother cell membrane, assemble helical arcs and foci, and are predicted to contain one transmembrane domain and a large extracellular domain. Although these proteins are in separate cells, it is possible that their extracellular domains directly interact within the septal space to mediate SpoIIIAH localization. To test this possibility, we used a yeast two-hybrid system (Finley and Brent 1994), fusing the hydrophilic C terminus of each protein (amino acids 25-218 for SpoIIIAH, amino acids 41-283 for SpoIIQ) to either LexA (a DNA-binding protein) or to the transcriptional activation domain of AB. Interaction between AB and LexA fusion proteins is indicated by increased β-galactosidase activity in a yeast strain carrying lacZ preceded by eight LexA-binding sites. When SpoIIQ was fused to LexA and SpoIIIAH was fused to AB, the yeast strain produce ∼25-fold more β-galactosidase activity than negative controls (Table 1). In the reciprocal experiment (SpoIIIAH-LexA and SpoIIQ-AB), a small but reproducible increase in β-galactosidase activity was observed.
Table 1.
Two-hybrid interaction between SpoIIIAH and SpoIIQ
| β-Galactosidase activity in yeast strains producing the indicated fusion proteins LexA fusion
|
|||
|---|---|---|---|
| AB fusion | Empty | spoIIIAH | spoIIQ |
| Empty | ND | 3.5 | 2.3 |
| spoIIIAH | 2.2 | ND | 58.8 |
| spoIIQ | 1.7 | 6.1 | ND |
The numbers shown are the average Miller Units produced in two independent experiments.
We used affinity chromatography to further investigate the interaction between the extracellular domains of SpoIIQ and SpoIIIAH. We first constructed a plasmid encoding a GST-SpoIIQ fusion protein, and bound GSTSpoIIQ or GST (as a negative control) to GST trap columns. Extracts from sporulating B. subtilis cells expressing either SpoIIIAH-Flag or FtsH-Flag (as a negative control) or a mixture of the two were loaded onto these columns and washed, and the eluates were subjected to immunoblot analysis to determine if SpoIIIAH-Flag was retained on the column. These experiments demonstrated that SpoIIIAH-Flag but not FtsH-Flag interacted with GST-SpoIIQ but not with GST itself (Fig. 3A). Importantly, the eluates included only a few bands (Fig. 3A), one the same size as GST-SpoIIQ. Together with the yeast two-hybrid data described above, these experiments support a direct interaction between the extracellular domains of SpoIIIAH and SpoIIQ.
Figure 3.
Biochemical evidence for an interaction between SpoIIQ and SpoIIIAH. (A) Affinity chromatography. Lysates were prepared from B. subtilis strains expressing either SpoIIIAH-Flag (lanes 1-3,5,10-14) or FtsH-Flag (lanes 4,6,7) or a mixture of the two (lanes 8,9) and loaded onto columns with GST-SpoIIQ (lanes 2,3,6-9,13,14), or GST only (lanes 11,12), washed, and eluted. Total protein was detected by Coomassie staining (lanes 1-3), and the presence of Flag-containing fusion proteins by immunoblots (lanes 4-14). The arrowhead indicates the position of GST-SpoIIQ, the asterisk unknown SpoIIQ-binding proteins. Lane 1 contains extract from 16 μL of original culture, lanes 4 and 5 from 166 μL of cells, while lanes 3, 7, and 9 contain the binding proteins from between 1.8 and 3.6 mL of original culture volume. The decreased amount of protein loaded and the presence of Triton in the lane 5 sample could explain the decreased level of SpoIIIAH-Flag compared to lane 9. (Lanes 11,12) SpoIIIAH-Flag was not retained on GST columns. (B) Coimmunoprecipitation. Membrane fractions were prepared from strains KP845 (GFP-spoIIQ; lanes 1,5,9,13), KP856 (spoIIIAH-Flag; lanes 2,6,10,14), KP867 (spoIIIAH-Flag, GFP-spoIIQ; lanes 3,7,11,15), or KP895 (spoIIIAH-Flag, PspoIIQ-malF-GFP; lanes 4,8,12,16), and immunoprecipitated with anti-Flag affinity gel. Total protein in lysates and eluates after immunoprecipitation (from 0.3 or 1.2 mL of culture, respectively) were detected by Coomassie staining, and GFP fusion proteins by immunoblot analysis. Forespore expressed MalF-GFP did not copurify with SpoIIIAH (lanes 12,16), but the larger of two GFPSpoIIQ bands (labeled SpoIIQ) almost quantitatively copurified with SpoIIIAH-Flag (lanes 11,15); a band of similar size was visible after Coomassie staining (lane 7, double arrowhead). The smaller band (SpoIIQ⋆) could be a degradation product. During elution, a small amount of antibody heavy and light chain (arrowheads) and SpoIIIAH-Flag (⋆) was released.
SpoIIQ and SpoIIIAH are present in the same membrane-bound complex in B. subtilis
We used coimmmunoprecipitation to demonstrate that SpoIIQ and SpoIIIAH interacted in living cells. Strains were constructed that expressed either GFP-SpoIIQ or MalF-GFP with SpoIIIAH-Flag, and membrane fractions were prepared and solubilized with 0.5% NP-40 (see Materials and Methods) and incubated with anti-Flag affinity gel. The eluates were subject to SDS-PAGE followed by Coomassie staining and Western blot analysis, probing the blots with GFP-specific antibodies. The results demonstrated that GFP-SpoIIQ but not forespore-expressed MalF-GFP copurified with SpoIIIAH-Flag (Fig. 3B). A comparison of the total amount of GFP-SpoIIQ in the membrane fraction prior to purification demonstrated that the larger of the two GFP-SpoIIQ bands was almost quantitatively recovered from the affinity gel, indicating that this protein, but not the shorter product, was completely bound to SpoIIIAH. The presence of SpoIIIAH-Flag in the eluate (Fig. 3B, asterisk) was confirmed by immunoblot with Flag-specific antibodies (data not shown). Thus, SpoIIQ and SpoIIIAH are present in the same membrane-bound complex in B. subtilis cells. Importantly, the shared topology of these two proteins (N terminus in, C terminus out) and the fact that they are synthesized in different cells means that this complex reaches from the mother cell cytoplasm across the septal space (where their C termini interact) to the forespore cytoplasm.
SpoIIIAH is not the primary tether for SpoIIQ
Retention of SpoIIQ at the septum requires an unidentified mother cell protein, since in the absence of mother cell gene expression, SpoIIQ moves around the forespore ahead of the engulfing mother cell membrane (Rubio and Pogliano 2004). To test if SpoIIIAH was this tether, we introduced GFP-SpoIIQ into a ΔspoIIIAG-AH strain. In wild type, GFP-SpoIIQ tracks the engulfing mother cell membrane, often in multiple foci similar to those seen for SpoIIIAH-Flag (Fig. 4A, arrow), while later in sporulation it is degraded to release cytoplasmic GFP (Fig. 4F; Rubio and Pogliano 2004). Early in sporulation, 31% of ΔspoIIIAG-AH sporangia showed normal SpoIIQ localization (Fig. 4B, arrow, double arrowhead), although 69% showed a small amount of GFP-SpoIIQ ahead of the engulfing membranes (Fig. 4B [arrowhead], G; Supplementary Table 1). This defect in early SpoIIQ localization is much less severe than occurs in mutants lacking all mother cell gene expression (spoIIGB), when SpoIIQ is uniformly distributed throughout the forespore membrane, moving ahead of the engulfing mother cell membrane (Figs. 4E [arrow], 2H). Thus, SpoIIIAH is not the primary tether for SpoIIQ.
Figure 4.
Localization of the forespore protein SpoIIQ. In all strains shown here, GFP-SpoIIQ (green) was expressed in strains lacking wild-type SpoIIQ protein and images collected from live cells at t2 of sporulation. Membranes were stained with mitotracker red (A-E) or FM 4-64 (red; I,J). (A) GFP-SpoIIQ localization in wild type (KP845). Bar, 2 μm. (B) GFP-SpoIIQ localization in ΔspoIIIAG—AH (KP872). (C) GFP-SpoIIQ localization in spoIIIJ (KP873). (D) GFP-SpoIIQ localization in ΔspoIIIAG-AH spoIIIJ double mutant (KP874). (E) GFP-SpoIIQ localization in the strain lacking all mother cell gene expression (spoIIGB; KP854). GFP-SpoIIQ was uniformly distributed throughout the forespore membrane in the absence of engulfment (arrow), although sporangia with very faint GFP fluorescence show enrichment at the septum (arrowhead), as does any forespore expressed nonlocalized protein (i.e., MalF) (Rubio and Pogliano 2004). (F) Cartoon showing SpoIIQ localization (blue circles) throughout engulfment, culminating in the degradation of SpoIIQ to release cytoplasmic GFP. (G) GFP-SpoIIQ localization in the absence of SpoIIIAG-AH; some SpoIIQ moves ahead of the engulfment membrane. (H) In the absence of mother cell gene expression, SpoIIQ is distributed throughout the forespore membrane and engulfment is blocked. (I) Mother-cell-expressed GFP-SpoIIQ (KP897) localizes exclusively to the second division site in the mother cell (arrow), which is activated, leading to partial septa opposite the forespore (arrowhead). The dotted line indicates sporangia added to the field to include more rare partial septa. (J) In a spoIIIAA-AH mutant (KP898), mother-cell-expressed GFP-SpoIIQ is only slightly enriched at some partial septa (arrow), and is rapidly degraded to yield cytoplasmic GFP fluorescence. (K) Depiction of SpoIIQ (blue) and SpoIIIAH (green) expressed in the same cell. (L) We propose that the binding sites are oriented such that the two proteins interact only at membrane invaginations. (M) Mother-cell-expressed GFP-SpoIIQ fails to localize without SpoIIIAH.
The effect of spoIIIA mutations on GFP-SpoIIQ localization could be increased by either inactivating spoIIIJ or using strains that also expressed wild-type SpoIIQ. While the spoIIIJ mutation had no discernible effect on GFP-SpoIIQ localization (Fig. 4C), spoIIIA spoIIIJ double-mutant sporangia showed increased GFP-SpoIIQ fluorescence ahead of the engulfing membranes compared to spoIIIA single mutants (Fig. 4D, arrowhead). We conclude that the absence of SpoIIIAH only slightly reduces the affinity of SpoIIQ for the sporulation septum, and that another mother cell protein is the primary SpoIIQ tether.
SpoIIIAH mediates localization of mother-cell-expressed SpoIIQ
If two extracellular protein domains interact only in opposing membranes rather than in the same membrane, their interaction could mediate localization to active division sites in symmetrically dividing cells. To determine if the interaction between SpoIIQ and SpoIIIAH could mediate septal localization when each protein was made in the same cell, GFP-SpoIIQ was expressed only in the mother cell, where it failed to complement the null mutation. In strains also expressing SpoIIIAH, GFPSpoIIQ localized as a ring at the partial septa formed when division initiates at the second potential site of polar septation (Fig. 4I,K). However, in strains lacking SpoIIIAH, GFP-SpoIIQ was delocalized, with a slight enrichment at the second division site of some sporangia and strong cytoplasmic fluorescence (Fig. 4J,M). Thus, SpoIIIAH can mediate localization of mother-cell-expressed SpoIIQ to active sites of cell division and protect it from degradation.
Colocalization of SpoIIIAH and SpoIIQ
If the extracellular domains of SpoIIQ and SpoIIIAH interact, they should colocalize at least until after engulfment, when SpoIIQ is cleaved. To test this, we used immunofluorescence microscopy to colocalize SpoIIQ-myc and SpoIIIAH-Flag. During engulfment, SpoIIQ-myc substantially colocalized with SpoIIIAH-Flag, and both formed foci along the engulfing membrane (Fig. 5A, arrow). After engulfment, many SpoIIQ-myc foci colocalized with SpoIIIAH-Flag foci (Fig. 5B, arrow), although some foci did not colocalize (Fig. 5B, arrowhead), perhaps due to loss of cell structure during preparation of the cells for immunofluorescence (which requires fixation and lysozyme treatment to permeabilize the cells). Some sporangia showed greatly reduced SpoIIQ-myc immunostaining and clear SpoIIIAH-Flag foci (Fig. 5B, double arrowhead), likely because SpoIIQ-myc (or the myc epitope) is unstable during sporulation (Rubio and Pogliano 2004). Thus, the extracellular domains of SpoIIIAH and SpoIIQ colocalize to a significant extent, although SpoIIIAH appears to persist during sporulation, whereas the extracellular domain of SpoIIQ-myc is less stable.
Figure 5.
Colocalization of SpoIIIAH-Flag and SpoIIQ-myc. Immunofluorescence microscopy was used to colocalize epitope fusions to the C termini of SpoIIQ (myc, green) and SpoIIIAH (Flag; false-colored red; strain KP868), and the membranes stained with FM 4-64 (false-colored blue), at t2.5 (A) and t3.5 (B). In late sporangia with α/β SASP/DNA rings (data not shown), SpoIIQ immunostaining was greatly reduced, while SpoIIIAH remained in foci (double arrowhead). (C) Observed colocalization pattern of the C termini of SpoIIQ (blue circles) and SpoIIIAH (green circles). Bar, 2 μm.
SpoIIIAH localization is inhibited by septal peptidoglycan
SpoIIIAH and SpoIIQ are ideally suited to couple σG activity to engulfment, as they interact across the septal space and move around the forespore during engulfment. If so, then engulfment mutants might alter SpoIIIAH localization by preventing its interaction with SpoIIQ. Three mother cell proteins (SpoIID, SpoIIM, and SpoIIP) are required for septal thinning, when peptidoglycan is degraded from the septal midpoint to the edges (Fig. 1, step i), and for membrane migration (Abanes-De Mello et al. 2002). In the absence of SpoIID, SpoIIM, or SpoIIP, septal thinning is incomplete, and the growing forespore pushes through the septum, and bulges into the mother cell cytoplasm (Fig. 6K). Septal thinning is also defective in spoIIB spoVG strains (Margolis et al. 1993; Perez et al. 2000). Electron micrographs show that in septal thinning mutants, the septum but not the bulge contains peptidoglycan (Illing and Errington 1991; Margolis et al. 1993; Smith et al. 1993; Frandsen and Stragier 1995; Perez et al. 2000).
Figure 6.
Effect of septal thinning mutants on SpoIIIAH-Flag localization. Immunolocalization of SpoIIIAH-Flag in samples harvested early (t2.5 [A,C]; t2 [E,G,I]) or late in sporulation (t3.5 [B,D]; t3 [F,H,J]). PY79 strains sporulated somewhat more rapidly, with similar engulfment seen 30 min later in MB24-derivatives. In engulfment mutants with forespore bulges, the double arrowheads indicate the bulge, and the arrowhead indicates the forespore. (A,B) The wild-type strain KP875. Bar, 2 μm. (C,D) The spoIIB spoVG strain KP878. (E,F) The spoIID strain KP861. (G,H) The spoIIP strain KP862. The dotted line indicates a sporangium that was pasted into the field. (I,J) The spoIID spoIIP double-mutant strain KP863 fails to form bulges, and SpoIIIAH remained in the mother cell cytoplasmic membrane late in sporulation. (K) Localization of SpoIIIAH (green circles) in the engulfment mutants before (left) and after (right) bulge formation.
Most spoIIB spoVG double-mutant sporangia with no bulge showed little or no localization of SpoIIIAH-Flag (Fig. 6C, arrow; scored in Supplementary Table 2). Once the bulge had formed (Fig. 6C,D [double arrowheads], K), SpoIIIAH-Flag localized to the bulge (Fig. 6C,D; Supplementary Table 2). The SpoIIIAH assemblage in the bulge looked similar to that seen in wild type, as it was spherical and had multiple foci (Fig. 6D, double arrowhead); however, the sphere was restricted to the bulge, rather than enclosing the forespore. Thus, SpoIIIAH localization spatially correlated with the absence of peptidoglycan, as in the spoIIB spoVG mutant, it localized specifically to bulges, which have little peptidoglycan.
The spoIID- and spoIIP-null mutants also had clear localization defects early in sporulation, with increased immunostaining in the mother cell cytoplasmic membrane (Fig. 6E,G, arrows; Supplementary Table 2). However, later in sporulation, this nonlocalized material decreased, and SpoIIIAH localized mostly to the bulge (Fig. 6F-H, double arrowheads; Supplementary Table 2). The spoIID spoIIP double mutant eliminates bulge formation, perhaps by decreasing residual septal thinning (Pogliano et al. 1999). It also showed a more persistent SpoIIIAH localization defect, with immunostaining remaining in the mother cell cytoplasmic membrane even late in sporulation (Fig. 6I,J, arrow). Together with the preferential localization of SpoIIIAH to the bulge, these results suggest that a peptidoglycan-free region of the septum facilitates the assembly of the SpoIIQ-SpoIIIAH complex.
Discussion
Most bacterial cells contain one membrane system, the cytoplasmic membrane, with localized membrane proteins being restricted to a particular region of the membrane, such as the cell pole or division site. Within such a cell, diffusion of these proteins away from their appropriate cellular location must be prevented by either a local reduction in membrane fluidity or interaction with another macromolecule that tethers the protein to the appropriate location. The cell wall provides an attractive candidate for tethering cell surface proteins, because it is stationary and encodes positional information. Indeed, the cell-wall-binding domain of Listeria monocytogenes murein hydrolase Ply118 mediates its localization to the septum and cell poles (Loessner et al. 2002). Here we describe a second simple mechanism to restrict membrane proteins to their appropriate subcellular address: an interaction between extracellular domains of proteins in adjacent daughter cells, in this case the mother cell protein SpoIIIAH and the forespore protein SpoIIQ (Fig. 7A-D). This interaction localizes SpoIIIAH to the sporulation septum, and without SpoIIQ, SpoIIIAH is uniformly distributed throughout the mother cell membrane. Curiously, although mother cell gene expression is essential for SpoIIQ localization (Rubio and Pogliano 2004), the absence of SpoIIIAH only slightly affects SpoIIQ tethering, indicating that another mother cell protein must be the primary tether for SpoIIQ. This interaction between proteins in adjacent cells allows each protein to remain localized to the septum during engulfment. We also propose that such interactions could mediate localization to division sites in growing cells (Fig. 7E), since when SpoIIQ and SpoIIIAH are made in the same cell, their interaction localizes SpoIIQ to invaginating septa (Fig. 4K). We propose that the binding sites of these proteins are positioned such that interaction can occur only at site of membrane invaginations (Fig. 4L).
Figure 7.
Model for engulfment-dependent localization of SpoIIQ and SpoIIIAH. (A) SpoIIQ (blue circles) is initially localized to the septum, while SpoIIIAH (green circles) is initially randomly distributed throughout the mother cell cytoplasmic membrane. (B) Septal thinning allows SpoIIIAH to interact with SpoIIQ. (C) The two proteins colocalize in foci throughout engulfment. (D) After the final step of engulfment, membrane fusion, SpoIIQ is degraded to yield a cytoplasmic N-terminal product (blue shading), while SpoIIIAH remains in foci and σG is activated. (E) Interaction between extracellular domains could also localize proteins in symmetrically dividing cells, if the proteins only interact in opposing membranes (such as those formed during division) but not when adjacent to one another in the cytoplasmic membrane. (F) Enlarged view of the septum prior to septal thinning (as in the cell shown in A), showing the peptidoglycan (PG) between the forespore membrane (FM) and the mother cell membrane (MM). Interaction between SpoIIQ (“IIQ”, blue) and SpoIIIAH (“IIIAH”, green) is inhibited by septal peptidoglycan. (G) After septal thinning the proteins interact, mediating SpoIIIAH localization (as in B,C). (H) After membrane fusion (as in D), SpoIIQ is degraded by a protease (red pacman) (Rubio and Pogliano 2004) and σG is activated.
The initial membrane insertion of localized proteins could either occur at the site where the proteins will ultimately reside (direct insertion) or throughout the membrane (random insertion) with proteins later diffusing to the appropriate location. In the forespore, random membrane proteins, such as Escherichia coli MalF, are initially localized to the septum, later diffusing throughout the forespore membrane, consistent with the direct insertion of forespore proteins into the septum (Rubio and Pogliano 2004). In the mother cell, we and others have demonstrated that sporulation-specific membrane proteins can reach the septum from a presynthesized and nonlocalized pool, (Rudner et al. 2002; van Ooij and Losick 2003), as would be required if membrane protein insertion occurred randomly. However, because membrane protein localization in bacteria requires either a tether, a diffusion barrier, or degradation of nonlocalized proteins, and because nonlocalized membrane proteins freely diffuse, even a protein normally directly inserted should be capable of localization by diffusion from a nonlocalized pool. Distinguishing between directed and random insertion will ultimately require localizing the site at which individual proteins engage the membrane insertion apparatus.
SpoIIIAH and SpoIIQ are both required to activate the late forespore transcription factor, σG, which is synthesized by the early forespore transcription factor, σF but activated only after engulfment (for review, see Rudner and Losick 2001; Errington 2003). Once active, σG directs its own synthesis, resulting in a positive feedback loop that allows high levels of σG activity. Multiple levels of regulation are therefore required to keep σG activity in check during vegetative growth and sporulation. Expression of spoIIIG (which encodes σG) is likely subject to post-transcriptional control, since there are differences in the ability of transcriptional and translational spoIIIG-lacZ fusions to be expressed by σF in vivo and in vitro (Sun et al. 1991). Expression of spoIIIG also requires both mother cell gene expression and SpoIIQ (Partridge and Errington 1993; Sun et al. 2000), suggesting that signaling between the forespore and mother cell is required for σG synthesis. In contrast, spoIIIA mutants, and mutants that fail to complete engulfment, show almost normal levels of spoIIIG expression (Kellner et al. 1996; Sun et al. 2000), indicating that the SpoIIIA proteins are more specifically required for σG activation.
SpoIIIAH and SpoIIQ coassemble multiple foci and arcs that ultimately enclose the forespore in an icosahedron-like structure (Rubio and Pogliano 2004). Our data suggests that this complex is ideally suited to couple activation of late transcription factors to engulfment. First, assembly of the SpoIIIAH-SpoIIQ complex is inhibited by septal peptidoglycan (Fig. 7F-H), which is removed during the first step of engulfment (Abanes-De Mello et al. 2002). Second, given the interaction between the extracellular domains of SpoIIQ and SpoIIIAH, at least until SpoIIQ is degraded (Rubio and Pogliano 2004), the complex reaches from the mother cell cytoplasm across the septum to the forespore cytoplasm, and is thus well positioned to transmit signals from one cell to the other. It will be interesting to determine more precisely the structure of the SpoIIIAH-SpoIIQ complex, and if other proteins involved in cell-cell communication localize to this structure.
Materials and methods
Bacterial strains, genetic manipulations, and growth conditions
B. subtilis strains (Table 2) were constructed by transformation (Dubnau and Davidoff-Abelson 1971) from PY79 (Youngman et al. 1984) and MB24 (Kellner et al. 1996). KP867 was constructed by transforming KP856 with KP845 DNA, selecting spectinomycin resistance (100 μg/mL); Spo+ transformants also acquired amyE::PspoIIQ-gfp-spoIIQΩcat. KP868 was constructed by transforming KP890 with KP889 DNA, selecting MlsR (1 μg/mL erythromycin and 25 μg/mL lincomycin); Spo+ recombinants also acquired spoIIIAH-FlagΩcm. Sporulation was induced by resuspension (Sterlini and Mandelstam 1969) or nutrient exhaustion (Schaeffer et al. 1965). Plasmids were constructed by PCR (using primers described in Supplemental Material), propagated in E. coli DH5α, TG1, or KJ622 (pcnB), and sequenced.
Table 2.
Strains used in this study
| Strain | Genotype | Reference | |
|---|---|---|---|
| PY79 | Prototrophic | Youngman et al. 1984 | |
| KP856 | spoIIIAH-flagΩcm | This study | |
| KP857 | spoIIIAH-flagΩcm, ΔspoIIQ::spec | This study | |
| KP858 | spoIIIAH-flagΩcm, spoIIIAA::EZ::TNΩkan | This study | |
| KP861 | spoIIIAH-flagΩcm, spoIID::Tn917Ωmls | This study | |
| KP862 | spoIIIAH-flagΩcm, ΔspoIIP::tet | This study | |
| KP863 | spoIIIAH-flagΩcm, spoIID::Tn917Ωmls, ΔspoIIP::tet | This study | |
| KP864 | spoIIIAH-flagΩcm, ΔspoIIA::spec, amy E::PspoIIE-spoIIRΩneo | This study | |
| KP865 | spoIIQ-mycΩcm | Rubio and Pogliano 2004 | |
| KP866 | amyE::PspoIIQ-gfp-spoIIQΩcm | This study | |
| KP867 | spoIIIAH-flagΩcm, amy E::PspoIIQ-gfp-spoIIQΩcm, ΔspoIIQ::spec | This study | |
| KP868 | spoIIQ-mycΩcm, spoIIIAH-flagΩcm, thrC::cotD-lacZΩmls | This study | |
| KP845 | amyE::PspoIIQ-gfp-spoIIQΩcm, ΔspoIIQ::spec | Rubio and Pogliano 2004 | |
| KP854 | amyE::PspoIIQ-gfp-spoIIQΩcm, ΔspoIIQ::spec, spoIIGB::erm | Rubio and Pogliano 2004 | |
| KP872 | amyE::PspoIIQ-gfp-spoIIQΩcm, ΔspoIIQ::spec, ΔspoIIAG-AH::kan | This study | |
| KP873 | amyE::PspoIIQ-gfp-spoIIQΩcm, ΔspoIIQ::spec, ΔspoIIIJ-jag::cm::tet | This study | |
| KP874 | amyE::PspoIIQ-gfp-spoIIQΩcm, ΔspoIIQ::spec, ΔspoIIIAG-AH::Kan, ΔspoIII-jag::cm::tet | This study | |
| KP889 | thrC::cotD-lacZΩmls, spoIIIAH-flagΩcm | This study | |
| KP890 | ΔspoIIIAG-AH::kan, spoIIQ-mycΩcm | This study | |
| KP891 | spoIIIAA::EZ::TNΩkan | This study | |
| KP892 | ΔspoIIIJ-jag::cm | This study | |
| KP893 | amyE::PspoIIQ-gfp-spoIIQΩcm, ΔspoIIQ::spec, sspB-lacZ::cm::tet | This study | |
| KP894 | spoIIIAH-flagΩcm, spoIIIJ::EZ::TNΩkan | This study | |
| KP895 | amyE::PspoIIQ-ma1F12-gfpΩcm, spoIIIAH-flagΩcm, thrC::cotD-lacZΩmls | This study | |
| KP896 | ΔspoIIIAG-AH::kan | This study | |
| KP897 | amyE::PspoIID-gfp-spoIIQΩcm, ΔspoIIQ::spec | This study | |
| KP898 | amyE::PspoIID-gfp-spoIIQΩcm, ΔspoIIQ::spec, ΔspoIIIAA-AH::kan | This study | |
| MB24 | trpC2, metC3 | Kellner et al. 1996 | |
| KP875 | trpC2, metC3, spoIIAH-flagΩcm | This study | |
| KP878 | trpC2, phe, spoIIAH-flagΩcm, spoIIB131, spoVG | This study | |
| KP879 | trpC2, metC3, spoIIAH-flagΩcm, spoIID298 | This study | |
| KP881 | trpC2, metC3, spoIIAH-flagΩcm, spoIID298, ΔspoIIP::tet | This study | |
| KP884 | trpC2, metC3, ΔspoIIIAH, amyE::PspoIIQ-spoIIQΩcm | This study | |
| KP885 | trpC2, metC3, ΔspoIIIAH, amyE::PspoIIQ-gfp-spoIIQΩcm, ΔspoIIQ::spec | This study | |
| KP886 | trpC2, metC3, amyE::Pxyl-spoIIIAH-flagΩcm | This study | |
| Yeast two hybridization strains | |||
| 1 | pEG202 (lexA) pJG4-5 (spoIIQ-ab) | This study | |
| 2 | pEG202 (lexA) pJG4-5 (spoIIIAH-ab) | This study | |
| 3 | pEG202 (spoIIQ-lexA) pJG4-5 (ab) | This study | |
| 4 | pEG202 (spoIIIAH-lexA) pJG4-5 (ab) | This study | |
| 5 | pEG202 (spoIIIAH-lexA) pJG4-5 (spoIIQ-ab) | This study | |
| 6 | pEG202 (spoIIQ-lexA) pJG4-5 (spoIIIAH-ab) | This study | |
Microscopy
Live cells were stained with 4′,6′-diamidino-2-phenylindole (DAPI) (0.2 μg/mL; Molecular Probes) and Mitotracker Red (0.1 μg/mL; Molecular Probes) or FM 4-64 (5 μg/mL; Molecular Probes) (Sharp and Pogliano 1999), and images were collected with an Applied Precision Spectris optical sectioning microscope. Immunofluorescence microscopy (Perez et al. 2000) used 0.5 μg/mL mouse monoclonal anti-myc antibodies (Sigma), and rabbit polyclonal (0.2 μg/mL; Sigma) or mouse monoclonal (1.0 μg/mL; Sigma) anti-Flag antibodies.
spoIIIAG-H and spoIIIJ-jag deletions
PCR amplification used primers O1 and O2 (726-bp fragment upstream of spoIIIAG) and O3 and O4 (705-bp fragment downstream of spoIIIAH); the fragments were digested and cloned to 5′ and 3′ of the pXJ52 kan gene. The plasmid was transformed into PY79, integrated by double recombination, replacing spoIIIAG and spoIIIAH with kan, yielding KP896. PCR amplification using primers O5 and O6 (524-bp fragment upstream of spoIIIJ) and O7 and O8 (498-bp fragment downstream of jag); the fragments were cloned to 5′ and 3′ of the pXJ3 chloramphenicol cassette (cat). The plasmid was transformed into PY79, integrated by double recombination, replacing spoIIIJ and jag coding regions with cat, yielding KP892.
Isolation of EZ::TN(kan) insertions in spoIIIAA and spoIIIJ
For mutagenesis, a kanamycin drug cassette (kan) from pH1 (Trieu-Cout and Courvalin 1983) was cloned into pMOD-2 (Epicentre) and EZ::TN(kan) isolated by PCR. In vitro transposition was carried out, mixing equimolar amount of target DNA (KP893 chromosomal DNA) and EZ::TN(kan) using transposase and buffers (Epicentre). Products were purified (QIAGEN QiaexII), and single-stranded gaps repaired by T4 DNA polymerase and T4 DNA ligase (Pelicic et al. 2000). Mutated DNA was transformed into KP893, plating on DSM X-gal (60 μg/mL) kanamycin (10 μg/mL). White colonies were isolated and screened by microscopy to identify two mutants that blocked GFPSpoIIQ degradation. These were sequenced by random primed PCR (Jacobs et al. 2003). The insertions were between codon Arg 115 and Ile 116 of spoIIIAA, and between codon Gly 8 and Leu 9 of spoIIIJ.
Construction of spoIIIAH-Flag, ftsH-Flag, and Pxyl-spoIIIAH-Flag
SpoIIIAH and FtsH were Flag epitope tagged by amplifying the 3′-end of each gene (spoIIIAH-O9 and O10; ftsH-O21 and O22) to insert the Flag epitope before the stop codon of each gene. PCR products were cloned into pCR2.1 (Invitrogen) and the cat cassette from pMS38 (Zilhao et al. 2004) excised with BamHI and BglII and cloned into BamHI-digested pCR2.1. Transformation of the resulting plasmid into MB24 integrated the plasmids via a single recombination event at spoIIIAH or ftsH.
amyE::Pxyl-spoIIIAH-Flag was constructed by PCR amplification of spoIIIAH using primers O11 and O12 and the 3′-end of spoIIIAH cloned adjacent to the BamHI site on pCR2.1. The gene was excised using BamHI, and inserted into pX (Kim et al. 1996), placing the 5′-end of spoIIIAH adjacent to Pxyl. The plasmid was transformed into MB24 amyE::erm and integrated at the amyE gene by double recombination, replacing amyE::erm with amyE::Pxyl-spoIIIAH-Flag. SpoIIIAH-Flag synthesis was induced by adding 1% xylose to the growth medium.
Construction of mother-cell-expressed GFP-SpoIIQ
The spoIIQ coding region was amplified (using the primers described in Rubio and Pogliano 2004) and cloned into EagI-digested pMDS14 (Sharp and Pogliano 2002), and the resulting plasmid was transformed into the spoIIQ-null mutant strain KP575.
Functionality of GFP-SpoIIQ
Although GFP-SpoIIQ supports efficient engulfment and spore formation, and localizes identically to native SpoIIQ using immunofluorescence, expressing GFP-SpoIIQ together with wild-type SpoIIQ eliminated the ability of GFP-SpoIIQ to form foci (Supplementary Fig. 2). In a strain also deleted for spoIIIAH, 84% of sporangia showed faint GFP-SpoIIQ fluorescence ahead of the engulfing membranes (Supplementary Fig. 2; Supplementary Table 1). We suggest that GFP-SpoIIQ is slightly less functional than the wild-type protein, which out-competes GFPSpoIIQ for assembly into foci and interaction with the SpoIIQ tether.
Yeast two-hybrid analysis
Yeast strains were grown on YPD media. Yeast strain EGY48 (ura3 trp1 his3 3LexA-operator-leu2) was used with reporter pSH18-34 (ura3 8LexA-operator-lacZ). spoIIIAH was amplified from B. subtilis DNA with primers O15 and O16, and spoIIQ was similarly amplified with primers O17 and O18. These fragments were cloned into vectors pEG202 (his3 bla lexA) and pJG4-5 (trp1 bla ab) using EcoRI and XhoI sites. The plasmids were transformed using Yeastmaker transformation system (Clontech) and β-galactosidase assays performed (Reynolds et al. 1997).
Affinity chromatography
A plasmid encoding GST-SpoIIQ was constructed using primers O19 and O20 to amplify the region of spoIIQ encoding the extracellular domain, and cloning this fragment into the EcoRI and XhoI sites of the GST-encoding pGEX-4T-3 (Amersham). DH5α carrying this plasmid was grown to OD600 0.5-0.8 and induced with 1 mM IPTG. One-hundred milliliters of culture was concentrated 25-fold in PBS, passed through a French press, and centrifuged at 4000g for 10 min. The cleared lysate was loaded onto GSTrap columns (Amersham) and washed with PBS. B. subtilis strains expressing either SpoIIIAH-Flag of FtsH-Flag were sporulated by resuspension, and 100 mL of the cultures was concentrated 25-fold in PBS plus 1% Triton X-100 and 0.1 mg/mL lysozyme and passed through a French press twice. The lysates were centrifuged at 4000g for 10 min and loaded, in three column volumes, onto GST trap columns prebound with either GST or GST-SpoIIQ and incubated for 90 min. The columns were washed with PBS plus 1% Triton X-100 and eluted with 50 mM Tris and 10 mM glutathione (pH 8), and 0.5-mL fractions were collected. For immunoblot analysis in Figure 3, 6 μL of eluate was loaded on the gel (corresponding to 1.8-3.6 mL of culture). Extract corresponding to 166 μL of culture was loaded for immunoblot analysis of whole cell proteins, 16.6 μL for Coomassie blue staining.
Western blot analysis
Western blot samples were prepared (Pogliano et al. 1997), heated 10 min at 50°C, loaded on 12.5% SDS-polyacrylamide gel, transferred to PVDF (Perez et al. 2000), and probed with 0.4 μg/mL mouse monoclonal anti-GFP antibodies (Roche), using 1:1,500 horseradish peroxidase-labeled anti-mouse antibodies, with enhanced chemiluminescence (ECL) used for detection (Amersham).
Coimmunoprecipitation
The appropriate B. subtilis strains were induced to sporulate by resuspension and fractionated; 25 mL of culture was harvested at t2.5, and fractionated essentially as described (Rudner and Losick 2002), except the lysate was ultracentrifuged at 100,000g for 1 h, and the membrane pellet was resuspended in 1 mL of Buffer B (50 mM Tris at pH 8.0, 150 mM KCl, and 10% glycerol). A final concentration of 0.5% NP-40 was added, and the samples were incubated on ice for 1 h with gentle mixing every 10 min. The samples were centrifuged at 4°C at 16,100g for 15 min, and 0.5 mL of supernatant was added to 20 μL of anti-Flag M2 affinity gel (Sigma) pre-equilibrated in Buffer B. Samples were incubated overnight at 4°C with gentle rolling, and washed three times with 0.5 mL of Buffer B plus 0.5% NP-40 at 4°C. The bound proteins were eluted with 250 μL of 1× SDS-loading buffer (without reducing agent) at room temperature. βME was added to 0.355 M, and 25 μL of eluate was loaded for SDS-PAGE and Western blot analysis. Mouse anti-GFP monoclonal antibody (Roche) was used to detect GFP-SpoIIQ on Western blots.
Acknowledgments
We thank D. Kalman (Emory University) for help with microscopy, P. Stragier for strains, and S. Chiba (UCSD) for advice on the coimmunoprecipitation experiments. This work was supported by the National Science Foundation (NSF 0135955 to K.P.) and the National Institutes of Health (GM 054395 to C.P.M.). A.R. was supported by an NSF Postdoctoral Fellowship (DBI-0109229).
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1252704.
References
- Abanes-De Mello A., Sun, Y.L., Aung, S., and Pogliano, K. 2002. A cytoskeleton-like role for the bacterial cell wall during engulfment of the Bacillus subtilis forespore. Genes & Dev. 16: 3253-3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arigoni F., Pogliano, K., Webb, C.D., Stragier, P., and Losick, R. 1995. Localization of protein implicated in establishment of cell type to sites of asymmetric division [see comments]. Science 270: 637-640. [DOI] [PubMed] [Google Scholar]
- Autret S. and Errington, J. 2003. A role for division-site-selection protein MinD in regulation of internucleoid jumping of Soj (ParA) protein in Bacillus subtilis. Mol. Microbiol. 47: 159-169. [DOI] [PubMed] [Google Scholar]
- Carballido-Lopez R. and Errington, J. 2003a. The bacterial cytoskeleton: In vivo dynamics of the actin-like protein Mbl of Bacillus subtilis. Dev. Cell 4: 19-28. [DOI] [PubMed] [Google Scholar]
- ____. 2003b. A dynamic bacterial cytoskeleton. Trends Cell Biol. 13: 577-583. [DOI] [PubMed] [Google Scholar]
- Driks A., Roels, S., Beall, B., Moran Jr., C.P., and Losick, R. 1994. Subcellular localization of proteins involved in the assembly of the spore coat of Bacillus subtilis. Genes & Dev. 8: 234-244. [DOI] [PubMed] [Google Scholar]
- Dubnau D. and Davidoff-Abelson, R. 1971. Fate of transforming DNA following uptake by competent Bacillus subtilis. I. Formation and properties of the donor-recipient complex. J. Mol. Biol. 56: 209-221. [DOI] [PubMed] [Google Scholar]
- Errington J. 2003. Regulation of endospore formation in Bacillus subtilis. Nat. Rev. Microbiol. 1: 117-126. [DOI] [PubMed] [Google Scholar]
- Errington J., Daniel, R.A., and Scheffers, D.J. 2003. Cytokinesis in bacteria. Microbiol. Mol. Biol. Rev. 67: 52-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett P., Melnikov, A., and Youngman, P. 1998. The Bacillus SpoIIGA protein is targeted to sites of spore septum formation in a SpoIIE-independent manner. Mol. Microbiol. 28: 931-943. [DOI] [PubMed] [Google Scholar]
- Finley R.L. and Brent, R. 1994. Interaction mating reveals binary and ternary connections between Drosophila cell cycle regulators. Proc. Natl. Acad. Sci. 91: 12980-12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frandsen N. and Stragier, P. 1995. Identification and characterization of the Bacillus subtilis spoIIP locus. J. Bacteriol. 177: 716-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K., Moller-Jensen, J., Ebersbach, G., Kruse, T., and Nordstrom, K. 2004. Bacterial mitotic machineries. Cell 116: 359-366. [DOI] [PubMed] [Google Scholar]
- Glaser P., Sharpe, M.E., Raether, B., Perego, M., Ohlsen, K., and Errington, J. 1997. Dynamic, mitotic-like behavior of a bacterial protein required for accurate chromosome partitioning. Genes & Dev. 11: 1160-1168. [DOI] [PubMed] [Google Scholar]
- Hofmann K. and Stoffel, W. 1993. TMbase—A database of membrane spanning proteins segments. Biol. Chem. HoppeSeyler 374: 166. [Google Scholar]
- Holtzendorff J., Hung, D., Brende, P., Reisenauer, A., Viollier, P.H., McAdams, H.H., and Shapiro, L. 2004. Oscillating global regulators control the genetic circuit driving a bacterial cell cycle. Science 304: 983-987. [DOI] [PubMed] [Google Scholar]
- Hu Z. and Lutkenhaus, J. 1999. Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Mol. Microbiol. 34: 82-90. [DOI] [PubMed] [Google Scholar]
- Illing N. and Errington, J. 1991. Genetic regulation of morphogenesis in Bacillus subtilis: Roles of σ E and σ F in prespore engulfment. J. Bacteriol. 173: 3159-3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs M.A., Alwood, A., Thaipisuttikul, I., Spencer, D., Haugen, E., Ernst, S., Will, O., Kaul, R., Raymond, C., Levy, R., et al. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. 100: 14339-14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs-Wagner C. 2004. Regulatory proteins with a sense of direction: Cell cycle signalling network in Caulobacter. Mol. Microbiol. 51: 7-13. [DOI] [PubMed] [Google Scholar]
- Kellner E.M., Decatur, A., and Moran Jr., C.P. 1996. Two-stage regulation of an anti-σ factor determines developmental fate during bacterial endospore formation. Mol. Microbiol. 21: 913-924. [DOI] [PubMed] [Google Scholar]
- Kim L., Mogk, A., and Schumann, W. 1996. A xylose-inducible Bacillus subtilis integration vector and its application. Gene 181: 71-76. [DOI] [PubMed] [Google Scholar]
- King N., Dreesen, O., Stragier, P., Pogliano, K., and Losick, R. 1999. Septation, dephosphorylation, and the activation of σF during sporulation in Bacillus subtilis. Genes & Dev. 13: 1156-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroos L. and Yu, Y.T. 2000. Regulation of σ factor activity during Bacillus subtilis development. Curr. Opin. Microbiol. 3: 553-560. [DOI] [PubMed] [Google Scholar]
- Lin D.C.H., Levin, P.A., and Grossman, A.D. 1997. Bipolar localization of a chromosome partition protein in Bacillus subtilis. Proc. Natl. Acad. Sci. 94: 4721-4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loessner M.J., Kramer, K., Ebel, F., and Scherer, S. 2002. C-terminal domains of Listeria monocytogenes bacteriophage murein hydrolases determine specific recognition and high-affinity binding to bacterial cell wall carbohydrates. Mol. Microbiol. 44: 335-349. [DOI] [PubMed] [Google Scholar]
- Londoño-Vallejo J.A., Frehel, C., and Stragier, P. 1997. SpoIIQ, a forespore-expressed gene required for engulfment in Bacillus subtilis. Mol. Microbiol. 24: 29-39. [DOI] [PubMed] [Google Scholar]
- Lybarger S.R. and Maddock, J.R. 2001. Polarity in action: Asymmetric protein localization in bacteria. J. Bacteriol. 183: 3261-3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis P.S., Driks, A., and Losick, R. 1993. Sporulation gene spoIIB from Bacillus subtilis. J. Bacteriol. 175: 528-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAdams H.H. and Shapiro, L. 2003. A bacterial cell-cycle regulatory network operating in time and space. Science 301: 1874-1877. [DOI] [PubMed] [Google Scholar]
- Partridge S.R. and Errington, J. 1993. The importance of morphological events and intercellular interactions in the regulation of prespore-specific gene expression during sporulation in Bacillus subtilis. Mol. Microbiol. 8: 945-955. [DOI] [PubMed] [Google Scholar]
- Pelicic V., Morelle, S., Lampe, D., and Nassif, X. 2000. Mutagenesis of Neisseria meningitidis by in vitro transposition of Himar1 mariner. J. Bacteriol. 182: 5391-5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez A.R., Abanes-De Mello, A., and Pogliano, K. 2000. SpoIIB localizes to active sites of septal biogenesis and spatially regulates septal thinning during engulfment in bacillus subtilis. J. Bacteriol. 182: 1096-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogliano K., Hofmeister, A.E., and Losick, R. 1997. Disappearance of the σ E transcription factor from the forespore and the SpoIIE phosphatase from the mother cell contributes to establishment of cell-specific gene expression during sporulation in Bacillus subtilis. J. Bacteriol. 179: 3331-3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogliano J., Osborne, N., Sharp, M.D., Abanes-De Mello, A., Perez, A., Sun, Y.L., and Pogliano, K. 1999. A vital stain for studying membrane dynamics in bacteria: A novel mechanism controlling septation during Bacillus subtilis sporulation. Mol. Microbiol. 31: 1149-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quardokus E.M. and Brun, Y.V. 2003. Cell cycle timing and developmental checkpoints in Caulobacter crescentus. Curr. Opin. Microbiol. 6: 541-549. [DOI] [PubMed] [Google Scholar]
- Raskin D.M. and de Boer, P.A. 1999a. MinDE-dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J. Bacteriol. 181: 6419-6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 1999b. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc. Natl. Acad. Sci. 96: 4971-4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnekov O., Alper, S., and Losick, R. 1996. Subcellular localization of proteins governing the proteolytic activation of a developmental transcription factor in Bacillus subtilis. Genes Cells 1: 529-542. [DOI] [PubMed] [Google Scholar]
- Reynolds A., Lundblad, V., Dorris, D., and Keaveney, M. 1997. Yeast vectors and assays for expression of cloned genes. In Current protocols in molecular biology, 39th Edition, pp. 13.6.1-13.6.6. Wiley, New York. [DOI] [PubMed]
- Rubio A. and Pogliano, K. 2004. Septal localization of forespore membrane proteins during engulfment in Bacillus subtilis. EMBO J. 23: 1636-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner D.Z. and Losick, R. 2001. Morphological coupling in development: Lessons from prokaryotes. Dev. Cell 1: 733-742. [DOI] [PubMed] [Google Scholar]
- ____. 2002. A sporulation membrane protein tethers the pro-σK processing enzyme to its inhibitor and dictates its subcellular localization. Genes & Dev. 16: 1007-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner D.Z., Pan, Q., and Losick, R.M. 2002. Evidence that subcellular localization of a bacterial membrane protein is achieved by diffusion and capture. Proc. Natl. Acad. Sci. 99: 8701-8706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K.R. and Shapiro, L. 2003. Temporal and spatial regulation in prokaryotic cell cycle progression and development. Annu. Rev. Biochem. 72: 367-394. [DOI] [PubMed] [Google Scholar]
- Schaeffer P., Millet, J., and Aubert, J. 1965. Catabolite repression of bacterial sporulation. Proc. Natl. Acad. Sci. 54: 704-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro L., McAdams, H.H., and Losick, R. 2002. Generating and exploiting polarity in bacteria. Science 298: 1942-1946. [DOI] [PubMed] [Google Scholar]
- Sharp M.D. and Pogliano, K. 1999. An in vivo membrane fusion assay implicates SpoIIIE in the final stages of engulfment during Bacillus subtilis sporulation. Proc. Natl. Acad. Sci. 96: 14553-14558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2002. Role of cell-specific SpoIIIE assembly in polarity of DNA transfer. Science 295: 137-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih Y.L., Le, T., and Rothfield, L. 2003. Division site selection in Escherichia coli involves dynamic redistribution of Min proteins within coiled structures that extend between the two cell poles. Proc. Natl. Acad. Sci. 100: 7865-7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerker J.M. and Laub, M.T. 2004. Cell-cycle progression and the generation of asymmetry in Caulobacter crescentus. Nat. Rev. Microbiol. 2: 325-337. [DOI] [PubMed] [Google Scholar]
- Smith K., Bayer, M.E., and Youngman, P. 1993. Physical and functional characterization of the Bacillus subtilis spoIIM gene. J. Bacteriol. 175: 3607-3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterlini J.M. and Mandelstam, J. 1969. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem. J. 113: 29-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stragier P. 2002. A gene odyssey: Exploring the genomes of endospore-forming bacteria. In Bacillus subtilis and its relatives: From genes to cells (eds. A.L. Sonenshein et al.), pp. 519-526. American Society for Microbiology, Washington, DC.
- Sun D., Cabrera-Martinez, R.M., and Setlow, P. 1991. Control of transcription of the Bacillus subtilis spoIIIG gene, which codes for the forespore specific transcription factor σG. J. Bacteriol. 173: 2977-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y.L., Sharp, M.D., and Pogliano, K. 2000. A dispensable role for forespore-specific gene expression in engulfment of the forespore during sporulation of Bacillus subtilis. J. Bacteriol. 182: 2919-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieu-Cout P. and Courvalin, P. 1983. Nucleotide sequence of the Streptococcus faecalis plasmid gene encoding the 3′5″-aminoglycoside phosphotransferase type III. Gene 23: 331-341. [DOI] [PubMed] [Google Scholar]
- Tusnady G.E. and Simon, I. 2001. The HMMTOP transmembrane topology prediction server. Bioinformatics 17: 849-850. [DOI] [PubMed] [Google Scholar]
- van Ooij C. and Losick, R. 2003. Subcellular localization of a small sporulation protein in Bacillus subtilis. J. Bacteriol. 185: 1391-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngman P., Perkins, J.B., and Losick, R. 1984. A novel method for the rapid cloning in Escherichia coli of Bacillus subtilis chromosomal DNA adjacent to Tn917 insertions. Mol. Gen. Genet. 195: 424-433. [DOI] [PubMed] [Google Scholar]
- Zilhao R., Serrano, M., Isticato, R., Ricca, E., Moran Jr., C.P., and Henriques, A.O. 2004. Interactions among CotB, CotG, and CotH during assembly of the Bacillus subtilis spore coat. J. Bacteriol. 186: 1110-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]