Abstract
The Drosophila Myb complex has roles in both activating and repressing developmentally regulated DNA replication. To further understand biochemically the functions of the Myb complex, we fractionated Drosophila embryo extracts relying upon affinity chromatography. We found that E2F2, DP, RBF1, RBF2, and the Drosophila homolog of LIN-52, a class B synthetic multivulva (synMuv) protein, copurify with the Myb complex components to form the Myb-MuvB complex. In addition, we found that the transcriptional repressor protein, lethal (3) malignant brain tumor protein, L(3)MBT, and the histone deacetylase, Rpd3, associated with the Myb-MuvB complex. Members of the Myb-MuvB complex were localized to promoters and were shown to corepress transcription of developmentally regulated genes. These and other data now link together the Myb and E2F2 complexes in higher-order assembly to specific chromosomal sites for the regulation of transcription.
Keywords: Drosophila, transcription, Myb, E2F, RB, synMuv
The Myb families of proteins are recognized as important regulators of nuclear activity, and model eukaryote organisms provide approaches to dissect the functions of such factors. The Drosophila melanogaster homolog of the vertebrate Myb proto-oncoprotein (Katzen et al. 1985) forms a stable complex with four additional proteins, Mip130, Mip120, Mip40, and Caf1/p55 (Beall et al. 2002). This five-subunit complex was originally identified as an activity present in Drosophila extracts that specifically recognizes two critical control elements (ACE-3 and ori-β) required for chorion gene DNA replication-mediated amplification in the follicle cells surrounding the developing oocyte (Orr-Weaver et al. 1989; Beall et al. 2002). Mutations in the Myb- and Mip120-binding sites in ACE-3-containing transgenes result in reduced amplification of such reporters. Moreover, somatic follicle cell clones devoid of Myb are defective for chorion gene amplification (Beall et al. 2002). Together, these data demonstrated that binding by Myb is a critical determinant for site-specific DNA replication at the locus.
Genetic studies have uncovered a more general role for the Myb complex during Drosophila development. Homozygous Drosophila myb temperature-sensitive and null mutations are lethal (Katzen et al. 1998; Manak et al. 2002), and Myb steady-state levels are dependent upon the integrity of the complex (Beall et al. 2004). Females harboring homozygous null mutations in the largest subunit of the Myb complex, mip130, are sterile and lay eggs with thin eggshells due to reduced chorion gene amplification (Beall et al. 2004). mip130 mutants incorporate bromodeoxyuridine (BrdU) throughout the follicle cell nucleus in stages normally undergoing site-specific amplification, suggesting that Mip130 acts as an inhibitor of replication in genomic regions normally not targeted for replication. Therefore, the Myb complex may serve dual functions in both the activation and repression of DNA replication that may depend upon the presence or absence of other factors at a given chromosomal location and/or developmental context. Surprisingly, myb mip130 double mutants are viable and display the same phenotypes as do mip130 mutants. Clearly, for normal eggshell development, both functions of the Myb complex are required. We have previously suggested that the myb mutant lethality in Drosophila results from the inability to counteract the repressive components of the Myb complex and/or associated factors in unknown developmental pathways (Beall et al. 2004). Thus, in the absence of both the repressive and activating functions of the Myb complex (as in myb mip130 double mutants), the animals have abnormalities but are viable.
To better understand biochemically the repressive activity of the Myb complex, we employed a purification scheme to reveal interacting proteins that might substantiate our previous genetic studies and provide a mechanistic insight into such negative regulation. Here we show that the Myb complex subunits are also components in a larger complex that represses expression of a number of developmentally regulated genes.
Results
Previously we discovered the Myb complex in Drosophila tissue culture cell nuclear extracts by using DNase protection assays with ACE3 and ori-β and fractions derived from high-density charged chromatography resins. We suspected that these methods might have disrupted interactions between the Myb complex and other less tightly associated factors. We thus employed different chromatography schemes and used immunoblot analysis to follow the members of the Myb complex to determine if the complex purified with any additional proteins. We arrived at two different purification schemes that resulted in the isolation of an interesting complex from Drosophila embryo nuclear extracts.
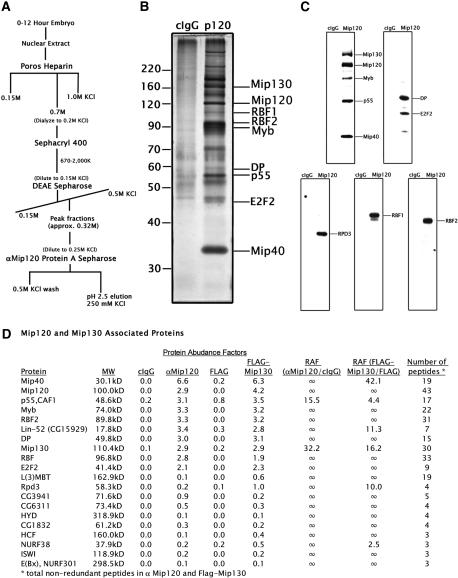
In the first approach, we employed a final affinity column using anti-Mip120 antibodies (Fig. 1A). As a control, a mock purification was performed by using nonspecific rabbit IgG. After eluting with low pH, many polypeptides in addition to those previously identified in the Myb complex were found associated specifically with Mip120 (Fig. 1B). The presence of the Myb complex members was confirmed by immunoblot analysis (Fig. 1C).
Figure 1.
The Drosophila Myb complex copurifies with E2F2/RBF. (A) The fractionation scheme used to purify Mip120-associated proteins from Drosophila embryo nuclear extract is shown. An anti-Mip120 affinity column was used as the final step in the purification. (B) SDS-PAGE and silver stain analysis of the anti-Mip120 (p120) or nonspecific immunoglobulin (cIgG) affinity column eluates. Proteins from the Mip120 affinity column eluate were identified by direct analysis of large protein complexes (DALPC) mass spectrometry (Link et al. 1999) and are indicated at the right. (C) Verification of the proteins identified in the anti-Mip120 or the nonspecific immunoglobulin (cIgG) eluate. Shown are immunoblots performed with the antibodies indicated on the right of each panel confirming that all members of the Myb and E2F2 complex were present in the supercomplex. (D) Data from duplicate control (cIgG and Flag) and experimental (Mip120 affinity and Flag-Mip130) samples were analyzed using mass spectroscopy. Briefly, a protein abundance factor (PAF) for each protein was expressed as the total number of nonredundant peptide hits normalized to the molecular weight of the cognate protein. An average PAF for each protein found in duplicate samples was then calculated. Average PAF values from experimental samples were normalized to that from the control samples to determine a relative abundance factor (RAF), shown as the ratio of the experimental/control samples.
By using a mass spectrometry approach termed direct analysis of large protein complexes (DALPC) (Link et al. 1999), we identified all of the Mip120-associated proteins (Fig. 1D), including the Drosophila transcriptional repressor protein E2F2, its dimerization partner DP, and the Drosophila retinoblastoma protein homologs RBF1 and RBF2. The presence of these proteins was confirmed by immunoblot analysis (Fig. 1C). A novel Drosophila protein (CG15929), which shares 34% identity with the Caenorhabditis elegans protein, LIN-52 (Thomas et al. 2003), was also identified by mass spectrometry and is also present in the gels with higher amounts of the eluate (Fig. 2; data not shown). The lethal (3) malignant brain tumor protein, L(3)MBT, was also identified from the mass spectrometry data. Loss-of-function alleles of L(3)MBT gene cause malignant growth of the adult optic neuroblasts in the larval brain in addition to imaginal disc overgrowth (Gateff et al. 1993). Additionally, the human L(3)MBT homolog represses transcription in transient transfection assays (Boccuni et al. 2003). The Drosophila homolog of the histone deacetylase, Rpd3, was present in the Mip120 affinity column fraction, and like L(3)MBT, perhaps in substoichiometric quantities compared with E2F2 and the RBFs. The presence of Rpd3 was not unexpected, as prior data showed that immunoprecipitates of the Myb complex contained an associated deacetylase activity (Fig. 5A, below).
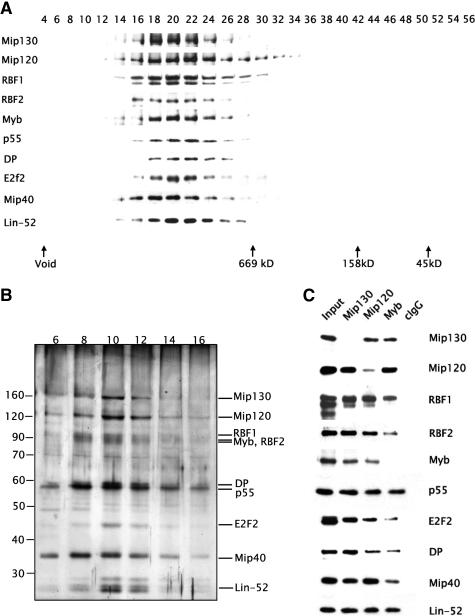
Figure 2.
The Mip130-associated proteins form a large stable complex. (A) Shown are immunoblot analyses of sephacryl S-400 gel filtration column fractions using antibodies against the proteins listed on the left. The eluate from the affinity-purified Flag-Mip130 column was used as the starting material for the column profile. (B) Shown is a silver-stained gradient SDS-PAGE gel of fractions derived from glycerol gradient centrifugation of the anti-Flag-Mip130 eluate. Approximately 6 pmol of Flag-Mip130 protein was loaded onto a 15%-40% gradient and the sample centrifuged at 150,000g for 12 h. The protein components of the peak fractions are indicated on the right. (C) Partially clarified embryo nuclear extracts were immunoprecipitated using the antibodies indicated on the top. The precipitates were washed with buffer containing 0.5M KCl, and the coimmunoprecipitated proteins were eluted with 0.4% sarcosyl. All immunoprecipitations were performed in the presence of 50 μg/mL ethidium bromide. Shown are immunoblot analyses of the immunoprecipitated material using antibodies directed against each of the super-complex members as indicated on the right.
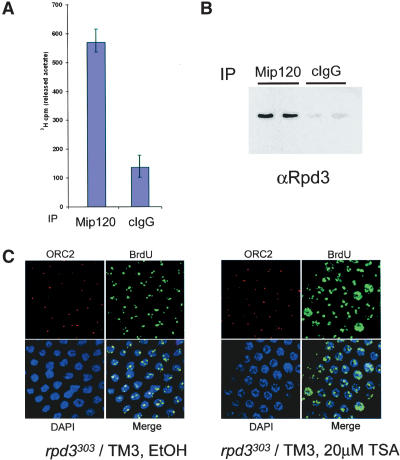
Figure 5.
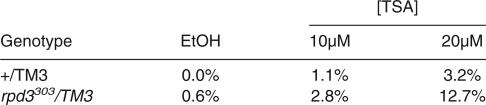
The Drosophila Myb complex interacts with the histone deacetylase, Rpd3. (A) Immunoprecipitates using antibodies against either Mip120 or nonspecific IgG (cIgG) and Drosophila 0-12-h embryo nuclear extract were incubated with 3H-acetylated core Drosophila histones. Released 3H-acetic acid was measured by scintillation counting, and the average and standard deviations from three independent experiments are indicated by the bar graph. (B) Immunoprecipitations as in A were analyzed for the presence of Rpd3 following immunoblot analysis. Specific enrichment for Rpd3 was detected in the anti-Mip120 IP versus the control IgG. (C) The histone deacetylase inhibitor, TSA, resulted in abnormal BrdU incorporation in follicle cell nuclei. Both wild-type (TM3) and Rpd3303 heterozygous mutant flies were grown in the presence of various concentrations of TSA as indicated. Shown are confocal images taken of stage-10 egg chambers from heterozygous Rpd3303 females treated either with ethanol alone (left) or with 20 μM TSA in ethanol (right) and stained for anti DmOrc2 (red), anti-BrdU (green), and DAPI (blue). The percentage of egg chambers displaying one or more follicle cells exhibiting overall genomic BrdU incorporation for each sample is indicated at the bottom. The total number of egg chambers scored are as follows: for wild type: EtOH alone, 149; 10 μM TSA, 267; and 20 μM TSA, 279; for Rpd3303: EtOH alone, 160; 10 μM TSA, 289; and 20 μM TSA, 310.
In a separate approach, we constructed a P element vector for expressing an N-terminal Flag-tagged Mip130 protein. The Flag-tagged protein fully rescued the mip130 female-sterile phenotype of homozygous mip130 null mutants (data not shown). Flag-Mip130-containing embryo extracts were fractionated as in Figure 1A through the heparin step, followed by an anti-Flag affinity column. All washes were performed in high salt followed by incubation with micrococcal nuclease and later ethidium bromide to ensure the removal of any contaminating DNA. After washing, the bound material was eluted with an excess of Flag peptide. The mass spectroscopy of the anti-Flag-Mip130 eluate revealed the same set of factors previously listed for the Mip120-associated proteins (Fig. 1D). Both gel filtration chromatography (Fig. 2A) and sedimentation fractionation through a glycerol-gradient (Fig. 2B) of the anti-Flag-Mip130 eluate confirmed that E2F2, DP, RBF1, RBF2, and dLin-52 are tightly associated with Myb and the Myb-interacting proteins. These data also show that the five previously identified members of the Myb complex interact with the E2F2, DP, RBF, and dLin-52 proteins to form the core of a larger complex. It is important to note that a very substantial amount (>50%) of the previously identified Myb complex can be accounted for in this new higher-order assembly, but that in different experiments between 10% and 50% of the Mips and Myb are found in subassemblies.
In addition to the putative LIN-52 homolog, we note that Mip130, Mip120, p55/Caf1, E2F2, DP, RBF, L(3)MBT, and Rpd3 are all homologs of the C. elegans synthetic multivulva (synMuv) class B genes (Table 1). In C. elegans the synMuv genes ensure the proper anatomy of the organism by repressing the vulval developmental pathway. Loss-of-function mutations in these genes thus result in a multivulva (Muv) phenotype as a result of the ectopic expression of those genes required for the development of a vulva. The C. elegans synMuv genes form three classes, A, B, and C, that function most likely through repression of transcription in hypodermal blast cells (Ferguson and Horvitz 1989; Hsieh et al. 1999; Ceol and Horvitz 2004). From the synthetic nature of the genetic penetrance, one infers that either one of two overlapping or redundant mechanisms are sufficient for repression. Animals with a class A and a class B mutation have a Muv phenotype, while animals having one or more mutations of the same class have a wild-type vulval phenotype. Thus, our data suggest that many of the class B synMuv genes required for proper developmental regulation of the nematode vulva execute functions through a single complex related to the one characterized in this study. Because of this likely conservation and the presence of the Drosophila Myb protein, we have named this purified unit the Myb-MuvB complex.
Table 1.
Comparison of Myb-MuvB complex subunits and associated proteins to C. elegans homologs
| Drosophila | C. elegans | Homology | synMuv phenotype (class B) |
|---|---|---|---|
| 1 Mip 130 | LIN-9 | 48% | yes |
| 2 Mip120 | LIN-54 | 57% | yes |
| 3 Myb | GEI-11 | 48% | ? |
| 4 p55/Caf1 | LIN-53 | 85% | yes |
| 5 Mip40 | — | — | — |
| 6 E2F2 | EFL-1 | 57% | yes |
| 7 DP | DPL-1 | 67% | yes |
| 8 RBF1 | LIN-35 | 40% | yes |
| 9 RBF2 | LIN-35 | 43% | yes |
| 10 dLin52 | LIN-52 | 64% | yes |
| 11 Rpd3 | HDA-1 | 79% | yes |
| 12 L(3)MBT | LIN-61 | 40% | yes |
BLAST analyses of the Drosophila Mby-MuvB complex sub-units were conducted to identify the C. elegans homologs. The genes as identified in the NCBI database are given. The core Myb-MuvB complex (labeled 1-10) and the associated proteins (labeled 11 and 12) have homology to synMuv C. elegans class B proteins. No apparent Mip40 homolog exists in C. elegans. The putative C. elegans Myb homolog, gei-11, has not been identified as having a synMuv phenotype. The references for the C. elegans genes are: lin-54 (Owen et al. 2003; C. Ceol and H.R. Horvitz, pers. comm.), lin-53 (Lu and Horvitz 1998), lin-35 (Lu and Horvitz 1998), efl-1 (Ceol and Horvitz 2001; Page et al. 2001), dpl-1 (Ceol and Horvitz 2001; Page et al. 2001), lin-9 (Beitel et al. 2000), gei-11 (Tsuboi et al. 2002), hda-1 (Lu and Horvitz 1998), lin-61 (M. Harrison, X. Lu, and H.R. Horvitz, pers. comm.), lin-52 (Thomas et al. 2003).
To confirm the presence of the Myb-MuvB complex in unfractionated extracts, we performed immunoprecipitations with antisera specific to several members of the Myb complex. By using embryo extracts, immunoprecipitations with antisera specific to Myb, Mip130, or Mip120 resulted in the coimmunoprecipitation of E2F2, DP, RBF1, RBF2, and Lin-52 (Fig. 2C). All immunoprecipitations were performed in the presence of ethidium bromide in order to disrupt protein-DNA interactions.
Genetic studies of the gene-amplification process in Drosophila follicle cells support our biochemical studies linking the Myb and E2F2 complexes: Females containing mutant alleles of e2f2, dp, and rbf1 lay thinly shelled eggs and exhibit reduced fertility (Royzman et al. 1999; Bosco et al. 2001; Cayirlioglu et al. 2001). Moreover, as with mip130 mutants, e2f2 null or female sterile rbf1 mutant females exhibit delocalization of the origin recognition complex (ORC) and promiscuous genomic BrdU incorporation (Bosco et al. 2001; Cayirlioglu et al. 2001; Beall et al. 2004). In recent work, we suggested that the phenotypes of the mip130 and myb mutants could be understood if the complex was involved in chromatin-associated pathways that might affect both transcription and DNA replication. Therefore, we sought to provide direct evidence that the Myb-MuvB complex described above functions in transcriptional repression.
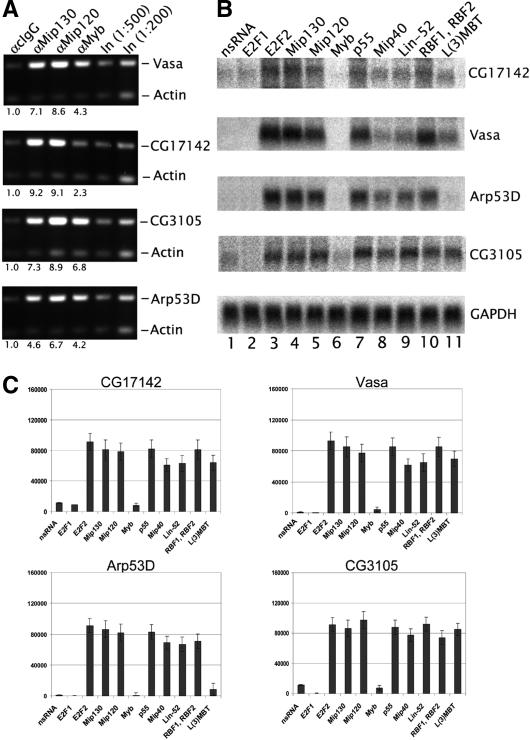
The Drosophila E2F2/RBF proteins repress transcription of both cell cycle and developmentally regulated genes. Microarray data from RNAi-treated tissue culture cells have established a list of some 47 genes that are part of the E2F2 repressive network as present in a cell culture system (Dimova et al. 2003). Chromatin immunoprecipitation experiments also indicate a direct role for E2F2 and the RBFs at a few key promoters (Dimova et al. 2003). Thus, we chose four specific sites where E2F2, RBF1, and RBF2 were localized and had a regulatory role in repression, to ask if the E2F2 repression might function through the Myb-MuvB complex. We performed chromatin immunoprecipitation using antibodies against Mip130, Mip120, or Myb and did find specific enrichment for promoter-derived fragments from the Vasa, CG17142, CG3105, and Arp53D genes (Fig. 3A). Moreover, we found that elimination of Mip130, Mip120, p55/Caf1, Mip40, dLin52, or L(3)MBT using RNAi in Drosophila tissue culture cells resulted in dramatically increased RNA levels of each of these genes as measured by Northern blot analysis (Fig. 3B). Both RBF1 and RBF2 have previously been shown to aid E2F2 in repression of transcription (Dimova et al. 2003). Because of the apparent functional redundancy among the RBF proteins in tissue culture cells, we used RNAi against both RBF1 and RBF2 for our Northern blot analysis.
Figure 3.
The Myb-MuvB complex is involved in transcriptional corepression. (A) Chromatin immunoprecipitation (ChIP) was performed using the antibodies indicated at the top, and the isolated DNA was analyzed using PCR for the presence of promoter fragments derived from the E2F2 regulated genes indicated on the right. Shown is an ethidium bromide-stained gel of PCR reactions performed with both gene-specific and actin primers simultaneously. The ratio of intensities of the particular gene promoter to the actin PCR signal in the ChIP and input DNA lanes were used to calculate the relative fold enrichment, as indicated below each lane. (B) RNAi was used to specifically eliminate members of the Myb-MuvB complexes as indicated on the top. Total RNA isolated from the RNAi-treated samples was analyzed for the abundance of E2F2 regulated transcripts using probes specific for the genes indicated on the right. nsRNA indicates RNAi performed using a double-stranded RNA against a region of the plasmid, pBSKS(+). GADPH was used as a loading control. (C) Signals derived from three independent Northern blots for the genes indicated were quantitated using PhosphorImager analysis and compared to the signal obtained for GADPH. Shown are histograms and standard deviations for the average normalized signal.
Previously we showed that RNAi against each individual subunit of the Myb complex member subunit effectively eliminated the targeted protein without affecting the nontargeted subunit mRNAs (Beall et al. 2004). Quantitation of the RNA levels (Fig. 3C) showed that de-repression occurred to the same extent whether E2F2, RBF, Mip130, Mip120, or p55/Caf1 was lost. RNAi directed at Mip40, Lin-52, and L(3)MBT also exhibited loss of repression, though somewhat less dramatic than for what was measured with loss of other complex components, and at one promoter (Arp53D), L(3)MBT appeared to have little effect on repression. In some cases, targeting of proteins simultaneously had a small additive effect, but we speculate that repression of transcription requires the fully intact complex (Fig. 4). Despite the presence of the Myb protein at the promoter loci for these genes, when Myb was targeted, there was no de-repression of any of the genes examined (Fig. 3B), suggesting that Myb is not required for the observed transcriptional repression. We note that RNAi ablation of Myb mRNA leads to a loss of >99% of the pool of Myb, and therefore after four cell doublings, the actual number of doublings achieved during the RNAi procedure, essentially no Myb protein should be bound to the DNA of daughter cells.
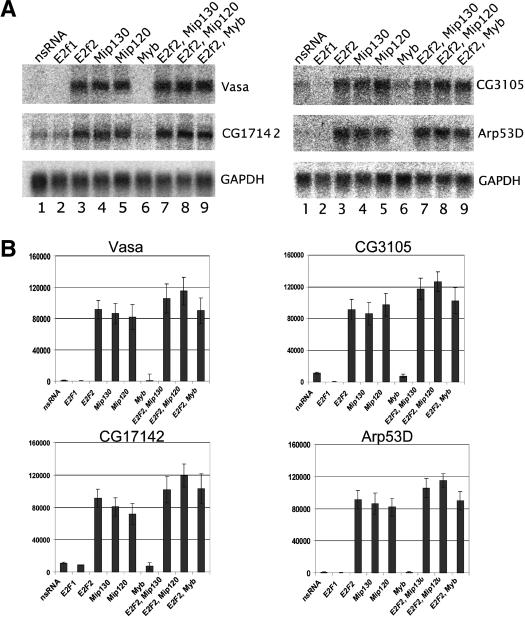
Figure 4.
Simultaneous double RNAi directed at Myb-MuvB complex members show some additive repressive effects. (A) RNAi directed at E2F2 and Mip130 or E2F2 and Mip120 displayed a small additive effect of derepression on the four genes we tested as mentioned in Figure 3. (B) Signals derived from five independent Northern blots for the genes indicated were quantitated using PhosphorImager analysis and compared to the signal obtained for GADPH. Shown are histograms and standard deviations for the average normalized signal.
The presence of the Myb protein at these repressed promoters suggests that Myb is a “silent” member of the repressive Myb-MuvB complex. We suspect that Myb is latent at these promoters, poised to participate as a transcriptional activator in specialized tissues where appropriate developmental signals may lead to activation. Evidence does exist to support Drosophila Myb's proposed role as a transcriptional activator (Hou et al. 1997; Okada et al. 2002). These ideas parallel what was suggested for the control of DNA replication in follicle cells: Specifically at the ACE-3 locus, Myb is thought to be maintained as a silent repressive partner that can later serve to switch the repressive complex to an activated complex for gene amplification (Beall et al. 2004). As we have shown here, Mip130 does contribute to the repressive activity of the Myb-MuvB complex and supports the idea that suppression of the lethality of myb mutants by mutations in mip130 (Beall et al. 2004) may involve this switch from repression to activation. Therefore, it is likely that the lethality of myb mutant animals is due to the unchecked transcriptional repressive activity of Mip130, a situation where the switch from repression to activation cannot occur. The gene expression profile in the absence of both activator and repressor activities (myb, mip130 double mutant) is probably more compatible with viability than is the situation when only the activator (myb mutant) is absent.
We posit that Rpd3 is recruited, perhaps transiently, to repress promoters by the Myb-MuvB complex in some cell-type-specific contexts. In mammalian systems, HDAC1, the Rpd3 homolog, is recruited to Rb-repressed promoters through a C-terminal LXCXE motif (Brehm et al. 1998). We note that the Drosophila and C. elegans Rpd3 homologs do not contain the LXCXE motif required for Rb binding (Brehm et al. 1998; Chan et al. 2001), suggesting that targeting of Rpd3 may occur through members of the Myb complex. When grown in the presence of histone deacetylase inhibitor, trichostatin A (TSA), wild-type Drosophila females displayed the same phenotypes observed with loss-of-function alleles of mip130, e2f2, dp, and rbf1: BrdU incorporation into follicle cell nuclei showed inappropriate genome-wide DNA replication. The penetrance of the inhibitor was, however, not as great as that observed with mutant animals. The effect of TSA was markedly exacerbated in rpd3 heterozygous mutant flies (Fig. 5C), suggesting that Rpd3 deacetylase function is necessary for proper replication in egg chamber follicle cell nuclei. Similar results were obtained in a recent study where rpd3 null follicle cell patches exhibited increased genome-wide acetylation and BrdU incorporation (Aggarwal and Calvi 2004). It is germane to our study to point out that the complex we have identified is also associated with the components of the NURF complex [Caf1/p55, ISWI, E(Bx), and NURF38] (Fig. 1D). Thus, some of the functions of the ensemble are likely directed toward chromatin remodeling and modification (Mizuguchi et al. 1997; Wysocka et al. 2003). These chromatin remodeling activities may be important for the initiation of repression in certain tissues, and accessing the roles of many of the Myb-MuvB complex proteins in vivo will require further extensive analysis.
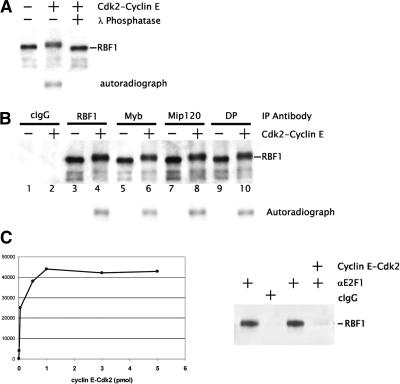
The genes we analyzed that are repressed by the Myb-MuvB complex are not required for normal cell cycle progression, and repression is stable through cell divisions in cell culture (Dimova et al. 2003). Moreover, both RBF1 and RBF2 are required for the repression at these sites (Du et al. 1996). RBF1 is a known target for Cyclin E-Cdk activity (Du et al. 1996; Frolov et al. 2003), and phosphorylation of human Rb by Cyclin E-Cdk causes dissociation from E2F1:DP and subsequent activation of transcription (Knudsen and Wang 1997; Harbour et al. 1999). We were therefore curious to determine if the RBF1 present in the Myb-MuvB complex would dissociate from the complex upon cyclin-Cdk phosphorylation. We found that treatment of the purified complex with Drosophila Cyclin E-Cdk2 in vitro yielded a hyperphosphorylated RBF1 (Fig. 6A), and this activity could be titrated to a plateau (Fig. 6C). However, the hyperphosphorylated RBF1 remained associated with the Myb-MuvB complex (Fig. 6B). As a control, we found that phosphorylation by Cyclin E-Cdk2 of immunoprecipitated E2F1/RBF1 from embryo extract resulted in the dissociation of RBF1 from E2F1 (Fig. 6D). That RBF1 in the Myb-MuvB complex did not dissociate upon phosphorylation suggests that the transcriptional repression mediated by RBF1 may be refractory to cell cycle cues mediated by the cyclin-Cdks. Previous data hinted that some E2F2/RBF1 complexes appear to be stable and persist in S phase, when in contrast RBF1 seems to be released from E2F1 bound to cell cycle-regulated promoters (Dimova et al. 2003). Our data show that activation of G1/S Cdks is likely not sufficient to disrupt all E2F2/RBF repressor complexes, suggesting that additional factors dictate repressor complex activity. Further, a switch to activation may involve an allosteric change in the Myb-MuvB complex, leading to the association of a coactivator rather than the conventional Rb release mechanism for activation (Weintraub et al. 1992; Cam and Dynlacht 2003). Mammalian E2F4/p107 complexes are known to persist in S phase, suggesting that the mechanism for escaping S-phase-Cdk-mediated inactivation may be conserved (Moberg et al. 1996).
Figure 6.
Cdk phosphorylated RBF1 does not dissociate from the Myb-MuvB complex. (A) Flag-Mip130-purified proteins were incubated with purified recombinant Cyclin E-Cdk2. Immunoblot analysis using anti-RBF1 antibodies as indicated showed that the mobility of RBF1 was retarded in SDS-PAGE upon phosphorylation. Incubation with λ-phosphatase restored the normal mobility of the RBF1 protein. Incubation with 32P-autoradiographs indicated that RBF1 was phosphorylated in the presence of cyclin-Cdk. (B) Immunoprecipitations of Flag-Mip130 proteins were performed after mock (odd lanes) or Cyclin E-Cdk2 phosphorylation (even lanes). Shown is an anti-RBF1 immunoblot analysis performed after immunoprecipitation with the various antibodies indicated on the top. The presence of phosphorylated RBF1 is indicated by the autoradiograph. (C) A titration of the Cyclin E-Cdk2 levels was performed using a constant amount of purified Myb-MuvB complex. PhosphorImager analysis indicated that the RBF1 phosphorylation levels plateaued at the kinase concentration used in the experiments (3 pmol Cyclin E-Cdk2). (D) E2F1/RBF1 was immunoprecipitated from 0- to 12-h Drosophila embryo nuclear extract. The immunoprecipitated complex was washed and equilibrated with the kinase reaction buffer. Following immunoprecipitation using either anti-E2F1 or control nonspecific IgG, the pellet was washed and incubated for 30 min at 30°C with Cyclin E-Cdk2. Shown is an immunoblot analysis using anti-RBF1 antibodies in which Cylin E-Cdk2 treatment dirupts the interaction of E2F1 with RBF1.
The experiments presented above raise interesting questions about how the RBF proteins are anchored to the repressor complex. The structural and molecular biological basis for the binding of E2F proteins to Rb is well documented in mammalian systems (Nevins et al. 1997; Lee et al. 2002). All E2F homologs in humans and other metazoans except for hE2F6 and hE2F7 contain an RB-interacting domain (Ogawa et al. 2002; Logan et al. 2004). It is noteworthy that the complex we have purified contains both RBF1 and RBF2 and that the primary structure of Drosophila E2F2 predicts that only one Rb moiety may be bound by E2F2. Therefore, it is plausible that some other member of the Myb-MuvB complex is associating with the second RBF subunit. We find it intriguing that Mip130 contains an LXCXE motif, which may bind either RBF1 or RBF2 perhaps with less avidity than E2F2. Alternatively a second RBF may interact directly with Myb as data from human proteins suggest. Human B-Myb, the Myb protein most similar to Drosophila Myb, functions to regulate the G1/S transition through a direct contact with the Rb related protein, p107, as B-Myb is required to overcome a G1 arrest mediated by p107 in human tumor cells (Joaquin et al. 2002). These data point to the possibility that a single complex may coordinate two different RBF proteins. Immunoprecipitations using antibodies specific for RBF1 or RBF2 showed that the majority of the purified Myb-MuvB complex contained one RBF protein, either RBF1 or RBF2. However, in these experiments a small amount of RBF1 coimmunoprecipitated with RBF2, and vice versa, suggesting that both RBF proteins may be present in the same complex (data not shown). Although this two RBF-containing complex is a small proportion of the total Myb-MuvB complex, it may be required for a specialized repressive function.
Discussion
The discovery that the Myb complex proteins are needed to repress developmentally regulated genes raises the possibility that some of the phenotypes observed in mip130 mutant animals may be due in part to the inappropriate expression of differentiation factors. The number of such target genes regulated by the repressive Myb-MuvB complex identified here is likely quite large. Multiple site-specific DNA-binding proteins contained together in one complex, such as Myb, Mip120, and E2F2:DP, increase the potential diversity for DNA sites that may be bound. Thus, at certain enhancers, the E2F2 site in combination with Mip120 may target the assembly, while at other sites the Myb DNA-binding activity may be important. Furthermore, although the majority of the Myb complex subunits in embryo extracts are present in the Myb-MuvB complex, it seems likely that the Myb and E2F2 proteins function independently at some chromosomal positions. We also posit that the Myb complex may be modified in such a way as to provide a signal for activation rather than repression. In work to be presented elsewhere, by using microarray analysis and genomic localization of the Myb complex, we have found that a family of transcripts is indeed dependent upon the Myb complex for expression. Thus a network of chromosomal domains may be independently regulated by these factors. In the context of such a network, it is intriguing that as an activator of DNA amplification, it appears as if Myb plays an active role in targeting the Mips and associated activities to the ACE-3 site. In contrast for the transcriptional repression studied in cell culture, Myb does not seem important for such targeting. Clearly understanding the DNA sequence context and associated factors in activation may shed some light on this difference.
A conserved complex for repression
We have called the Drosophila repressor characterized in this work the Myb-Muv B complex because of the striking resemblance of its protein composition to those encoded by the synMuv class B genes of C. elegans. The elegant genetic screens described by Horvitz and his colleagues (Horvitz and Sulston 1980; Ferguson and Horvitz 1989) have defined a regulatory pathway essential for vulval development that is entirely consistent with a model in which these synMuv proteins are individual members of a large complex. Our biochemical data together with the known expression patterns of the proteins associated with the Drosophila Myb-MuvB complex and phenotypes of the mutants for many of the factors of the complex argue for a general role for the repressor in many tissue types. Phenotypic differences between the putative nematode complex and the Drosophila counterpart may ultimately be ascribed to subunit composition or perhaps other more complex differences in the actual developmental programs between the two organisms. To highlight these in vivo differences, it is worthwhile to briefly review the SynMuv mutant phenotypes.
Wild-type C. elegans hermaphrodites contain a single vulva organ, while synMuv mutants may posses multiple vulva. In the wild-type organism, the activity of the synMuv genes antagonize the effects of the basal activity of the RTK/Ras pathway by repressing transcription of vulval genes (Lu and Horvitz 1998). The class B synMuv genes likely inhibit vulval induction by a conserved mechanism whereby the class B synMuv proteins form a repressive complex with the sequence-specific transcription factor EFL-1/E2F protein, and recruit corepressor proteins to inhibit the transcription of vulval specification genes via EFL-1/E2F-binding sites. As a result, those cells adopt the nonvulval fate. However, in the key vulval precursor cells, the antagonistic action of the synMuv genes is inactivated or can be overcome by the activated RTK/Ras pathway, thereby permitting downstream activation and transcription of keys genes for vulval fate. Some of the findings from our studies on the biochemical properties of the components of the Myb-MuvB complex may be relevant to a putative nematode complex. The Mip120 protein binds specifically to the ACE-3 and ori-β sequence (Beall et al. 2002) and is probably involved in sequence-specific interactions for the Myb-MuvB complex. It is therefore possible that the C. elegans Mip120 homolog, LIN-54 (C. Ceol and H.R. Horvitz, pers. comm.), is also a sequence-specific DNA-binding protein that helps direct the class B gene complex to specific promoters for repression of vulval genes.
L(3)MBT, a homolog of LIN-61 (M. Harrison and H.R. Horvitz, pers. comm.), is similar to the Drosophila polycomb group protein Sex Combs on Midleg (SCM), which is a member of the PRC1 complex. PRC1 is thought to primarily repress gene expression through blocking the nucleosome remodeling activity of SWI/SNF (Shao et al. 1999). As shown here, RNAi directed against L(3)MBT indicates that it is required for transcriptional repression at many of the sites coordinately repressed by E2F2 and the Myb-associated proteins. The L(3)MBT protein appears substoichiometric relative to core Myb-MuvB complex subunits. Like L(3)MBT in the Myb-MuvB complex, the SCM protein is present in substoichiometric quantities relative to the other subunits of the human and Drosophila hPRC-H and PRC1 complexes (Levine et al. 2002). Both L(3)MBT and LIN-61 contain multiple MBT repeats that are evolutionarily conserved domains found throughout metazoa. The X-ray crystal structure of the MBT repeats (Wang et al. 2003) provides some hints as to how both LIN-61 and the Drosophila L(3)MBT protein may function. The MBT repeat consists of a five-stranded β-barrel core domain that shares structural similarity to the Tudor and chromodomains. The Tudor domain interacts with methylated arginine residues (Sprangers et al. 2003), and chromodomains interact with methylated lysine residues of histone H3 (Bannister et al. 2001; Lachner et al. 2001; Nakayama et al. 2001; Cao et al. 2002). Consistent with the speculation that this domain is critical for function of the MBT family and that the proteins bind modified histones, several hypomorphic mutations in Drosophila SCM map to residues within in the putative ligand-binding pocket (Bornemann et al. 1998; Wang et al. 2003). The MBT domains in LIN-61 and L(3)MBT may maintain a repressed chromatin domain through interaction with histone tails methylated at specific lysine residues on neighboring nucleosomes, thus hindering the nucleosome mobility by chromatin remodeling factors. For maximum repression, genes regulated by the Myb-MuvB or the putative nematode complex may require additional mechanisms of repression such as histone modification and thus the association of the deacetylase Rpd3.
Like the Myb-MuvB complex the putative C elegans complex may also play a wide role in repression in different tissues. For example, several C. elegans class B synMuv genes, including the homologs of Mip130, E2F2, DP, and RBF, have been shown to function independently of synMuv A genes for regulation of the G1/S transition (Boxem and van den Heuvel 2002).
Activation and repression
The particular genes regulated by the Myb-MuvB complex are likely determined in a tissue-specific and cell-type-specific manner. In Drosophila tissue culture cells, E2F2 appears to function primarily for repression of developmentally regulated genes (Dimova et al. 2003), while E2F1/RBF1 complexes are involved in regulating genes involved in cell cycle progression. However, microarray studies performed in e2f2 and rbf1 mutant follicle cells indicate that both E2F2 and RBF1 are involved in the repression of several S-phase genes, including CDT1 and the ORC and MCM complex subunits (Cayirlioglu et al. 2003). Therefore, it is likely that the set of genes regulated by the Myb-MuvB complex may change depending on the developmental context.
In the embryo extracts that we have fractionated, the repressive form of the Myb complex seems to predominate. However, other much less abundant complexes between the previously identified Myb complex and activators should also be found. Certainly at ACE3, E2F1 and the Myb complex cooperate for amplification and proper ORC localization (Bosco et al. 2001; Beall et al. 2002), and we have proposed that a switch between E2F2 and E2F1 at ACE3 may be epistatic to activation (Beall et al. 2004). Other RNAi studies using Drosophila cell lines (Dimova et al. 2003; Frolov et al. 2003) indicate that E2F1 and E2F2 primarily occupy and regulate the expression of a non-overlapping set of genes, and the work presented here implies that this non-overlapping control may be dictated by other proteins associated with the well-studied E2F proteins. As drawn in Figure 7, the Myb complex might assemble with either activators or repressors of the E2F family to regulate either transcription or DNA replication in response to appropriate developmental cues. In future work it will be important to understand how in a given cell type, the cis-acting DNA sites and chromosomal context determine a region for either repression or activation.
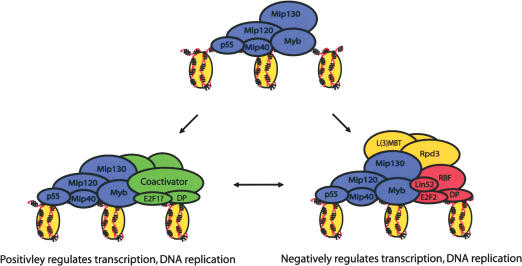
Figure 7.
A model for Drosophila Myb complex activity. The Myb complex may serve dual functions in both the activation and repression of transcription and DNA replication that may depend upon the presence or absence of other factors at a given chromosomal location and/or developmental context. The conversion between activator and repressor functions may alter the subunit composition of the Myb complex, favoring either coactivator or corepressor interactions. Drosophila E2F1 may preferentially interact with the Myb complex to promote activation of both transcription and replication. Ultimately, the complex acts through recruitment of chromatin modifying enzymes, such as the histone deacetylase, Rpd3, to alter local chromatin architecture in order to influence both origin usage and promoter activity.
Materials and methods
Purification of the Myb-MuvB complex
Embryos (0-12 h) were harvested and stored at 4°C. Nuclear extracts were prepared as previously described (Kamakaka et al. 1991) with some modification. Embryos were homogenized by one pass through a Yamato LH-21 homogenizer at 1500 rpm in buffer (0.35 M sucrose, 15 mM HEPES at pH 7.7, 10 mM KCl, 5 mM MgCl2, 0.5 mM EGTA, 0.1 mM EDTA, 10% glycerol, 1 mM NaMBS, 1 mM Benzamidine, 1 mM PMSF, 10 mM 2-mercaptoethanol) at a concentration of 4 mL buffer/gram of embryo. The homogenate was filtered through a layer of Miracloth, and the filtrate was centrifuged in GSA bottles for 10 min at 10,000 rpm. The pellets were resuspended in lysis buffer (15 mM HEPES at pH 7.6, 0.1 M KCl, 3 mM MgCl2, 0.1 mM EDTA, 10% glycerol with protease inhibitor cocktail; Roche) at a concentration of 1 mL buffer/gram of embryos in Ti45 tubes. A 1:10 volume of a saturated ammonium sulfate solution (pH 7.9) was added, and the tubes were rotated for 30 min. Following centrifugation for 120 min at 35,000 rpm, the supernatant was precipitated by addition of solid ammonium sulfate to a final concentration of 30% saturation followed by centrifugation for 20 min at 10,000 rpm. The pellet was resuspended in 20 mM HEPES (pH 7.7), 0.13 M KCl, 0.5 mM EDTA, 0.01% NP-40, 10% glycerol, 0.5 mM PMSF, and 5 mM 2-mercaptoethanol, and loaded onto a Poros-Heparin Column. Proteins from the 0.7 M KCl step elution were dialyzed to 0.2 M KCl and loaded onto a Sephacryl S-400 column. Peak fractions for the Myb complex were pooled, diluted to 0.15 M KCl, and loaded onto DEAE Sepharose column. Peak fractions of the Myb complex were pooled and diluted to 0.25 M KCl. The pooled fractions were added to Protein A Sepharose beads with rabbit IgGs (Sigma) cross-linked followed by Protein A Sepharose beads with cross-linked anti-Mip120 antibodies. The fractions were recirculated over the beads for 4 h. The cIgG or Mip120 beads were washed extensively with 0.5M KCl buffer and later eluted with 50 mM glycine (pH 2.5) and 0.25 M KCl.
Mip130-Flag complexes were purified from mip1301-36/mip1301-36;Mip130-Flag/Mip130-Flag 0-12-h embryos. The embryos were processed as stated above. The 0.7 M fraction from the Poros-Heparin column was dialyzed in 20 mM HEPES (pH 7.7), 0.3 M KCl, 0.5 mM EDTA, 0.01% NP-40, 10% glycerol, 0.5 mM PMSF, and 5 mM 2-mercaptoethanol and rotated with M2 anti-Flag resin (Sigma). The beads were extensively washed with 0.5 M KCl buffer supplemented with 50 μg/mL ethidium bromide. The beads were washed with 0.3 M KCl buffer supplemented with 2 mM CaCl2. The beads were incubated with Micrococcal nuclease (Roche; 1 U/μL) for 1 h at 4°C. The beads were washed with 0.3 M KCl buffer and eluted with 0.4 mg/mL Flag peptide.
DALPC
Eluates from the affinity or control columns (aliqutos containing ∼20 μg total protein) were reduced by adding DTT to 5 mM and incubating for 15 min at 65°C. Reduced samples were then alkylated by adding iodoacetamide to 10 mM and incubating for 30 min at 30°C. After alkylation, the samples were adjusted to pH 8.0 using 1 M Tris (pH 8.0), and CaCl2 was added to 1 mM and acetonitrile to 10%. One microgram modified sequencing-grade Trypsin (Promega) was added, and the samples were digested overnight at 37°C. After the digest, any precipitate was removed by centrifugation at 20,000g for 5 m. DALPC was used to identify proteins copurifying with p120 and p130 as described previously (Sanders et al. 2002). Acquired tandem mass spectral data obtained on an LCQ Deca Plus (ThermoFinnigan) were searched against a Drosophila subset of the RefSeq database. Data processing of the SEQUEST output files into a list of proteins copurifying with p120 or p130 was performed as described previously (Link et al. 1999).
RNAi and Northern blot
Double-stranded RNA synthesis, RNA transfection, and total cellular RNA purification for Mip130, Mip120, Myb, E2F2, and Lin-52 were performed as previously described (Beall et al. 2004).
Chromatin immunoprecipitation
Drosophila KC tissue culture cells were grown to a density of 2 to 3 × 106 cells/mL. Cells were harvested, washed with PBS and 1 mM MgCl2, and subsequently resuspended in a hypotonic buffer (50 mM HEPES at pH 7.5, 10 mM KCl, 0.5 mM EDTA, 1 mM MgCl2, and 0.5 mM PMSF). Cells were broken by dounce homogenization, and nuclei were collected and resuspended in a lysis buffer (50 mM HEPES at pH 7.5, 10 mM KCl, 0.5 mM EDTA, 1 mM MgCl2, 0.4 mM PMSF, 1% Triton X-100). Nuclei were cross-linked with 1% formaldehyde for 10 min and sonicated.
Growth of Drosophila, antibody statining, and BrdU incorporation
The effects of TSA on BrdU incorporation in follicle cells. Wild-type (W1118) or Rpd3 null (Rpd3303) (Mottus et al. 2000) flies were grown on media containing TSA or ethanol. Adult flies were fed yeast paste with TSA or ethanol prior to ovary dissection. Ovaries were fixed and processed as described (Beall et al. 2002, 2004)
Cyclin E:Cdk2 kinase reaction and immunoprecipitation
Recombinant His-tagged Drosophila Cyclin E:Cdk2 was incubated with 300 ng of Flag-Mip130-purified proteins in buffer (20 mM HEPES at pH 7.7, 1 mM MgCl2, 0.1 mM EDTA, 2 mM ATP, 1 μCi 32Pγ-ATP; Amersham). Immunoprecipitations were performed as described previously (Beall et al. 2002), except for the wash buffer (20 mM HEPES at pH 7.7, 0.3 M KCl, 0.5 mM EDTA, 0.1% NP40, 0.4 mM PMSF). All kinase reactions were performed in 1 mM ATP with 3 pmol of recombinant cyclin E-Cdk2.
Gel filtration and coimmunoprecipitations
Purified Myb complex using M2 anti-Flag beads (Sigma) was further purified by Sephacryl S-400 HR (Amersham Pharmacia) gel filtration chromatography; 200 μL Myb complex was loaded onto a sephacryl S-400 HR column in 20 mM HEPES (pH 7.7), 0.5 mM EDTA, 10% glycerol, 0.01% NP40, 0.4 mM PMSF, 2 mM 2-mercaptoethanol, and 100 μg/mL Arg-insulin (Sigma). The column was calibrated with protein markers with high and low relative molecular mass (Amersham Pharmacia). Coimmunoprecipitations were performed as previously described (Beall et al. 2002).
Immunoprecipitation and HDAC assay
Rabbit polyclonal antibodies against Mip120 were affinity-purified from serum. For immunoprecipitation experiments, antibodies were performed as described using affinity-purified rabbit IgG (Sigma) as a control. Where indicated, 50 μg/mL ethidium bromide was included in the extracts before immunoprecipitation.
HDAC assays were performed as described by (Taunton et al. 1996) with minor changes. Purified Drosophila core histones were purified as described (Kamakaka et al. 1993). Purified core histones were acetylated in vitro using recombinant Drosophila MOF and the HAT1-p55 complex with 3H-acetyl CoA. Core histones were repurified on hydroxyapeptite resin (Bio-Rad) and dialyzed. 3H acetylated histones (10,000 dpm) were incubated with immunoprecipitated material for 45 min at 30°C. The reaction was quenched with 1 M HCl and 0.16 M acetic acid. Released 3H acetic acid was extracted with ethyl acetate and quantified by scintillation counting.
Acknowledgments
We thank Robert Duronio, Nick Dyson, Peter Harte, Giovanni Bosco, and Alex Brehm for antibodies, and Randy Mottus for the Rpd3303 strain. We also thank an anonymous reviewer for providing the name of the complex. Dirk Remus provided the recombinant Drosophila Cyclin E:Cdk2. E.L.B was supported by NIH training grant (CA 09041) to the Cancer Research Laboratory of the University of California at Berkeley. T.C.F. is supported by NIH training grant T32 AI49824. D.G. is supported by a fellowship from the Belgian American Educational Foundation (B.A.E.F.-75). A.J.L. is supported by NIH grants GM64779, HL68744, NS43952, ES11993, and CA098131. This work was supported by NIH grant CA 30490 to M.B.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1255204.
References
- Aggarwal B.D. and Calvi, B.R. 2004. Chromatin regulates origin activity in Drosophila follicle cells. Nature 430: 372-376. [DOI] [PubMed] [Google Scholar]
- Bannister A.J., Zegerman, P., Partridge, J.F., Miska, E.A., Thomas, J.O., Allshire, R.C., and Kouzarides, T. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120-124. [DOI] [PubMed] [Google Scholar]
- Beall E.L., Manak, J.R., Zhou, M. Bell, S., Lipsick, J.S., and Botchan, M.R. 2002. Role for a Drosophila Myb-containing protein complex in site-specific DNA replication. Nature 420: 833-837. [DOI] [PubMed] [Google Scholar]
- Beall E.L., Bell, M., Georlette, D., and Botchan, M.R. 2004. Dm-myb mutant lethality in Drosophila is dependent upon mip130: Positive and negative regulation of DNA replication. Genes & Dev. 18: 1667-1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beitel G.J., Lambie, E.J., and Horvitz, H.R. 2000. The C. elegans gene lin-9, which acts in an Rb-related pathway, is required for gonadal sheath cell development and encodes a novel protein. Gene 254: 253-263. [DOI] [PubMed] [Google Scholar]
- Boccuni P., MacGrogan, D., Scandura, J.M., and Nimer, S.D. 2003. The human L(3)MBT polycomb group protein is a transcriptional repressor and interacts physically and functionally with TEL (ETV6). J. Biol. Chem. 278: 15412-15420. [DOI] [PubMed] [Google Scholar]
- Bornemann D., Miller, E., and Simon, J. 1998. Expression and properties of wild-type and mutant forms of the Drosophila sex comb on midleg (SCM) repressor protein. Genetics 150: 675-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco G., Du, W., and Orr-Weaver, T.L. 2001. DNA replication control through interaction of E2F-RB and the origin recognition complex. Nat. Cell Biol. 3: 289-295. [DOI] [PubMed] [Google Scholar]
- Boxem M. and van den Heuvel, S. 2002. C. elegans class B synthetic multivulva genes act in G1 regulation. Curr. Biol. 12: 906-911. [DOI] [PubMed] [Google Scholar]
- Brehm A., Miska, E.A., McCance, D.J., Reid, J.L., Bannister, A.J., and Kouzarides, T. 1998. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature 391: 597-601. [DOI] [PubMed] [Google Scholar]
- Cam H. and Dynlacht, B.D. 2003. Emerging roles for E2F: Beyond the G1/S transition and DNA replication. Cancer Cell 3: 311-316. [DOI] [PubMed] [Google Scholar]
- Cao R., Wang, L., Wang, H., Xia, L., Erdjument-Bromage, H., Tempst, P., Jones, R.S., and Zhang, Y. 2002. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298: 1039-1043. [DOI] [PubMed] [Google Scholar]
- Cayirlioglu P., Bonnette, P.C., Dickson, M.R., and Duronio, R.J. 2001. Drosophila E2f2 promotes the conversion from genomic DNA replication to gene amplification in ovarian follicle cells. Development 128: 5085-5098. [DOI] [PubMed] [Google Scholar]
- Cayirlioglu P., Ward, W.O., Silver Key, S.C., and Duronio, R.J. 2003. Transcriptional repressor functions of Drosophila E2F1 and E2F2 cooperate to inhibit genomic DNA synthesis in ovarian follicle cells. Mol. Cell Biol. 23: 2123-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceol C.J. and Horvitz, H.R. 2001. dpl-1 DP and efl-1 E2F act with lin-35 Rb to antagonize Ras signaling in C. elegans vulval development. Mol. Cell 7: 461-473. [DOI] [PubMed] [Google Scholar]
- ____. 2004. A new class of C. elegans synMuv genes implicates a Tip60/NuA4-like HAT complex as a negative regulator of Ras signaling. Dev. Cell 6: 563-576. [DOI] [PubMed] [Google Scholar]
- Chan H.M., Smith, L., and La Thangue, N.B. 2001. Role of LXCXE motif-dependent interactions in the activity of the retinoblastoma protein. Oncogene 20: 6152-6163. [DOI] [PubMed] [Google Scholar]
- Dimova D.K., Stevaux, O., Frolov, M.V., and Dyson, N.J. 2003. Cell cycle-dependent and cell cycle-independent control of transcription by the Drosophila E2F/RB pathway. Genes & Dev. 17: 2308-2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W., Vidal, M., Xie, J.E., and Dyson, N. 1996. RBF, a novel RB-related gene that regulates E2F activity and interacts with cyclin E in Drosophila. Genes & Dev. 10: 1206-1218. [DOI] [PubMed] [Google Scholar]
- Ferguson E.L. and Horvitz, H.R. 1989. The multivulva phenotype of certain Caenorhabditis elegans mutants results from defects in two functionally redundant pathways. Genetics 123: 109-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolov M.V., Stevaux, O., Moon, N.S., Dimova, D., Kwon, E.J., Morris, E.J., and Dyson, N.J. 2003. G1 cyclin-dependent kinases are insufficient to reverse dE2F2-mediated repression. Genes & Dev. 17: 723-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gateff E., Loffler, T., and Wismar, J. 1993. A temperature-sensitive brain tumor suppressor mutation of Drosophila melanogaster: Developmental studies and molecular localization of the gene. Mech. Dev. 41: 15-31. [DOI] [PubMed] [Google Scholar]
- Harbour J.W., Luo, R.X., Dei Santi, A., Postigo, A.A., and Dean, D.C. 1999. Cdk phosphorylation triggers sequential intra-molecular interactions that progressively block Rb functions as cells move through G1. Cell 98: 859-869. [DOI] [PubMed] [Google Scholar]
- Horvitz H.R. and Sulston, J.E. 1980. Isolation and genetic characterization of cell-lineage mutants of the nematode Caenorhabditis elegans. Genetics 96: 435-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou D.X., Akimaru, H., and Ishii, S. 1997. Trans-activation by the Drosophila myb gene product requires a Drosophila homologue of CBP. FEBS Lett. 413: 60-64. [DOI] [PubMed] [Google Scholar]
- Hsieh J., Liu, J., Kostas, S.A., Chang, C., Sternberg, P.W., and Fire, A. 1999. The RING finger/B-box factor TAM-1 and a retinoblastoma-like protein LIN-35 modulate context-dependent gene silencing in Caenorhabditis elegans. Genes & Dev. 13: 2958-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joaquin M., Bessa, M., Saville, M.K., and Watson, R.J. 2002. B-Myb overcomes a p107-mediated cell proliferation block by interacting with an N-terminal domain of p107. Oncogene 21: 7923-7932. [DOI] [PubMed] [Google Scholar]
- Kamakaka R.T., Tyree, C.M., and Kadonaga, J.T. 1991. Accurate and efficient RNA polymerase II transcription with a soluble nuclear fraction derived from Drosophila embryos. Proc. Natl. Acad. Sci. 88: 1024-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamakaka R.T., Bulger, M., and Kadonaga, J.T. 1993. Potentiation of RNA polymerase II transcription by Gal4-VP16 during but not after DNA replication and chromatin assembly. Genes & Dev. 7: 1779-1795. [DOI] [PubMed] [Google Scholar]
- Katzen A.L., Kornberg, T.B., and Bishop, J.M. 1985. Isolation of the proto-oncogene c-myb from D. melanogaster. Cell 41: 449-456. [DOI] [PubMed] [Google Scholar]
- Katzen A.L., Jackson, J., Harmon, B.P., Fung, S.M., Ramsay, G., and Bishop, J.M. 1998. Drosophila myb is required for the G2/M transition and maintenance of diploidy. Genes & Dev. 12: 831-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen E.S. and Wang, J.Y. 1997. Dual mechanisms for the inhibition of E2F binding to RB by cyclin-dependent kinase-mediated RB phosphorylation. Mol. Cell. Biol. 17: 5771-5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M., O'Carroll, D., Rea, S., Mechtler, K., and Jenuwein, T. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410: 116-120. [DOI] [PubMed] [Google Scholar]
- Lee C., Chang, J.H., Lee, H.S., and Cho, Y. 2002. Structural basis for the recognition of the E2F transactivation domain by the retinoblastoma tumor suppressor. Genes & Dev. 16: 3199-3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine S.S., A. Weiss, A., Erdjument-Bromage, H., Shao, Z., Tempst, P., and Kingston, R.E. 2002. The core of the polycomb repressive complex is compositionally and functionally conserved in flies and humans. Mol. Cell. Biol. 22: 6070-6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link A.J., Eng, J., Schieltz, D.M., Carmack, E., Mize, G.J., Morris, D.R., Garvik, B.M., and Yates III, J.R. 1999. Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 17: 676-682. [DOI] [PubMed] [Google Scholar]
- Logan N., Delavaine, L., Graham, A., Reilly, C., Wilson, J., Brummelkamp, T.R., Hijmans, E.M., Bernards, R., and La Thangue, N.B. 2004. E2F-7: A distinctive E2F family member with an unusual organization of DNA-binding domains. Oncogene 23: 5138-5150. [DOI] [PubMed] [Google Scholar]
- Lu X. and Horvitz, H.R. 1998. lin-35 and lin-53, two genes that antagonize a C. elegans Ras pathway, encode proteins similar to Rb and its binding protein RbAp48. Cell 95: 981-991. [DOI] [PubMed] [Google Scholar]
- Manak J.R., Mitiku, N., and Lipsick, J.S. 2002. Mutation of the Drosophila homologue of the Myb protooncogene causes genomic instability. Proc. Natl. Acad. Sci. 99: 7438-7443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G., Tsukiyama, T., Wisniewski, J., and Wu, C. 1997. Role of nucleosome remodeling factor NURF in transcriptional activation of chromatin. Mol. Cell 1: 141-150. [DOI] [PubMed] [Google Scholar]
- Moberg K., Starz, M.A., and Lees, J.A. 1996. E2F-4 switches from p130 to p107 and pRB in response to cell cycle reentry. Mol. Cell. Biol. 16: 1436-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottus R., Sobel, R.E., and Grigliatti, T.A. 2000. Mutational analysis of a histone deacetylase in Drosophila melanogaster: Missense mutations suppress gene silencing associated with position effect variegation. Genetics 154: 657-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J., Rice, J.C., Strahl, B.D., Allis, C.D., and Grewal, S.I. 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292: 110-113. [DOI] [PubMed] [Google Scholar]
- Nevins J.R., Leone, G., DeGregori, J., and Jakoi, L. 1997. Role of the Rb/E2F pathway in cell growth control. J. Cell. Physiol. 173: 233-236. [DOI] [PubMed] [Google Scholar]
- Ogawa H., Ishiguro, K., Gaubatz, S., Livingston, D.M., and Nakatani, Y. 2002. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science 296: 1132-1136. [DOI] [PubMed] [Google Scholar]
- Okada M., Akimaru, H., Hou, D.X., Takahashi, T., and Ishii, S. 2002. Myb controls G2/M progression by inducing cyclin B expression in the Drosophila eye imaginal disc. EMBO J. 21: 675-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver T.L., Johnston, C.G., and Spradling, A.C. 1989. The role of ACE3 in Drosophila chorion gene amplification. EMBO J. 8: 4153-4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen A.B., Stuart, J., Mach, K., Villeneuve, A.M., and Kim, S. 2003. A gene recommender algorithm to identify coexpressed genes in C. elegans. Genome Res. 13: 1828-1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page B.D., Guedes, S., Waring, D., and Priess, J.R. 2001. The C. elegans E2F- and DP-related proteins are required for embryonic asymmetry and negatively regulate Ras/MAPK signaling. Mol. Cell 7: 451-460. [DOI] [PubMed] [Google Scholar]
- Royzman I., Austin, R.J., Bosco, G., Bell, S.P., and Orr-Weaver, T.L. 1999. ORC localization in Drosophila follicle cells and the effects of mutations in dE2F and dDP. Genes & Dev. 13: 827-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders S.L., Jennings, J., Canutescu, A., Link, A.J., and Weil, P.A. 2002. Proteomics of the eukaryotic transcription machinery: Identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry. Mol. Cell Biol. 22: 4723-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z., Raible, F., Mollaaghababa, R., Guyon, J.R., Wu, C.T., Bender, W., and Kingston, R.E. 1999. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell 98: 37-46. [DOI] [PubMed] [Google Scholar]
- Sprangers R., Groves, M.R., Sinning, I., and Sattler, M. 2003. High-resolution X-ray and NMR structures of the SMN Tudor domain: Conformational variation in the binding site for symmetrically dimethylated arginine residues. J. Mol. Biol. 327: 507-520. [DOI] [PubMed] [Google Scholar]
- Taunton J., Hassig, C.A., and Schreiber, S.L. 1996. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 272: 408-411. [DOI] [PubMed] [Google Scholar]
- Thomas J.H., Ceol, C.J., Schwartz, H.T., and Horvitz, H.R. 2003. New genes that interact with lin-35 Rb to negatively regulate the let-60 ras pathway in Caenorhabditis elegans. Genetics 164: 135-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi D., Qadota, H., Kasuya, K., Amano, M., and Kaibuchi, K. 2002. Isolation of the interacting molecules with GEX-3 by a novel functional screening. Biochem. Biophys. Res. Commun. 292: 697-701. [DOI] [PubMed] [Google Scholar]
- Wang W.K., Tereshko, V., Boccuni, P., MacGrogan, D., Nimer, S.D., and Patel, D.J. 2003. Malignant brain tumor repeats: A three-leaved propeller architecture with ligand/peptide binding pockets. Structure (Camb) 11: 775-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub S.J., Prater, C.A., and Dean, D.C. 1992. Retinoblastoma protein switches the E2F site from positive to negative element. Nature 358: 259-261. [DOI] [PubMed] [Google Scholar]
- Wysocka J., Myers, M.P., Laherty, C.D., Eisenman, R.N., and Herr, W. 2003. Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes & Dev. 17: 896-911. [DOI] [PMC free article] [PubMed] [Google Scholar]