Abstract
We have conducted a survey of RNA editing in human brain by comparing sequences of clones from a human brain cDNA library to the reference human genome sequence and to genomic DNA from the same individual. In the RNA sample from which the library was constructed, ∼1:2000 nucleotides were edited out of >3 Mb surveyed. All edits were adenosine to inosine (A→I) and were predominantly in intronic and in intergenic RNAs. No edits were found in translated exons and few in untranslated exons. Most edits were in high-copy-number repeats, usually Alus. Analysis of the genome in the vicinity of edited sequences strongly supports the idea that formation of intramolecular double-stranded RNA with an inverted copy underlies most A→I editing. The likelihood of editing is increased by the presence of two inverted copies of a sequence within the same intron, proximity of the two sequences to each other (preferably within 2 kb), and by a high density of inverted copies in the vicinity. Editing exhibits sequence preferences and is less likely at an adenosine 3′ to a guanosine and more likely at an adenosine 5′ to a guanosine. Simulation by BLAST alignment of the double-stranded RNA molecules that underlie known edits indicates that there is a greater likelihood of A→I editing at A:C mismatches than editing at other mismatches or at A:U matches. However, because A:U matches in double-stranded RNA are more common than all mismatches, overall the likely effect of editing is to increase the number of mismatches in double-stranded RNA.
RNA editing is a widespread biological process that occurs in prokaryotes, plants, and animals. However, the patterns and extent of RNA editing differ markedly. In humans, several different classes of RNA editing have been described (for review, see Gott and Emeson 2000; Keegan et al. 2001; Bass 2002; Blanc and Davidson 2003). There is strong evidence for widespread adenosine-to-inosine (A→I) editing and for a small number of cytidine-to-uridine (C→U) edits. In addition, examples of T→G (Nutt et al. 1994), G→A (Nutt et al. 1994), U→A (Novo et al. 1995), U→C (Sharma et al. 1994), and deletion (van Leeuwen et al. 1998) editing have been reported in human tissues.
A→I RNA editing in humans is carried out by adenosine deaminases that act on RNA (ADARs). These enzymes catalyze the hydrolytic deamination of adenosine to inosine in double-stranded RNA. Inosine undergoes noncanonical base-pairing with cytidine, and is interpreted by the translational machinery as a guanosine. A→I edits have been reported in translated mRNA, where they alter the sequences and functions of glutamate receptors, serotonin receptors, and hepatitis delta antigen (Higuchi et al. 1993; Lomeli et al. 1994; Casey and Gerin 1995; Herb et al. 1996; Polson et al. 1996; Burns et al. 1997). A→I editing has also been reported in 5′- and 3′-untranslated regions (UTRs) of spliced mRNA and in intronic RNA (Morse et al. 2002). Double-stranded RNA molecules (hairpins) formed by inverted copies are common substrates for A→I editing. These are often formed between common repeat sequences such as Alus (Morse et al. 2002). Many potential A→I edits of this type were identified from novel large human cDNA sequences (Kikuno et al. 2002).
Only three C→U edits have been described in human RNA (Keegan et al. 2001; Blanc and Davidson 2003; Kondo et al. 2004). C→U editing in APOB in the small intestine results in a truncated form of the protein with different functional characteristics. C→U editing in NF1 also causes truncation of the encoded protein and in IL12R causes a missense substitution. C→U editing is carried out at specific RNA target sequences by a family of cytidine deaminases in a complex with other proteins.
The functions of RNA editing are not fully understood. Some RNA edits clearly change the function of proteins. Others may alter RNA splicing patterns or RNA retention in the nucleus (Rueter et al. 1999; Zhang and Carmichael 2001). Some may modulate entry into the RNA interference pathway (Tonkin and Bass 2003). A description of the patterns of RNA editing derived from large numbers of systematically acquired edits would facilitate understanding of its functions. However, our current knowledge of editing in human tissues is based on a relatively limited amount of information. Many of these known edits have been encountered serendipitously in the course of other studies, although some A→I edits have been discovered through more systematic approaches (Morse et al. 2002; Hoopengardner et al. 2003). Moreover, the numbers of edits of each class (other than A→I) that have been reported are very small, and the prevalence and distribution of RNA edits has not been fully established. The advent of the finished human genome sequence provides a new tool with which patterns of editing can be established and analyzed. In this study, we have identified large numbers of RNA edits and analyzed their distribution in the human genome in order to understand the features of sequence structure that influence the likelihood of editing.
Results
A total of 3,049,060 bp of cDNA sequence (Table 1) derived from 6768 cDNA clones was analyzed for sequence differences from the reference human genome sequence. The average size of cDNA inserts was ∼800 bp, and their distribution across the genome (by BLAT alignment) shows very close correspondence to the distribution of known genes, indicating that the cDNA library is predominantly composed of transcribed sequences (data not shown). SNPs found on dbSNP were excluded from further analyses.
Table 1.
RNA edits by class of sequence
| Bases sequenced | RNA edits | Edits/Mb | |
|---|---|---|---|
| Intergenic | 346,113 | 333 | 962 |
| Exonic (translated) | 541,772 | 0 | 0 |
| Intronic | 1,486,947 | 1214 | 816 |
| Exonic (5′-untranslated) | 50,129 | 0 | 0 |
| Exonic (3′-untranslated) | 310,551 | 9 | 29 |
| Unknown | 313,548 | 171 | 545 |
| Total | 3,049,060 | 1727 | 566 |
To distinguish RNA edits from other causes of sequence variation (including novel SNPs and sequence artifacts), we analyzed genomic DNA from the individual from whom the cDNA library was constructed and compared it to cDNA sequences. Since there were a large number of potential edits (>4000), which would have required extensive PCR-based genomic and cDNA sequencing for complete assessment, we implemented a parsimonious, two-stage evaluation of these variants.
First, we searched for putative multiply edited transcripts, all of which turned out to show multiple A→GorT→C variants. We chose a random sample of 12 cDNA sequences from 297 sequences with three or more A→G or T→C variants. Sequence analysis of genomic DNA from the individual from whom the library was constructed demonstrated that none of the variants observed in these 12 sequences were SNPs. Moreover, in 11 out of 12 (99/102 variants) they were confirmed as RNA edits by RT-PCR of the relevant sequence segments from total cerebral RNA followed by sequencing of the PCR product. Since almost all A→G or T→C variants in this class of sequence appeared to be RNA edits, all cDNA sequences with three or more A→G or T→C variants that passed the criteria described in Methods were included in the subsequent analyses without further confirmation (1665 edits). However, it should be noted that a small proportion of these 1665 presumed A→I edits (<5%) may not be correct.
Second, we evaluated a subset of the 1824 variants (potential edits) from sequences containing fewer than three A→G or T→C variants. Of these, 503 (from 374 different PCR fragments) were successfully amplified from genomic DNA and, if the variants were shown not to be SNPs, were evaluated by RT-PCR and sequencing of total brain RNA. Of 185 A→G/T→C variants in these experiments, 62 were confirmed as RNA edits. Of 285 other base substitution variants and 33 insertion/deletion variants, all were either SNPs or artifacts. The results demonstrate that A→I edits are common compared to all other classes of potential edit. However, further analysis may be required to investigate more exhaustively the precise prevalence of other types of editing.
The 1665 edits from the first stage of evaluation were combined with the 62 confirmed edits from the second stage of evaluation and included in the analyses described below (a total of 1727 edits of which 161 [9%] were directly confirmed by RT-PCR and sequencing of brain RNA). Because we were only able to evaluate 503 of the 1824 potential edits that were present in sequences with fewer than three variants, we have underrepresented A→I edits that occur in such sequences in the final 1727. However, identification of the remainder by sequencing would have increased the total number of A→I edits by <10%. Moreover, subsequent analyses indicate that these show similar patterns to A→I edits from multiply edited sequences. (A→I edited cDNA sequences and the genomic coordinates of A→I edits are available as Supplemental material).
In all, 541,777 bp of translated exon sequence formed part of these analyses. Using the lower quality score threshold (described in Methods), 286 sequence variants were detected (one per 1.9 kb). Of these variants, 125 failed the higher quality score threshold, and 19 out of these 125 were known SNPs, leaving 106 potentially novel variants, 22 of which were successfully evaluated further. Two were novel SNPs, 20 were artifacts, and none were RNA edits. In all, 161 out of 286 translated sequence variants passed the higher quality score threshold (one per 3.3 kb), of which 93 were known SNPs, and 33/68 of the remaining sequence variants were successfully evaluated further and were shown either to be SNPs or artifacts (including 13 out of the 17 potential nonsynonymous coding variants that were present in the set of 68). Although only 167 out of 286 variants from the 541,777-bp translated exon sequence have been directly investigated and categorized, none out of the 55 that were not previously known SNPs turned out to be RNA edits. This suggests that very few of the remaining 119 are likely to be edits and therefore that the total number of edits in the 541,777-bp translated exon sequence is very small.
A total of 1727 RNA edits, all A→G, were identified (Table 1). Most edits were in intronic RNA (1214, presumably from unprocessed or partially processed transcripts) or in intergenic RNAs (333). A relatively small number of edits was observed in 3′- and 5′-UTRs (compared to intronic and intergenic transcript RNA). No edits were found in translated exon sequence. (cDNA sequences were obtained from the glutamate receptor, which is known to be edited in brain. However, these did not overlap the region of the gene that is edited. We confirmed by RT-PCR of total RNA that editing of glutamate receptor coding sequences is present in the cerebral cortex RNA from which the library was constructed [data not shown].) No edits were found in mitochondrial RNA.
Most edits were in high-copy-number repeats (Table 2). Of these, Alus accounted for the large majority and showed more edits per base sequenced than other repeat classes. Among the subfamilies of Alus, the number of edits per base analyzed did not differ markedly. Threefold greater numbers of edits were observed in Free Right Arm Monomers (FRAMs) than in Free Left Arm Monomers (FLAMs). However, the FRAM-derived component of complete Alus was not edited to a higher frequency than the FLAM component (data not shown). There was considerable variation in the extent of editing of individual Alus. The Alu with the greatest number of edits had 20 over 529 bp sequenced. A small number of RNA edits were not obviously in repeat sequences (Other Sequences in Table 1). However, on further inspection, all of these turned out to be in close proximity to high- or low-copy-number repeats.
Table 2.
RNA edits by repeat class and subclass
| Repeat | Bases sequenced | Repeats sequenced | Repeats edited | Edits | Edits/Mb |
|---|---|---|---|---|---|
| SINE/Alu (All) | 339,546 | 2151 | 302 | 1548 | 4559 |
| AluJ | 83,801 | 519 | 79 | 367 | 4379 |
| AluS | 196,178 | 1197 | 164 | 900 | 4588 |
| AluY | 45,628 | 283 | 43 | 231 | 5063 |
| FLAM | 9256 | 99 | 8 | 23 | 2485 |
| FRAM | 3114 | 34 | 8 | 27 | 8671 |
| Alu (MISC) | 1569 | 19 | 0 | 0 | 0 |
| SINE/MIR | 49,704 | 455 | 1 | 5 | 101 |
| LINE/L1 | 269,044 | 1258 | 18 | 116 | 431 |
| LINE/L2 | 71,420 | 456 | 0 | 0 | 0 |
| Simple | 21,191 | 497 | 6 | 11 | 519 |
| Low complexity | 18,502 | 471 | 0 | 0 | 0 |
| DNA | 54,155 | 398 | 2 | 6 | 111 |
| LTR | 103,375 | 505 | 4 | 7 | 68 |
| Other repeats | 10,743 | 69 | 0 | 0 | 0 |
| Other sequences | 2,111,380 | 11,041 | 20 | 35 | 17 |
Previously published results indicate that double-stranded RNA formed by inverted copies of a sequence on the same transcript is a major substrate for A→I editing enzymes. We therefore investigated the role of double-stranded RNA hairpin formation in A→I RNA editing by comparing the characteristics of the genome surrounding edited and unedited sequences. Since edits in Alus account for most RNA edits, the results shown are predominantly from Alus.
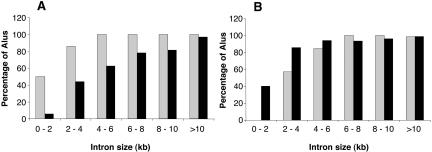
Several analyses confirm that the presence of an inverted Alu copy is an important determinant of the likelihood of editing. For example, approximately half of both edited and unedited Alus within introns <2 kb have another Alu within the same intron (Fig. 1A,B). However, all additional Alus within introns <2 kb occupied by edited Alus are inverted (antisense) copies with respect to the edited Alu. Conversely, almost all additional Alus within introns <2 kb occupied by unedited Alus are same sense to the unedited Alu (Fig. 1A,B).
Figure 1.
Proportion of edited and unedited Alus with additional Alus in the same intron. All Alus aligning to the introns of known genes, and for which ≥80% of the genomic extent of the Alu was sequenced, were included in this analysis. The proportion of edited Alus (gray bars) and the proportion of unedited Alus (black bars) having an antisense Alu (A) or a same sense Alu (B) in the same intron are shown for different intron sizes.
We next investigated whether absence of an intron boundary between the two inverted Alu copies is important, by comparing the sizes of introns containing edited and unedited Alus. We found 3% edited Alus in introns <2 kb compared to 9% of unedited Alus. There is no difference in the frequency of edited and unedited Alus in introns >2 kb. This suggests that inverted Alu copies separated by an intron-exon boundary are less likely to form double-stranded RNA and become edited. It should be noted, however, that some edited Alus do not have an inverted copy within the same intron.
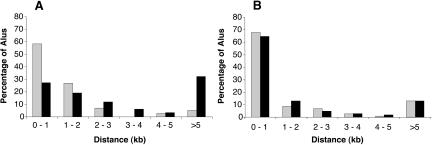
We then evaluated whether proximity of an inverted Alu copy within an intron is associated with the likelihood of an Alu being edited. Even in introns >10 kb, edited Alus are more likely than unedited Alus to have an inverted copy within 1 kb (data not shown). Similarly, edited Alus are more frequently close to an inverted copy within the same intron than unedited Alus (Fig. 2A). This effect is most marked at distances up to 2 kb and is weaker at greater distances. No association with likelihood of Alu editing is observed for proximity of same-sense Alus (Fig. 2B).
Figure 2.
Distance from edited and unedited Alus to the nearest Alu in the same intron. The Alu sequences included in this analysis are the same as in Figure 1. The proportion of edited Alus (gray bars) and the proportion of unedited Alus (black bars) at different distances from the nearest antisense Alu (A) or same sense Alu (B) are shown.
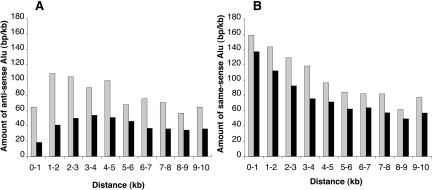
We next investigated whether the amount of inverted Alu copy sequence in the vicinity influences the likelihood of editing. Edited Alus have more inverted Alu copy sequences than unedited Alus at all distances up to 10 kb, although the effect is strongest within 2 kb of the Alu (Fig. 3A). A similar effect, of lesser magnitude, is observed for same-sense Alus (Fig. 3B).
Figure 3.
Amount of flanking Alu sequence at different distances from edited and unedited Alus. All Alus for which ≥80% of the genomic extent of the Alu was sequenced were included in this analysis. For each Alu, the amount of flanking Alu sequence in the opposite orientation (A) or same orientation (B) in successive 1-kb windows was recorded. For each distance, the flanking Alu sequences in the 1-kb window 5′ and 3′ of the reference Alu were combined. The data presented are the average amount of Alu sequence flanking all edited Alus (gray bars) or unedited Alus (black bars).
The amount of information for repeat classes other than Alus was limited. However, the only repeat class in which there clearly did not seem to be a relationship between editing and the presence of a nearby inverted copy was simple AT repeats, which can form double-stranded RNA molecules internally (data not shown).
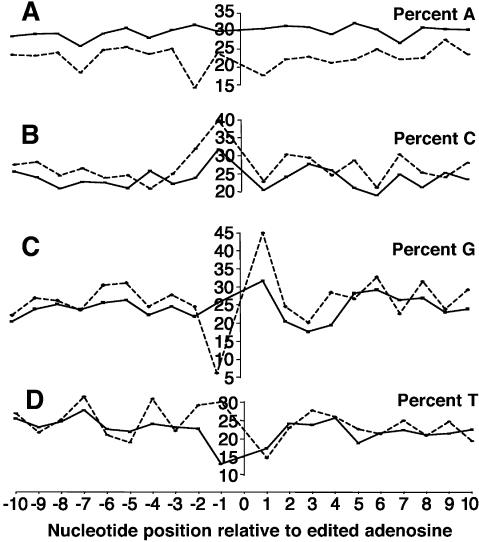
We assessed the role of sequence context in RNA editing by selecting edited Alu sequences, identifying the bases at positions up to 10 bp 5′ and up to 10 bp 3′ to edited adenosines and comparing these to the bases up to 10 bp 5′ and up to 10 bp 3′ to unedited adenosines. The results show that there is a marked deficit of G at the 5′ position to an edited A, with a compensatory increase of U (and to a lesser extent C) (Fig. 4). There is also an excess of G at the 3′ position to an edited A with minor compensatory fluctuations of the other bases. At all positions 5′ and 3′ to the edited adenosine, edited bases show fewer adenosines than unedited bases. This seems to be attributable mainly to complete absence of editing of the FRAM associated poly(A) tail of Alus (data not shown).
Figure 4.
Sequence context of adenosines in edited Alu sequences. The sequence context of all edited adenosines and all unedited adenosines from all edited Alu sequences was compared. For each of the 10 bases either side of edited adenosines (dashed lines) and unedited adenosines (solid lines), the proportion of bases that were A, C, G, or T (A-D, respectively) at that position was calculated.
To investigate further the factors that determine whether a particular base is edited and to assess the overall effect of RNA editing, we formed hypothetical double-stranded RNA molecules by BLAST alignments between edited Alus and the nearest inverted repeat copy. We then identified the mismatches and matches in each hypothetical double-stranded RNA molecule, and by superimposing the observed edits, assessed the likelihood of A→I editing at each class of mismatch and match (Table 3). The results indicate that A→I editing at an A:C mismatch (which will generate an I:C matched base pair) is more likely than editing at other types of base pair. Similar results were obtained by examining edited sequences for which there is only a single inverted copy in the same intron (Table 3). This data set (although smaller) is probably a more accurate simulation of the in vivo situation.
Table 3.
A→I editing at different RNA base pairings
| Match/mismatch | Subset of Alu | Total (bp) | Edits | Edited % |
|---|---|---|---|---|
| A:U matches | All Alus | 5839 | 465 | 8 |
| Alus with single inverted copy | 581 | 44 | 8 | |
| A:G mismatches | All Alus | 217 | 13 | 6 |
| Alus with single inverted copy | 23 | 0 | 0 | |
| A:C mismatches | All Alus | 1166 | 249 | 21 |
| Alus with single inverted copy | 113 | 24 | 21 | |
| A:A mismatches | All Alus | 264 | 11 | 4 |
| Alus with single inverted copy | 24 | 1 | 4 | |
| Total matches | All Alus | 25,363 | 465 | 1.8 |
| Alus with single inverted copy | 2400 | 44 | 1.8 | |
| Total mismatches | All Alus | 8368 | 273 | 3.3 |
| Alus with single inverted copy | 769 | 25 | 3.1 |
Each edited Alu has been BLAST-aligned to the nearest inverted Alu copy in the same transcript to form a hypothetical double-stranded RNA molecule. The numbers of adenines that are matched (A:U) and mismatched (A:A, A:C, A:G) and the numbers of each class of match/mismatch that are edited have been calculated. The calculations have been performed for all edited Alus and separately for the subset of these that have only a single inverted copy in the same intron. The results were from 159 alignments and 738 edits (all Alus) and 14 alignments and 69 edits (Alus with a single inverted copy in the same intron).
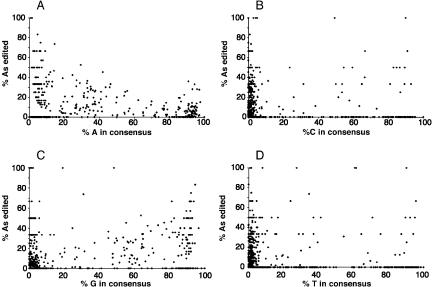
We also investigated the effect of match/mismatch on the likelihood of editing by aligning every edited Alu to all other edited Alus and then correlating the proportion of the adenosines edited at each position in the alignment with the proportion of each nucleotide at that position. The scatter graphs (Fig. 5) show that a high proportion of adenosines at a particular position in the alignment (which would be uracil in the antisense strand forming A:U matches in double-stranded RNA) is correlated with a low frequency of editing, whereas a high proportion of guanosines at a particular position in the alignment (which would be cytidine in the antisense strand forming A:C mismatches in double-stranded RNA) is correlated with a high frequency of editing.
Figure 5.
Effect of sequence composition on the likelihood of RNA editing. A multiple alignment of all edited Alu sequences was prepared using CLUSTALW. At each position in the alignment, the proportion of edited adenosines was calculated from the number of sequenced edited adenosines and the total number of sequenced adenosines. The sequence composition at each position was calculated from all Alus. For each position in the alignment, the proportion of edited adenosines is compared to the proportion of A, C, G, or T at that position (A-D, respectively).
Since A→I editing may result in matching base pairs being formed from mismatched base pairs (A:C→I:C), mismatches being formed from matches (A:U→I:U) and mismatches from mismatches (A:A→I:A and A:G→I:G), we next evaluated the overall effect of editing on the balance of matched base-pairing in the hypothetical double-stranded RNA molecules formed by an edited Alu BLAST-aligned to its nearest inverted copy. There is a net increase in mismatches of ∼2.6% (from 8368 to 8584), resulting, on balance, in an additional 0.6% (216 out of 33,731) of bases in double-stranded RNAs becoming mismatched after editing. (Since we can only evaluate editing of one of the RNA strands in the double-stranded molecule, it is likely that the number of additional mismatched base pairs is twice this estimate, i.e., 1.2%.) It should be noted, however, that in a minority of individual simulated double-stranded RNA molecules there was on balance an apparent increase in matches (data not shown). We also examined hypothetical double-stranded RNA molecules between repeats that have only a single inverted copy within the same intron. Following editing in this set there is a 2.5% (from 796 to 816) increase in mismatches resulting, on balance, in an additional 0.6% of bases (20 out of 3196) becoming mismatched after editing (1.2% taking into account both strands). However, one out of the 14 double-stranded RNA molecules included in this analysis still would appear slightly better matched after editing (six matches to mismatches and seven mismatches to matches) (data not shown). Finally, we evaluated the balance of editing using the alignments of all edited Alus to all other edited Alus. The average nucleotide composition at 1539 edited adenosines from 301 multiply aligned Alus confirms that 57% of editing reactions create a mismatch (I:U) from a match (A:U), 28% create a match (I:C) from a mismatch (A:C), and 15% create a mismatch from a mismatch.
Discussion
We have conducted a survey of RNA editing in human brain. A substantial number of A→I edits and no examples of any other type of edit were detected. We have not sequenced exhaustively enough to completely exclude the existence of edits other than A→I. However, they are clearly very rare compared to A→I edits.
All edits that could be classified are in intronic, intergenic, or untranslated RNA. No edits in mitochondrial RNA or in translated exon sequences were found. Nonsynonymous A→I coding edits in human brain have previously been described. Our data indicate that coding edits represent a very small proportion of the total number of edits and that a small fraction of coding bases, compared to noncoding bases, are edited. However, our evaluation of coding edits is relatively limited, and more exhaustive investigation directed at translated exon sequences may still be warranted to detect rare, functionally important coding edits.
In the brain mRNA sample from which the cDNA library used in these experiments was made, approximately one in two thousand nucleotides are A→I edited. This is ∼10-fold higher than a previous estimate of one in 17,000 nucleotides based upon analyses of rat brain (Paul and Bass 1998). However, the proportion of bases edited is clearly dependent on the degree to which the mRNA has been processed. Since most edits are in intronic RNA, the proportion of edited bases in completely spliced cytoplasmic mRNA will be much lower. Conversely, because our cDNA library is enriched in spliced mRNAs, the proportion of edited bases in unspliced nuclear mRNA is likely to be even higher. In intronic and intergenic RNA approximately 1:1000 bp is edited (Table 1), and this may represent a plausible estimate for total unspliced brain mRNA.
The large majority of A→I edited sequences are high-copy-number repeats, particularly Alus. Therefore, most of our further analyses have been conducted on this class of repeat. Analysis of the finished human genome sequence in the vicinity of edited sequences strongly confirms that the potential for double-stranded RNA formation is an important factor in determining whether a sequence is edited. The likelihood of a sequence being edited is increased in proportion to the amount and proximity of inverted copy sequence (which can potentially serve as a partner in double-stranded RNA formation) with the strongest effects observed when the two copies are within 2 kb of each other.
The likelihood of a sequence being edited also appears to be dependent on the two inverted copies being within the same intron. Thus, edited Alus are observed less frequently than unedited Alus within small introns (<2 kb), presumably because of the preference for an inverted copy within the restricted space. These data suggest that inverted copies can form double-stranded RNA and become edited if they are within the same loop (lariat) of RNA that is removed during RNA splicing, but are much less likely to do so if they are in different loops. Interestingly, there is 30-fold less editing of 3′-untranslated RNA compared to intronic or intergenic mRNAs, despite only twofold to threefold fewer Alus (12.4% of intronic sequence is Alu compared to 4.5% of 3′-untranslated sequence). The low prevalence of RNA edits in 3′-untranslated sequence is probably attributable to two factors. The last exon of a gene (average size ∼700 bp) is unlikely to contain two inverted copies of an Alu sequence and there is low likelihood of double-stranded RNA formation with the nearest inverted Alu in the last intron because of the presence of an intervening intron-exon junction.
The presence of inverted copies at distances >2 kb appears to have less influence on the likelihood of a transcript being edited. Nevertheless, the frequency of inverted repeats up to 10 kb distant (and even further distant) (data not shown) is higher for edited sequences than unedited sequences. Although this may in part be due to a direct biological interaction between two distant inverted copies to form double-stranded RNA, the effect (although less marked) is observed for same-sense sequences as well. These longer distance associations of repeat copy density with likelihood of editing may be a reflection of the existence of large Alu-rich genomic domains. Edited Alus are more likely to be in Alu-rich domains because this will be associated with a higher frequency of Alus in close proximity, which in turn will influence the likelihood of editing by influencing the rate of double-stranded RNA formation.
If the likelihood of editing is increased by the proximity of inverted sequence copies, it is conceivable that same-sense copies might reduce the likelihood of editing, perhaps by competing for nearby inverted copies in the formation of double-stranded RNA. The results suggest, however, that the presence of a same-sense Alu in the vicinity is not associated with a decrease in the likelihood of editing (except in small introns, where they occupy the space that might be taken by an inverted copy). Indeed, there is a slightly higher frequency of same-sense Alus at all distances up to 10 kb (and, indeed, up to 50 kb) (data not shown) from edited sequences compared to unedited sequences (perhaps because of the existence of large Alu-rich domains in which both sense and antisense Alus are more common; see above).
The most commonly edited repeats are Alus. A much smaller proportion of MIRs, LINEs, and other repeats are edited. The lower frequency of editing of repeats other than Alus may simply be a consequence of a lower likelihood of nearby inverted copies that would be available for double-stranded RNA formation. That is, 71% of Alus have an inverted Alu within 5 kb that overlaps by at least 50 bp and with which it might therefore form double-stranded RNA. Conversely, only 2% of L1LINEs have an inverted L1LINE within 5 kb that overlaps by at least 50 bp. In contrast, although Alu subfamilies vary substantially in their genomic copy number, there seems to be little difference in the frequency of editing of these subfamilies. This would suggest that members of Alu subfamilies do not discriminate between each other in the formation of double-stranded mRNA, that is, that a member of one subfamily is as likely to form double-stranded RNA and be edited with a member of its own subfamily as with a member of another subfamily.
Overall, the data are consistent with a model in which the likelihood of A→I editing is largely determined by the likelihood of double-stranded RNA formation, which, in turn, is predominantly determined by the proximity and amount of inverted copy sequence, particularly in the same intron. The results also indicate that most edited double-stranded RNAs are formed by intramolecular RNA base-pairing. The observations are, therefore, broadly consistent with previous reports on smaller data sets using different strategies for identifying RNA edits (see reviews and Morse et al. 2002).
There are, however, edited Alus for which no inverted copy within the same intron can currently be identified. Some of these may be due to anomalies in gene annotation. Alternatively, double-stranded mRNA formation with independent mRNA molecules such as antisense transcripts, double-stranded mRNA formation with an inverted copy in an adjacent intron before the splicing machinery separates the two copies, or conceivably an editing process that does not rely on double-stranded mRNA may be responsible.
Despite the evidence that the presence of adjacent inverted sequence copies increases the likelihood of editing, many sequences with nearby inverted repeat copies do not appear to be edited. In reality, some of these may actually be edited to a small extent, and hence only a small proportion of randomly selected cDNA clones from the library exhibit evidence of editing. However, we have confirmed by RT-PCR and sequencing of total RNA from human brain that most sequences that appear unedited in cDNA clone sequencing are not edited to a detectable extent (data not shown). The results, therefore, suggest that, in addition to the presence of a nearby inverted copy within the same intron, other factors influence the likelihood of editing. One of these may simply be whether a transcript is predominantly expressed in a cell type(s) that has low levels of editing. Previous data (and our unpublished results) show that the extent of RNA editing is highly variable between tissues (Paul and Bass 1998). Brain is a heterogeneous tissue composed of several constituent cell types including nerve cells, astrocytes, oligodendrocytes, endothelial cells, and microglia. The expression pattern among these different cell types of most of the transcripts we have sequenced and the extent of editing in each of these cell types is unknown. Therefore, unedited brain transcripts may simply be expressed exclusively in cells with no editing activity. Similarly, fully edited transcripts may be expressed only in cells with high editing activity. It is therefore currently difficult to evaluate the role of other, unknown factors that influence the likelihood of editing.
In vitro analyses have suggested that editing of double-stranded RNA shows some sequence preferences. Editing by Xenopus ADAR1 takes place preferentially at adenosines that are immediately 3′ to U = A > C > G and by ADAR2 at adenosines 3′ to U = A > C = G (Polson and Bass 1994; Lehmann and Bass 2000). It has also been suggested that for ADAR2 there is an influence of the base 3′ to the edited adenosine, with the preference U = G > C = A. Analyses of a small number of edited adenosines in ADAR2 itself were broadly concordant with these patterns (Dawson et al. 2004). Our results indicate that at the immediately 5′ position to an edited adenosine there is a relative deficit of G and compensatory increase in U and C, consistent with the previously reported patterns associated with ADAR1 and ADAR2. At the immediately 3′ position to an edited adenosine there is a relative excess of guanosine with compensatory decrease mainly of adenosine. This is consistent with the pattern previously proposed for ADAR2, but not forADAR1 (Polson and Bass 1994) and may possibly reflect a predominant role of ADAR2 in editing of brain mRNA.
We have evaluated the impact of RNA editing on the extent of matching in double-stranded RNA molecules by simulating, in several ways, double-stranded RNA molecules. First, we have made the assumption that the double-stranded RNA that was the in vivo substrate for editing enzymes was formed with the closest inverted repeat copy. Although this assumption is unlikely to be correct for all sequences, the overall results of this study indicate that it is often likely to be the case. The advantage of invoking this assumption is that it allows use of most available information. Second, we have examined edited sequences that have only a single inverted copy in the same intron. While these represent a fraction of the available information, our results would indicate that these are likely to be more accurate simulations of the in vivo substrate. Third, we have aligned each Alu that we have shown to be edited to all other edited Alus. In the three types of analysis, the hypothetical double-stranded RNA molecules generated are dependent on the parameters used to generate the alignments and are unlikely to completely replicate the biological conditions present in vivo. Moreover, our results only provide information on editing of one strand of the double-stranded RNA molecule. Editing on the other strand (probably at an equivalent rate) is likely, but we cannot evaluate it.
Double-stranded RNA formed between a sequence and an inverted copy usually includes several base mispairings. We have therefore investigated whether editing is equally likely at mismatches and matches. The likelihood of editing at A:C mismatches in double-stranded RNA appears to be higher than at A:G or A:A mismatches or at A:U matches. Since an A:C mismatch is converted into an I:C base pair by A→I editing, the enzymatic configuration of the editing machinery seems to favor the creation of fully matched double-stranded RNA. These observations are consistent with previous in vitro experiments that indicate that editing at A:C mismatches is more efficient than at A:U matches or other mismatches (Wong et al. 2001).
The frequency of A:U matches in most RNA duplexes formed by inverted copies is, however, much higher than the number of A:C mismatches. Therefore, despite the higher likelihood of editing at A:C mismatches, the overall effect of RNA editing may be to increase the number of mismatches in double-stranded RNA molecules. This appears to be the prediction of all three types of analysis. Overall, the results suggest that the effect of editing is to increase the number of mismatches in double-stranded RNA molecules, albeit by a relatively modest amount (in edited sequences, an additional 1%-2% of base pairs become mismatched after editing).
The functions of RNA editing in mammals are still being investigated. On the basis of previously reported evidence, a small number of edits alter the coding sequence and activities of certain proteins. An additional small number have direct effects on mRNA splicing, by altering transcript sequence at consensus splice sites. However, the function of the large majority of RNA edits, which are of intronic or of intergenic high-copy-number repeats, is not known. One possibility is that they have no function at all. They may simply be the collateral damage of an enzyme system that uses double-stranded RNA as a template and that therefore generates large numbers of edits of high-copy-number repeat elements. According to this hypothesis, the important functional consequences for the cell reside in the small number of coding, splice site, and other functional edits. This would be a system of remarkable metabolic profligacy because <1% (and probably <0.1%) edits would be functional.
Alternatively, editing of intronic and intergenic high-copy-number repeats may have a function. One possibility is that RNA editing influences other cellular responses to double-stranded RNA (or the product of processing of double-stranded RNA) that are deleterious to cellular function, including activation of 2′,5′-oligoA synthetase/RNaseL resulting in RNA degradation, activation of the dsRNA-dependent Protein kinase (PKR) resulting in suppression of protein synthesis, activation of interferon response leading to apoptosis, or gene silencing via the RNAi pathway (for review, see McManus and Sharp 2002; Yelin et al. 2003). Previous studies in Caenorhabditis elegans support the idea that RNA editing abrogates RNAi-dependent toxic effects of endogenous double-stranded RNAs (Tonkin and Bass 2003). An increased number of mismatches generated by editing of double-stranded RNA molecules may limit their deleterious RNAi-dependent effects by destabilizing the hairpin, by reducing the efficiency of processing (perhaps by retention in the nucleus) (Zhang and Carmichael 2001), by generating products that are less effective in mediating the effects of RNAi (e.g., by interrupting long, perfectly matched stretches of base pairing) or by other, currently obscure, mechanisms.
While our paper was under review, two additional articles reporting A→I edits in transcribed Alu sequences were published (Kim et al. 2004; Levanon et al. 2004). In both studies, A→I edits were identified by computational analyses of publicly available EST and cDNA sequences. The patterns of A→I editing of Alu sequences identified in these studies were broadly consistent with our results.
Methods
Library construction and sequencing
All procedures were approved by Cambridge Addenbrookes Local Research Ethics Committee. A cDNA library was constructed in pUC19 by random hexamer priming of twice poly(A) selected RNA from the cerebral cortex of a 67-yr-old male who had died following cardiac failure and a chest infection. The library was amplified. Clones were picked and sequenced using fluorescent dideoxysequencing in one direction on ABI3700 DNA sequencers using the M13 forward primer. A single sequencing run was taken from each clone. Differences between cDNA sequences and the genome reference sequence were further evaluated to assess whether they were germ-line polymorphisms or RNA edits by amplification of genomic DNA of the individual from whom the cDNA library had been constructed and by RT-PCR of whole RNA from the cerebral cortex sample from which the library had been constructed. Sequences from genomic DNA and cDNA were aligned and viewed in a GAP4 database.
cDNA sequence alignment and variant detection
Custom Perl programs were used extensively in the analysis of cDNA clone sequences. ABI Sequence trace files were base scored and quality scored using PHRED. Vector sequence was masked using Cross_match. Sequences were quality scored and were aligned to the reference human genome sequence (NCBI 34 assembly) using BLAT (http://genome.ucsc.edu/cgi-bin/hgBlat). In order to identify alignments that showed differences from the reference genome sequence due to RNA edits, but at the same time to discard sequences that showed differences from the reference because of poor sequence quality or false alignments (e.g., to pseudogenes), the following algorithm was adopted. Sequences that aligned with an identity of <95% to the reference genome sequence were discarded. These are likely to be poor quality. To exclude sequences that align with a high degree of similarity to more than one region of the genome (e.g., to a gene and a pseudogene), we removed sequences for which the product of the BLAT score and the percentage identity of the best alignment was >95% that of the product of the BLAT score and the percentage identity of the second best BLAT alignment, where the second alignment had greater sequence identity than the first. Having taken measures to exclude sequences that might generate false sequence variants when aligned against the reference genome, we then retrieved sequences in which there were multiple differences from the reference genome that were likely to be due to RNA editing. To do this, we recovered sequences that had more than three variants of a single type (e.g., A→G, T→C, or C→T, G→A) that accounted for >75% of all variants. Examination of 20 sequences processed according to these criteria demonstrated that all rejected sequences were poor quality or false alignments, and all likely edited sequences were retained. For each cDNA clone, the start and end of each region of alignment and the position of each variant were recorded in genomic coordinates. Variant quality was assessed by reference to the PHRED quality score: only variants with a quality score of 20 or more flanked on either side by five bases of quality score 15 or more were used in the analyses. Variants with a quality score of 30 or more with two preceding bases of quality score 30 or more and a following base of score 20 or more were selected for resequencing as the highest quality candidate novel RNA edits.
cDNA sequence annotation and determination of editing frequency
Custom Perl programs incorporating the EnsEMBL application-programming interface (API) were used to query EnsEMBL (version 19.34b.1; http://www.ensembl.org/). Known single nucleotide polymorphisms (SNPs) were excluded by reference to dbSNP (http://www.ncbi.nlm.nih.gov/SNP/). The genomic coordinates of each clone were compared with the genomic coordinates of overlapping transcripts. For clones that could be aligned unambiguously to a single transcript, the gene name, the genomic coordinates of any intron/exon boundaries, and the genomic coordinates of translated region/untranslated region boundaries were recorded. For all clones, the start and end position of repeat elements was recorded in genomic coordinates. Genomic coordinates were used to calculate the amount of editing by sequence class (Table 1) and repeat class (Table 2).
Analysis of repeat distributions
Full-length Alu sequences corresponding to repeats sequenced as part of cDNA clones were obtained from EnsEMBL. For all studies of edited and unedited Alus (Figs. 1, 2, 3), only Alus for which at least 80% of their genomic extent was sequenced as part of a cDNA clone were used as reference Alus in the analyses. For studies of the patterns of Alu elements in the same intron as edited and unedited Alus (Figs. 1 and 2), only Alu elements from cDNA clones that aligned to the introns of EnsEMBL known genes were used as reference Alus in the analyses. Intron sizes and the orientation and genomic coordinates of flanking Alus were obtained from the EnsEMBL genome annotation database using the genomic coordinates of reference Alus as queries.
BLAST simulation of RNA duplexes
Reference Alus were aligned to neighboring Alus using BLAST (http://www.ncbi.nlm.nih.gov/blast/bl2seq/) (Table 3). The positions of mismatches in the alignments were recorded and compared with the positions of edited bases in the reference sequence. BLAST is not generally considered an algorithm for simulating RNA duplexes. However, we compared the base-pairing produced by BLAST to that generated by MFOLD, a program designed to simulate RNA secondary structure and found that for the 32 edited bases evaluated, the predicted base-pairing was identical using the two methods. We therefore used BLAST for this purpose.
Alu multiple sequence alignments
Multiple alignments were constructed from all edited Alu sequences using CLUSTALW. Information from all sequences was used to calculate the percent nucleotide composition at each position in the alignment. Only bases sequenced in this study were used to calculate the proportion of adenosines edited at each position in the alignment.
Acknowledgments
We thank other members of the Cancer Genome Project for their support and Nav Navaratnam for advice. M.B. is a Wellcome Trust Prize Ph.D. student.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2951204. Article published online ahead of print in November 2004.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Bass, B.L. 2002. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 71: 817-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc, V. and Davidson, N.O. 2003. C-to-U RNA editing: Mechanisms leading to genetic diversity. J. Biol. Chem. 278: 1395-1398. [DOI] [PubMed] [Google Scholar]
- Burns, C.M., Chu, H., Rueter, S.M., Hutchinson, L.K., Canton, H., Sanders-Bush, E., and Emeson, R.B. 1997. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature 387: 303-308. [DOI] [PubMed] [Google Scholar]
- Casey, J.L. and Gerin, J.L. 1995. Hepatitis D virus RNA editing: Specific modification of adenosine in the antigenomic RNA. J. Virol. 69: 7593-7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson, T.R., Sansam, C.L., and Emeson, R.B. 2004. Structure and sequence determinants required for the RNA editing of ADAR2 substrates. J. Biol. Chem. 279: 4941-4951. [DOI] [PubMed] [Google Scholar]
- Gott, J.M. and Emeson, R.B. 2000. Functions and mechanisms of RNA editing. Annu. Rev. Genet. 34: 499-531. [DOI] [PubMed] [Google Scholar]
- Herb, A., Higuchi, M., Sprengel, R., and Seeburg, P.H. 1996. Q/R site editing in kainate receptor GluR5 and GluR6 pre-mRNAs requires distant intronic sequences. Proc. Natl. Acad. Sci. 93: 1875-1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi, M., Single, F.N., Kohler, M., Sommer, B., Sprengel, R., and Seeburg, P.H. 1993. RNA editing of AMPA receptor subunit GluR-B: A base-paired intron-exon structure determines position and efficiency. Cell 75: 1361-1370. [DOI] [PubMed] [Google Scholar]
- Hoopengardner, B., Bhalla, T., Staber, C., and Reenan, R. 2003. Nervous system targets of RNA editing identified by comparative genomics. Science 301: 832-836. [DOI] [PubMed] [Google Scholar]
- Keegan, L.P., Gallo, A., and O'Connell, M.A. 2001. The many roles of an RNA editor. Nat. Rev. Genet. 2: 869-878. [DOI] [PubMed] [Google Scholar]
- Kikuno, R., Nagase, T., Waki, M., and Ohara, O. 2002. HUGE: A database for human large proteins identified in the Kazusa cDNA sequencing project. Nucleic Acids Res. 30: 166-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D.D., Kim, T.T., Walsh, T., Kobayashi, Y., Matise, T.C., Buyske, S., and Gabriel, A. 2004. Widespread RNA editing of embedded Alu elements in the human transcriptome Genome Res. 14: 1719-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo, N., Matsui, E., Kaneko, H., Aoki, M., Kato, Z., Fukao, T., Kasahara, K., and Morimoto, N. 2004. RNA editing of interleukin-12 receptor β2, 2451 C-to-U (Ala 604 Val) conversion, associated with atopy. Clin. Exp. Allergy 34: 363-368. [DOI] [PubMed] [Google Scholar]
- Lehmann, K.A. and Bass, B.L. 2000. Double-stranded RNA adenosine deaminases ADAR1 and ADAR2 have overlapping specificities. Biochemistry 39: 12875-12884. [DOI] [PubMed] [Google Scholar]
- Levanon, E.Y., Eisenberg, E., Yelin, R., Nemzer, S., Hallegger, M., Shemesh, R., Fligelman, Z.Y., Shoshan, A., Pollock, S.R., Sztybel, D., et al. 2004. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 22: 1001-1005. [DOI] [PubMed] [Google Scholar]
- Lomeli, H., Mosbacher, J., Melcher, T., Hoger, T., Geiger, J.R., Kuner, T., Monyer, H., Higuchi, M., Bach, A., and Seeburg, P.H. 1994. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science 266: 1709-1713. [DOI] [PubMed] [Google Scholar]
- McManus, M.T. and Sharp, P.A. 2002. Gene silencing in mammals by small interfering RNAs. Nat. Rev. Genet. 3: 737-747. [DOI] [PubMed] [Google Scholar]
- Morse, D.P., Aruscavage, P.J., and Bass, B.L. 2002. RNA hairpins in noncoding regions of human brain and Caenorhabditis elegans mRNA are edited by adenosine deaminases that act on RNA. Proc. Natl. Acad. Sci. 99: 7906-7911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novo, F.J., Kruszewski, A., MacDermot, K.D., Goldspink, G., and Gorecki, D.C. 1995. Editing of human α-galactosidase RNA resulting in a pyrimidine to purine conversion. Nucleic Acids Res. 23: 2636-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt, S.L., Hoo, K.H., Rampersad, V., Deverill, R.M., Elliott, C.E., Fletcher, E.J., Adams, S.L., Korczak, B., Foldes, R.L., and Kamboj, R.K. 1994. Molecular characterization of the human EAA5 (GluR7) receptor: A high-affinity kainate receptor with novel potential RNA editing sites. Receptors Channels 2: 315-326. [PubMed] [Google Scholar]
- Paul, M.S. and Bass, B.L. 1998. Inosine exists in mRNA at tissue-specific levels and is most abundant in brain mRNA. EMBO J. 17: 1120-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polson, A.G. and Bass, B.L. 1994. Preferential selection of adenosines for modification by double-stranded RNA adenosine deaminase. EMBO J. 13: 5701-5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polson, A.G., Bass, B.L., and Casey, J.L. 1996. RNA editing of hepatitis delta virus antigenome by dsRNA-adenosine deaminase. Nature 380: 454-456. [DOI] [PubMed] [Google Scholar]
- Rueter, S.M., Dawson, T.R., and Emeson, R.B. 1999. Regulation of alternative splicing by RNA editing. Nature 399: 75-80. [DOI] [PubMed] [Google Scholar]
- Sharma, P.M., Bowman, M., Madden, S.L., Rauscher III, F.J., and Sukumar, S. 1994. RNA editing in the Wilms' tumor susceptibility gene, WT1. Genes & Dev. 8: 720-731. [DOI] [PubMed] [Google Scholar]
- Tonkin, L.A. and Bass, B.L. 2003. Mutations in RNAi rescue aberrant chemotaxis of ADAR mutants. Science 302: 1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen, F.W., de Kleijn, D.P, van den Hurk, H.H., Neubauer, A., Sonnemans, M.A., Sluijs, J.A., Koycu, S., Ramdjielal, R.D., Salehi, A., Martens, G.J., et al. 1998. Frameshift mutants of β amyloid precursor protein and ubiquitin-B in Alzheimer's and Down patients. Science 279: 242-247. [DOI] [PubMed] [Google Scholar]
- Wong, S.K., Sato, S., and Lazinski, D.W. 2001. Substrate recognition by ADAR1 and ADAR2. RNA 7: 846-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelin, R., Dahary, D., Sorek, R., Levanon, E.Y., Goldstein, O., Shoshan, A., Diber, A., Biton, S., Tamir, Y., Khosravi, R., et al. 2003. Widespread occurrence of antisense transcription in the human genome. Nat. Biotechnol. 21: 379-386. [DOI] [PubMed] [Google Scholar]
- Zhang, Z. and Carmichael, G.G. 2001. The fate of dsRNA in the nucleus: A p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell 106: 465-475. [DOI] [PubMed] [Google Scholar]
Web site references
- http://genome.ucsc.edu/cgi-bin/hgBlat; BLAT.
- http://www.ensembl.org/; EnsEMBL.
- http://www.ncbi.nlm.nih.gov/blast/bl2seq/; BLAST.
- http://www.ncbi.nlm.nih.gov/SNP/; dbSNP.