Abstract
The 156 breeds of registered dogs in the United States offer a unique opportunity to map genes important in disease susceptibility, morphology, and behavior. Linkage disequilibrium (LD) is of current interest for its application in whole genome association mapping, since the extent of LD determines the feasibility of such studies. We have measured LD at five genomic intervals, each 5 Mb in length and composed of five clusters of sequence variants spaced 800 kb-1.6 Mb apart. These intervals are located on canine chromosomes 1, 2, 3, 34, and 37, and none is under obvious selective pressure. Approximately 20 unrelated dogs were assayed from each of five breeds: Akita, Bernese Mountain Dog, Golden Retriever, Labrador Retriever, and Pekingese. At each genomic interval, SNPs and indels were discovered and typed by resequencing. Strikingly, LD in canines is much more extensive than in humans: D′ falls to 0.5 at 400-700 kb in Golden Retriever and Labrador Retriever, 2.4 Mb in Akita, and 3-3.2 Mb in Bernese Mountain Dog and Pekingese. LD in dog breeds is up to 100× more extensive than in humans, suggesting that a correspondingly smaller number of markers will be required for association mapping studies in dogs compared to humans. We also report low haplotype diversity within regions of high LD, with 80% of chromosomes in a breed carrying two to four haplotypes, as well as a high degree of haplotype sharing among breeds.
Whole genome association mapping is emerging as a promising tool in the effort to understand genetic and phenotypic variation. Mapping by association requires only a population-based sample rather than the multigenerational families that are needed for linkage mapping. However, the feasibility of whole genome association mapping depends strongly on the level of linkage disequilibrium (LD), since only a limited number of markers can be genotyped. Typed markers are used to draw inferences about surrounding regions of a locus through the non-random association between markers known as LD. In a population where LD extends over relatively short distances, comparatively more markers must be typed, as each marker allows inference about relatively small segments of flanking DNA. In a population with more extensive LD, fewer markers may be typed and the workload is concomitantly lower.
Purebred dogs have several features that make them attractive for mapping traits of interest in both companion animal health and human health and biology (Galibert et al. 1998; Ostrander and Kruglyak 2000; Patterson 2000; Galibert and Andre 2002). First, dogs share many of the most frequently occurring diseases that plague humans, including cancer, heart disease, deafness, blindness, and joint disease (for reviews, see Patterson 1980; Patterson et al. 1982; Ostrander and Giniger 2001; Ostrander and Comstock 2004). In addition, individual breeds often exhibit breed-specific patterns of disease susceptibility (Patterson et al. 1982; Galibert et al. 1998; Ostrander et al. 2000), consistent with individual breeds carrying a subset of the possible alleles responsible for canine diseases. Diseases that are prevalent in one or a small number of breeds either are not observed in others or are observed infrequently. This observation is expected; as a result of differential selection for traits important in behavior and appearance, dog breeds have developed into closed breeding populations with a high degree of genetic differentiation (Parker et al. 2004).
Key to the success of such studies is an understanding of the extent to which LD is observed across the dog genome. While one might expect extensive LD due to admixture, the use of popular sires, and population bottlenecks (Ostrander and Kruglyak 2000), little empirical data have been available to date. The precise values of LD in any particular breed will depend not only on the above considerations but also on additional factors, such as the number of initial founders and relative popularity of the breed. Hyun et al. (2003) used a set of microsatellite markers to measure LD around the copper toxicosis locus on chromosome 10q26 and reported that it extended for 33.3 cM in the Bedlington Terrier. This locus is unusual, however, since at least a third of Bedlington Terriers were reportedly carriers of the copper toxicosis mutation in 1997 (Yuzbasiyan-Gurkan et al. 1997), and as recently as the late 1970s, nearly two thirds of Bedlington Terriers in the United States were affected with the disease (Hardy and Stevens 1978). This suggests that the gene lies near another gene that is under selection or that a very limited number of popular sires are responsible for most of the gene pool. The only other study of LD in dogs focused on a purpose-bred pedigree of Labrador Retrievers and Greyhounds managed at Cornell Veterinary College. This cross was designed to identify genes for hip dysplasia (Lou et al. 2003). In that study, again using microsatellites, LD was observed more frequently between pairs of markers separated by <40 cM than those separated by >40 cM. Neither of the above study designs is useful for predicting how well LD mapping will work in purebred dogs that are not part of unusual breeds or purpose-bred pedigrees, nor do they predict the number of SNPs needed for a canine SNP map.
We recently completed a phylogenetic analysis of 85 of the most popular breeds registered by the American Kennel Club (AKC). Phylogenetic analysis initially separated a dozen breeds with ancient origins, and of Asian origin, from the remaining breeds with modern European origins. Then, using genetic cluster analysis, we demonstrated that all remaining breeds could be assigned to one of four main populations, reflecting similarities in morphology and geographic origin (Parker et al. 2004).
With that information in hand, we sought to determine the extent of LD in the dog genome, as well as the degree of haplotype sharing among domestic dog breeds, in a representative set of selected breeds. We chose five breeds with distinctive phylogenetic origins and population histories: Akita, Bernese Mountain Dog, Golden Retriever, Labrador Retriever, and Pekingese, which together represent the four groups described by Parker et al. (2004). In addition, each breed features a distinct population history, providing a good estimate for the range of LD extent likely to be present among different breeds. To analyze each breed, we discovered and genotyped single nucleotide polymorphisms (SNPs) and insertion/deletions (indels) by resequencing in up to 20 unrelated dogs from each breed. In this report we describe extensive LD in all five dog breeds; D′ falls to 0.5 at marker pair distances ranging from 0.4 to 3.2 Mb. This is dramatically longer than the <100-kb range that has been reported for human populations (Moffatt et al. 2000; Daly et al. 2001; Patil et al. 2001; Reich et al. 2001; Stephens et al. 2001a; Dawson et al. 2002; Gabriel et al. 2002; Weiss and Clark 2002; Varilo et al. 2003; Ke et al. 2004). We conclude that whole genome association scans are more tractable in dog breeds than in human populations. In addition, we report a high degree of haplotype sharing among phylogenetically divergent breeds.
Results
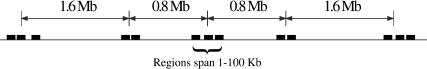
Sequence reads at each locus were clustered in five regions: a central region, and regions 0.8 and 2.4 Mb distant from the center in each direction along the chromosome (Fig. 1). Each region was composed of one to five sequence reads (average = 2.8) and spanned 775 bp to 96 kb, with 15 of the 25 regions spanning at least 50 kb. This ∼50-kb scale yields markers separated at distances comparable to that at which LD breaks down in human populations (Abecasis et al. 2001; Reich et al. 2001; Varilo et al. 2003; Ke et al. 2004), and in addition, our experimental design also allowed us to measure LD over much longer distances of 0.8-5 Mb. Pilot studies in the Golden Retriever and Pekingese breeds at two loci on chromosome 12 suggested that the extent of LD in dog breeds would likely exceed that seen in human populations (data not shown). Finally, by resequencing the entire data set of 97 dogs from all five breeds, we have minimized ascertainment bias related to rare SNPs and/or SNPs “private” to one or a subset of breeds (Bonnen et al. 2000; Akey et al. 2002; Carlson et al. 2003), thus providing a representative set of polymorphisms with which to compare the five breeds.
Figure 1.
Schematic design of resequencing strategy for each of five loci, one each on canine chromosomes 1, 2, 3, 34, and 37. At each locus sequence reads were clustered into five “regions.” Each region is composed of one to five sequence reads (average, 2.8). Regions are separated by ∼0.8 or ∼1.6 Mb.
All biallelic markers were discovered in a set of 95 dogs (20 from each of four breeds, plus 15 Akitas; two more Akitas were added to the data set at a later point). In 53,088 resequenced bases, we discovered and genotyped 220 markers, including 200 biallelic SNPs, one triallelic SNP, and 19 indels. The ratio of indels to SNPs found in the dog is ∼1:10, similar to what is reported for the human genome (Clark et al. 1998; Cambien et al. 1999; Crawford et al. 2004). For 126 of the 220 markers, the minor allele frequency (MAF) over all chromosomes was <0.2, and for 52 markers the minor allele was observed in only one of the breeds. We observed large differences in allele frequencies among the five breeds, and Akita was least similar to the other breeds. The correlation coefficients between the overall allele frequencies and those of Akita, Bernese Mountain Dog, Golden Retriever, Labrador Retriever, and Pekingese were 0.52, 0.69, 0.70, 0.72, and 0.64, respectively.
For the LD analysis, we used only SNPs for which the MAF was at least 0.2 in the breed under consideration. By removing rare SNPs, we reduced the upward bias in the D′ statistic expected due to the limited sample size in each breed. Using this criterion, on average, each breed had at least two SNPs in 63% of regions.
We first analyzed patterns of LD within regions. Most pairs of SNPs separated by 1-100 kb were in LD. Across all breeds and averaging over all regions with multiple SNPs, we found that 71% of intra-region SNP pairs were in significant LD with D′ > 0.8 (Table 1). By breed, a higher proportion of intra-region SNP pairs were in LD in Pekingese (88%) and Bernese Mountain Dog (87%) than in Golden Retriever (68%), Labrador Retriever (58%), and Akita (53%). These differences occur despite broadly similar numbers of SNP pair comparisons in each breed (217, 185, 137, 155, and 240, respectively). Thus, the extent of LD in each of five dog breeds exceeds the extent of LD reported for human populations (Reich et al. 2001; Weiss and Clark 2002), even those that are comparatively isolated (Mohlke et al. 2001; Service et al. 2001; Hall et al. 2002; Varilo et al. 2003; Tenesa et al. 2004). Values for the extent of LD in human populations vary with different experimental designs but are on the order of 10s of kb.
Table 1.
Fraction of SNP pairs <100 kb apart and in significant linkage disequilibrium (D′ > 0.8; P < 0.01)
|
Dog breed
|
||||||
|---|---|---|---|---|---|---|
| Locus on chromosome | AK | BM | GR | LB | PK | All breeds |
| 1 | 0.50 | 1.00 | 1.00 | 0.50 | 0.85 | 0.66 |
| 2 | 0.82 | 0.95 | 0.86 | 1.00 | 1.00 | 0.93 |
| 3 | 0.57 | 0.84 | 0.77 | 0.64 | 1.00 | 0.78 |
| 34 | 0.41 | 0.96 | 0.39 | 0.29 | 0.64 | 0.51 |
| 37 | 0.48 | 0.81 | 0.77 | 0.71 | 1.00 | 0.72 |
| All loci | 0.53 | 0.87 | 0.68 | 0.58 | 0.88 | 0.71 |
(AK) Akita; (BM) Bernese Mountain Dog; (GR) Golden Retriever; (LB) Labrador Retriever; (PK) Pekingese.
We next looked for evidence of more extensive LD and found that many SNP pairs spaced between 0.8 and 5 Mb were in LD (Table 2; Fig. 2A, red squares). In fact, we detected significant LD with D′ > 0.8 for one-fifth of all pairs of SNPs occurring in different regions but at the same locus (i.e., the SNPs are separated by ∼0.8, 1.6, 2.4, 3.2, or 5 Mb, see schematic in Fig. 1). The values across the five loci and five breeds varied from 0% of SNP pairs (observed in the Golden Retriever at three loci and Akita and Labrador Retriever at one each) to 91% of SNP pairs in LD (in Pekingese on CFA37). In all breeds, we observed at least two loci containing SNPs that are in LD and separated by >2.4 Mb.
Table 2.
Fraction of SNP pairs 0.8–5 Mb apart and in significant linkage disequilibrium (D′ > 0.8; P < 0.01)
|
Dog breed
|
||||||
|---|---|---|---|---|---|---|
| Locus on chromosome | AK | BM | GR | LB | PK | All breeds |
| 1 | 0.12 | 0.75 | 0.00 | 0.03 | 0.55 | 0.33 |
| 2 | 0.00 | 0.49 | 0.00 | 0.08 | 0.33 | 0.23 |
| 3 | 0.13 | 0.10 | 0.04 | 0.16 | 0.10 | 0.10 |
| 34 | 0.19 | 0.09 | 0.00 | 0.00 | 0.17 | 0.11 |
| 37 | 0.28 | 0.61 | 0.17 | 0.05 | 0.91 | 0.30 |
| All loci | 0.17 | 0.27 | 0.05 | 0.08 | 0.31 | 0.19 |
| Unlinked | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
(AK) Akita; (BM) Bernese Mountain Dog; (GR) Golden Retriever; (LB) Labrador Retriever; (PK) Pekingese. (Unlinked) SNP pairs on separate chromosomes.
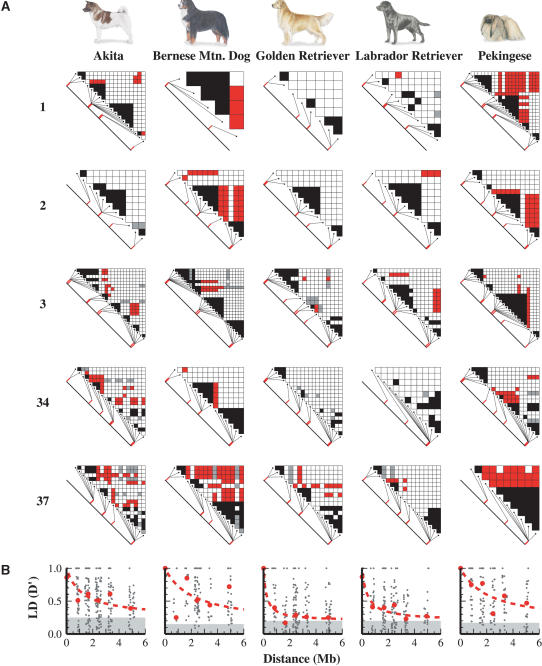
Figure 2.
(A) Triangle plots of LD between common SNPs (MAF > 0.2) for each chromosome and dog breed. Black squares represent SNP pairs in significant LD (D′ > 0.8; P < 0.01) and <100 kb apart. Red squares indicate significant LD between markers at least 0.8 Mb apart. Gray squares represent high LD values (D′ > 0.8) that are less significant (P > 0.01). (B) LD (D′) decay plots for common SNPs (MAF > 0.2) in each dog breed corresponding to the triangle plots above. Red circles show the median LD values within each bin of marker pair distances, and the dashed line is a best-fitting curve to the median values. Gray shading indicates the background LD level between unlinked markers. The top of the gray rectangle shows the median of D′ values between markers on different chromosomes.
As expected, we observed variable patterns of LD across the five loci surveyed as well as between breeds. In particular, the difference between the retriever breeds and the other three breeds was apparent at most loci. For example, at the locus on CFA1 the Golden Retriever and Labrador Retriever had almost no SNP pairs in LD except for those within regions (<100Kb). In stark contrast, SNPs located 3.2 Mb apart were in LD in the Bernese Mountain Dog, and SNPs 5 Mb apart were in LD in the Akita and, frequently, in the Pekingese. At CFA2 the Golden Retriever and Akita showed little evidence for LD extending over megabases, whereas many SNP pairs spaced 0.8 and 2.4 Mb apart in Pekingese and Bernese Mountain Dogs were in LD. CFA3 had a more uniform pattern across breeds: Every breed had SNP pairs in LD that were separated by 0.8-2.4 Mb, although the Golden Retriever had the fewest. At the CFA34 locus, there were no SNP pairs spaced 0.8-5 Mb apart in LD in either the Golden Retriever or Labrador Retriever. This contrasts with SNP pairs located >2.4 Mb apart that were in LD in the other three breeds. Interestingly, the CFA37 locus showed the most extensive LD, >0.8 Mb in all five breeds. Indeed, SNP pairs 3-5 Mb apart were found in LD in the Akita, Bernese Mountain Dog, Pekingese, and Golden Retriever.
Our results showing the extent of LD are summarized in Figure 2B, where the D′ values for all loci together are plotted for each breed. D′ falls to half its maximum value at 370 kb in Golden Retrievers, 680 kb in Labrador Retrievers, 2.4 Mb in Akitas, 3.0 Mb in Bernese Mountain Dogs, and 3.3 Mb in Pekingese. Thus, examination of only five dog breeds yields an approximately 10-fold range in the extent of LD in the dog genome.
We next sought to describe haplotype diversity and sharing among these breeds. We inferred haplotypes for all regions with at least four SNPs of 0.2 MAF or greater in all breeds, and in which the SNPs spanned a distance of at least 9 kb. Ten of the 25 regions met these criteria and had an average of 6.8 SNPs per region (range = 4 to 9). All five chromosomal loci were represented, with two, one, three, two, and two regions from CFA1, 2, 3, 34, and 37 loci, respectively.
When all five breeds were considered simultaneously, we found an average of 12.4 haplotypes per region (median = 10.5). The smallest number of haplotypes was six, at a region with the minimum four markers. The highest number of haplotypes was 30, at one of the two regions with nine markers. This represents an outlier, as the second-highest number of haplotypes was just 15. On average, there were approximately five haplotypes total at each region within each individual breed, with the Labrador Retriever displaying the highest average (6.4) and the Pekingese the lowest (4.1).
We were also interested in describing haplotype diversity in terms of the number of haplotypes carried by the majority of chromosomes in a breed. We therefore counted the number of chromosomes carrying each haplotype, and then determined the fewest number of haplotypes needed to account for at least 80% of chromosomes. For example, Table 3 shows that for region 1 Akita had four different haplotypes and they were carried by 41%, 38%, 12%, and 9% of Akita chromosomes sampled. Therefore, 80% of Akita chromosomes can be explained by three haplotypes. At the same region, the Bernese Mountain Dog, Golden Retriever, Labrador Retriever, and Pekingese require one, two, four, and three haplotypes, respectively, to account for 80% of their chromosomes. Averaging over all breeds and regions, 2.7 haplotypes account for 80% of chromosomes. By breed, on average, 2.9, 2.3, 2.9, 3.4, and 2.1 haplotypes explain 80% of chromosomes in the Akita, Bernese Mountain Dog, Golden Retriever, Labrador Retriever, and Pekingese, respectively. Using this method of counting, the Labrador retriever has 50% more haplotypes than does the Pekingese. Counting haplotypes across all dog chromosomes together and ignoring breed indication, an average of 4.5 haplotypes are contained in 80% of chromosomes at the 10 regions (range = 2 to 10). Thus, two to three haplotypes account for most chromosomes at each region in any single breed, and four to five haplotypes account for most chromosomes in all breeds surveyed.
Table 3.
Haplotypes inferred at each region and the fraction of chromosomes carrying each haplotype
|
Frequency
|
||||||
|---|---|---|---|---|---|---|
| Region | Haplotype | AK | BM | GR | LB | PK |
| 1 | GGTICAC | 0.09 | — | 0.78 | 0.33 | 0.20 |
| AGTICAC | — | 0.83 | 0.10 | 0.25 | 0.08 | |
| GGCDTTT | 0.12 | — | 0.10 | — | 0.58 | |
| GCTICAC | 0.38 | 0.18 | 0.03 | 0.20 | — | |
| GCCDTTT | 0.41 | — | — | — | — | |
| 2 | GCTCTTC | — | 0.08 | 0.30 | 0.53 | 0.60 |
| GGTCTTT | 0.12 | — | 0.60 | 0.23 | 0.10 | |
| ACATCCT | — | 0.78 | — | — | — | |
| ATATCCT | 0.68 | — | — | — | 0.15 | |
| ACTCTTC | — | 0.03 | 0.10 | 0.08 | 0.05 | |
| 3 | CATITC | 0.65 | 0.28 | 0.73 | 0.50 | 0.80 |
| GGCDGT | 0.24 | 0.73 | 0.28 | 0.28 | 0.20 | |
| 4 | CCTTATC | 0.47 | 0.38 | 0.20 | 0.13 | 0.55 |
| CCCGTCT | 0.09 | 0.15 | 0.05 | 0.38 | 0.30 | |
| AACGTCT | — | 0.13 | 0.65 | 0.40 | — | |
| CCCGACT | 0.26 | — | — | — | 0.15 | |
| AATTATC | — | 0.35 | — | — | — | |
| 5 | CADCAATAG | 0.09 | 0.45 | 0.15 | 0.28 | 0.60 |
| CAITATCCA | 0.35 | 0.05 | 0.40 | 0.25 | — | |
| GGITATCCA | 0.03 | 0.43 | 0.23 | 0.30 | — | |
| GGITCTCCA | 0.38 | — | — | — | 0.30 | |
| CAITCTCCA | 0.09 | 0.03 | 0.23 | 0.03 | 0.10 | |
| 6 | TCGIGCTT | — | 0.55 | — | — | 0.55 |
| CTADATTT | — | 0.33 | 0.40 | 0.28 | — | |
| CCGIGCTT | 0.62 | — | — | — | — | |
| CTADATCC | 0.12 | — | 0.10 | 0.30 | — | |
| CTAIGCTT | 0.06 | — | 0.05 | — | 0.30 | |
| CTAIGCTC | — | — | 0.30 | — | 0.08 | |
| TCGDATCC | — | — | 0.05 | 0.28 | — | |
| CCGIGCCC | 0.18 | 0.10 | — | — | — | |
| 7 | GCAAGATTA | — | 0.50 | 0.35 | 0.15 | — |
| GCGGGGCCG | 0.56 | — | — | — | 0.25 | |
| AGAGCACTA | — | 0.33 | 0.23 | — | — | |
| GCAGGGCCG | 0.06 | — | 0.10 | 0.28 | — | |
| AGAGCACCG | — | — | — | 0.03 | 0.35 | |
| GCGGGACTA | — | — | — | — | 0.28 | |
| AGGGCACCG | — | 0.10 | 0.03 | 0.10 | 0.03 | |
| 8 | GIGA | 0.59 | — | 0.40 | 0.75 | 0.70 |
| GDAG | 0.32 | 0.65 | 0.13 | — | 0.08 | |
| AIGA | — | — | 0.20 | 0.08 | 0.23 | |
| AIGG | — | 0.33 | — | — | — | |
| 9 | CCAATGG | 0.53 | 0.33 | 0.30 | 0.28 | 0.55 |
| TATGCTC | — | 0.20 | 0.45 | 0.43 | 0.33 | |
| TCTGCGC | 0.18 | — | 0.10 | 0.05 | 0.05 | |
| TATGCGC | — | 0.23 | — | 0.10 | — | |
| CCTGCGC | — | 0.23 | 0.03 | — | 0.05 | |
| CATGCGC | 0.18 | — | 0.05 | 0.08 | — | |
| 10 | ADTT | 0.32 | 0.18 | 0.18 | 0.28 | 0.88 |
| ADTG | 0.18 | 0.38 | 0.38 | 0.30 | — | |
| GIAT | 0.06 | 0.28 | 0.25 | 0.20 | 0.05 | |
| GDAT | 0.12 | 0.13 | 0.15 | 0.20 | 0.08 | |
Haplotypes that are observed on <5% of all chromosomes are not shown. Alleles for insertion/deletion polymorphisms are: (I) inserted; (D) deleted sequence. Note that the second marker in region 2 is a triallelic SNP. Haplotypes were inferred for these 10 regions since they contain at least four SNPs of ≥0.2 MAF in all breeds, and the SNPs span a distance of at least 9 kb. (AK) Akita; (BM) Bernese Mountain Dog; (GR) Golden Retriever; (LB) Labrador Retriever; (PK) Pekingese.
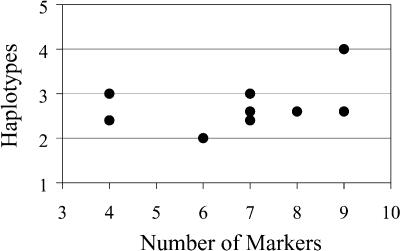
For the 10 regions in which haplotypes were analyzed, the number of markers within a region did not affect the conclusions about haplotype diversity. Two of the 10 regions contained the minimum of just four markers each, but by breed, the average number of haplotypes in these two regions was 3, 2.5, 3.5, 3, and 1.5 haplotypes for Akita, Bernese Mountain Dog, Golden Retriever, Labrador Retriever, and Pekingese, respectively. These values are essentially the same as the averages by breed over all 10 regions together (Fig. 3).
Figure 3.
Relationship between the number of markers and the number of haplotypes observed for 10 regions selected for haplotype analysis. This count of haplotypes is the average over the five breeds for the number of haplotypes carried by 80% of chromosomes within the breed. Two regions with four markers have the same average number of haplotypes as regions with more markers.
For each breed pair, we next considered how many chromosomes on a per breed basis carry shared haplotypes. Over the 10 breed pairings, an average of 63% of the chromosomes within each breed in a given pair carried haplotypes shared between both breeds in the pair. The highest sharing occurred in the Golden Retriever and Labrador Retriever pair (84% of chromosomes carried shared haplotypes), while the lowest occurred between the Akita and Bernese Mountain Dog pair (46%) (Table 4). In fact, looking over all breed pairings, each breed's highest degree of sharing was with the Golden Retriever, with the exception of Akita. Although this sharing is only moderately higher than the sharing among other breeds, the Golden Retriever's creation from a rich mixture of breeds places this finding in context (see Discussion). In contrast, sharing between Akita and other breeds was low; the lowest degree of Bernese Mountain Dog, Golden Retriever, and Labrador Retriever sharing occurred with the Akita. Interestingly, the degree of sharing between Pekingese and Akita was average, not low, when compared with Pekingese sharing with the other breeds (Table 4).
Table 4.
Haplotype sharing between pairs of dog breeds
| BM | LB | GR | PK | |
|---|---|---|---|---|
| AK | 0.46 | 0.55 | 0.60 | 0.65 |
| BM | 0.62 | 0.69 | 0.57 | |
| LB | 0.84 | 0.62 | ||
| GR | 0.69 |
The table entry indicates the proportion of breed chromosomes carrying a haplotype shared between the two breeds (row and column). The value here is the average of the proportion in each of the two breeds of the given pair, and is also averaged over all 10 regions in which haplotypes were analyzed. Breeds: (AK) Akita; (BM) Bernese Mountain Dog; (GR) Golden Retriever; (LB) Labrador Retriever; (PK) Pekingese.
We next asked whether the majority of haplotypes were shared among all five breeds. Of the total 124 observed haplotypes, just 10 were identified in all five breeds (Table 5). In the five regions in which they occurred, there is an average of 6.6 markers, very similar to the overall average of 6.8 markers. For each breed, the number of haplotypes shared by all breeds, by definition, must be equal. However, such haplotypes may be common or rare in different breeds. Surprisingly, the proportion of chromosomes carrying such haplotypes was nearly equal across four of the five breeds, with 26%, 29%, 25%, 25%, and 41% of Akita, Bernese Mountain Dog, Golden Retriever, Labrador Retriever, and Pekingese chromosomes, respectively, carrying these haplotypes (Table 5).
Table 5.
Number of chromosomes in each breed carrying haplotypes shared with zero to four other breeds
|
Chromosomes carrying haplotypes (and number of haplotypes) shared with zero to four other dog breeds
|
||||||
|---|---|---|---|---|---|---|
| Breed | 0 | 1 | 2 | 3 | 4 | Total |
| AK | 63 (14) | 79 (11) | 32 (9) | 76 (9) | 90 (10) | 340 (53) |
| BM | 69 (12) | 49 (5) | 50 (7) | 116 (10) | 116 (10) | 400 (44) |
| LB | 52 (23) | 32 (9) | 64 (9) | 151 (13) | 101 (10) | 400 (64) |
| GR | 15 (8) | 35 (9) | 90 (13) | 160 (14) | 100 (10) | 400 (54) |
| PK | 23 (7) | 79 (10) | 46 (4) | 88 (10) | 164 (10) | 400 (41) |
In parentheses are the number of haplotypes shared. The numbers of chromosomes and haplotypes for all 10 regions are added together, so, for example, the total of 400 chromosomes for four of the breeds comes from 40 chromosomes (20 dogs) assayed at each of 10 regions. Thirty-four chromosomes were assayed in Akita. Breeds: (AK) Akita; (BM) Bernese Mountain Dog; (GR) Golden Retriever; (LB) Labrador Retriever; (PK) Pekingese.
Discussion
Because of the closed breeding pools, detailed population records, and large variation in morphology, disease susceptibility, and behavior in purebred domestic dogs, we and others have hypothesized that complex genes could be localized in the dog by using LD-based mapping approaches (Ostrander and Kruglyak 2000; Hyun et al. 2003). In recent years, resources and knowledge have advanced to the point where this is now feasible. We and colleagues have produced meiotic linkage (Mellersh et al. 1997), radiation hybrid (Breen et al. 2001; Guyon et al. 2003; Breen et al. 2004), and comparative maps (Breen et al. 1999; Yang et al. 1999; Sargan et al. 2000) of the dog genome. In addition, high-quality genome sequence now exists for dog, with the availability of 1.5× sequence of a Standard Poodle (Kirkness et al. 2003) and 7.8× assembled Boxer sequence (www.genome.ucsc.edu/). Together, these efforts will describe many thousands of canine SNPs for the first time. Concomitantly, recent work by Parker et al. (2004) suggests that, while each dog breed is a distinct and identifiable genetic entity, most breeds may nevertheless be grouped into a limited number of genetic clusters.
Motivated by this recent progress, we sought to describe LD in dog breeds in order to determine the relationship between breed history and extent of LD and to estimate the number of SNPs that would be needed for association studies. We determined that dog breeds feature extensive LD, particularly when compared with humans. Purebred dogs, therefore, hold strong promise for both simple and complex trait mapping studies in the near future.
In dogs, the trend toward genetic differentiation accelerated 150 years ago with the establishment of dog breeding clubs in Europe and the emerging concept of a “pure” breed. Today the “breed barrier rule” is firmly established: No dog may be a member of an AKC-registered breed unless its dam and sire are registered members of the same breed (AKC 1998). The genetic isolation implied by application of this rule has profound implications for the utility of purebred dog populations in mapping studies. First and foremost, we would predict that only a few of the many potentially contributing genes for a complex trait will actually be segregating a causative variant in any single breed. This reduction in locus heterogeneity is crucial to the success of both classical linkage mapping and whole genome association studies and represents a large part of the rationale for the study of dog populations and their diseases. Second, closely related breeds will frequently share a common ancestral mutation responsible for traits shared by these breeds, and unique meiotic recombination events occurring in popular sires and breed founders will have reduced the relevant haplotype to a small region of sharing across the breeds.
In this study we show, first, that five dog breeds have LD that extends ∼10 to 100 times farther than does LD in even the most isolated human populations (Hall et al. 2002; Varilo et al. 2003; Tenesa et al. 2004). Large and outbred populations are predicted to have LD of minimal extent, and this is indeed seen in human populations (Reich et al. 2001). Depending on how LD is measured, it extends ∼10 kb in African populations and up to 50-100 kb in European populations (Reich et al. 2001; Weiss and Clark 2002). It has been estimated that several hundred thousand markers must be typed in a typical whole genome association study in human populations (Kruglyak 1999; HapMap Consortium 2003). Our results show, in contrast, that only ∼5 to 30,000 markers would be needed for an experiment of similar design in dogs.
Our findings of extensive LD in the dog is comparable with that observed in other domesticated species. In five distinct pig populations, LD extended for ∼5 to 10 cM (measured as the distance at which D′ falls to 0.5), but at least one of the two loci surveyed is almost certainly under selection (Nsengimana et al. 2004). In cattle and sheep, similar results were observed (Farnir et al. 2000; McRae et al. 2002; Tenesa et al. 2003). In these situations, extensive LD is perhaps not surprising, as the use of popular sires dominates domestic farm animal breeding programs. In one extreme case, the effective population size of Japanese black cattle was estimated to be ∼20 animals during the 1990s (Nomura et al. 2001).
The dog breeds chosen for this study were carefully selected to represent a range of population histories. The Labrador and Golden Retrievers have been among the two most popular breeds in the United States for the past century. The Labrador was originally described as a small water dog from Newfoundland used for fowling (AKC 1998) and was established as a breed in England during the 19th century. Today, distinct lines are bred for field and obedience trials, drug detection and cadaver recovery, and as guide dogs for the blind (Fogel 1995; Wilcox and Walkowicz 1995). It isn't surprising that the extent of LD is comparatively lower in this breed. The Golden Retriever was deliberately created in the 1850s from a combination of Bloodhound, small Newfoundland, Labrador, Irish Setter, and the now-extinct Tweed Water Spaniel, and its similarly low levels of LD likely reflect the lack of a single founding population (Wilcox and Walkowicz 1995; AKC 1998). In addition, both breeds enjoy enormous popularity today and account together for ∼22% of newly registered dogs each year (www.AKC.org), even though the AKC recognizes >150 unique breeds.
By comparison, while the Pekingese breed is somewhat popular today, with 4700 registrations in 2003, it is believed to derive from a small number of founders (∼20) that came from China during the second half of the 19th century (Wilcox and Walkowicz 1995; AKC 1998). The United States population specifically derives from similarly small numbers of animals imported from Great Britain. The Akita and Bernese Mountain Dog also represent population dynamics typical of rare breeds. The Akita population was severely reduced in Japan during World War II, and the current United States population was founded by a small number of animals imported to the United States from Japan following the war (Rogers and Brace 1995; Wilcox and Walkowicz 1995; AKC 1998). The Akita is neither popular nor rare today; 3200 dogs were newly registered by the AKC in 2003. Like the Akita, the Bernese Mountain Dog population underwent a tight population bottleneck during World War II (Rogers and Brace 1995; AKC 1998), but it is a breed of modest popularity today, with 3100 registrations in 2003. The known history for these three breeds, with large reductions in effective population size, fits well with the long extent of LD in each. We predict that many other breeds should also feature LD at least this extensive based on their rarity.
We also analyzed haplotype diversity and sharing. We find that between two and three haplotypes account for 80% of the chromosomes in each breed, and the variation in this number fits with what is known about the histories of the different breeds. For example, the Pekingese has the fewest haplotypes and is known to have undergone tight population bottlenecks. By comparison, the continuous popularity of the Labrador Retriever and the selection for lines with distinct behaviors argue for greater haplotype diversity, as we observed. The observed degree of haplotype sharing by pairs of breeds is consistent with breed membership in genetic clusters as described in Parker et al. (2004); for instance, Akita, the most divergent breed, showed the least average sharing with other breeds, but shared the most with Pekingese, the other breed with partial membership in the Asian/Ancient cluster (Parker et al. 2004).
This pattern of haplotype diversity has important implications for association mapping. The large extent of LD limits the number of markers needed for genome-wide studies, as discussed above. The limited haplotype diversity within regions of high LD means that a small number of markers (haplotype-tagging SNPs or tagSNPs) (Daly et al. 2001; Johnson et al. 2001; Patil et al. 2001; Gabriel et al. 2002; Carlson et al. 2003) is needed to distinguish among the haplotypes, further reducing the necessary marker density. The high degree of haplotype sharing among breeds suggests that it may be possible to develop an efficient universal set of tagSNPs for studies in many different breeds.
While association studies in humans are predicted to require hundreds of thousands of SNPs (Kruglyak 1999; HapMap Consortium 2003) we show here the same traits can be mapped in dogs using almost two orders of magnitude fewer SNPs. But the long extent of LD may prove less welcome when attempting to fine map and ultimately identify a causal mutation. Our data do provide evidence that LD is not complete over ∼100-kb distances (Table 1), with breakdown of LD with distance at least in some breeds at some regions, although our experimental design does not allow us to quantify this precisely. To further address this situation during fine mapping, we propose to genotype sets of breeds that all carry the same trait or disease susceptibility. The fact that we observe the same haplotypes in multiple breeds argues favorably for this strategy. The breeds selected for any given study can be picked based on the phenotype in question as well as on their genetic relatedness as determined by our recent studies of population structure (Parker et al. 2004). Closely related breeds are more likely to share ancestral chromosomes, and hence ancestral mutations responsible for the same traits of interest, and recombination events in multiple breeds, including ones with comparatively short extent of LD, can be used to narrow the regions of interest (Ostrander and Kruglyak 2000).
At present we are witnessing the fortuitous confluence of exciting experimental findings combined with the maturation of vital genomic tools, including radiation hybrid (RH) (Breen et al. 1999; Guyon et al. 2003, 2004) and comparative maps (Breen et al. 2004), two genomic sequences (Kirkness et al. 2003; www.genome.ucsc.edu), and dense marker sets (Clark et al. 2004) suitable for genome-wide scans. We have suggested previously that the large size of canine pedigrees, together with the reduction in genetic complexity caused by closed breeding programs, would eventually allow for rapid identification via linkage mapping of loci of interest in dogs and then in humans (Patterson et al. 1988; Galibert et al. 1998; Ostrander et al. 2000). We believe dogs hold promise for association mapping as well, and we can for the first time begin to validate that claim by showing that markers separated by megabases are in LD. This work sets the stage for association studies aimed at understanding the genetics not only of disease susceptibility but also of morphology and behavior. In summary, the findings presented here and by Parker et al. (2004), together with the 7.8× canine genome sequence, officially end the era of canine genomics as we first defined it in 1992 (Ostrander et al. 1992) and initiate a new era of canine functional genomics, at the end of which we may finally understand how DNA variation causes diversity and distinction.
Methods
Sample collection and selection
DNA was extracted from whole blood samples collected from adult Akitas, Pekingese, Golden Retrievers, and Bernese Mountain Dogs using a standard phenol-chloroform-based purification protocol. For Labrador Retrievers, samples were collected from cheek swabs using cytobrushes (Medical Packaging Corporation) and DNA was purified using Qiagen Mini Blood DNA Kits according to the manufacturer's protocol. The Fred Hutchinson Cancer Research Center (FHCRC) Institutional Animal Care and Use Committee approved all protocols. To best represent the multiple lineages within breeds, we selected dogs such that no dog in the sample set shared any grandparent with any other dog in the set. For all studies reported here we used the same set of dogs for each breed: 17 Akitas, 20 for each of the other four breeds. Our nomenclature for breed names follows that of the AKC (AKC 1998) and www.akc.org.
Selection of loci
Sequence reads from the 1.5× shotgun sequence of a standard poodle were aligned to finished human genome sequence (National Center for Biotechnology Information [NCBI] build 31, November 2002, http://www.ncbi.nlm.nih.gov/) to define segments of ordered synteny between dog and human. Approximately 50% of the 1.5× sequence data could be assigned to these segments on the basis of sequence conservation (Kirkness et al. 2003). One syntenic block was selected from each of five canine chromosomes: Canis familiaris (CFA) 1, 2, 3, 34, and 37. Syntenic blocks were chosen based on length of the alignment available, and gene content was ignored. A series of canine sequence reads (Supplemental Table 2) were then chosen that spanned up to 100 kb of human DNA within five individual regions at each of the five loci. At each locus, the central region is flanked by two regions on either side, at 1-Mb and 3-Mb distance in the human sequence from the center (Fig. 1). Because the physical size of the dog genome is estimated to be 15%-20% smaller than that of the human genome (Kirkness et al. 2003), the true distances in the dog genome are shorter. To make this relationship precise, we utilized the recently assembled 7.8× Boxer sequence (NCBI build 1.1, July 2004, http://www.ncbi.nlm.nih.gov/; K. Lindblad-Toh, pers. comm.) to define positions for our markers. Our loci were designed to span 6 Mb in the human sequence and are found to span, on average, 5 Mb in the Boxer sequence. All marker positions reported here are derived from the Boxer sequence. Canine reads were masked by RepeatMasker (www.repeatmasker.org/), and primers for each region were designed by primer3 software (www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi). At each region, between one and five (average = 2.8) sequence reads were resequenced in the full set of 97 dog samples.
SNP discovery and resequencing
All SNPs and indels were discovered and genotyped by resequencing in 95 dogs, including 20 Bernese Mountain Dogs, Golden Retrievers, Labrador Retrievers, and Pekingese, and 15 of the 17 Akitas. Dog genomic DNA was arrayed using a BIOMEK FX in ∼10-ng aliquots to 96-well microtiter plates and then frozen for storage until later use. DNA from each 96-well plate was amplified using 17.5 U Biolase (BIOLINE), 115.2 μL 10× PCR buffer, 34.6 μL MgCl2 (50 mM), and 115.2 μL dNTP (1 mM dNTP). For both forward and reverse primers (Invitrogen) 5.8 μL of 100μΜ concentration solutions were used per 96-well plate. Amplification conditions were as follows: 1 min at 95°C denaturing, followed by 35 cycles of 94°C for 30 sec, 62°C at 30 sec, 72°Cat 30 sec, and a final 7-min hold at 72°C followed by a hold at 4°C.
For each sequence read, the presence of PCR amplicons was tested by dotting 2 μL PCR product and 2 μL 10 ng/mL ethidium bromide solution. These were visualized on an Eagle Eye 2 UV transilluminator (Stratagene). If successful, a 3-μL aliquot from each original PCR reaction was then cleaned for sequencing reactions by treatment with 5.7 U Exonuclease I (EPICENTER) and 0.6 U shrimp alkaline phosphatase (USB). Samples were incubated for 30 min at 37°C and then for 15 min at 80°C to inactivate the enzymes. Prepared template was then split for use in two sequencing reactions, utilizing both the forward and reverse PCR primers as sequencing primers. ABI BigDye 3.0 and 3.1 was used for all sequencing reactions according to the manufacturer's specifications. Sequencing reactions were cleaned on Sephadex G-50 Fine (Amersham Biosciences) columns using Millipore MultiScreen-HV plates. All sequence data were collected using an ABI 3730 sequencer.
SNP genotyping
Raw sequence reads from forward and reverse reactions for all samples were analyzed and aligned using Polyphred (Nickerson et al. 1997) and Phrap (Ewing and Green 1998; Ewing et al. 1998) and visualized using the Consed editor (Gordon et al. 1998). All Polyphred genotype calls were individually quality checked by viewing the forward and reverse chromatograms in Consed. After the first pass, data collection for missing data points (<3% of data set) was attempted a second time in the same way with a single PCR reaction and two sequencing reactions, yielding a final data set with only 1% missing data.
LD and haplotype analysis
Nucleotide positions were assigned to all SNPs and indels based on the 7.8× Boxer sequence (NCBI build 1.1, July 2004, http://www.ncbi.nlm.nih.gov/). For LD calculations SNPs and indels were included if the MAF within the given breed was at least 0.2. In addition, one triallelic SNP (BLA12-253) (see Supplemental Table 1) was excluded from LD analysis, and an unlikely apparent third allele for indel BLC15-148 was coded as missing data. The apparent third allele was homozygous in a single dog, not observed in any other dogs, and based on a single PCR reaction.
The LD statistic D′ and significance levels (Hartl and Clark 1997) were calculated for all possible marker pairs. Significance levels were calculated from χ2 tests on the two-by-two table of the estimated haplotype frequencies (Hill 1974). For haplotype analysis, SNPs and indels were included if the MAF was at least 0.2 when the entire data set of 97 dogs was considered together. Haplotypes were inferred independently within each breed using the program PHASE (Stephens et al. 2001b). This program has been demonstrated to possess increased accuracy when estimating haplotypes when compared to parsimony algorithms (Clark 1990) and standard EM algorithms (Fallin and Schork 2000).
Acknowledgments
We acknowledge the support of the Burroughs Wellcome Innovation Award (E.A.O. and L.K.), grants from the AKC-Canine Health Foundation (E.A.O.), National Institutes of Health training grant T32 HG00035 (H.G.P.), and a postdoctoral fellowship from the Waltham Foundation (N.B.S.). E.A.O. also acknowledges support from K05 CA90754 and L.K. from R37 MH59520. L.K. is a James S. McDonnell Centennial Fellow. Special thanks to members of Local and National Clubs for the Akita, Labrador Retriever, Bernese Mountain Dog, Pekingese, and Golden Retriever breeds for their enthusiasm as well as willingness to provide samples and pedigree information.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3147604. Article published online ahead of print in November 2004.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Abecasis, G.R., Noguchi, E., Heinzmann, A., Traherne, J.A., Bhattacharyya, S., Leaves, N.I., Anderson, G.G., Zhang, Y., Lench, N.J., Carey, A., et al. 2001. Extent and distribution of linkage disequilibrium in three genomic regions. Am. J. Hum. Genet. 68: 191-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akey, J.M., Zhang, G., Zhang, K., Jin, L., and Shriver, M.D. 2002. Interrogating a high-density SNP map for signatures of natural selection. Genome Res. 12: 1805-1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The American Kennel Club (AKC). 1998. The complete dog book. Howell Book House, New York.
- Bonnen, P.E., Story, M.D., Ashorn, C.L., Buchholz, T.A., Weil, M.M., and Nelson, D.L. 2000. Haplotypes at ATM identify coding-sequence variation and indicate a region of extensive linkage disequilibrium. Am. J. Hum. Genet. 67: 1437-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breen, M., Thomas, R., Binns, M.M., Carter, N.P., and Langford, C.F. 1999. Reciprocal chromosome painting reveals detailed regions of conserved synteny between the karyotypes of the domestic dog (Canis familiaris) and human. Genomics 61: 145-155. [DOI] [PubMed] [Google Scholar]
- Breen, M., Jouquand, S., Renier, C., Mellersh, C.S., Hitte, C., Holmes, N.G., Cheron, A., Suter, N., Vignaux, F., Bristow, A.E., et al. 2001. Chromosome-specific single-locus FISH probes allow anchorage of an 1800-marker integrated radiation-hybrid/linkage map of the domestic dog genome to all chromosomes. Genome Res. 11: 1784-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breen, M., Hitte, C., Lorentzen, T.D., Thomas, R., Cadieu, E., Sabacan, L., Scott, A., Evanno, G., Parker, H.G., Kirkness, E., et al. 2004. An integrated 4249 marker FISH/RH map of the canine genome. BMC Genomics 5: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambien, F., Poirier, O., Nicaud, V., Herrmann, S.M., Mallet, C., Ricard, S., Behague, I., Hallet, V., Blanc, H., Loukaci, V., et al. 1999. Sequence diversity in 36 candidate genes for cardiovascular disorders. Am. J. Hum. Genet. 65: 183-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson, C.S., Eberle, M.A., Rieder, M.J., Smith, J.D., Kruglyak, L., and Nickerson, D.A. 2003. Additional SNPs and linkage-disequilibrium analyses are necessary for whole-genome association studies in humans. Nat. Genet. 33: 518-521. [DOI] [PubMed] [Google Scholar]
- Clark, A.G. 1990. Inference of haplotypes from PCR-amplified samples of diploid populations. Mol. Biol. Evol. 7: 111-122. [DOI] [PubMed] [Google Scholar]
- Clark, A.G., Weiss, K.M., Nickerson, D.A., Taylor, S.L., Buchanan, A., Stengard, J., Salomaa, V., Vartiainen, E., Perola, M., Boerwinkle, E., et al. 1998. Haplotype structure and population genetic inferences from nucleotide-sequence variation in human lipoprotein lipase. Am. J. Hum. Genet. 63: 595-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, L.A., Tsai, K.L., Steiner, J.M., Williams, D.A., Guerra, T., Ostrander, E.A., Galibert, F., and Murphy, K.E. 2004. Chromosome-specific microsatellite multiplex sets for linkage studies in the domestic dog. Genomics 85: 550-554. [DOI] [PubMed] [Google Scholar]
- Crawford, D.C., Carlson, C.S., Rieder, M.J., Carrington, D.P., Yi, Q., Smith, J.D., Eberle, M.A., Kruglyak, L., and Nickerson, D.A. 2004. Haplotype diversity across 100 candidate genes for inflammation, lipid metabolism, and blood pressure regulation in two populations. Am. J. Hum. Genet. 74: 610-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly, M.J., Rioux, J.D., Schaffner, S.F., Hudson, T.J., and Lander, E.S. 2001. High-resolution haplotype structure in the human genome. Nat. Genet. 29: 229-232. [DOI] [PubMed] [Google Scholar]
- Dawson, E., Abecasis, G.R., Bumpstead, S., Chen, Y., Hunt, S., Beare, D.M., Pabial, J., Dibling, T., Tinsley, E., Kirby, S., et al. 2002. A first-generation linkage disequilibrium map of human chromosome 22. Nature 418: 544-548. [DOI] [PubMed] [Google Scholar]
- Ewing, B. and Green, P. 1998. Base-calling of automated sequencer traces using phred, II: Error probabilities. Genome Res. 8: 186-194. [PubMed] [Google Scholar]
- Ewing, B., Hillier, L., Wendl, M.C., and Green, P. 1998. Base-calling of automated sequencer traces using phred, I: Accuracy assessment. Genome Res. 8: 175-185. [DOI] [PubMed] [Google Scholar]
- Fallin, D. and Schork, N.J. 2000. Accuracy of haplotype frequency estimation for biallelic loci, via the expectation-maximization algorithm for unphased diploid genotype data. Am. J. Hum. Genet. 67: 947-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnir, F., Coppieters, W., Arranz, J.J., Berzi, P., Cambisano, N., Grisart, B., Karim, L., Marcq, F., Moreau, L., Mni, M., et al. 2000. Extensive genome-wide linkage disequilibrium in cattle. Genome Res. 10: 220-227. [DOI] [PubMed] [Google Scholar]
- Fogel, B. 1995. The encyclopedia of the dog. DK Publishing, New York.
- Gabriel, S.B., Schaffner, S.F., Nguyen, H., Moore, J.M., Roy, J., Blumenstiel, B., Higgins, J., DeFelice, M., Lochner, A., Faggart, M., et al. 2002. The structure of haplotype blocks in the human genome. Science 296: 2225-2229. [DOI] [PubMed] [Google Scholar]
- Galibert, F. and Andre, C. 2002. The canine genome: Alternative model for the functional analysis of mammalian genes. Bull. Acad. Natl. Med. 186: 1489-1499; discussion 1499-1502. [PubMed] [Google Scholar]
- Galibert, F., Andre, C., Cheron, A., Chuat, J.C., Hitte, C., Jiang, Z., Jouquand, S., Priat, C., Renier, C., and Vignaux, F. 1998. The importance of the canine model in medical genetics. Bull. Acad. Natl. Med. 182: 811-821. [PubMed] [Google Scholar]
- Gordon, D., Abajian, C., and Green, P. 1998. Consed: A graphical tool for sequence finishing. Genome Res. 8: 195-202. [DOI] [PubMed] [Google Scholar]
- Guyon, R., Lorentzen, T.D., Hitte, C., Kim, L., Cadieu, E., Parker, H.G., Quignon, P., Lowe, J.K., Renier, C., Gelfenbeyn, B., et al. 2003. A 1 Mb resolution radiation hybrid map of the canine genome. Proc. Natl. Acad. Sci. 100: 5296-5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyon, R., Kirkness, E.F., Lorentzen, T.D., Hitte, C., Comstock, K.E., Quignon, P., Derrien, T., Andréa, C., Fraser, C.M., Galibert, F., et al. 2003. Building comparative maps using 1.5× sequence coverage: Human chromosome 1p and the canine genome. Cold Spring Harbor Symp. Quant. Biol. 68: 171-177. [DOI] [PubMed] [Google Scholar]
- Hall, D., Wijsman, E.M., Roos, J.L., Gogos, J.A., and Karayiorgou, M. 2002. Extended intermarker linkage disequilibrium in the Afrikaners. Genome Res. 12: 956-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HapMap Consortium. 2003. The International HapMap Project. Nature 426: 789-796. [DOI] [PubMed] [Google Scholar]
- Hardy, R. and Stevens, J. 1978. Chronic progressive hepatitis in Bedlington terriers. Proc. Am. Anim. Hosp. Assoc. (AAHA) 45: 187-190. [Google Scholar]
- Hartl, D. and Clark, A. 1997. Principles of population genetics. Sinauer Associates, Sunderland, MA.
- Hill, W.G. 1974. Estimation of linkage disequilibrium in randomly mating populations. Heredity 33: 229-239. [DOI] [PubMed] [Google Scholar]
- Hyun, C., Filippich, L.J., Lea, R.A., Shepherd, G., Hughes, I.P., and Griffiths, L.R. 2003. Prospects for whole genome linkage disequilibrium mapping in domestic dog breeds. Mamm. Genome 14: 640-649. [DOI] [PubMed] [Google Scholar]
- Johnson, G.C., Esposito, L., Barratt, B.J., Smith, A.N., Heward, J., Di Genova, G., Ueda, H., Cordell, H.J., Eaves, I.A., Dudbridge, F., et al. 2001. Haplotype tagging for the identification of common disease genes. Nat. Genet. 29: 233-237. [DOI] [PubMed] [Google Scholar]
- Ke, X., Hunt, S., Tapper, W., Lawrence, R., Stavrides, G., Ghori, J., Whittaker, P., Collins, A., Morris, A.P., Bentley, D., et al. 2004. The impact of SNP density on fine-scale patterns of linkage disequilibrium. Hum. Mol. Genet. 13: 577-588. [DOI] [PubMed] [Google Scholar]
- Kirkness, E.F., Bafna, V., Halpern, A.L., Levy, S., Remington, K., Rusch, D.B., Delcher, A.L., Pop, M., Wang, W., Fraser, C.M., et al. 2003. The dog genome: Survey sequencing and comparative analysis. Science 301: 1898-1903. [DOI] [PubMed] [Google Scholar]
- Kruglyak, L. 1999. Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nat. Genet. 22: 139-144. [DOI] [PubMed] [Google Scholar]
- Lou, X.Y., Todhunter, R.J., Lin, M., Lu, Q., Liu, T., Wang, Z., Bliss, S.P., Casella, G., Acland, G.M., Lust, G., et al. 2003. The extent and distribution of linkage disequilibrium in a multi-hierarchic outbred canine pedigree. Mamm. Genome 14: 555-564. [DOI] [PubMed] [Google Scholar]
- McRae, A.F., McEwan, J.C., Dodds, K.G., Wilson, T., Crawford, A.M., and Slate, J. 2002. Linkage disequilibrium in domestic sheep. Genetics 160: 1113-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellersh, C.S., Langston, A.A., Acland, G.M., Fleming, M.A., Ray, K., Wiegand, N.A., Francisco, L.V., Gibbs, M., Aguirre, G.D., and Ostrander, E.A. 1997. A linkage map of the canine genome. Genomics 46: 326-336. [DOI] [PubMed] [Google Scholar]
- Moffatt, M.F., Traherne, J.A., Abecasis, G.R., and Cookson, W.O. 2000. Single nucleotide polymorphism and linkage disequilibrium within the TCR α/δ locus. Hum. Mol. Genet. 9: 1011-1019. [DOI] [PubMed] [Google Scholar]
- Mohlke, K.L., Lange, E.M., Valle, T.T., Ghosh, S., Magnuson, V.L., Silander, K., Watanabe, R.M., Chines, P.S., Bergman, R.N., Tuomilehto, J., et al. 2001. Linkage disequilibrium between microsatellite markers extends beyond 1 cM on chromosome 20 in Finns. Genome Res. 11: 1221-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson, D.A., Tobe, V.O., and Taylor, S.L. 1997. PolyPhred: Automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res. 25: 2745-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura, T., Honda, T., and Mukai, F. 2001. Inbreeding and effective population size of Japanese Black cattle. J. Anim. Sci. 79: 366-370. [DOI] [PubMed] [Google Scholar]
- Nsengimana, J., Baret, P., Haley, C.S., and Visscher, P.M. 2004. Linkage disequilibrium in the domesticated pig. Genetics 166: 1395-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrander, E.A. and Comstock, K.E. 2004. The domestic dog genome. Curr. Biol. 14: R98-R99. [PubMed] [Google Scholar]
- Ostrander, E.A. and Giniger, E. 2001. Semper Fidelis: What man's best friend can teach us about human biology and disease. Am. J. Hum. Genet. 61: 475-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrander, E.A. and Kruglyak, L. 2000. Unleashing the canine genome. Genome Res. 10: 1271-1274. [DOI] [PubMed] [Google Scholar]
- Ostrander, E.A., Jong, P.M., Rine, J., and Duyk, G. 1992. Construction of small-insert genomic DNA libraries highly enriched for microsatellite repeat sequences. Proc. Natl. Acad. Sci. 89: 3419-3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrander, E.A., Galibert, F., and Patterson, D.F. 2000. Canine genetics comes of age. Trends Genet. 16: 117-123. [DOI] [PubMed] [Google Scholar]
- Parker, H.G., Kim, L.V., Sutter, N.B., Carlson, S., Lorentzen, T.D., Malek, T.B., Johnson, G.S., DeFrance, H.B., Ostrander, E.A., and Kruglyak, L. 2004. Genetic structure of the purebred domestic dog. Science 304: 1160-1164. [DOI] [PubMed] [Google Scholar]
- Patil, N., Berno, A.J., Hinds, D.A., Barrett, W.A., Doshi, J.M., Hacker, C.R., Kautzer, C.R., Lee, D.H., Marjoribanks, C., McDonough, D.P., et al. 2001. Blocks of limited haplotype diversity revealed by high-resolution scanning of human chromosome 21. Science 294: 1719-1723. [DOI] [PubMed] [Google Scholar]
- Patterson, D.F. 1980. A catalogue of genetic disorders of the dog. In Current veterinary therapy, vol. VII. WB Saunders, Philadelphia, PA.
- ———. 2000. Companion animal medicine in the age of medical genetics. J. Vet. Intern. Med. 14: 1-9. [PubMed] [Google Scholar]
- Patterson, D.F., Haskins, M.E., and Jezyk, P.F. 1982. Models of human genetic disease in domestic animals. Adv. Hum. Genet. 12: 263-339. [DOI] [PubMed] [Google Scholar]
- Patterson, D.F., Haskins, M.E., Jezyk, P.F., Giger, U., Meyers-Wallen, V.N., Aguirre, G., Fyfe, J.C., and Wolfe, J.H. 1988. Research on genetic diseases: Reciprocal benefits to animals and man. J. Am. Vet. Med. Assoc. 193: 1131-1144. [PubMed] [Google Scholar]
- Reich, D.E., Cargill, M., Bolk, S., Ireland, J., Sabeti, P.C., Richter, D.J., Lavery, T., Kouyoumjian, R., Farhadian, S.F., Ward, R., et al. 2001. Linkage disequilibrium in the human genome. Nature 411: 199-204. [DOI] [PubMed] [Google Scholar]
- Rogers, C.A. and Brace, A.H. 1995. The international encyclopedia of dogs. Howell Book House, New York.
- Sargan, D.R., Yang, F., Squire, M., Milne, B.S., O'Brien, P.C., and Ferguson-Smith, M.A. 2000. Use of flow-sorted canine chromosomes in the assignment of canine linkage, radiation hybrid, and syntenic groups to chromosomes: Refinement and verification of the comparative chromosome map for dog and human. Genomics 69: 182-195. [DOI] [PubMed] [Google Scholar]
- Service, S.K., Ophoff, R.A., and Freimer, N.B. 2001. The genome-wide distribution of background linkage disequilibrium in a population isolate. Hum. Mol. Genet. 10: 545-551. [DOI] [PubMed] [Google Scholar]
- Stephens, J.C., Schneider, J.A., Tanguay, D.A., Choi, J., Acharya, T., Stanley, S.E., Jiang, R., Messer, C.J., Chew, A., Han, J.H., et al. 2001a. Haplotype variation and linkage disequilibrium in 313 human genes. Science 293: 489-493. [DOI] [PubMed] [Google Scholar]
- Stephens, M., Smith, N.J., and Donnelly, P. 2001b. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 68: 978-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenesa, A., Knott, S.A., Ward, D., Smith, D., Williams, J.L., and Visscher, P.M. 2003. Estimation of linkage disequilibrium in a sample of the United Kingdom dairy cattle population using unphased genotypes. J. Anim. Sci. 81: 617-623. [DOI] [PubMed] [Google Scholar]
- Tenesa, A., Wright, A.F., Knott, S.A., Carothers, A.D., Hayward, C., Angius, A., Persico, I., Maestrale, G., Hastie, N.D., Pirastu, M., et al. 2004. Extent of linkage disequilibrium in a Sardinian sub-isolate: Sampling and methodological considerations. Hum. Mol. Genet. 13: 25-33. [DOI] [PubMed] [Google Scholar]
- Varilo, T., Paunio, T., Parker, A., Perola, M., Meyer, J., Terwilliger, J.D., and Peltonen, L. 2003. The interval of linkage disequilibrium (LD) detected with microsatellite and SNP markers in chromosomes of Finnish populations with different histories. Hum. Mol. Genet. 12: 51-59. [DOI] [PubMed] [Google Scholar]
- Weiss, K.M. and Clark, A.G. 2002. Linkage disequilibrium and the mapping of complex human traits. Trends Genet. 18: 19-24. [DOI] [PubMed] [Google Scholar]
- Wilcox, B. and Walkowicz, C. 1995. Atlas of dog breeds of the world. T.F.H. Publications, Neptune City, NJ.
- Yang, F., O'Brien, P.C., Milne, B.S., Graphodatsky, A.S., Solanky, N., Trifonov, V., Rens, W., Sargan, D., and Ferguson-Smith, M.A. 1999. A complete comparative chromosome map for the dog, red fox, and human and its integration with canine genetic maps. Genomics 62: 189-202. [DOI] [PubMed] [Google Scholar]
- Yuzbasiyan-Gurkan, V., Blanton, S.H., Cao, V., Ferguson, P., Li, J., Venta, P.J., and Brewer, G.J. 1997. Linkage of a microsatellite marker to the canine copper toxicosis locus in Bedlington terriers. Am. J. Vet. Res. 58: 23-27. [PubMed] [Google Scholar]
Web site references
- http://www.genome.ucsc.edu/; UCSC Genome Browser.
- http://www.AKC.org; American Kennel Club.
- http://www.ncbi.nlm.nih.gov/; NCBI, National Center for Biotechnology Information.
- http://www.repeatmasker.org/; RepeatMasker online tool.
- http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi; Primer3 online tool.