Abstract
Analysis of sequence variation among members of a single species offers a potential approach to identify functional DNA elements responsible for biological features unique to that species. Due to its high rate of allelic polymorphism and ease of genetic manipulability, we chose the sea squirt, Ciona intestinalis, to explore intraspecies sequence comparisons for genome annotation. A large number of C. intestinalis specimens were collected from four continents, and a set of genomic intervals were amplified, resequenced, and analyzed to determine the mutation rates at each nucleotide in the sequence. We found that regions with low mutation rates efficiently demarcated functionally constrained sequences: these include a set of noncoding elements, which we showed in C. intestinalis transgenic assays to act as tissue-specific enhancers, as well as the location of coding sequences. This illustrates that comparisons of multiple members of a species can be used for genome annotation, suggesting a path for the annotation of the sequenced genomes of organisms occupying uncharacterized phylogenetic branches of the animal kingdom. It also raises the possibility that the resequencing of a large number of Homo sapiens individuals might be used to annotate the human genome and identify sequences defining traits unique to our species.
Sequence comparisons between the genomes of organisms separated by a varying degree of evolutionary distances currently serve as an essential means to identify genes as well as gene regulatory elements (Ansari-Lari et al. 1998; Nobrega et al. 2003; Thomas et al. 2003). These comparisons are based on the well established molecular evolution principle that negative selection reduces the accumulation of sequence differences in functional sequences of related species (Hartl and Clark 1997; Hardison 2003). An important limitation of interspecies comparisons is that they can only be used to identify sequences underlying biological traits shared by the species examined. Although recently described approaches, leveraging the sequences of many closely related species to increase the total evolutionary branch length of the comparisons, have begun to address this issue (Boffelli et al. 2003), they are nevertheless ill-suited to uncover species-specific features. Intraspecies comparisons of the genomes of numerous members of the same species offer a theoretical strategy to tackle this problem. Although this approach is clearly expected to require the sequences of a very large number of individuals of the same species, the progressive lowering of the barrier to large-scale resequencing made possible by advances in sequencing technology now provides the opportunity to determine whether the theoretical advantages of intraspecies comparisons for genome annotation can be supported by experimental data.
The sequencing of the ascidian Ciona intestinalis (sea squirt) genome recently revealed this organism to be a particularly attractive candidate target for testing the feasibility of resequencing for annotating genomes. An important attribute is its very high allelic polymorphism, with an average 1.2% of the nucleotides differing between chromosome pairs of a single individual (Dehal et al. 2002). This high degree of allelic variation, more than 15-fold that noted in humans, is probably a consequence of the large effective population size of C. intestinalis. In addition, the genetic manipulability of the sea squirts offers a rigorous in vivo experimental system to test the functional activity of identified candidate regulatory elements (Satoh 2003).
In this study, we determined the extent of sequence polymorphism in several C. intestinalis subpopulations collected at multiple locations worldwide. We exploited sequence variation within C. intestinalis to computationally identify regions subjected to fast and slow rates of evolution, and we experimentally characterized their functional roles. These studies illustrate that slowly evolving regions correspond to protein-coding or enhancer regions, indicating the feasibility of using intraspecies polymorphism to annotate a species' genome.
Results
Amplification of genomic targets and phylogenetic analysis
C. intestinalis specimens were collected from several coastal locations in North America, Europe, Eastern Asia, and Oceania. Samples were defined as C. intestinalis based on characteristic morphological features supported by sequence analysis. Although the formal proof that all the samples analyzed are members of the same species would require successful sexual mating, the sequence difference among the samples examined indicated that they are several-fold more similar to each other than to their closest intra-genus relative, C. savignyi. Any two pairs of C. intestinalis analyzed in this study were at least 85% identical, a value within the range of allelic polymorphism reported for a single C. intestinalis individual (Dehal et al. 2002). Conversely, BLAST comparisons between C. intestinalis and C. savignyi, carried out at the relaxed threshold of e = -1, revealed sequence alignments for less than 10% of the sequences. Four genomic intervals, each ∼4 kb long, were chosen for analysis in this study based on previous knowledge of genes and gene regulatory elements located within these intervals. Target regions included exons 18–25 of patched homolog, exons 1–4 of col5a1, and the 5′ sequences of forkhead and snail. The targeted coding regions all had strong gene structure predictions supported by EST sequences, while several 5′ tissue-specific enhancers for forkhead and the promoter of snail had been previously defined and characterized (Erives et al. 1998; Di Gregorio et al. 2001).
Using the consensus sequence of an individual collected from a West Coast location for PCR primer design, amplification was attempted for each of the four genomic target regions from 140 animals. Successful PCR reactions yielded unique bands in most cases. When two bands were obtained, they were both sequenced independently and in all instances resulted in heterozygous allelic variants due to insertion/deletion events. Even though all four target regions were unique in the current C. intestinalis assembly, its draft status raises the possibility that amplicons from different individuals represent paralogous, rather than allelic, copies. This is unlikely, since all the sequences for the same target locus had stretches of perfect identity longer than 100 base pairs, which are not normally observed between paralogous loci.
We were able to obtain amplified genomic targets from ∼50% of the individuals collected from West Coast locations, 20% of the individuals from New Zealand and Japan, and less than 20% of the individuals collected from coastal locations on the Atlantic Ocean or the Mediterranean Sea (Table 1). The difficulty in amplifying C. intestinalis genomic samples, explained by the high levels of polymorphism of this species, suggests that only a reduced subset of the polymorphism present in C. intestinalis was likely captured by this study.
Table 1.
Summary of PCR amplification of forkhead, snail, col5a1 and patched
| forkhead | snail | col5a1 | patched | |
|---|---|---|---|---|
| WC | 16 (.48) | 16 (.48) | 17 (.52) | 19 (.58) |
| JP | 5 (.25) | 6 (.30) | 4 (.20) | 4 (.20) |
| NZ | 3 (.21) | 1 (.7) | 5 (.36) | 0 (.0) |
| EC | 8 (.17) | 8 (.17) | 5 (.11) | 0 (.0) |
| FR | 1 (.7) | 4 (.27) | 3 (.20) | 0 (.0) |
| IT | 0 (.0) | 2 (.17) | 2 (.17) | 0 (.0) |
Number of individuals amplified from each subpopulation for each of the four target regions analyzed. The numbers in parentheses indicate the fraction of individuals that yielded successful amplification out of all the amplifications attempted from that subpopulation. WC: samples from three collection locations on the Pacific coast of the United States. JP: samples from two collection locations in western Japan. NZ: samples from one collection location in New Zealand. FR: samples from one collection location on the Atlantic coast of France. EC: samples from three collection locations on the northern Atlantic coast of the United States. IT: samples from two collection locations in Italy.
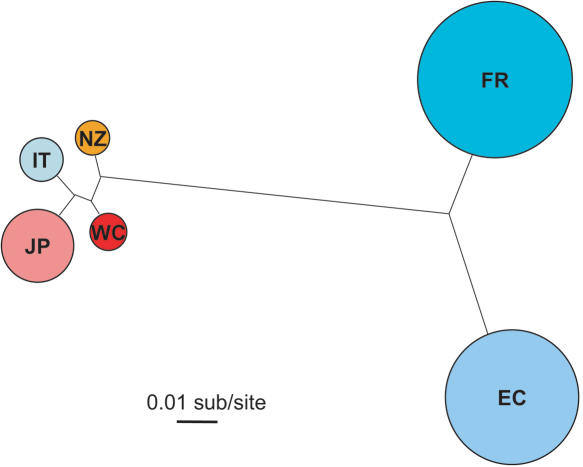
Successfully amplified target regions were fully sequenced using custom primers designed ∼every 250 bp on each strand, for a total of 40 sequencing reads for each amplified target region. This level of coverage ensured that each base was read at least four times. Analysis of the sequences revealed that specimens from the same collection location clustered nearly exclusively close to each other as estimated by their degree of sequence similarity, supporting the conjecture that they are members of largely isolated subpopulations (Fig. 1). Surprisingly, individuals from Mediterranean locations appeared more related to individuals collected from the Pacific rather than Atlantic Ocean. In addition, subpopulations collected from locations on the Atlantic Ocean showed much higher heterozygosity than subpopulations from locations on the Pacific Ocean, as reflected by the size of the circles in Figure 1. C. intestinalis is an invasive species reported to be spread in ship bilges along shipping routes. This is supported by the clustering of samples from the Pacific Ocean, reflecting the greater shipping activity between California, Japan, and New Zealand ports. The lower heterozygosity among those samples also suggests that the Pacific Ocean was colonized after the Atlantic Ocean.
Figure 1.
Phylogenetic relationships of C. intestinalis subpopulations. Consensus sequences for the col5a1 interval, obtained for each of the six subpopulations analyzed in this study, were used to calculate the population tree. Subpopulations are defined by their collection locations, as in Table 1. The size of the circle surrounding each subpopulation is proportional to the heterozygosity of that subpopulation.
The phylogenetic tree of the individuals sequenced in each genomic interval was obtained in two steps. First, phylogenetic relationships between subpopulations were calculated from the consensus sequences for each subpopulation, defined by their collection locations (see legend, Table 1). Phylogenetic relationships for individuals from the same subpopulation were then estimated from the average distance of all members of that subpopulation, since the degree of sequence similarity among individuals from the same subpopulation did not allow the computation of statistically significant trees within subpopulations. The resulting composite trees were used to calculate the likelihood that each nucleotide site in the multiple sequence alignment is mutating at a high or at a low rate (Boffelli et al. 2003). Variation profiles of the genomic intervals analyzed were displayed through likelihood ratio curves to identify regions undergoing the slowest mutation rates relative to the rate of their surrounding regions.
Identification of regulatory elements of forkhead and snail
The identification of gene regulatory elements in the proximity of two early-development genes, forkhead and snail, was sought using C. intestinalis sequence comparisons. Both genes are expressed in the larval stage of C. intestinalis development and are therefore amenable to in vivo assessments in transgenic C. intestinalis tadpoles. The mutation rate ratio plot for the forkhead 5′ regulatory region revealed five distinct minima representing the sequences likely under the strongest selective constraints (Fig. 2A, regions 1, 2, 4, 5, and 7). We explored the ability of these five regions to function as enhancers in vivo using reporter constructs assayed in transgenic C. intestinalis tadpoles. Constructs which reproducibly drove expression in a tissue-specific manner included that for region 1 in the notochord and endoderm, region 4 in the notochord, endoderm, and neural tube, region 5 in the notochord, and region 7 in the neural tube, endoderm, and notochord (Fig. 2B). These patterns are consistent with the endogenous forkhead expression characteristics (Corbo et al. 1997a). As putative negative controls, we examined two regions with high mutation rates (regions 3 and 6, Fig. 2A). Consistent with the expectation that fast-evolving regions likely lack gene regulatory activity, these two regions failed to drive gene expression in this assay (data not shown).
Figure 2.
(A) Mutation rate analysis of the genomic interval containing the 5′ region of the forkhead gene. The x-axis represents the position in the multiple alignment consensus sequence, the y-axis the log likelihood ratio for a fast- over a slow-mutation regime at that position. The plot is smoothed using a 20%-trimmed mean over the 24-base window centered at each aligned site. A lower ratio indicates a low mutation rate. The sequence of 33 individuals (total tree length = 0.28) was used to generate this plot. The blue bar labeled “P” indicates the position of the forkhead promoter; the red and purple bars indicate the positions of low- and high-mutation rate intervals, respectively, that were functionally analyzed in this study. (B) Transgenic analysis of intervals identified by mutation rate analysis of the 5′ region of the forkhead gene. C. intestinalis larvae were electroporated with a reporter construct containing the genomic fragments 1, 2, 4, 5, and 7, respectively and the expression was visualized by histochemical staining with X-gal. Constructs for region 2 never yielded LacZ expression, and the position marked on the plot corresponds to a segment previously analyzed (Di Gregorio et al. 2001). Red arrows indicate expression in the neural tube, yellow arrows that in the notochord, and green arrows in the endoderm. Constructs for region 2 failed to yield tissue-specific expression.
While the slow-evolving region 2 failed to show tissue-specific expression in this assay, a previous study, analyzing a reporter construct containing the forkhead 5′ region through sequential deletions, had shown that removal of a segment containing region 2 abolished neural tube expression in the C. intestinalis tadpole (Di Gregorio et al. 2001). This suggests that this functional element might require interactions with nearby enhancers for driving gene expression in the neural tube. The same previous study also demonstrated that deletions corresponding to regions 1 and 3 resulted in a loss of neural tube, endoderm, and notochord expression, in a manner consistent with our results.
We applied a similar analysis to the genomic interval containing snail's 5′ region. One 5′ noncoding region characterized by a mutation rate similar to that of forkhead enhancers was also investigated using the transgenic C. intestinalis tadpole assay (region 2, Fig. 3). The reporter construct for this region drove expression in the neuronal lineage (inset, Fig. 3). The other 5′ noncoding region showing a similarly low variation rate (region 1, Fig. 3) corresponded exactly to the previously described minimal promoter/mesodermal enhancer of snail (Erives et al. 1998). The mutation rate ratio plot for this interval also correctly identified the first exon of snail (region E, Fig. 3).
Figure 3.
Mutation rate analysis of the genomic interval containing the 5′ region and the first exon of the snail gene. The plot was drawn as described in the Figure 2 legend. The sequence of 37 individuals (total tree length = 0.52) was used to generate this plot. The position of the first exon is indicated by the green bar labeled “E”; region 1 is snail's promoter, and region 2 is a constrained interval upstream of snail. The inset shows the transgenic analysis of region 2. C. intestinalis larvae were electroporated with a reporter construct containing region 2, and the expression was visualized by histochemical staining with X-gal. The red arrow indicates expression in the neural tube.
Identification of coding exons of col5a1 and patched
To further test our ability to predict the location of exonic sequences through intraspecies sequence comparisons, we investigated two genomic intervals containing several exons of the col5a1 and patched genes. Both intervals revealed a similarly uneven distribution of mutation rates, with several regions characterized by a low rate interspersed among high mutation rate regions (Fig. 4). Of the six regions with low mutation in the col5a1 interval, four corresponded to the exon-coding regions annotated by gene prediction programs and EST sequences available for this interval (1–4, Fig. 4A). The two additional regions of very low mutation rate revealed by the variation plot were not functionally investigated in this study but could represent additional col5a1 regulatory elements. Consistent with the results for the col5a1 interval, the position of the eight patched exons were all coincident with regions of low mutation rate (Fig. 4B). Overall sequence diversity was very low in the patched interval (total tree length = 0.10 substitutions per site), likely due to the successful amplification of DNA from only 20 individuals exclusively from California or Japanese locations, but was nevertheless sufficient to identify regions encompassing the exon locations.
Figure 4.
Mutation rate analysis of the genomic interval containing the 5′ region of the col5a1 (A) and patched (B) genes. The plot was drawn as described in the Figure 2 legend. The sequence of 36 and 22 individuals was used to generate the col5a1 and patched plots (total tree lengths were 0.69 and 0.10), respectively. The blue bar labeled “P” indicates the position of col5a1's promoter; the numbered green bars indicate the position of exons 1–4 of col5a1 and exons 18–25 of patched.
Discussion
In contrast to all previous comparative genomic studies, which used evolutionarily fixed sequence differences between two or more species to estimate mutation rates, we are here able to show that intraspecies sequence polymorphism can be effectively used to identify gene regulatory elements and exons through a combination of phylogenetic analysis and polymorphism diversity. Intraspecies sequence polymorphism has been extensively used by population geneticists to study the action of selection on protein coding sequences and by biologists to detect linkage disequilibrium and associate deleterious allelic variants with disease. Our results extend the interest in using polymorphism beyond those applications and suggest that intraspecies sequence comparisons may be useful for identifying functional regions in a genome.
Intraspecies comparisons fill several important niches in the comparative genomics analysis of sequenced genomes. First, they have the potential to identify sequences underlying biological traits unique to a single species. They are also useful when a species under investigation is too far away from its nearest evolutionary neighbor for useful pair-wise comparisons. For example, C. intestinalis shared a last common ancestor with its sister species, C. savignyi, ∼100 million years ago. Consequently, these two species show limited sequence similarity that, while often sufficient to identify many highly constrained sequences, might lead to failure in identifying many other functional sequences, which can otherwise be detected by the approach described here. Finally, intraspecies comparisons suggest a path for the annotation of organisms occupying previously uncharacterized phylogenetic branches of the animal kingdom, including the green alga C. reinhardtii, the diatom T. pseudonana, and human malaria parasite P. falciparum, whose sequences are now becoming available.
Because of the high rate of allelic polymorphism of C. intestinalis, the sequences from as few as 30 individuals were required to achieve sufficient total sequence variation to identify intervals evolving significantly more slowly than their surrounding regions. Applying the same intraspecies approach to human is made more difficult by its low rate of polymorphism (Sachidanandam et al. 2001). The complexity of human population dynamics and haplotype structure complicates estimating the number of individuals required for such a study. Nonetheless, making several simplifications and assumptions, extrapolation of the data from Yu et al. (2002) suggests that sequencing of a few thousand individuals would yield 0.3 single nucleotide polymorphism (SNP)/site, a number comparable to the total tree length of the sequences used in the analysis of forkhead 5′ region in this study. Irregardless of the exact number of humans required for such a study, with the increase in human genome resequencing predicted for the near future (Collins et al. 2003; Shendure et al. 2004), intra-Homo sapiens comparisons exploiting the approaches described here for C. intestinalis may prove fruitful for the identification of functional sequences shared with other species as well as human-specific ones.
Methods
Collection of C. intestinalis specimens
Specimens were collected from the following coastal locations: South San Francisco, California; Half-Moon Bay, California; Santa Barbara, California; Cobscook Bay, Maine; Darling Marine Center, Maine; Woods Hole, Massachussetts; Kobe, Japan; Kochi, Japan; Marlborough Sounds, New Zealand; Reading, England; Roscoff, France; Grevelingen, Netherlands; Den Hosse, Netherlands; Viaggio Coppola, Italy; Fusaro, Italy.
Direct resequencing of target regions
Genomic DNA was isolated from C. intestinalis muscle tissue using the Puregene kit (Gentra Systems). Target regions were amplified by PCR using the Elongase systems (Invitrogen) following the manufacturer's recommendations, with primers described in the Supplemental information. Single-band PCR products were gel-purified (SNAP UV-free Gel purification Kit, Invitrogen) and subjected to direct microsequencing using custom primers designed every 250 bp on each strand. Fluorescence automated DNA sequencing was carried out using BygDye chemistry in an ABI3700 Sequencer (Applied Biosystems). Both the (+) and (-) strands were sequenced at least twice. Base calling, quality assessment, and assembly were carried out using the phred, Phrap, Consed software suite developed by Phil Green (www.phrap.org). All the sequences generated in this study have been submitted to GenBank.
Data analysis
Sequences were aligned using MAVID (Bray and Pachter 2003) (baboon.math.berkeley.edu/mavid) or ClustalW (www.ebi.ac.uk/clustalw/index.html) and the alignments were manually verified. Consensus sequences for individuals collected from California, Japan, New Zealand, East Coast, and Europe were derived from the multiple alignment. Maximum likelihood phylogenetic trees for the consensus sequences were reconstructed using FastDNAml. In order to identify conserved sequences, a likelihood ratio (conserved vs. nonconserved) was calculated for each position in the multiple alignment of the individuals (McAuliffe et al. 2004) (http://bonaire.lbl.gov/newshadower/). To explicitly account for both the phylogenetic relationships between the consensus sequences, as well as the polymorphic diversity between individuals from the same subpopulation, a phylogenetic likelihood computation was coupled with a nucleotide diversity calculation for both fast and slow rates (Pamilo et al. 1987). Although nucleotide diversity is typically computed with a Jukes-Cantor correction, it can be viewed as a phylogenetic likelihood computation on a star tree and can be based on any evolutionary model. We matched the model to that used for the phylogenetic computations on the consensus sequences (Boffelli et al. 2003), and by conditioning on the consensus sequences we obtain a probabilistic model for modeling phylogenetic relationships between groups as well as polymorphic differences among same-population individuals. The rates for the slow (conserved) and fast (nonconserved) computations were based on previous estimates. Regions characterized by the slowest rates were defined relative to the rates of their surroundings, implicitly accounting for the local rate of mutation.
Maintenance of C. intestinalis colony
Adult C. intestinalis were collected from several locations in Northern California, purchased from Woods Hole, Massachusetts, and Long Beach, Southern California. The animals were kept at 18°C in recirculating artificial seawater.
Construction of plasmids and electroporation
All constructs were made with the pCES (plasmid Ciona enhancer screen) vector previously reported (Harafuji et al. 2002). Different fragments from the Ci-fkh 5′ and Ci-snail 5′ flanking regions were amplified by PCR using the Ciona genomic DNA isolated from Northern California animals, with primers described in the Supplemental information. The DNA fragments were then ligated into the BamHI site of the pCES vector. Electroporations, fixation, and staining reactions were done as described by Corbo et al. (1997b). Aliquots containing 100 μg of purified plasmid were used in each electroporation. Each transgene was tested at least twice. The number of positive embryos scored in each experiment ranged ∼between 70 and 200.
GenBank accession numbers
Forkhead region: AY667314–AY667347. Snail region: AY667371–AY667407. Col5a1 region: AY667278–AY667313. Patched region: AY667348–AY667370.
Acknowledgments
We thank Shigeki Fujiwara, Arjan Gittenberger, Kevin Heasman, Helene Huelvan, Di Jiang, Shungo Kano, Aimee Phillippi, Andy Sexton, and Seb Shimeld for providing C. intestinalis samples. Research was conducted at the E.O. Lawrence Berkeley National Laboratory and at the Joint Genome Institute, with support by grants from the Programs for Genomic Application, NHLBI (E.M.R.) and NIH (L.P.), and performed under Dept. of Energy Contract DE-AC0378SF00098, Univ. of California.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3199704. Article published online ahead of print in November 2004.
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study were submitted to GenBank under accession nos. AY667278–AY667407. The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: S. Fujiwara, A. Gittenberger, K. Heasman, H. Huelvan, D. Jiang, S. Kano, A. Phillippi, A. Sexton, and S. Shimeld.]
References
- Ansari-Lari, M.A., Oeltjen, J.C., Schwartz, S., Zhang, Z., Muzny, D.M., Lu, J., Gorrell, J.H., Chinault, A.C., Belmont, J.W., Miller, W., et al. 1998. Comparative sequence analysis of a gene-rich cluster at human chromosome 12p13 and its syntenic region in mouse chromosome 6. Genome Res. 8: 29-40. [PubMed] [Google Scholar]
- Boffelli, D., McAuliffe, J., Ovcharenko, D., Lewis, K.D., Ovcharenko, I., Pachter, L., and Rubin, E.M. 2003. Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science 299: 1391-1394. [DOI] [PubMed] [Google Scholar]
- Bray, N. and Pachter, L. 2003. MAVID multiple alignment server. Nucleic Acids Res. 31: 3525-3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, F.S., Green, E.D., Guttmacher, A.E., and Guyer, M.S. 2003. A vision for the future of genomics research. Nature 422: 835-847. [DOI] [PubMed] [Google Scholar]
- Corbo, J.C., Erives, A., Di Gregorio, A., Chang, A., and Levine, M. 1997a. Dorsoventral patterning of the vertebrate neural tube is conserved in a protochordate. Development 124: 2335-2344. [DOI] [PubMed] [Google Scholar]
- Corbo, J.C., Levine, M., and Zeller, R.W. 1997b. Characterization of a notochord-specific enhancer from the Brachyury promoter region of the ascidian, Ciona intestinalis. Development 124: 589-602. [DOI] [PubMed] [Google Scholar]
- Dehal, P., Satou, Y., Campbell, R.K., Chapman, J., Degnan, B., De Tomaso, A., Davidson, B., Di Gregorio, A., Gelpke, M., Goodstein, D.M., et al. 2002. The draft genome of Ciona intestinalis: Insights into chordate and vertebrate origins. Science 298: 2157-2167. [DOI] [PubMed] [Google Scholar]
- Di Gregorio, A., Corbo, J.C., and Levine, M. 2001. The regulation of forkhead/HNF-3β expression in the Ciona embryo. Dev. Biol. 229: 31-43. [DOI] [PubMed] [Google Scholar]
- Erives, A., Corbo, J.C., and Levine, M. 1998. Lineage-specific regulation of the Ciona snail gene in the embryonic mesoderm and neuroectoderm. Dev. Biol. 194: 213-225. [DOI] [PubMed] [Google Scholar]
- Harafuji, N., Keys, D.N., and Levine, M. 2002. Genome-wide identification of tissue-specific enhancers in the Ciona tadpole. Proc. Natl. Acad. Sci. 99: 6802-6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardison, R.C. 2003. Comparative genomics. PLoS Biol. 1: E58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl, D.L. and Clark, A.G. 1997. Principles of population genetics. Sinauer Associates, Sunderland, MA.
- McAuliffe, J.D., Pachter, L., and Jordan, M.I. 2004. Multiple-sequence functional annotation and the generalized hidden Markov phylogeny. Bioinformatics 12: 1850-1860. [DOI] [PubMed] [Google Scholar]
- Nobrega, M.A., Ovcharenko, I., Afzal, V., and Rubin, E.M. 2003. Scanning human gene deserts for long-range enhancers. Science 302: 413. [DOI] [PubMed] [Google Scholar]
- Pamilo, P., Nei, M., and Li, W.H. 1987. Accumulation of mutations in sexual and asexual populations. Genet. Res. 49: 135-146. [DOI] [PubMed] [Google Scholar]
- Sachidanandam, R., Weissman, D., Schmidt, S.C., Kakol, J.M., Stein, L.D., Marth, G., Sherry, S., Mullikin, J.C., Mortimore, B.J., Willey, D.L., et al. 2001. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 409: 928-933. [DOI] [PubMed] [Google Scholar]
- Satoh, N. 2003. The ascidian tadpole larva: Comparative molecular development and genomics. Nat. Rev. Genet. 4: 285-295. [DOI] [PubMed] [Google Scholar]
- Shendure, J., Mitra, R., Varma, C., and Church, G. 2004. Advanced sequencing technologies: Methods and goals. Nat. Rev. Genet. 5: 335-343. [DOI] [PubMed] [Google Scholar]
- Thomas, J.W., Touchman, J.W., Blakesley, R.W., Bouffard, G.G., Beckstrom-Sternberg, S.M., Margulies, E.H., Blanchette, M., Siepel, A.C., Thomas, P.J., McDowell, J.C., et al. 2003. Comparative analyses of multi-species sequences from targeted genomic regions. Nature 424: 788-793. [DOI] [PubMed] [Google Scholar]
- Yu, N., Chen, F.C., Ota, S., Jorde, L.B., Pamilo, P., Patthy, L., Ramsay, M., Jenkins, T., Shyue, S.K., and Li, W.H. 2002. Larger genetic differences within Africans than between Africans and Eurasians. Genetics 161: 269-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
Web site references
- http://bonaire.lbl.gov/newshadower/; phylogenetic shadowing.
- www.phrap.org; consed suite.
- www.ebi.ac.uk/clustalw/index.html; multiple sequence alignment.
- baboon.math.berkeley.edu/mavid; multiple sequence alignment.