Abstract
The completion of the mouse and other mammalian genome sequences will provide necessary, but not sufficient, knowledge for an understanding of much of mouse biology at the molecular level. As a requisite next step in this process, the genes in mouse and their structure must be elucidated. In particular, knowledge of the transcriptional start site of these genes will be necessary for further study of their regulatory regions. To assess the current state of mouse genome annotation to support this activity, we identified several hundred gene predictions in mouse with varying levels of supporting evidence and tested them using RACE–PCR. Modifications were made to the procedure allowing pooling of RNA samples, resulting in a scaleable procedure. The results illustrate potential errors or omissions in the current 5′ end annotations in 58% of the genes detected. In testing experimentally unsupported gene predictions, we were able to identify 58 that are not usually annotated as genes but produced spliced transcripts (∼25% success rate). In addition, in many genes we were able to detect novel exons not predicted by any gene prediction algorithms. In 19.8% of the genes detected in this study, multiple transcript species were observed. These data show an urgent need to provide direct experimental validation of gene annotations. Moreover, these results show that direct validation using RACE–PCR can be an important component of genome-wide validation. This approach can be a useful tool in the ongoing efforts to increase the quality of gene annotations, especially transcriptional start sites, in complex genomes.
The sequence of the human genome has been completed, and the mouse genome is rapidly nearing completion (Lander et al. 2001; Venter et al. 2001; Waterston et al. 2002). An important next step is defining the mammalian gene set and accurate annotation of gene structures. There are currently two major sets of mammalian gene annotations found at Ensembl and National Center for Biotechnology Information (NCBI).
A canonical set of mouse genes is rapidly coalescing, and similar progress is also being made on other species. Programs contributing to this progress, at both Ensembl and NCBI, make use of full-length cDNAs and add the genes predicted by ab initio algorithms such as Genscan (Burge and Karlin 1997) and Twinscan (Yeh et al. 2001). However, these predictions are further filtered by Ensembl, which requires additional support such as multiple EST matches or similarity to genes in another species before pure predictions rise to the status of confirmed genes. Thus, the sequencing of full-length cDNAs by the Mammalian Gene Collection (MGC) and the RIKEN group (Carninci et al. 2002, 2003; Waterston et al. 2002) has greatly improved the annotation quality of the mammalian genome. However, these approaches can miss the genes expressed in a restricted manner or at very low levels. This conservative approach results in a very low false-positive rate. However, its sensitivity, the probability of missing real genes, is less clear and is one of the questions that we set out to address.
Current gene predictions and annotation also focus largely on predicting and confirming coding regions. Transcriptional start sites (TSSs) have been identified for some genes by full-length cDNA approaches. Other efforts such as the MGC (http://mgc.nci.nih.gov/) do not attempt to identify the TSS, but have as their end goal confirming and providing the coding region of the gene. Even if some percentage of the genes in the MGC were to actually contain the TSS, since the collection is not designed to contain the TSS one could not know a priori which were in fact complete. In fact, at this point it is not even known what percentage of the MGC contains the TSS. Hence annotations are particularly incomplete in describing the TSS of many genes.
An important facet of the annotation of any genome is the determination of the TSSs of both coding and noncoding RNA genes. Even for many well-characterized genes, TSSs are often unknown. This is of critical importance due to the association between the TSS and the cis-acting sites that regulate transcription. Information about the start sites of all the genes is ultimately required for large-scale computational analysis of sequence patterns that are associated with transcriptional regulatory themes. A specific example will perhaps clarify this point. If a set of microarray data indicates that there are several hundred genes that are expressed together in a pattern, it would be of interest to search the putative promoters of all of these genes for common structural motifs. We cannot do this now because of a lack of a confirmed TSSs for many, perhaps even most, genes.
Approximately 40% of the known human genes have completely noncoding first exons (Davuluri et al. 2000). Although a similar study in mouse has not, to our knowledge, been reported, one might expect a similar percentage to apply. Due to difficulty in their purely computational prediction, experimental data are presently required to determine the location of the promoter or the 5′ end of a transcript.
Here we describe results from a systematic study in which gene discovery and the understanding of gene structure is approached by integrating existing gene models and ab initio gene predictions into an experimental pipeline geared both to identify the TSSs of known and novel genes and to develop more accurate gene models. Three hundred genes with varying degrees of associated experimental evidence were chosen to test the approach. The structure of the 5′ end of a sizable number of both previously known and unknown genes was established by using the methods described here. Based on our results, this approach seems ideal for validation of a large number of gene predictions, as well as better annotation of the TSSs in known genes, in a rapid yet reliable manner.
Results
Gene categorization
The 300 gene predictions or annotated genes selected for analysis were grouped into five different categories, based on the quality and quantity of supporting evidence for the gene model (for the selection process, see Methods and ftp://ftp.cshl.org/pub/sequences/mouse/data_for_paper/). These gene categories included well-characterized genes in the Eukaryotic Promoter Database (EPD category), genes from the RefSeq gene set (RefSeq category), automated gene predictions that are linked to multiple ESTs/mRNAs, (category B), as well as computational predictions with a single (category C) or no extant empirical support (category D). It should be noted that our goal for inclusion of category C and D was to experimentally verify these gene prediction with much less emphasis on comparison of our experimentally derived transcript structure with the predicted gene model since these models are based on limited to no experimental evidence and therefore will be invariably incomplete at the 5′ end. The category names, definitions, and the number of genes tested in each are shown in Table 1. A smaller number of genes were assigned to the EPD and RefSeq categories since they were originally intended solely as controls. However, as described below, interesting data were obtained from their analysis.
Table 1.
Description of gene categories
| Category | Definition | No. of genes in category | No. of genes successfully amplified by 5′ RACE (%) | No. of RACE sequences that differed from the 5′ gene annotation (%) |
|---|---|---|---|---|
| EPD | Genes in the Eukaryotic Promoter Database having experimentally verified transcriptional start sites | 13 | 13 (100%) | 4 (31%) |
| RefSeq | NCBI's curated non-redundant gene set | 27 | 20 (74%) | 8 (40%) |
| B | Automated NCBI predictions covered by multiple ESTs | 23 | 15 (65%) | 7 (47%) |
| C | Gene predictions which are covered by a single EST only and do not overlap any mRNA, cDNA, ENSEMBLE or GENIE evidence | 169 | 40 (24%) | 30 (75%) |
| D | Gene predictions that do not overlap any EST, mRNA, cDNA, ENSEMBLE, or GENIE evidence | 68 | 18 (26%) | 12 (67%) |
| Total | 300 | 106 (35%) | 61 (58%) |
Three hundred mouse genes or gene predictions were classified into five categories based on the quality of associated evidence. The definition column describes the basis for the classification. Genes in the EPD category have the highest quality evidence and were used as internal positive control for all experiments. Genes in category D were considered to be based on evidence with least amount of confidence. 5′ RACE–PCR was performed on 15 mouse tissues/stages as described in Methods. The number of genes successfully amplified in each category and satisfying the criteria described in the Methods section are listed. The number of 5′ RACE sequences where the reference sequence annotation was found to be incomplete at the 5′ end is shown for each category.
Amplification of 5′ end of transcripts by RACE–PCR
The result of 5′ rapid amplification of cDNA ends (RACE)–PCR on a set of 15 mouse tissues/stages is shown in Table 1. The RACE–PCR fragments were cloned, and eight clones for each amplification were sequenced. Of the 300 genes in all the categories, 106 were successfully amplified. The genes in the EPD set served as our internal positive control. All 13 genes in the EPD category were detected. For all of these genes, at least one splice variant was detected that agreed with the annotated first exon for the corresponding gene in the EPD. We amplified from the 5′ cap of the transcript to an internal exon so that the presence of a spliced product would rule out the possibility of genomic contamination. Spliced products for a majority of the well-characterized and curated genes (100% in the EPD and 74% in the RefSeq categories) were successfully detected.
Fewer of the category C (24%) and D (26%) genes were detected, which was expected based upon a number of possible hypotheses. For example, the predicted gene may not be expressed in the tissues/stages tested—which may also be the case for the 26% of the RefSeq genes as well as the 35% of category B genes that were also not detected. Alternatively, the gene model may have an incorrect structure. Finally, the predicted gene may be a false positive of the prediction algorithm and not truly be an expressed gene.
Annotation at the 5′ ends of genes is incomplete
To assess the quality of current annotations of 5′ ends of genes in the mouse genome, the sequences obtained by 5′RACE–PCR were compared to the corresponding gene annotation/prediction. Sequences were filtered using a set of stringent criteria as described in the Methods section. Table 1 shows the number of genes in the different categories whose 5′ RACE sequences differed from the gene annotation. Overall, the RACE–PCR method detected 43 first exons that were unannotated/not predicted. Fourteen of these 43 exons did not have any matching experimental evidence in GenBank (and hence were termed as novel first exons).
Of the 106 genes successfully detected in this study, 61 (58%) produced 5′ RACE–PCR sequences that were longer than the annotation. We analyzed these sequences in relation to their alignment to their associated gene model or prediction (Fig. 1A). In 20 of these 61 genes, we found at least one additional exon upstream of the existing annotation/prediction (Fig. 1B). We extended the annotated first exon of two genes (15%) in the EPD and six genes (30%) in the RefSeq categories by an average of 33 and 48 transcribed bases, respectively. EST or similar evidence in GenBank supported our results concerning all eight of these well-characterized genes whose annotation was in disagreement with our results.
Figure 1.
Comparison of current 5′ annotation with 5′ RACE–PCR results. (A) Schematic representation of the possible alignments of the first exon suggested by 5′ RACE sequence relative to the annotated exons (ex) and introns (int). (B) Percentage of genes detected in each category whose 5′ RACE sequence exhibits the alignments as follows: first exon is upstream of the annotated first exon (black), first exon overlaps with annotated first exon but extends it by at least 20 bp (blue), and first exon lies completely in the annotated first intronic region or aligns to the second annotated exon and extends into the first intronic region (light blue).
Additionally, for two other genes in the EPD category, alternative TSSs defining an exon located in the annotated first intron were detected by RACE–PCR (Fig. 1B). The 5′ RACE–PCR sequence obtained for the c-myc oncogene (GenBank accession no. NM_010849) identifies an exon that has supporting evidence in the form of one EST, and our mapped TSS has previously been reported as an alternate transcription start site in the EPD database (ID nos. EP14066 and EP14067). However, this exon is not represented in the RefSeq annotation. In the second case, sequence obtained for the myb oncogene (GenBank accession no. NM_010848) indicates an alternate first exon, which has no experimental support in GenBank or EPD (see Supplemental Fig. 3). Interestingly, this exon sequence is conserved in human but not in Takifugu rubripues. In the case of the RefSeq category, four genes were observed in which the first exon was located in the first intronic region. Altogether, we detected 23 cases in all the categories (Fig. 1B) in which the 5′ RACE sequence aligned to the first intronic region. These findings show the limitations of current annotation, which must collate various empirical data sources (such as ESTs) and computer predictions in an attempt to determine the TSS of a gene.
Comparison of 5′ RACE–PCR sequences with full-length RIKEN cDNA
The RACE–PCR results were compared to full-length RIKEN cDNAs. Of the 106 genes successfully detected in this study, 54 genes were represented in the RIKEN full-length cDNA collection. For each of these 54 genes, at least one RACE–PCR-derived sequence agreed with the first exon in the corresponding RIKEN cDNA (although the precise TSSs differed in several cases). However, for six genes the RACE–PCR approach detected additional first exon variants that lie in the annotated first intron. These exons are not represented in the RIKEN collection. This is most likely due to the presence of alternate first exons, which can be more easily identified by a directed approach such as RACE–PCR rather than a sampling approach such as cDNA library construction.
Detection of novel genes
Large-scale studies, including those undertaken by the RIKEN group and the Mammalian Gene Consortium, have been extremely valuable in detecting genes (Carninci et al. 2002, 2003; Waterston et al. 2002). These random-sampling methods suffer, over time, from decreasing efficiency and increasing costs for novel gene discovery. To assess the feasibility of using the RACE–PCR approach to discover novel genes, 237 genes predicted by Twinscan (Yeh et al. 2001) and GenomeScan (Korf et al. 2001) programs were tested (genes in categories C and D; Table 1). We detected products for 58 genes in categories C and D. It is important to note that the category C and D genes are not presently considered as annotated genes, suggesting that a targeted analysis of predicted genes has the potential to yield a large increase in the mammalian gene set.
After the initiation of this study, at least one full-length cDNA was submitted by the RIKEN group for 28 of the 237 genes tested in category C and D. Of the 58 category C and D genes that were detected in this study, nine overlapped with Riken clones. The TSS suggested by RACE–PCR agreed with that of the RIKEN clone for all nine genes (data not shown). In addition, eight of the 237 genes tested in category C and D have subsequently been annotated as RefSeq genes. However, the RefSeq annotation of the start sites of four of these eight genes is indicated to be incomplete by our data.
First exon variation
Identification of variation in first exons is of great importance as it can potentially help understand gene regulation based on usage of alternate promoters and tissue-specific expression (Zhang et al. 2004). First exon variation is also likely to be involved in the utilization of alternate splice sites (Zavolan et al. 2002) and therefore influence the production of different protein isoforms, particularly at the amino terminus. In this study, 21 of the 106 genes (∼20%) successfully detected exhibited variation in transcription start sites (Fig. 1B). This percentage is similar to that observed by Zavolan and colleagues wherein 30% of mouse genes in their set exhibited first exon variation.
CpG island association with first exons
Approximately 50% of genes in mammals are associated with CpG islands (Antequera and Bird 1993), which have been used as markers to predict promoter regions. We determined the relationship between CpG islands and the first exons detected by RACE–PCR. The overall percentage of first exons associated with CpG islands (∼53%) is similar to the previously reported observation. The first exons for a significantly higher percentage of genes in the well-characterized set (∼70% in both EPD and RefSeq) are CpG linked. In contrast, <50% of the first exons of genes in the other categories were associated with CpG islands (Supplemental Fig. 4). This finding may be due to the “types” of genes that are in the different categories. Specifically, the EPD and RefSeq categories may consist of a large percentage of constitutively expressed or housekeeping genes that are known to be associated with CpG islands at a high frequency (Larsen et al. 1992), while the other categories may consist of tissue-specific or developmental stage–specific genes that may have a lower frequency of association with CpG islands.
Discussion
We have carried out a study both to determine the TSSs of a number of mouse genes and to assess the accuracy of current mouse genome annotation. The results show that there are considerable deficiencies in the current annotation of TSSs. For 61 of the 106 genes (58%) successfully amplified (Table 1), 5′ RACE–PCR detected transcripts had incomplete TSS annotation. This is not surprising, since in the absence of full-length cDNA information, the annotation of transcription start sites must collate and evaluate data from disparate sources, assessing their relative quality in making a “final” determination of the TSS. We would contend that this is an extremely difficult task. Since a substantial number of genes are not represented in the publicly available full-length cDNA sets, the occurrence of misannotated TSSs could be expected to be very common. Our results suggest that a viable way to address this issue rigorously is with a systematic program of directed RACE–PCR based on known gene structures and/or gene predictions that will aid in “correcting” the existing gene annotation. Such corrections will be to either capture previously misannotated structures or, alternately and importantly, alternate TSSs for correctly annotated genes. Such variation in start sites undoubtedly exists for many genes, and a directed approach is ultimately probably the only way to capture it. Only with such data in hand will we be able to begin carrying out large-scale computational studies on likely motifs used in the coregulation of expression of large sets of genes.
In addition, we found that the conservative nature of gene annotation in general leads to a very low false-positive rate but a surprisingly high number of missed genes. It is interesting to note that the current estimates of gene numbers in mammalian genomes is lower than previously thought (Lander et al. 2001; Venter et al. 2001; Waterston et al. 2002). This runs contrary to experimental evidence accumulating in the literature (Shoemaker et al. 2001; Kapranov et al. 2002; Rinn et al. 2003; Kampa et al. 2004;) as well as the data reported in this study. It is possible that the emphasis on database consistency and high specificity is leading to a substantial underestimate of gene numbers. As with TSS determination, an effective way to address this problem is with a systematic experimental confirmation of several types of gene predictions, including those with low confidence and stringency.
We chose RACE–PCR rather than reverse transcriptase (RT)–PCR to carry out confirmation of gene predictions for two major reasons. First, in addition to confirming a transcript, this approach would provide information about the TSS. Second, the work of Vidal and colleagues in experimental validation of gene predictions in C. elegans (Reboul et al. 2001, 2003) showed that a common cause of failing to recover a transcript from a predicted gene was that the primer(s) picked from predicted exons were slightly outside the actual exons. The modified 5′RACE–PCR approach described in this report allows us to have a known primer (based on the oligo adapter at the 5′ end), and the second primer in an internal exon, which is more reliably predicted using current methodologies. We reasoned the overall procedure would be more robust than using exon-specific primers on both ends.
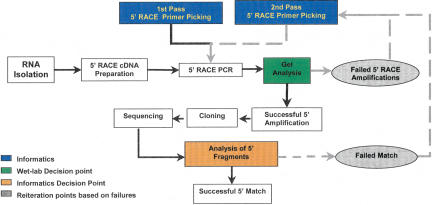
The optimized 5′ RACE–PCR protocol used to obtain the present data was designed to be performed in a high-throughput format. Figure 2 provides an overview of the workflow that was used in this study. Other than the production of 5′ RACE cDNA from specific tissues, all other steps were carried out in 96-well format with liquid handling performed by the Biomek FX robotic workstation or a multichannel digital pipettor. Based on the procedures and workflow we have optimized in the course of this work, we estimate that two people can obtain 500–1000 5′ RACE–PCR sequences per week.
Figure 2.
Overview of high-throughput workflow. The initiation of this workflow begins with the selection of genes for experimental verification. The gene models associated with these genes are used to select 5′ RACE–PCR primers (1st pass) (for more details, see Supplemental information). The primers are used to amplify RACE templates that have been generated in parallel. Failed amplifications are identified via agarose gel analysis, and new primers are picked for those genes (2nd pass). The products of the successful amplification are ligated into a plasmid vector. Eight clones from every amplification are sequenced and analyzed by comparison to the existing gene model. Amplified fragments that do not match their associated gene model are evaluated, and new primers are picked for the associated gene model (2nd pass). Successful matches are further evaluated in terms of whether they modify the current gene model, and the sequence is submitted to GenBank.
There are ∼6000 Twinscan and GenomeScan predictions matching the criteria that we have used for assigning gene predictions category C and D (ftp://ftp.cshl.org/pub/sequences/mouse/data_for_paper/). By extrapolation of our gene combined detection rate in these categories (∼25%), we estimate that there are likely to be at least 1450 additional genes in the mouse genome that are not part of the current annotated gene set. Our results clearly suggest that a significant fraction of the transcriptome may go undetected if gene verification strategies are restricted to testing only those gene predictions that have highly conserved structures or previous EST matches. It has previously been reported that three-quarters of all human genes (and by extrapolation, mouse genes) can be recognized in the Fugu genome (Aparicio et al. 2002). In contrast, only ∼21% of the 58 novel genes detected in this study have identifiable homologs in the sequenced Fugu genome. This may show circularity in current gene annotation efforts that are having the effect of pushing gene estimates unrealistically downward.
Methods
Three hundred annotated genes or gene predictions were chosen for analysis. Some of these were selected to allow an evaluation of the quality of the annotation describing the TSS of well-characterized genes, while others represent predictions chosen to allow an estimate of the unannotated genes that might exist in the genome. Table 1 describes the criteria for categorizing the genes into different groups based on the quality and confidence level of information associated with each known gene or gene prediction (for data sources, and detailed information about gene selection, see Supplemental information; the complete gene set is available at ftp://ftp.cshl.org/pub/sequences/mouse/data_for_paper/). Briefly, all known (RefSeq and mRNA) and predicted (Ensembl, GenomeScan, TwinScan) transcripts were mapped to the mouse genome. For genes in category C and D, the following steps were taken: (1) genic regions that are only mapped by GenomeScan and/or TwinScan transcripts (novel gene set) were selected; (2) from this set, only those predictions were retained that have at least one FirstEF (Davuluri et al. 2001) predicted promoter located <20 kb upstream of the 5′-boundary; and (3) all mouse ESTs were mapped to the mouse genome. Predictions from step 2, which overlap only a single EST, were placed into category C. Predictions from step 2 that do not overlap any EST were placed into category D. (4) Only those genes from step 3 that have at least one human–mouse conserved sequence element (hm-CSE) in the exonic regions were retained, and (5) the genes in each category were then sorted based on the distance from the FirstEF predicted TSS to the transcript-mapped gene 5′-end. Two hundred of the category C genes and 100 of the category D genes with shortest distance were then used for primer selection. Primers were generated for 237 of these genes based on the primer picking criteria described below.
Primer design
First exon boundaries were determined by aligning the predicted sequence to the genome using BLAT (Kent 2002). Primers were selected within exons other than the first exon to obtain spliced products. Two primers were chosen for each gene, both in internal exons, one in the middle of the gene and one flanking the known or predicted first exon (Supplemental Fig. 5). Perl scripts were written to design primers using the primer3 software (Rozen 2000). Primers were checked for uniqueness by querying against a customized database of all mRNA and Riken full-length cDNA collection. Primers matching sequences other than the corresponding predicted or known mRNA sequences in the customized database with >70% identity were discarded. For 3′RACE–PCR experiments, two additional primers were picked for each gene. These primers lie upstream of the two primers picked for 5′ RACE–PCR experiments and were designed based on the sequences obtained from 5′ RACE–PCR experiments for each gene. Primers were synthesized in 96-well plate format by Illumina or on site using the Mermade IV oligo-synthesizer.
RACE protocol
Total cytosolic mouse RNA from the following tissues/stages was obtained from BD Biosciences: 7-, 11-, 15-, and 17-d-total embryo, whole brain, eye, kidney, liver, lung, prostate, submaxilary gland, smooth muscle, spleen, testes, and uterus. The RACE protocol was adapted from the RNA-ligase–mediated RACE (RLM–RACE) system from Ambion. The following modifications were made to the protocol to increase the robustness and efficiency of transcript amplification. RNA samples were treated with DNAse I and purified (Qiagen) prior to the procedure. Following isolation, 10 μg of total RNA was dephosphorylated for 60 min at 37°C in a 10-μL reaction with 10 U shrimp alkaline phosphatase (SAP; Roche Diagnostics). RNA was then phenol/chloroform extracted, precipitated, and resuspended in water; 4.5 μg of dephosphorylated RNA was digested for 60 min at 37°C in a 10-μL reaction with 10 U tobacco acid pyrophosphatase (TAP; Ambion). Nine hundred nanograms of TAP-digested RNA was then incubated for 60 min at 37°C with 1 U T4 RNA ligase (Ambion) and 17 μM of an RNA adapter (5′GCUGAUGGCGAUGAAUGAACACUGCGUU UGCUGGCUUUGAUGAAA-3′, Ambion). After ligation, 180 ng of RNA was incubated for 2 min at 75°C in the presence of 5 μM random decamers (Ambion) in RT buffer. The sample was allowed to cool slowly to room temperature to allow the random decamers to anneal and prevent the refolding of RNA. Single-stranded cDNA was generated by the addition of 100 U M-MLV RT (Ambion) and incubation at 42°C for 60 min.
RACE–PCR amplification
PCR amplification was carried out in 96-well plate format. Amplification of 5′ RACE cDNA was carried out using nested gene-specific primers and adapter specific primers. Primers and cycling conditions are described in Supplemental information.
Cloning and sequencing of RACE–PCR products
After amplification, 5′ RACE–PCR products were cloned into pCR-TOPO2.1 vector (Invitrogen). Briefly, 2 μL PCR reaction was incubated with 0.1 ng of vector for 30 min at room temperature in a 96-well PCR plate (Robbins). The ligated samples were then incubated on ice with 20 μL TOP10 competent cells (Invitrogen) for 30 min and then heat shocked for 30 sec at 42°C. Cells were then transferred to 100 μL SOC in a 96-deep well block (Beckman-Coulter) and incubated for 1 h at 37°C with shaking. Eight clones from each transformation were then inoculated in LB media and grown overnight in a 96-well deep-well plate. DNA was isolated from the cultures using an automated alkaline lysis prep. The cloned products were then sequenced using flanking -21 M13 forward and reverse primers in 1/16th Big Dye Terminator v3.0 reactions (ABI). After precipitation, samples were resuspended in water and separated/detected on an ABI 3700 DNA sequencer.
Sequence analysis
Gene sequences and sequences obtained using the RACE–PCR method were aligned to the mouse genome (Oct.2003 build) using BLAT (Kent 2002). For genes in EPD and RefSeq categories, the latest annotation (RefSeq release 3, January 2004) was used as the gene sequence. For genes in other categories, the original gene sequence (corresponding to RefSeq release 2) was used. For cases in which a corresponding gene structure from RefSeq RNA was not available, the predicted gene structure was used as the reference annotation. A RACE–PCR sequence was counted as a positive hit if it satisfied the following criteria: (1) in the case of multi-exon genes, if alignment indicated a spliced product; (2) if the product mapped to the same region of the genome as the gene sequence; (3) if the product could be mapped uniquely to the genome with >98% identity over >95% of the sequence; (4) if at least two clones were obtained with similar exon structure; and (5) if the sequence contained the RACE-specific primer sequence. Sequences for 15 genes (in categories C and D) did not produce spliced products but agreed with all the other criteria mentioned above. Analysis indicated that for all these cases, the primer sequences were present in the first exon. For two of these cases, spliced products were obtained using additional primers and amplifying the 3′ end of the gene using 3′ RACE–PCR. These were therefore counted as positive hits. The genomic sequence corresponding to the gene and a flanking 15-kb sequence on each side was extracted. This sequence was used as the reference sequence to align gene sequence and sequences obtained using the RACE–PCR method using Sim4 (Florea et al. 1998).
CpG island analysis
Coordinates of CpG islands were obtained from the UCSC (University of California–Santa Cruz) Genome database. A first exon was considered to be CpG associated if a CpG island overlapped with a region that lies within 200 bp upstream of the transcription start and the first donor sites suggested by RACE–PCR sequence.
Acknowledgments
This work was supported by grant 3 U54 HG02135 from the National Human Genome Research Institute. We thank Damon J. Kelly for assistance in programming PERL modules used in primer selection. We would also like to thank William Tansey and Lincoln Stein for critical reading of the manuscript.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3158304.
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to dbEST/GenBank under accession nos. CV303589–CV309218.]
References
- Antequera, F. and Bird, A. 1993. Number of CpG islands and genes in human and mouse. Proc. Natl. Acad. Sci. 90: 11995-11999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio, S., Chapman, J., Stupka, E., Putnam, N., Chia, J.M., Dehal, P., Christoffels, A., Rash, S., Hoon, S., Smit, A., et al. 2002. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297: 1301-1310. [DOI] [PubMed] [Google Scholar]
- Burge, C. and Karlin, S. 1997. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268: 78-94. [DOI] [PubMed] [Google Scholar]
- Carninci, P., Shiraki, T., Mizuno, Y., Muramatsu, M., and Hayashizaki, Y. 2002. Extra-long first-strand cDNA synthesis. Biotechniques 32: 984-985. [DOI] [PubMed] [Google Scholar]
- Carninci, P., Waki, K., Shiraki, T., Konno, H., Shibata, K., Itoh, M., Aizawa, K., Arakawa, T., Ishii, Y., Sasaki, D., et al. 2003. Targeting a complex transcriptome: The construction of the mouse full-length cDNA encyclopedia. Genome Res. 13: 1273-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davuluri, R.V., Suzuki, Y., Sugano, S., and Zhang, M.Q. 2000. CART classification of human 5′ UTR sequences. Genome Res. 10: 1807-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davuluri, R.V., Grosse, I., and Zhang, M.Q. 2001. Computational identification of promoters and first exons in the human genome. Nat. Genet. 29: 412-417. [DOI] [PubMed] [Google Scholar]
- Florea, L., Hartzell, G., Zhang, Z., Rubin, G.M., and Miller, W. 1998. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 8: 967-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampa, D., Cheng, J., Kapranov, P., Yamanaka, M., Brubaker, S., Cawley, S., Drenkow, J., Piccolboni, A., Bekiranov, S., Helt, G., et al. 2004. Novel RNAs identified from an in-depth analysis of the transcriptome of human chromosomes 21 and 22. Genome Res. 14: 331-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov, P., Cawley, S.E., Drenkow, J., Bekiranov, S., Strausberg, R.L., Fodor, S.P., and Gingeras, T.R. 2002. Large-scale transcriptional activity in chromosomes 21 and 22. Science 296: 916-919. [DOI] [PubMed] [Google Scholar]
- Kent, W.J. 2002. BLAT: The BLAST-like alignment tool. Genome Res. 12: 656-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf, I., Flicek, P., Duan, D., and Brent, M.R. 2001. Integrating genomic homology into gene structure prediction. Bioinformatics 17: S140-S148. [DOI] [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- Larsen, F., Gundersen, G., Lopez, R., and Prydz, H. 1992. CpG islands as gene markers in the human genome. Genomics 13: 1095-1107. [DOI] [PubMed] [Google Scholar]
- Reboul, J., Vaglio, P., Tzellas, N., Thierry-Mieg, N., Moore, T., Jackson, C., Shin-i, T., Kohara, Y., Thierry-Mieg, D., Thierry-Mieg, J., et al. 2001. Open-reading-frame sequence tags (OSTs) support the existence of at least 17,300 genes in C. elegans. Nat. Genet. 27: 332-336. [DOI] [PubMed] [Google Scholar]
- Reboul, J., Vaglio, P., Rual, J.F., Lamesch, P., Martinez, M., Armstrong, C.M., Li, S., Jacotot, L., Bertin, N., Janky, R., et al. 2003. C. elegans ORFeome version 1.1: Experimental verification of the genome annotation and resource for proteome-scale protein expression. Nat. Genet. 34: 35-41. [DOI] [PubMed] [Google Scholar]
- Rinn, J.L., Euskirchen, G., Bertone, P., Martone, R., Luscombe, N.M., Hartman, S., Harrison, P.M., Nelson, F.K., Miller, P., Gerstein, M., et al. 2003. The transcriptional activity of human chromosome 22. Genes & Dev. 17: 529-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen, S. and Skaletsky, H. 2000. Primer3 on the WWW for general users and for biological programmers. In Bioinformatics methods and protocols: Methods in molecular biology (eds. S. Krawetz and S. Misner), pp. 365-386. Humana Press, Totowa, NJ. [DOI] [PubMed]
- Shoemaker, D.D., Schadt, E.E., Armour, C.D., He, Y.D., Garrett-Engele, P., McDonagh, P.D., Loerch, P.M., Leonardson, A., Lum, P.Y., Cavet, G., et al. 2001. Experimental annotation of the human genome using microarray technology. Nature 409: 922-927. [DOI] [PubMed] [Google Scholar]
- Venter, J.C., Adams, M.D., Myers, E.W., Li, P.W., Mural, R.J., Sutton, G.G., Smith, H.O., Yandell, M., Evans, C.A., Holt, R.A., et al. 2001. The sequence of the human genome. Science 291: 1304-1351. [DOI] [PubMed] [Google Scholar]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
- Yeh, R.F., Lim, L.P., and Burge, C.B. 2001. Computational inference of homologous gene structures in the human genome. Genome Res. 11: 803-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavolan, M., van Nimwegen, E., and Gaasterland, T. 2002. Splice variation in mouse full-length cDNAs identified by mapping to the mouse genome. Genome Res. 12: 1377-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, T., Haws, P., and Wu, Q. 2004. Multiple variable first exons: A mechanism for cell- and tissue-specific gene regulation. Genome Res. 14: 79-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
Web site references
- http://mgc.nci.nih.gov/; Mammalian Gene Collection.
- ftp://ftp.cshl.org/pub/sequences/mouse/data_for_paper/; Author's additional mouse data.