Abstract
MSM/Ms is an inbred strain derived from the Japanese wild mouse, Mus musculus molossinus. It is believed that subspecies molossinus has contributed substantially to the genome constitution of common laboratory strains of mice, although the majority of their genome is derived from the west European M. m. domesticus. Information on the molossinus genome is thus essential not only for genetic studies involving molossinus but also for characterization of common laboratory strains. Here, we report the construction of an arrayed bacterial artificial chromosome (BAC) library from male MSM/Ms genomic DNA, covering ∼1× genome equivalent. Both ends of 176,256 BAC clone inserts were sequenced, and 62,988 BAC-end sequence (BES) pairs were mapped onto the C57BL/6J genome (NCBI mouse Build 30), covering 2,228,164 kbp or 89% of the total genome. Taking advantage of the BES map data, we established a computer-based clone screening system. Comparison of the MSM/Ms and C57BL/6J sequences revealed 489,200 candidate single nucleotide polymorphisms (SNPs) in 51,137,941 bp sequenced. The overall nucleotide substitution rate was as high as 0.0096. The distribution of SNPs along the C57BL/6J genome was not uniform: The majority of the genome showed a high SNP rate, and only 5.2% of the genome showed an extremely low SNP rate (percentage identity = 0.9997); these sequences are likely derived from the molossinus genome.
A number of inbred mouse strains characterized by different heritable traits have been established, and these strains are extensively used in biomedical research (Atchley and Fitch 1991; Beck et al. 2000). The distinct phenotypes manifested by these laboratory strains are attributable to differences in the genomic constitution carried by each strain. However, the gene pools of the commonly used laboratory mouse strains are probably small, as they were derived from a mixed, small number of fancy mice bred at the beginning of the 20th century (Morse III 1981; Silver 1995).
The house mouse, Mus musculus, is a complex species comprised of several subspecies. Results of our past studies (Yonekawa et al. 1980,1982; Moriwaki 1994) and those of others (Ferris et al. 1982; Bonhomme et al. 1987) suggest that commonly used laboratory strains were derived predominantly from the Mus musculus domesticus subspecies, which is indigenous to Western Europe and the Mediterranean Basin, and that there were some small contributions from the Japanese fancy mouse, which originated from the Mus musculus molossinus subspecies (Morse III 1981; Silver 1995). Thus, the laboratory strains can be considered a natural set of recombinant inbred strains derived from a small number of mice belonging to either M. m. domesticus or Asian subspecies (Wade et al. 2002). Different combinations of genomic segments contributed by the two major ancestral sources may have created the diverse phenotypic traits characteristic of each strain. Therefore, information as to how these inbred strains originated historically and how they are related to each ancestral subspecies is crucial in any genetic analyses of the phenotypes of interest.
Recent analyses using single nucleotide polymorphism (SNP) markers across the whole mouse genome have revealed uneven distributions of SNPs along the genomes of the laboratory strains (Wade et al. 2002; Wiltshire et al. 2003). From pairwise comparisons of inbred strains, Wade et al. (2002) found that the genomes of the laboratory strains have mosaic structures consisting of long segments of either low or high polymorphism rates, and they proposed that the high-SNP-rate regions are reflections of intersubspecific polymorphisms between M. m. domesticus and Asian subspecies, M. m. molossinus or M. m. musculus. However, this notion is based on rather indirect evidence obtained mainly from the genomic data on common laboratory strains, and the precise structures of such mosaic patterns in each strain have not been presented. Currently, sizable data on genomic sequences are available only for the common laboratory strains, and our knowledge of the genomic constitutions of other mouse subspecies is very limited. Such scarcity of genomic information hampers our better understanding of the genomic evolution of M. musculus subspecies and the origin of the common laboratory strains, as well as of the mosaic structures of variations among those strains.
We therefore attempted to gain genomic information from Asian subspecies. For this purpose, we chose MSM/Ms as a target strain, because there are several lines of evidence that M. m. molossinus predominantly contributed to the genomes of the common laboratory strains (Nagamine et al. 1992; Floyd et al. 2003). We constructed a bacterial artificial chromosome (BAC) genomic library, a fundamental resource for a variety of genomic research (Zhao et al. 2001) and subsequently sequenced the BAC-ends of ∼180,000 clones. The MSM/Ms strain was established from Japanese wild mice, M. m. molossinus, collected in 1978 in Mishima, Japan (Moriwaki 1994). The inbreeding generations of this strain had reached N71 at the end of 2003, and it is now established as a pure inbred strain. Genetic analysis using the MSM/Ms strain has several advantages: (1) MSM/Ms has unique characteristics not observed in the other laboratory strains, for example, extremely low incidence of tumor development (Miyashita and Moriwaki 1987) and characteristic behavioral phenotypes (Koide et al. 2000); (2) it shows high breeding performance (100–125 N2 progeny/pair/year); (3) it has a number of polymorphic DNA markers (more than 2000 SSLP markers are now available for the cross of MSM/Ms with the common laboratory strains; Kikkawa et al. 2001 and see Mouse Microsatellite Database of Japan [MMDBJ], http://www.shigen.nig.ac.jp/mouse/mmdbj/top.jsp); (4) a full series of consomic strains, in which each chromosome of MSM/Ms is introduced into the B6 background by repeated backcross, is established (T. Shiroishi, unpubl.); and (5) MSM/Ms has been selected as a target strain in the Mouse Phenome Project, and therefore accumulating data of phenotypes will be available (Paigen and Eppig 2000).
Here we describe the general characteristics of the MSM/Ms BAC library, which is the first, representative BAC library from M. m. molossinus, and the results of BAC-end sequence (BES) analysis of about 180,000 clones. We mapped the MSM/Ms BESs onto the C57BL/6J (B6) genome assembly (NCBI Build 30). Comparison of the MSM/Ms sequences with the B6 data provided a vast number of SNPs with defined genomic locations. Using this SNP information, we present the fine details of the genetic variation between M. m. molossinus and C57BL/6J, one of the most widely used laboratory strains.
Results
Construction of MSM BAC library
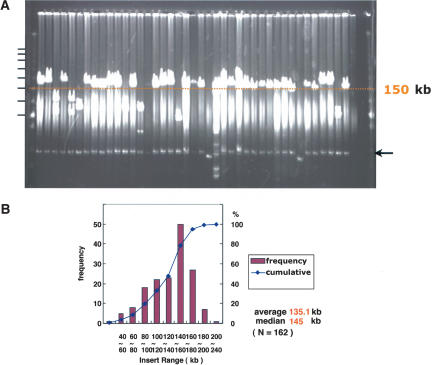
A BAC library was constructed from genomic DNA of male MSM/Ms (henceforth MSM), an inbred strain derived from mouse subspecies groups of M. m. molossinus (Moriwaki 1994). High-molecular-weight DNA isolated from MSM male mice was partially digested with a combination of EcoRI and EcoRI methylase and cloned into pBACe3.6 vector (Osoegawa et al. 1998). The library was composed of 210,432 BAC clones arrayed in 384-well microtiter plates. The average insert size of the library was 135.1 kb, as determined by pulsed-field gel electrophoresis analysis of 162 randomly selected clones (Fig. 1). Thus this library has ∼11.9 × coverage of the haploid mouse genome. This library can be screened by hybridizing probes with high-density colony blot or by performing PCR with DNA pools consisting of mixtures of the BAC DNA isolated from each of the library plates (data not shown).
Figure 1.
Estimation and distribution of insert sizes in MSM BAC clones. (A) Randomly chosen BACs were digested with NotI that flank the cloning site (EcoRI) in the pBACe3.6 vector. The digested DNAs were analyzed by pulsed-field gel electrophoresis on a 1% agarose gel in 0.5× TBE at 6V/cm, with 0.1 sec to 40 sec pulse time for 16 h at 14°C. The DNA markers are λ concatemers plus HindIII-digested λDNA. Horizontal black bars indicate positions of the concatemers. The arrow indicates the position of the vector. Distribution of insert size is shown in (B).
Derivation of BAC-end sequences and their mapping onto the mouse genome sequence
To exploit this genomic resource fully, we sequenced both ends of the BAC clones and mapped the BESs onto the B6 genome assembly (NCBI mouse Build 30). The precise assignment of each BAC clone position along the chromosomes by the BES mapping enabled computer-based screening of the library. BES mapping can also provide information on chromosomal rearrangements between the MSM and the B6 (Volik et al. 2003). More importantly, comparison of the MSM sequences with the B6 data will generate a vast amount of SNPs with defined genomic locations. The distribution patterns of the SNPs will reveal the fine details of the genetic variation between the two M. musculus subspecies (Wade et al. 2002).
Both ends of 176,256 BAC clone inserts were sequenced, corresponding roughly to 6.4% of the haploid mouse genome. After we had trimmed the vector sequences and masked any highly repetitive sequences, we performed a BLAST search of the BESs against the B6 genome to determine the genomic locations of these BAC clones. For this mapping, we considered the size of homologous sequences, percentage identity, sequence orientation, and the average size of the BAC inserts (<300 kb). If the BES hit multiple locations, the position of the region showing the highest BLAST score was assigned. The `paired-end' sequences were selected from 62,988 BACs. From the `paired-end' group, 38,611 clones with `unique paired-end' sequences were further selected; these were mapped unambiguously to one location in the genome. Coverage of the mouse genome by the MSM BAC clones was estimated on the basis of the results of the BES mapping. As shown in Table 1, 89% of the B6 genome sequence (i.e., 2,228,164 kbp) was covered by the `paired-end' clones. In other words, 62,988 × 2 = 125,976 BESs were interspersed across the 2,228,164 kbp genomic sequence (i.e., one BES was found in every 17.68 kb). The sum of the `paired-end' BES size was 51,137,941 bp (average, 406 bp per BES), corresponding to 2.29% of the total genome size. Coverage for each chromosome was similar, with the exception of the X chromosome (Table 1), which we would expect to be underrepresented in this library because male genomic DNA was used as a source material. BESs often carry highly repetitive sequences that prevent mapping onto the genome (Zhao et al. 2001). Clones with unique hit sequences at one end but unmapped sequences at the other and those with unmapped sequences at both ends were excluded from the analysis described above.
Table 1.
Coverage of the genome by the MSM BAC clones
| Chromosome | B6 genome sequenced (kb) | MSM covered (kb) | Coverage | No. of clones |
|---|---|---|---|---|
| chr1 | 191,720 | 172,683 | 0.90 | 4,597 |
| chr2 | 177,924 | 162,218 | 0.91 | 4,839 |
| chr3 | 156,874 | 141,439 | 0.90 | 4,056 |
| chr4 | 149,422 | 133,339 | 0.89 | 3,674 |
| chr5 | 145,320 | 130,549 | 0.90 | 3,706 |
| chr6 | 145,901 | 128,731 | 0.88 | 3,718 |
| chr7 | 128,702 | 112,502 | 0.87 | 3,056 |
| chr8 | 125,273 | 113,156 | 0.90 | 3,336 |
| chr9 | 120,867 | 109,326 | 0.90 | 3,057 |
| chr10 | 127,088 | 114,979 | 0.90 | 3,212 |
| chr11 | 119,563 | 111,820 | 0.94 | 3,402 |
| chr12 | 110,213 | 100,598 | 0.91 | 3,040 |
| chr13 | 112,393 | 104,410 | 0.93 | 2,948 |
| chr14 | 112,144 | 102,497 | 0.91 | 3,052 |
| chr15 | 100,912 | 94,315 | 0.93 | 2,659 |
| chr16 | 95,837 | 88,132 | 0.92 | 2,556 |
| chr17 | 89,430 | 79,658 | 0.89 | 2,226 |
| chr18 | 87,841 | 80,949 | 0.92 | 2,389 |
| chr19 | 57,643 | 53,088 | 0.92 | 1,653 |
| chrX | 145,596 | 93,775 | 0.64 | 1,452 |
| Total | 2,500,661 (kb) | 2,228,164 (kb) | 0.89 | 62,988 |
To show the effective coverage of the genome by paired-end clones in detail and to establish a convenient means of identifying BAC clones, we made a Web browser-based BAC library screening system (http://stt.gsc.riken.jp/msm/). In this system, it is possible to view genomic regions of interest covered by MSM BAC clones by either clicking on a region of interest on the chromosome map in the top menu or typing a gene symbol or a sequence ID in the search window (Fig. 2A,B). In this example, the genomic region covering H-2K gene of mouse major histocompatibility complex is shown (Fig. 2B). The BAC clones with a red triangle at both ends represent the `unique paired-end' BAC clones, whereas the pink triangle represents `ambiguous' BESs. Orange triangles indicate the positions of `one-ended' clones, of which only one BAC-end was unambiguously mapped. Including such clones, the coverage of total genomic regions by the MSM BAC clones was practically extended (data not shown).
Figure 2.
BAC contigs retrieved by the web-based clone screening system. By clicking a genomic region of interest on the chromosome map in the top menu, BAC contigs covering the corresponding genomic region can be viewed (A). The red triangles represent the `unique paired-end' BAC clones; the pink triangles represent `ambiguous' BESs. Locations of `one-ended' BAC clones are shown by the orange triangles (B). Clone MSMg01-114N13 is an example of such `one-ended' clones; its size is ∼150 kb long, extending toward the centromere, and the clone was found to contain the Rps28, Angptl4, Rab11b, and 9530046H09Rik genes.
Collection of SNPs from the BAC-end sequences and distribution of SNPs across the genome
The nucleotide substitutional difference between B6 and MSM was calculated by comparing the MSM BESs with the B6 genome sequences. Insertion or deletion of DNA sequences (so-called `indels') was excluded from this analysis (see Discussion). From the `paired-end' BESs, 489,200 SNPs were detected in 51,137,941 bp sequenced. For this SNP survey, we used BESs with PHRED (Ewing et al. 1998) values equal to or higher than 30 (PHRED ≥30). The calculation yielded a base substitution rate 0.0096. Of these nucleotide substitutions, there were 323,416 transition changes and 165,784 transversion changes. The ratio of transition changes to transversion changes was thus calculated to be 1.95. Similar figures for substitutional difference (0.0093) and transition versus transversion ratio (1.97) were obtained from `unique paired-end' data sets.
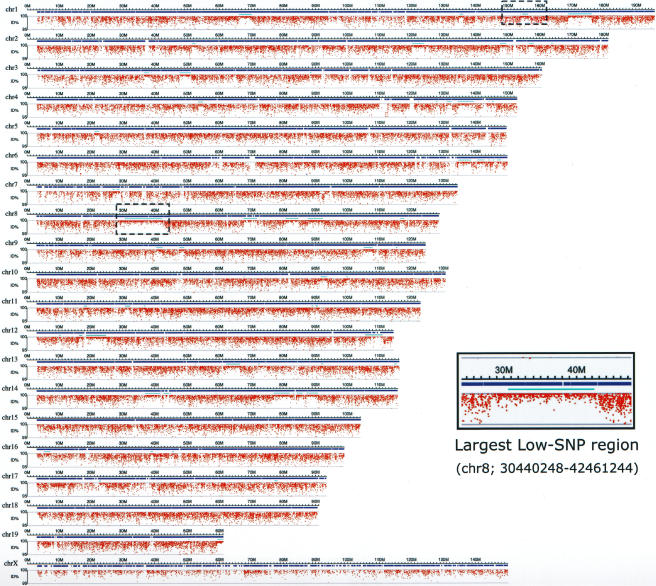
We next plotted the percentage identity of each of the MSM BESs in comparison with the B6 sequences along the chromosomes (Fig. 3). Although most of the genomic regions contained a high density of SNPs, as expected from the calculated substitutional differences, there were regions showing percentage identities close to 1.00 (average percentage identity = 0.9997), indicating that the distributions of the SNPs were not uniform. There were 66 such extremely low SNP regions across the whole genome. The size of the low-SNP-rate regions varied from ∼140 kb to 12 Mb, with an average size of 2 Mb (median = 1.37 Mb). The precise locations and sizes of the low-SNP-rate regions are summarized in Supplemental Table S1. The proportion of low-SNP segments versus high-SNP regions also varied for each chromosome. For example, 20% of chromosome 8 corresponded to low-SNP regions (Table 2), and the largest consecutive low-SNP segment (12 Mb) was also found on chromosome 8 (Fig. 3, inset). In contrast, there were no low-SNP regions on chromosomes 15 and 18 or on the X chromosome (Table 2). The overall occurrence of nucleotide substitutions did not vary significantly among the chromosomes. The X chromosome seemed to have a slightly lower substitutions than did the autosomes. Low-SNP segments occupied a total of 5.2% of the B6 genome (Table 2).
Figure 3.
Distribution of SNPs across the chromosomes; identification of low- and high-SNP regions. The percentage identity of each of the MSM BESs in comparison with the B6 sequences was plotted along the chromosomes. Red dots represent the MSM BESs, and positions of the dots along the y-axis indicate % identity of each BES. Examples of the low-SNP regions (chr8; 30440248–42461244) are enlarged and shown in the inset. Position of this enlarged region on chromosome 8 is indicated by a dashed rectangle. Another dashed rectangle on chromosome 1 indicates position of a BAC clone, MSMg01-122K03, which was completely sequenced (see Fig. 4). Horizontal bars represent chromosomes, and chromosome number is indicated at left side. Blue bars represent genomic regions covered by the MSM BAC clones. Light blue bars indicate the low-SNP regions.
Table 2.
Size of low-SNP-rate regions relative to each chromosome
| % of low SNP regions | Substitution rate | % of low SNP regions | Substitution rate | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| chr1 | 7.30% | 0.0096 | chr11 | 3.20% | 0.0096 | |||||
| chr2 | 4.90% | 0.0095 | chr12 | 11.30% | 0.0093 | |||||
| chr3 | 3.70% | 0.0104 | chr13 | 5.40% | 0.0102 | |||||
| chr4 | 6.60% | 0.0097 | chr14 | 15.10% | 0.0092 | |||||
| chr5 | 1.00% | 0.011 | chr15 | 0% | 0.0108 | |||||
| chr6 | 5.60% | 0.0098 | chr16 | 3.30% | 0.0105 | |||||
| chr7 | 1.10% | 0.011 | chr17 | 1.60% | 0.0107 | |||||
| chr8 | 20.00% | 0.0088 | chr18 | 0% | 0.011 | |||||
| chr9 | 5.30% | 0.01 | chr19 | 3.50% | 0.011 | |||||
| chr10 | 2.00% | 0.0108 | chrX | 0% | 0.0072 | |||||
| chrYa | ND | 0.0025 | ||||||||
| Total: 5.29%b (Sum of the Low SNP regions relative to the whole B6 genome) | ||||||||||
Based on NCBI build 32
801 genes out of 12,389 known genes (loci) are present in this 5.2% of the genome
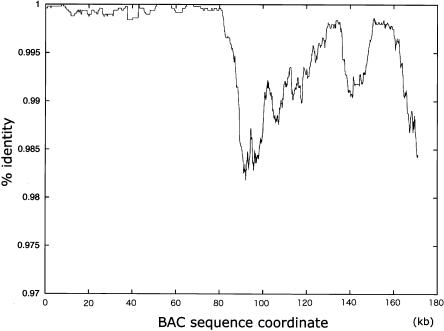
The B6 genome appeared to be composed of genomic segments with different SNP densities, according to the BES mapping data. To confirm this finding, we sequenced BAC clones that were on the borders of low-to-high SNP transitions and found that there were sharp transitions from low to high SNP frequency within those BAC finished sequences (Fig. 4). For example, in the clone MSMg01-122K03 (chr1_156195129_156370415), the percentage identity was close to 1.00 from bases 0 to 800, then abruptly decreased, reaching ∼0.985 within the region between bases ∼800 and ∼900. Although the percentage identity varied markedly in the next ∼80 kb, the average base substitutions remained high. An essentially similar result was also obtained from analysis of the clone MSMg01-275M02 (chr1_176213505_176378730) (data not shown). These results confirmed that the mosaic structures of low- and high-SNP regions revealed by the BES SNP distributions were true reflections of the nucleotide sequence variations between the B6 and MSM genomes, and that nucleotide substitutional differences between the two strains were in the range of ∼1% in the high-SNP-rate genomic segments.
Figure 4.
SNP distribution within the BAC clone MSMg01-122K03, on the border of low-to-high SNP regions. MSMg01-122K03 is mapped at chr1_156195129–156370415, which is on the border of low-to-high SNP transition. Complete sequence of this clone was determined, compared with the B6 sequences, and % identity (y-axis) was plotted along the entire sequence (x-axis).
Sixty-seven BESs were mapped onto the Y-chromosome sequence retrieved from the NCBI Build 32 data. The substitutional differences of the Y-derived BESs (0.0025) was much lower than those of the autosome and X chromosome sequences (Table 2). The ratio of transition versus transversion changes for the Y-SNPs was 0.7, the inverse of those obtained from other chromosomes.
As described above, an unexpectedly high incidence of nucleotide substitutions (i.e., 0.96%) was found between B6 and MSM. This nucleotide difference value was comparable with that of the genomic difference (i.e., 1.23%) between humans and chimpanzees, which were estimated to diverge more than five million years ago (Mya) (Fujiyama et al. 2002). Because the two mouse subspecies groups were considered to diverge ∼1 Mya (Moriwaki 1994; Silver 1995), mice (or rodents in general) should have a higher evolutionary rate than do hominoids. Alternatively (although unlikely), some biological constraints could exist on the BAC-end sequences, resulting in deviations from the normal evolutionary rate. To examine these possibilities, we performed comparative analyses of mouse and rat genomic sequences using two kinds of data set: one was the MSM BESs presented in this study and their orthologous sequences in the rat genome, and the other was the sequences of the protein coding regions of the mouse and their orthologous counterparts in the rat. Only fourfold degenerate (4D) synonymous sites were compared in the latter case (see Supplemental material for detailed methods). Nucleotide differences calculated from either the BES data or 4D synonymous sites of protein-coding region sequences were very similar and were estimated to be 13.66%∼14.61% (Supplemental Tables S2, S3).
This amount of divergence between mice and rat is compatible with that between MSM and B6 strains if we consider a reasonable range of divergence times (see Supplemental material and Discussion for details). This fact, in turn, suggests that the BES data from the MSM library are likely to comprehensively represent the neutral nucleotide sequences in the MSM genome, and implies that rodents have a higher evolutionary rate than do hominoids (see Discussion for details).
Discussion
We constructed a representative BAC genomic library from MSM/Ms. This is the first BAC library made from the M. m. molossinus subspecies. We sequenced both ends of 176,256 clone inserts and mapped the end sequences onto the C57BL/6J mouse genome assembly. Information on the genomic positions of the BAC clones allowed us to establish a browser-based clone screening system. Such computer-based screening has been possible only for the B6 BAC library until now. BES information is useful not only for clone mapping but also for identification of novel STSs and their corresponding EST markers (Zhao et al. 2001) or genomic rearrangements (Volik et al. 2003). Furthermore, to extend the utility of BESs, we compared MSM BESs with the B6 whole mouse genome sequences, generating a vast number of SNPs between the two strains. Thus the BAC library itself and the high-quality BES information obtained from this study will be valuable resources to the mouse research community.
Because we used BESs with PHRED values of ≥30 for this SNP survey, the incidence of “false-positives” in the 489,200 candidate SNPs was expected to be very small, if not negligible. This number of SNPs—nearly half a million—is much larger than those previously reported (Lindblad-Toh et al. 2000; Wade et al. 2002; Wiltshire et al. 2003). The SNP markers found every 17.68 kb across the genome should facilitate any type of genetic analysis involving B6 and MSM. Moreover, as it is highly likely that most of the MSM-associated SNPs are not MSM strain-specific but rather are shared by M. m. molossinus and M. m. musculus, they will be useful in crosses of these subspecies-derived strains.
It is generally believed that Asian mice have made substantial genetic contributions to the genomes of laboratory strains, although the majority of these genomes was derived from M. m. domesticus of Western European origin (Yonekawa et al. 1980, 1982; Moriwaki 1994). Wade et al. (2002), through SNP analyses, confirmed the notion that the genomes of laboratory strains are a mosaic of genetic segments derived from either domesticus or musculus (molossinus) ancestral sources, and suggested that the SNPs found among the laboratory strains are reflections of ancestral intersubspecific differences, not mutations specific to a particular strain. If this is the case, then most of the SNPs presented in this study can be used to type domesticus-molossinus differences. The unprecedented number of these SNPs, in conjunction with a high-throughput SNP typing method (e.g., Lindblad-Toh et al. 2000; Matsuzaki et al. 2004) should greatly facilitate genotyping, QTL mapping, modifier mapping, and the defining of SNP haplotype patterns in commonly used laboratory strains. We did not present details of indels, because (1) B6 genome assembly is not yet a `finished' sequence, and the BAC-end sequences used in this analysis are results of one-pass sequencing, and (2) long insertions may not be accurately assigned in the short BAC-end sequences, which are only ∼400 bp-long on average. Our analysis revealed ∼164,000 indels between B6 and MSM (H. Noguchi, unpubl.). Distribution of size of indels is depicted in Supplemental Figure S1. The majority of the indels were short, but a small peak at around 200 bp was detected. This peak likely was produced by the integrations of SINE, mainly B2 (Supplemental Fig. S1). A similar phenomenon due to Alu insertion was revealed by human–chimpanzee comparison (Watanabe et al. 2004). Information on such indels is potentially important for tracing evolutionary events after separation of two subspecies.
Wade et al. (2002) and Wiltshire et al. (2003) reported that the genomes of laboratory strains comprised blocks of low and high SNP occurrence, and that each strain appears to have a characteristic pattern of SNP haplotype. Estimates of the sizes of low and high regions, as well as of SNP densities, however, appeared to vary greatly in these studies and the present study. For example, about one-third (Wade et al. 2002) or about 40%–70% (Wiltshire et al. 2003) of the genome has high SNP densities, representing segments with intersubspecific difference, whereas in this study the low-SNP regions occupy only 5% of the genome in the B6-MSM comparison, representing molossinus-derived segments. These differences seem to be related to the extensiveness of the studies. For example, the sample sequence numbers and the sizes of the regions analyzed were different in the above studies. Moreover, differences in the methods used to make comparisons may explain the variations in data. In the previous studies, the B6 genome was compared with sequences from another common laboratory strain, whereas in our study the B6 genome was directly compared with BAC-end sequences of a molossinus-derived MSM strain. In our analysis, the low-SNP region (∼5% of the B6 genome) simply corresponded to the genomic segments inherited from M. m. molossinus. In the previous `indirect' analyses, however, the situation is more complicated. As shown in Supplemental Figure S2, the low- and high-SNP regions revealed by comparison of two laboratory strains rather reflect the composite patterns of SNP haplotypes from the two single strains. In the low-SNP regions, the two strains share a common subspecies origin (i.e., both domesticus or both molossinus). Conversely, the high-SNP regions are composites of segments from domesticus and molossinus, although we cannot distinguish which laboratory strain carries which ancestral source. Previous reports (Wade et al. 2002; Wiltshire et al. 2003) indicate that the distribution patterns of the low- and high-SNP regions vary in the comparison of different strain-pairs. Therefore, the sizes of the high-SNP regions revealed by comparison of any two laboratory strains will be usually larger than those revealed by comparison of one of the laboratory strains and an Asian wild mouse-derived strain (see Supplemental Figure S2). This would reconcile discrepancies in the extent of the contributions from the Asian mouse to the genomes of laboratory strains. In theory, our `direct' comparison of MSM-BES with the B6 genome should yield more straightforward interpretations, and our genome-wide, high-resolution data are effective in refining the structures of genetic variations in common laboratory strains. For example, our analysis identified relatively small (<1000-kb) regions of co-ancestry, which would be overlooked in other studies.
We estimated the base substitutional differences between MSM and B6 to be 0.0096, which is likely to represent the divergence rate between domesticus and molossinus. In a previous study (Wade et al. 2002), SNP rates were calculated on a different basis with different-sized data sets and were estimated to be lower than the value obtained in our study. Whether this difference is due to technical reasons or to genetic contributions from ancestral sources other than molossinus remains to be elucidated.
Knowledge of the fine structure of shared haplotype blocks will have a great impact on the design and interpretation of various genetical analyses (Wade et al. 2002; Wiltshire et al. 2003). In this context, one of the most important findings of our study is that we could define, for the first time, the precise structures of SNP haplotypes in the C57BL/6J genome, because B6 is one of the most widely used laboratory strains. It will be perhaps equally important to define two genomic segments with distant origins, which cannot coexist in the same genome. We found no molossinus-derived segments in the X chromosome of B6, consistent with the observation of Wiltshire et al. (2003) that the rate of polymorphism in the X chromosome is exceptionally low in the comparison of any strain combinations. This implies that interactions of X-linked genes of different evolutionary origins with autosomal or Y-linked genes would disturb the survival or fertility of individuals. In fact, such genetic interactions have been reported (Takagi et al. 1994; Oka et al. 2004).
In most of the present analyses, data from the Y chromosome were excluded, as the available data from this chromosome were still very limited. However, even the limited data on the Y-derived SNPs indicated that the Y chromosome of the B6 strain was much more similar to that of the MSM/Ms than was the case with the autosomes. This result is consistent with the previous findings that the Y chromosomes of the majority of laboratory strains, including B6, were derived from M. m. musculus (Bishop et al. 1985). More specifically, they are derived from the molossinus population, because most common laboratory strains have molossinus-specific polymorphisms of Y-linked genes such as SRY and Zfy (Nagamine et al. 1992, 1994). This can be further validated by the Y-SNPs from MSM/Ms. It is also notable that the ratio of transition versus transversion changes is quite different from the ones obtained in the autosomes and the X chromosome, implying that the mechanisms by which nucleotide substitutions are generated may be different in the Y chromosome (Skaletsky et al. 2003).
Data from comparative genomic analyses on the MSM BESs and rat sequences can be used for estimations of evolutionary rates of rodents. Evolutionary distance between mouse and rat was calculated to be 0.155 (Supplemental Table S2). This estimate is compatible with that obtained for the rat–mouse genomic comparison (Rat Genome Sequencing Project Consortium 2004). If we assume that mouse subspecies groups diverged from their ancestors ∼1 Mya, then mouse–rat speciation can be estimated to have occurred ∼16 (=0.155/0.0096 × ∼1) Mya, assuming that the evolutionary rate for rodents is constant during this period. This estimation of timescale for mouse–rat divergence appears to be consistent with a previous assumption that mice and rats diverged from a common ancestor 10–15 Mya (Jaeger et al. 1986).
If we use a formula, λ = d/2T (λ, evolutionary rate; d, evolutionary distance; T, divergence time) (Nei 1987), the evolutionary rate of nucleotide substitution in mouse was assumed to be 4.8 × 10-9 in the above consideration, where d = 0.0096 and T = 1 million years (Myr). Alternatively, we also estimated the evolutionary rate for rodents (between mouse and rat), λ, to be 2 × 10-9 ∼8 × 10-9, based on the evolutionary distance d = 0.155, and mouse–rat divergence time T = 10 or a maximum estimate of 40 Myr (Kumar and Hedges 1998). We therefore suggest that the estimated evolutionary rate in subfamily murinae is two to eight times larger than the maximum evolutionary rate (0.99 × 10-9) for humans and chimpanzees (Yi et al. 2002).
Methods
Construction of BAC library from MSM/Ms
The MSM/Ms BAC library was constructed in accordance with the methods described by Osoegawa et al. (1998). High-molecular-weight genomic DNA prepared from spleens and kidneys of MSM/Ms male mice was partially digested with a combination of EcoRI restriction endonuclease/EcoRI methylase (New England Biolabs) and size-fractionated by pulsed-field gel electrophoresis. The partially digested DNA was then ligated to EcoRI-digested and dephosphorylated pBACe3.6 vector and introduced into ElectroMAX DH10B competent cells (Invitrogen) by electroporation performed on a Gene Pulser (BioRad). Recombinant clones grown on agar plates containing 5% sucrose, and 20 μg/mL chloramphenicol were picked up with a Flexys colony picker (Genomic Solutions), and individual clones were arrayed into 384-well plates and stored at -80°C. A browserbased clone screening system will be available at these Web sites: http://stt.gsc.riken.jp/msm/ and http://shigen.lab.nig.ac.jp/mouse/polymorphism/ and http://analysis1.lab.nig.ac.jp/Mus_musculus/).
MSM BAC clones will be available from the RIKEN BioResource Center DNA bank (http://www.brc.riken.jp/lab/dna/).
BAC-end sequencing
The BAC library was rearrayed to 96-well plates with a Flexys system equipped with a gridding tool (GeneMachines). BAC clones were inoculated into a 96-deep-well plate and cultured overnight at 37°C in 1.5 mL 2× LB containing 12.5 μg/mL chloramphenicol. BAC DNA was isolated by using a Montage BAC96 Miniprep kit (Millipore) and dissolved in 35 μL of 10 mM Tris-HCl buffer (pH 8.0). Cycle sequencing reaction was carried out in a 15-μL reaction volume containing 10 μL BAC DNA (∼300 ng), 1.5 μL Big Dye terminator Ready Reaction mix (Applied Biosystems), and 1.5 μL sequence primer (3.2 pmol/μL). Custom-designed sequence primers at the T7 and SP6 ends were TGACAT TGTAGGACTATATTGC and ATCTGCCGTTTCGATCCTCC, respectively. PCR conditions were 95°C for 5 min, then 75 cycles of 95°C for 30 sec, 50°C for 10 sec, and 60°C for 4 min. The reaction mixture was cleaned up by isopropanol precipitation followed by 70% ethanol wash. The reaction products were loaded on ABI 3700 automated capillary DNA sequencers (Applied Biosystems).
Sequence processing and bioinformatics
Raw sequence data were base-called using the Phred program (Ewing and Green 1998; Ewing et al. 1998), and the vector sequences were filtered out. All sequence reads were submitted to the DNA Databank of Japan (DDBJ) under accession numbers AG275743–AG613213. They can also be viewed at our Web site (http://stt.gsc.riken.jp/msm/).
For BES analyses, repetitive sequences in the BESs were masked with RepeatMasker (ftp://ftp.genome.washington.edu), and then a similarity search was performed against NCBI Build 30 of the B6 genome assembly, using BLASTN (Altschul et al. 1997). Some BLAST hits were split because of young repetitive elements or other short indels. In such cases, those hits were merged into one hit, and sum of their bit-scores was assigned as a score of the merged hit. When a BAC-end was mapped to multiple locations in the genome, the hit with the highest score was selected as the position of the end, and the BES was labeled an `ambiguous hit'. BESs whose alignments were shorter than 50 bp were also treated as ambiguous hits. Once locations of BESs had been determined, detailed alignments were calculated without mask of repeats using dynamic programming. When both ends of a BAC clone were mapped with correct orientations and positions considering the possible insert size of BAC (<300 kb), the clone was regarded as the `paired-end' clone. For mapping of `one-ended' clones, we regarded end sequences as singletons if they were uniquely mapped onto B6 genome and had alignments longer than 100 bp.
The nucleotide substitutional differences between B6 and MSM was calculated from the alignments of unique paired-end sequences. For this calculation, we used nucleotide sequences with PHRED quality values of q ≥ 30.
Sequencing and data assembly
MSM BAC clones were sequenced by the conventional shotgun sequencing method. Briefly, shotgun sequencing was performed to provide 8× coverage of draft sequences. In addition, we constructed plasmid clone libraries from appropriate restriction fragments, and sequenced both ends of these clones to provide 2× additional coverage. After assembly of all the sequence data using Phred/Phrap/Consed (Ewing and Green 1998; Ewing et al. 1998; Gordon et al. 1998), gap-filling and resequencing of low-quality regions in the assembled data were performed by a nested deletion method (Hattori et al. 1997), primer-walking, and direct sequencing of the BAC clone. Accession numbers for the clones MSMg01-122K03 and MSMg01-275M02 are AP007207 and AP007208, respectively.
Rat–mouse comparative sequence analysis
Methods for the comparative analysis are described in the Supplemental material.
Acknowledgments
We thank C. Kawagoe, X. Son, and all technical staff of the Genome Sequencing Team (Human Genome Research Group, RIKEN Genomic Sciences Center, Japan) for their excellent sequencing work and support through computational data management; Dr. Hiromichi Yonekawa of The Tokyo Metropolitan Inst. of Medical Science for valuable discussion about mouse subspeciation; Dr. Satoshi Oota of Bioresource Information Division at RIKEN BRC for comments and discussion on mouse–rat speciation, and Drs. Kazutoyo Osoegawa and Pieter De Jong of Children's Hospital Oakland Research Inst. for advice on BAC library construction. This work was supported in part by Special Coordination Funds for Promoting Science and Technology from the Ministry of Education, Culture, Sports, Science and Technology of Japan to K.A. and T.S. and by the National Bio-Resources Project of the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2899304.
Footnotes
[Supplemental material is available online at www.genome.org, and the MSM BAC database is available at http://stt.gsc.riken.jp/msm/ or http://analysis1.lab.nig.ac.jp/Mus_musculus/.]
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atchley, W.R. and Fitch, W.M. 1991. Gene trees and the origins of inbred strains of mice. Science 254: 554-558. [DOI] [PubMed] [Google Scholar]
- Beck, J.A, Lloyd, S., Hafezparast, M., Lennon-Pierce, M., Eppig, J.T., Festing, M.F., and Fisher, E.M. 2000. Genealogies of mouse inbred strains. Nat. Genet. 24: 23-25. [DOI] [PubMed] [Google Scholar]
- Bishop, C.E., Boursot, P., Baron, B., Bonhomme, F., and Hatat, D. 1985. Most classical Mus musculus domesticus laboratory mouse strains carry a Mus musculus musculus Y chromosome. Nature 315: 70-72. [DOI] [PubMed] [Google Scholar]
- Bonhomme, F., Guenet J.-L., Dod, B., Moriwaki, K., and Bulfield, G. 1987. The polyphyletic origin of laboratory inbred mice and their rate of evolution. Biol. J. Linn. Soc. Lond. 30: 51-58. [Google Scholar]
- Ewing, B. and Green, P. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8: 186-194. [PubMed] [Google Scholar]
- Ewing, B., Hillier, L., Wendl, M.C., and Green, P. 1998. Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 8: 175-185. [DOI] [PubMed] [Google Scholar]
- Ferris, S.D., Sage, R.D., and Wilson, A.C. 1982. Evidence from mtDNA sequences that common laboratory strains of inbred mice are descended from a single female. Nature 295: 163-165. [DOI] [PubMed] [Google Scholar]
- Floyd, J.A., Gold, D.A., Concepcion, D., Poon, T.H., Wang, X., Keithley, E., Chen, D., Ward, E.J., Chinn, S.B., Friedman, R.A., et al. 2003. A natural allele of Nxf1 suppresses retrovirus insertional mutations. Nat. Genet. 35: 205-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiyama, A., Watanabe, H., Toyoda, A., Taylor, T.D., Itoh, T., Tsai, S.-F., Park, H.-S., Yaspo, M.-L. Lehrach, H., Chen, Z., et al. 2002. Construction and analysis of a human–chimpanzee comparative clone map. Science 295: 131-134. [DOI] [PubMed] [Google Scholar]
- Gordon, D., Abajian, C., and Green, P. 1998. Consed: A graphical tool for sequence finishing. Genome Res. 8: 195-202. [DOI] [PubMed] [Google Scholar]
- Hattori, M., Tsukahara, F., Furuhata, Y., Tanahashi, H., Hirose, M., Saito, M., Tsukuni, S., and Sakaki, Y. 1997. A novel method for making nested deletions and its application for sequencing of a 300 kb region of human APP locus. Nucleic Acids Res. 25: 1802-1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger, J.-J., Tong, H., and Denys, C. 1986. The age of Mus-Rattus divergence: Paleontological data compared with the molecular clock. C.R. Acad. Sci. Paris 302(ser. II): 917-922. [Google Scholar]
- Kikkawa, Y., Miura, I., Takahama, S., Wakana, S., Yamazaki, Y., Moriwaki, K., Shiroishi, T., and Yonekawa, H. 2001. Microsatellite database for MSM/Ms andJF1/Ms, molossinus-derived inbred strains. Mamm. Genome 12: 750-752. [DOI] [PubMed] [Google Scholar]
- Koide, T., Moriwaki, K., Ikeda, K., Niki, H., and Shiroishi, T. 2000. Multi-phenotype behavioral characterization of inbred strains derived from wild stocks of Mus musculus. Mamm. Genome 11: 664-670. [DOI] [PubMed] [Google Scholar]
- Kumar, S. and Hedges, S.B. 1998. A molecular timescale for vertebrate evolution. Nature 392: 917-920. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh, K., Winchester, E., Daly, M.J., Wang, D.G., Hirschhorn, J.N., Laviolette, J.P., Ardlie, K., Reich, D.E., Robinson, E., Sklar, P., et al. 2000. Large-scale discovery and genotyping of single-nucleotide polymorphisms in the mouse. Nat. Genet. 24: 381-386. [DOI] [PubMed] [Google Scholar]
- Matsuzaki, H., Loi, H., Dong, S., Tsai, Y.-Y., Fang, J., Law, J., Di, X., Liu, W.-M., Yang, G., Liu, G., et al. 2004. Parallel genotyping of over 10,000 SNPs using a one-primer assay on a high-density oligonucleotide array. Genome Res. 14: 414-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita, N. and Moriwaki, K. 1987. H-2-controlled genetic susceptibility to pulmonary adenomas induced by urethane and 4-nitroquinoline 1-oxide in A/Wy congenic strains. Jpn. J. Cancer Res. 78: 494-498. [PubMed] [Google Scholar]
- Moriwaki, K. 1994. Wild mouse from geneticist's viewpoint. In Genetics in wild mice: Its application to biomedical research (eds. K. Moriwaki, et al.), pp. xiii-xxiv, Japan Scientific Press/Karger, Tokyo.
- Morse III, H.C. 1981. The laboratory mouse—A historical perspective. In The mouse in biomedical research (eds. H.L. Foster, J.D. Small, and J.G. Fox), Vol. 1, pp. 1-16. Academic Press, New York. [Google Scholar]
- Nagamine, C.M., Nishioka, Y., Moriwaki, K., Boursot, P., Bonhomme, F., and Lau, Y.F. 1992. The musculus-type Y chromosome of the laboratory mouse is of Asian origin. Mamm. Genome 3: 84-91. [DOI] [PubMed] [Google Scholar]
- Nagamine, C.M., Shiroishi, T., Miyashita, N., Tsuchiya, K., Ikeda, H., Namikawa, T., Wu, X.-L., Jin, M.-L., Wang, F.-S., Kryukov, A.P., et al. 1994. Distribution of the molossinus allele of Sry, the testis-determining gene, in wild mice. Mol. Biol. Evol. 11: 864-874. [DOI] [PubMed] [Google Scholar]
- Nei, M. 1987. Molecular evolutionary genetics. Columbia University Press, New York.
- Oka, A., Mita, A., Sakurai-Yamatani, N., Yamamoto, H., Takagi, N., Takano-Shimizu, T., Toshimori, K., Moriwaki, K., and Shiroishi, T. 2004. Hybrid breakdown caused by substitution of the X chromosome between two mouse subspecies. Genetics 166: 913-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osoegawa, K., Woon, P.Y., Zhao, B., Frengen, E., Tateno, M., Catanese, J.J., and de Jong, P.J. 1998. An improved approach for construction of bacterial artificial chromosome libraries. Genomics 52: 1-8. [DOI] [PubMed] [Google Scholar]
- Paigen, K. and Eppig, J.T. 2000. A mouse phenome project. Mamm. Genome 11: 715-717. [DOI] [PubMed] [Google Scholar]
- Rat Genome Sequencing Project Consortium. 2004. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 428: 493-521. [DOI] [PubMed] [Google Scholar]
- Silver, L.M. 1995. Laboratory mice. In The mouse genetics, pp. 32-57. Oxford University Press, New York and Oxford, UK.
- Skaletsky, H., Kuroda-Kawaguchi, T., Minx, P.J., Cordum, H.S., Hillier, L., Brown, L.G., Repping, S., Pyntikova, T., Ali, J., Bieri, T., et al. 2003. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423: 825-837. [DOI] [PubMed] [Google Scholar]
- Takagi, N., Tada, M., Shoji, M., and Moriwaki, K. 1994. An X-linked gene governing sperm morphology revealed in laboratory mice consomic for X chromosome from Japanese house mouse, M. musculus molossinus. In Genetics in wild mice: Its application to biomedical research (eds. K. Moriwaki et al.), pp. 247-256. Japan Scientific Press/Karger, Tokyo.
- Volik, S., Zhao, S., Chin, K., Brebner, J.H., Herndon, D.R., Tao, Q., Kowbel, D., Huang, G., Lapuk, A., Kuo, W.L., et al. 2003. End-sequence profiling: Sequence-based analysis of aberrant genomes. Proc. Natl. Acad. Sci. 100: 7696-7701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade, C.M., Kulbokas III, E.J., Kirby, A.W., Zody, M.C., Mullikin, J.C., Lander, E.S., Lindblad-Toh, K., and Daly, M.J. 2002. The mosaic structure of variation in the laboratory mouse genome. Nature 420: 574-578. [DOI] [PubMed] [Google Scholar]
- Watanabe, H., Fujiyama, A., Hattori, M., Taylor, T.D., Toyoda, A., Kuroki, Y., Noguchi, H., BenKahla, A., Lehrach, H., Sudbrak, R. et al. 2004. DNA sequence and comparative analysis of chimpanzee chromosome 22. Nature 429: 382-388. [DOI] [PubMed] [Google Scholar]
- Wiltshire, T., Pletcher, M.T., Batalov, S., Barnes, S.W., Tarantino, L.M., Cooke, M.P., Wu, H., Smylie, K., Santrosyan, A., Copeland, N.G., et al. 2003. Genome-wide single-nucleotide polymorphism analysis defines haplotype patterns in mouse. Proc. Natl. Acad. Sci. 100: 3380-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, S., Ellsworth, D.L., and Li, W.-H. 2002. Slow molecular clocks in Old World monkeys, apes, and humans. Mol. Biol. Evol. 19: 2191-2198. [DOI] [PubMed] [Google Scholar]
- Yonekawa, H., Moriwaki, K., Gotoh, O., Hayashi, J.-I., Watanabe, J, Miyashita, N., Petras, M.L., and Tagashira, Y. 1980. Relationship between laboratory mice and the subspecies Mus Musculus domesticus based on restriction endonuclease cleavage patterns of mitochondrial DNA. Jpn. J. Genet. 55: 289-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekawa, H., Moriwaki, K., Gotoh, O., Miyashita, N., Migita, S., Bonhomme, F., Hjorth, J.P., Petras, M.L., and Tagashira, Y. 1982. Origins of laboratory mice deduced from restriction patterns of mitochondrial DNA. Differentiation 22: 222-226. [DOI] [PubMed] [Google Scholar]
- Zhao, S., Shatsman, S., Ayodeji, B., Geer, K., Tsegaye, G., Krol, M., Gebregeorgis, E., Shvartsbeyn, A., Russell, D., Overton, L., et al. 2001. Mouse BAC-ends quality assessment and sequence analyses. Genome Res. 11: 1736-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
Web site references
- http://www.shigen.nig.ac.jp/mouse/mmdbj/top.jsp; Mouse Microsatellite Database of Japan.
- http://stt.gsc.riken.jp/msm/; Web browser-based BAC library screening system.
- http://shigen.lab.nig.ac.jp/mouse/polymorphism/; a browser-based clone screening system site.
- http://analysis1.lab.nig.ac.jp/Mus_musculus/; the MSM BAC database, and a browser-based clone screening system site.
- http://www.brc.riken.jp/lab/dna/; the RIKEN BioResource Center DNA bank.
- ftp://ftp.genome.washington.edu; RepeatMasker.