Abstract
The genomic core concept has found several uses in comparative and evolutionary genomics. Defined as the set of all genes common to (ubiquitous among) all genomes in a phylogenetically coherent group, core size decreases as the number and phylogenetic diversity of the relevant group increases. Here, we focus on methods for defining the size and composition of the core of all genes shared by sequenced genomes of prokaryotes (Bacteria and Archaea). There are few (almost certainly less than 50) genes shared by all of the 147 genomes compared, surely insufficient to conduct all essential functions. Sequencing and annotation errors are responsible for the apparent absence of some genes, while very limited but genuine disappearances (from just one or a few genomes) can account for several others. Core size will continue to decrease as more genome sequences appear, unless the requirement for ubiquity is relaxed. Such relaxation seems consistent with any reasonable biological purpose for seeking a core, but it renders the problem of definition more problematic. We propose an alternative approach (the phylogenetically balanced core), which preserves some of the biological utility of the core concept. Cores, however delimited, preferentially contain informational rather than operational genes; we present a new hypothesis for why this might be so.
The concept of a genomic core plays a key role in the literature of evolutionary and comparative prokaryotic genomics (Makarova et al. 1999; Nesbø et al. 2001; Harris et al. 2003; Koonin 2003). Operationally, a core can be defined as the set of all genes shared as orthologs by all members of an evolutionarily coherent group (a species such as Escherichia coli, a phylum such as Proteobacteria, a domain such as Bacteria, or all of Life). Biologically, cores have been used for three purposes as follows: to help deduce the composition of ancestral genomes (Mushegian and Koonin 1996; Koonin 2003), to guide in the construction of minimal cells (Zimmer 2003), and to facilitate the reconstruction of organismal phylogenetic trees (Makarova et al. 1999; Nesbø et al. 2001; Daubin et al. 2002, 2003; Lerat et al. 2003). This last use involves the assumption that core genes, universally shared by all members of a taxon, are relatively unlikely to have experienced lateral gene transfer (LGT). Thus, in all three usages some biological— rather than simply statistical—meaning is attached to the size and composition of a core.
In prokaryotic species for which genomes of several different strains have been completely sequenced, orthologous genes of the species core can usually be identified easily; they are conserved in chromosomal position as well as in sequence. For deeper and more inclusive taxa, cores become progressively smaller and more elusive because of weak phylogenetic signals, genomic rearrangements, and problems in recognizing paralogy. Nevertheless, there is much interest in such deep cores, especially the universal (Bacteria + Archaea + Eukarya) core. Several investigators argue that this core's composition might reflect that of the genome of the Last Universal Common Ancestor (LUCA), while phylogenies of its genes, if congruent, could delineate the earliest branchings of the Tree of Life (Brown et al. 2001; Woese 2002; Koonin 2003).
Recent attempts to define the universal core have concluded that it contains very few genes. Harris et al. (2003), in a study of 34 genomes report 80 core genes, Koonin (2003) using about 100 genomes finds something like 60 core genes, Brown et al. (2001) with 45 genomes (and greater requirements for stringency) 23. Although the operational and largely arbitrary nature of the definition of this minimal set has been appreciated, several authors have attributed biological significance to the fact that the number of ubiquitous genes is so small. For instance, for Koonin (2003), “the important realization that comes from this type of analysis is the remarkable evolutionary plasticity of even the central, essential biological functions. Only a tiny group of genes (nearly all of them associated with translation and transcription) is truly ubiquitous among living things”. Woese (1987) asks, in the phylogenetic context, “What does it mean, then, to speak of an organismal genealogy when nearly all of the genes in the cell, genes that give it its general character, do not share a common history?”
Here, we describe a similarly motivated study, addressing a more extensive set of prokaryotic genomes (130 Bacteria and 17 Archaea) with a greater variety of methods of analysis. We, too, find a diminutive set of truly ubiquitous genes among prokaryotes. Our interest, however, is not so much in this number as in whether or not it might be a statistical illusion, what more useful algorithms for defining cores might be possible, and the difficult question of biological significance of core size and composition.
Results
Ubiquitous cores defined by reciprocal best matches
Table 1 summarizes our enumeration of genes shared by all members of selected prokaryotic taxa. Reported values are the means of all queries in which a search was launched with any one genome from a member of the taxon against all others from that taxon, requiring a reciprocal best match (RBM, see Methods) in each. Variation around these means is small, and reflects cases in which genes are variously disconnected from one another in the BLAST method. (By disconnection, we mean a situation in which an occasional gene X will be an above-threshold best-reciprocal match between genomes A and B and A and C, but not between B and C). The average prokaryotic genomic core defined in this way (genes shared by a launching genome and all other Bacteria and Archaea) has only 14.82 such ubiquitous genes. A more generous result, and one which actually produces a unique list of gene names, is obtained by taking the union of all such sets shared by all 147 genomes. The size of this union of RBM sets is 30 genes.
Table 1.
Number of genes strictly shared (as determined by reciprocal best match) within major taxa of prokaryotesa
| Prokaryotes: 14.82 (sd = 2.55), range 10-23, n = 147 |
| Archaea: 144.53 (sd = 8.00), range 128-156, n = 17 |
| Crenarchaeota: 587.75 (sd = 5.91), range 582-596, n = 4 |
| Euryarchaeotab: 174.00 (sd = 5.95), range 165-182, n = 12 |
| Bacteria: 61.53 (sd = 2.71), range 56-70, n = 130 |
| Bacteroidetes: 1219.00 (sd = 4.24), range 1216-1222, n = 2 |
| Chlamydia: 780.57 (sd = 1.81), range 778-784, n = 7 |
| Cyanobacteria: 820.50 (sd = 23.53), range 776-844, n =8 |
| High-G + C Firmicutes: 349.25 (sd = 11.99), range 327-363, n = 12 |
| Low-G + C Firmicutesc: 118.74 (sd = 3.56), range 109-125, n = 35 |
| Bacillus/Streptococcus group: 478.48 (sd = 11.86), range 446-491, n = 23 |
| Clostridium/Fusobacterium group: 466.20 (sd = 16.02), range 444-485, n = 5 |
| Mollicutesd: 236.83 (sd = 8.98), range 220-245, n = 6 |
| Proteobacteria: 131.47 (sd = 4.66), range 124-141, n = 58 |
| α: 456.54 (sd = 12.89), range 434-472, n = 11 |
| β + γ: 195.58 (sd = 7.41), range 185-213, n = 41 |
| ε: 787.40 (sd = 5.32), range 780-793, n = 5 |
| Spirochaetes: 321.00 (sd = 8.00), range 313-329, n = 3 |
| Clades were defined from a genomic phylogeny based on the mean normalized BLASTP similarity between pairs of genomes (Clarke et al. 2002; Charlebois et al. 2004), but with preferential weighting in favor of phylogenetically concordant sequences (Gophna et al. 2004). Reported are means: from the perspective of each genome in a clade, the size and composition of the shared set of genes is somewhat variable, owing to match threshold effects. For instance, the union of the set of genes shared by all 147 prokaryotic genomes (at the time of this analysis) has 30 members (see Table 3), despite the mean number of genes shared by all represented prokaryotes being only 14.8. |
Using the “Find ubiquitous genes” query within NGIBWS (Charlebois et al. 2003), and a BLASTP cutoff threshold value of 1.0e-5
Including Nanoarchaeum equitans, but excluding Halobacterium salinarum NRC-1 which branches basally to other Archaea in our phylogenies
Including Fusobacterium nucleatum
Excluding Phytoplasma asteris, which branches basally to the Bacillus/Streptococcus group in our phylogenies
It seems unlikely that this number is falsely large. Artifacts that could make it so would require extreme degrees of sequence convergence, and in any case, all or most of the genes identified in such an analysis are expected to be highly conserved on biological grounds (see below; Table 3, below). It could easily be falsely small, however, and the remaining analyses presented here variously address possible artifactual explanations for the diminutive size of the prokaryotic core.
Table 3.
The 34 consensus gene names found in all 147 prokaryotic genomes (from Figure 1)
| *argS (arginyl-tRNA synthetase) |
| dnaG (DNA primase) |
| *fusA (translation elongation factor EF-G) |
| *gcp (O-sialoglycoprotein endopeptidase) |
| *gltX (glutamyl-tRNA synthetase) |
| *hisS (histidyl-tRNA synthetase) |
| *infB (translation initiation factor IF-2) |
| ksgA (S-adenosylmethionine-6-N′,N′-adenosyl (rRNA) dimethyltransferase) |
| leuS (leucyl-tRNA synthetase) |
| lysS (lysyl-tRNA synthetase) |
| *metG (methionyl-tRNA synthetase) |
| nusA (transcription pausing, L factor) |
| nusG (involved in transcription antitermination) |
| *pheS (phenylalanyl-tRNA synthetase) |
| *proS (prolyl-tRNA synthetase) |
| *rplA (ribosomal protein L1) |
| *rplC (ribosomal protein L3) |
| *rplE (ribosomal protein L5) |
| rplF (ribosomal protein L6) |
| *rplK (ribosomal protein L11) |
| *rplN (ribosomal protein L14) |
| *rpoB (RNA polymerase, β subunit) |
| *rpsB (ribosomal protein S2) |
| *rpsC (ribosomal protein S3) |
| *rpsD (ribosomal protein S4) |
| *rpsG (ribosomal protein S7) |
| *rpsH (ribosomal protein S8) |
| secY (ATPase subunit of translocase) |
| *serS (seryl-tRNA synthetase) |
| *thrS (threonyl-tRNA synthetase) |
| *trpS (tryptophanyl-tRNA synthetase) |
| *tufA (translation elongation factor EF-Tu) |
| *valS (valyl-tRNA synthetase) |
| *ychF (GTP binding protein) |
| Core genes also recovered from the “strict-sharing” analysis (Table 1) are indicated by an asterisk. The latter analysis (30 genes in the set) also includes aspS (aspartyl-tRNA synthetase), dnaX (DNA polymerase III γ/τ, or replication factor C), ftsH (ATPase involved in cell division), and rpsl (ribosomal protein S9). |
Effects of BLAST parameters and genome size on apparent core size
One such possibility is that we have set cut-off values for BLASTP too stringently, so that legitimate orthologs have been overlooked. If this were so, the average number of genes with RBMs in all genomes should be exquisitely sensitive to that cut-off value. It is not, as Table 2 illustrates. There is remarkably little variation in average numbers of genes obtained for expectation values between 1.0e-3 and 1.0e-7. Nevertheless, a few more genes were added by combining the results of the union of shared RBMs at 1.0e-5 with the results of the consensus gene name, or CGN approach (see Methods), which might recover some cases of poorly or nonreciprocally matching orthologs. Both sets are listed in Table 3, and are largely overlapping (30 genes from the union of shared RBMs, 34 genes from CGNs, 26 in their intersection and 38 in their union).
Table 2.
Number of genes strictly shared by prokaryotes, by simple match and by reciprocal best match (RBM), at various BLASTP cutoff expectation values
|
147 Prokaryotes
|
130 Bacteria RBM
|
17 Archaea RBM
|
||
|---|---|---|---|---|
| E-value | Simple match | RBM | ||
| 1.0e-3 | 101.5, sd = 46.9 [28-267] | 18.0, sd = 2.8 [12-26] | 63.5 | 153.6 |
| 1.0e-4 | 93.6, sd = 45.6 [26-256] | 16.1, sd = 2.8 [11-24] | 62.6 | 150.1 |
| 1.0e-5 | 87.2, sd = 44.6 [25-248] | 14.8, sd = 2.6 [10-23] | 61.5 | 144.6 |
| 1.0e-7 | 78.1, sd = 42.5 [20-233] | 12.1, sd = 1.9 [7-17] | 59.5 | 133.6 |
| 1.0e-10 | 65.8, sd = 37.0 [15-204] | 10.0, sd = 1.8 [5-14] | 55.3 | 118.6 |
| 1.0e-15 | 35.2, sd = 17.1 [9-147] | 7.4, sd = 1.4 [4-11] | 46.6 | 99.9 |
| 1.0e-20 | 12.1, sd = 3.4 [5-26] | 5.5, sd = 1.1 [3-8] | 38.8 | 83.5 |
| 1.0e-30 | 4.7, sd = 1.5 [2-11] | 2.2, sd = 0.9 [0-4] | 24.5 | 58.8 |
| 1.0e-50 | 1.5, sd = 0.9 [0-5] | 9.9, sd = 0.7 [0-2] | 12.6 | 33.5 |
| 1.0e-100 | 0.0 | 0.0 | 5.9 | 10.5 |
Reported are means, standard deviations (sd), and ranges of estimates (in square brackets), from the perspective of different query genomes. See Table 1 for an explanation of why there is variance in such estimates.
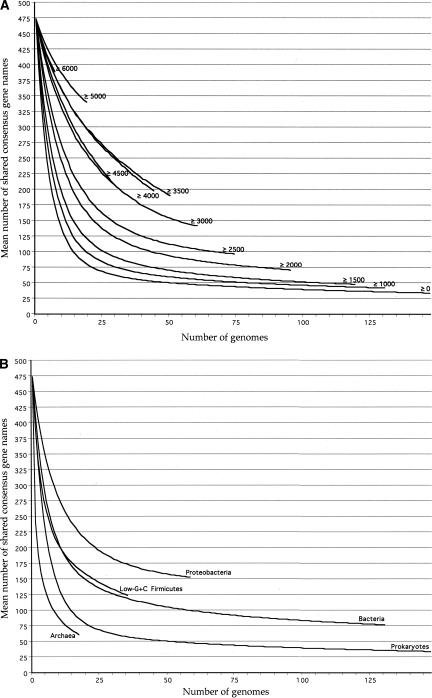
Another possible artifact (or misleading bias) could result from inclusion of the highly reduced genomes of endosymbionts or parasites, which can lack many genes required by free-living cells. To examine the effect of excluding small genomes, we performed the analysis shown in Figure 1A, using the CGN method. To limit errors due to inconsistent annotations of rare genes, we excluded any gene missing from more than 50 of the 147 genomes; this left 474 genes for analysis. The lower curve in Figure 1A plots the mean number of such genes found shared in 10,000 randomized comparisons using the indicated number of different genomes, selected randomly (without replacement) from the total set of 147. This method of representation mimics the general historical course of research in the definition of the universal core—each new survey involving more genomes has reduced the apparent number of genes, and some asymptotic low value less than 100 appears to be on the horizon. With our methods, restricting the comparisons to genomes of more than 1000 ORFs, 1500 ORFs, and so on (upper curves) raises the apparent asymptote, but the effect is not dramatic. Even with genomes of more than 2000 ORFs, we expect fewer than 75 ubiquitous genes.
Figure 1.
(A) Number of genes that are found (by having the same consensus gene name, see Methods) in at least two-thirds of prokaryotic genomes, and that are found in a random sample of x = 1 through x = 147 of these genomes. The point at x = 0 of y = 474 represents the number of genes found in at least 97 prokaryotes. For x > 0, means are reported for 10,000 random selections of x genomes. Small genomes were progressively deleted from the analysis in order to produce the series of curves shown. (B) As in A, but for selected clades of prokaryotes.
Figure 1B, which restricts comparisons to genomes in a single clade, shows additional, presumably biological, effects. At a given sample size, there are more genes shared between genomes when these are taken from the Proteobacteria than from the Bacteria generally, or prokaryotes more generally still. The proteobacterial core defined in this way is indeed larger than (and includes) the bacterial core. Archaea appear to comprise a generally less-coherent group. In part, this could be a genome size effect; the average sequenced archaeal genome is smaller than the average sequenced bacterial genome. The diminutive Nanoarchaeum equitans genome (only 563 ORFs and a seriously impoverished metabolic repertoire) may in particular exert an effect (Waters et al. 2003). By our measure, it shrunk the prior and presumed stabilized archaeal core (Makarova and Koonin 2003) by 41% (data not shown).
Core composition and the problem of missing genes
Table 3 lists the 30–38 largely identical genes that are found in the universal prokaryotic cores defined by either the union of RBMs (Table 1) or CGN methods (Fig. 1). (We note that the former method is likely vulnerable to false negatives, and the latter to false positives, but, nevertheless, that the agreement between them is quite strong.) These genes are generally included among those found by Koonin (2003) and Harris et al. (2003). The high fraction of translational components fits the generally popular theory that informational genes are less frequently transferred (Woese 1987, 1998, 2000, 2002; Jain et al. 2002), and the lack of genes for catabolic or anabolic pathways conforms to the view that these latter evolve though LGT, by a mix-and-match principle (Lake et al. 1999; Boucher et al. 2003; Koonin 2003). But, we are aware of no mixable and matchable alternatives for some of the genes that seem to be missing altogether from Table 3, in particular, RNA polymerase subunits other than RpoB and many ribosomal proteins.
As it happens, the distribution and conservation of ribosomal proteins has recently been subjected to a careful analysis by Lecompte et al. (2002). With 66 genomes (45 Bacteria, 14 Archaea, and seven Eukarya), they found 33 universal prokaryotic ribosomal proteins, after correcting for missed annotations. Extending their analysis to 147 prokaryotes (130 Bacteria and 17 Archaea), with a thorough tBLASTN search for each missing gene in each apparently deficient genome, we concluded that the status of these investigators' 15 ubiquitous small-subunit ribosomal proteins remains secure. However, four of the 18 then-ubiquitous large-subunit proteins can now be declared missing in at least one bacterial genomic sequence. The normally adjacent rplB and rplW genes cannot be located within the Streptococcus mutans UA159 genomic sequence, but rather the pair's neighbors (rpsS and rplD) overlap by 11 bp; and there is no sign of rpmC within the Wolinella succinogenes DSMZ 1740 genome. Whether these three absences represent legitimate losses or sequencing artifacts is impossible to tell. Finally, the rplM gene is annotated within the genome of Enterococcus faecalis V583 as having a frameshift, but is presumed nonfunctional (no protein sequence is described).
Some further losses clearly have occurred among several ribosomal protein genes described by Lecompte et al. (2002) as restricted to, but ubiquitous within Bacteria. The rplI gene cannot be located within the Mycoplasma penetrans HF-2 sequence; rpmB is annotated as a pseudogene in Mycobacterium leprae TN and appears to be absent from Pirellula sp. 1; rpmF cannot be found in Mycobacterium tuberculosis H37Rv; rpmH cannot be found in Pirellula sp. 1; rpmI cannot be found in Bdellovibrio bacteriovorus HD100; and rpmJ appears to be absent from both strains (C58 and C58 UWash) of Agrobacterium tumefaciens and from Corynebacterium glutamicum. All of the archaeal-specific ribosomal proteins that were ubiquitous in the study of Lecompte et al. (2002) are still ubiquitous with our slightly larger (17 vs. 14) archaeal genomic data set.
Our case-by-case searches described above revealed that the true ubiquitous core consists of 29 prokaryotic ribosomal protein genes, but our strictest measurement of the core using an automated analysis identified only 11 ribosomal protein genes (Table 3). Many of the false negatives in the automated search, which is necessarily based on annotated genes, were due to missed annotations (especially of small proteins not designated as ORFs in genome databases) and misannotations (usually where a larger overlapping ORF was selected at the expense of a smaller gene). Some of the false negatives were, however, attributable to deficiencies in our methods, where BLASTP thresholds were too strict or where alternate annotations confounded assignment of the CGN. Ribosomal proteins might, however, be worst-case examples both for annotation artifacts and algorithmic shortcomings; many ribosomal proteins are very small, and can easily be overlooked by annotators and by BLASTP alike. Furthermore, some of the apparently genuine absences affect very few genomes; four more genes could be added to the core if the ubiquity requirement were relaxed, even very slightly.
It will not be simple to extract from our analysis of ribosomal proteins any reliable estimate of how many genes in other functional categories have been excluded from Table 3 because of error, and how many are genuinely missing from at least one or a few genomes. Our focus, in any case, is not so much on the precise number or identity of core genes as on the methods best used to define them. In that regard, we infer that the requirement for ubiquity in defining genomic cores—as well as being completely unforgiving with respect to errors—might be standing in the way of our recognition of some biologically more significant collection of almost ubiquitous genes.
Relaxing the requirement for ubiquity
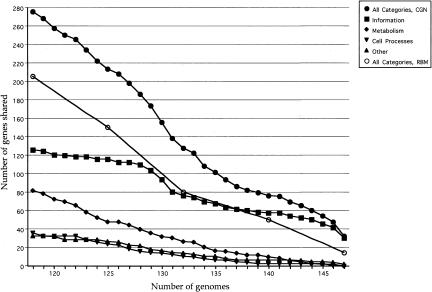
The analysis illustrated in Figure 2 was undertaken to test this inference. Here, we have asked not how many genes on average are shared by the unique sample of all 147 genomes, or by samples of smaller subsets of these genomes (as in Fig. 1, A and B), but instead how many genes are present in at least 147 genomes, or at least 146 or at least 145, and so forth. There is an inflection in the curve for informational genes (and because of this for total genes) at 130 genomes—expected because at this point genes limited to the 130 Bacteria but ubiquitous among them can first register. But, this inflection aside, there is little evidence for any discrete core. Emphasis here should be on the word `discrete'. If genomes comprised genes of two classes—variously dispensible genes whose genomic representation is normally distributed around some average, and core genes that are always present (although sometimes not detected)—we would have expected a curve more like that shown in Figure 1A.
Figure 2.
Number of genes that are shared by at least 80%–100% of prokaryotic genomes. The points at the extreme right of the All Categories curves represent the end points from Figure 1 (for consensus gene names, CGN), and from Table 1 (for reciprocal best matches, RBM), respectively. Toward the left are genes that are cumulatively shared by progressively fewer genomes. Also shown are consensus gene names by functional category, extrapolated from COG assignments (Tatusov et al. 1997).
A comparison of Figures 2 and 1 also illustrates a potential misunderstanding in the graphical representation of such analyses. Both achieve a terminal value of 34 for the number of consensus gene names found in all 147 of the genomes examined in this study. But at, say, 125 genomes, the number of shared genes has increased by only a few in Figure 1A and to more than 200 in Figure 2. This is because Figure 1A asks, for all possible samplings of 125 of the 147 genomes, what is the average number of genes commonly ubiquitous in these 125, while Figure 2 asks how many genes are independently ubiquitous in any sampling of 125 genomes. Thus, the second analysis differs from the first in that genes found in at least 125 genomes do not have to be found in the same 125 genomes (and manifestly are not). Figure 1A, we think, mimics the historical course of studies of the core. These have usually asked how many genes are present in all genomes sequenced at the time of analysis. Such studies quite quickly settled down to reporting fewer than 100 genes, an estimate that seems to be holding up well (Brown et al. 2001; Harris et al. 2003; Koonin 2003). This might easily be interpreted as evidence that we are asymptotically approaching some biologically meaningful number of core genes. But, in fact, this behavior (like Fig. 1A) is very much the consequence of the requirement that core genes be found in all genomes. If, with the current collection of genomes, we relax the requirement to be the presence in all genomes except one, we add 10 genes. If we allow as few as four of the 147 genomes to miss one of the core genes, we have already doubled its apparent size (Fig. 2). It does not seem at all unreasonable, in the quest for a biologically meaningful core, to relax the ubiquity requirement in this way, and define it as comprising genes present in the vast majority of genomes. Omissions due to error might be minimized in this way, as would the effects of very rare nonorthologous replacements. But then, the size of the core will depend very crucially on what we mean by vast majority.
It is appealing to propose a model in which each gene has a different and independent characteristic probability of going missing from a genome (Krylov et al. 2003), where even critical functions are not formally exempt from analogous replacement in evolution, and no gene is exempt from sequencing or annotation error. But, it would be very difficult to establish the parameters of such a model, not only the gene-specific loss propensities, but in the end, just what value of loss propensity is tolerable for inclusion in the core. In other words, although these gene-specific probabilities would have a biological significance, the core itself could be arbitrary or artifactual in two senses, depending on the number (and nature) of genomes examined and the cut-off value set for inclusion.
Toward a biologically more significant phylogenetically balanced core
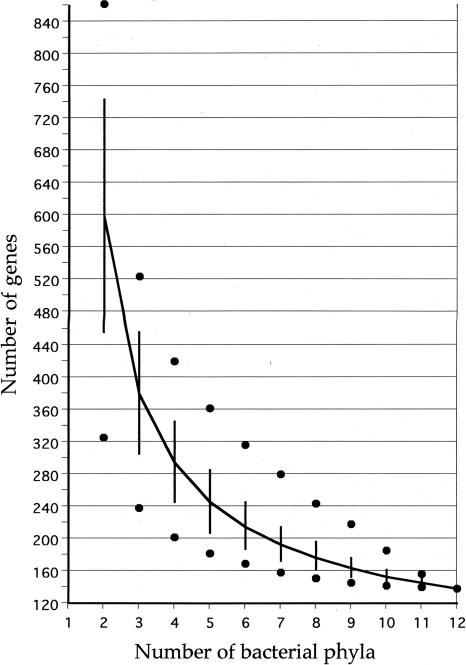
We reasoned that the biological goals of defining a prokaryotic core might be better achieved by methods that do not demand ubiquity or assert some arbitrary definition of ubiquity, but do retain the requirement that genes of the core be (1) very common, and (2) distributed as broadly as possible, phylogenetically. Table 4 and Figure 3 illustrate a computational experiment applied to bacterial genomes which suggests that such a phylogenetically balanced core (PBC) approach holds promise. For this analysis, all possible pairwise comparisons of any two genomes taken from two different bacterial phyla, three-way comparisons of any three genomes from three different phyla, and so forth, were performed. Mean, standard deviation, maximum, and minimum values for numbers of shared genes (detected as RBMs) are shown. (Again, the RBM method may identify different, albeit largely overlapping, sets of genes in different comparisons between any two, three, or more genomes, so Fig. 3 does not unequivocally identify 138 specific core genes, although there will be much overlap between sets.)
Table 4.
Mean number of orthologs (reciprocal best matches, RBM) shared among genomes from X different bacterial phyla (12 currently available*).
| Mean number shared (genome combinationsa) | n | Mean number shared (phylum combinationsb) | n | |
|---|---|---|---|---|
| X = 2 | 609 (sd 267) [169-1721] | 6.02e+3 | 599 (sd 143) [326-861] | 66 |
| X = 3 | 362 (sd 128) [132-835] | 1.30e+5 | 380 (sd 76) [237-523] | 220 |
| X = 4 | 272 (sd 82) [118-596] | 1.52e+6 | 294 (sd 52) [201-419] | 495 |
| X = 5 | 225 (sd 58) [109-477] | 1.04e+7 | 246 (sd 39) [182-360] | 792 |
| X = 6 | 197 (sd 44) [107-398] | 4.34e+7 | 215 (sd 29) [168-315] | 924 |
| X = 7 | 179 (sd 34) [105-357] | 1.14e+8 | 193 (sd 22) [157-280] | 792 |
| X = 8 | 166 (sd 28) [103-324] | 1.92e+8 | 177 (sd 17) [151-244] | 495 |
| X = 9 | 157 (sd 23) [102-297] | 2.06e+8 | 164 (sd 12) [145-217] | 220 |
| X = 10 | 149 (sd 20) [101-274] | 1.37e+8 | 153 (sd 8) [142-185] | 66 |
| X = 11 | 143 (sd 17) [100-253] | 5.10e+7 | 145 (sd 5) [139-155] | 12 |
| X = 12 | 138 (sd 15) [100-178] | 8.18e+6 | 138 (sd 0) [138-138] | 1 |
Bacterial phyla for which sequenced genomes were available at the time of constructing this table are as follows: Aquificales (1), Bacteroidetes (2), Chlamydiales (7), Chlorobi (1), Cyanobacteria (8), Thermus/Deinococcus group (1), High-G+C Firmicutes (12), Low-G+C Firmicutes (35), Planctomycetes (1), Proteobacteria (58), Spirochaetes (3), Thermotogales (1). (Fusobacterium nucleatum is included within the Low-G+C Firmicutes)
Computed as a global mean of all individual combinations of X genomes from X phyla
Means are first computed for each phylum combination (e.g., from the 174 genome combinations of Proteobacteria versus Spirochaetes), then a mean of these phylum means is computed. E.g., for X = 3, a genome from one phylum is compared with a genome from a second phylum and with a genome from a third phylum. The mean number of RBM for all such combinations is reported. (sd) Standard deviation, (square brackets) minimum and maximum values (n), number of combinations. See Figure 3 for a graphical representation of the phylum combinations data
Figure 3.
Graphical representation of Table 4. The x-axis denotes breadth of distribution amongst bacterial phyla, whereas the y-axis indicates the mean number of orthologs (reciprocal best matches) shared at that breadth. (Vertical bars) SD; (dots) minima and maxima.
This approach has at least three distinct advantages over ubiquity-requiring global analyses. First, for the mean values of multiphylum comparisons, genes that are missing (either truly or through error) from only a few genomes, will usually have less effect. Second, and for related reasons, highly reduced genomes have less impact on the size of the core—unless they are the only representatives of their phyla. The maximum and minimum values show the extent to which large and small genomes (endosymbiotic or parasitic) within well-sampled phyla influence our comparisons. Third, as more genomes are added within existing phyla, this estimate will become more precise (its standard error will decrease). Its value is not expected to diminish and will likely even increase, as larger genomes from phyla, so far poorly sampled, appear. The addition of new phyla will diminish the PBC core, but the list of such completely unsampled phyla is not, like that of unsequenced genomes, limitless.
Our ultimate interest here is in such a core for all prokaryotes, defined as genes present in some reasonable fraction of genomes in each and every one of 12 bacterial phyla and two archaeal phyla (Crenarchaeota and Euryarchaeota, including Nanoarchaeum equitans). In Table 5, we list all genes that are (1) present by consensus name in at least one genome in each of these 14 groups, and (2) within these groups, present in 100%, at least 90%, or at least 80% of genomes. Of course, the 100% gene set is identical to that shown in Table 3, except for minor differences arising from Table 5's more comprehensive data set; there is no phylogenetic balancing when ubiquity is required. (For constructing this table, we have used, in addition to the 147 prokaryotic genomes available in January 2004, 23 that have since appeared.) Interestingly, rpoB and rpoC, which are necessary and ubiquitous components of RNA polymerase, are only retrieved using our relaxed definition of the core. In some genomes, these genes are fused, and thus thwart the retrieval of rpoB and/or rpoC by RBM or CGN. This example helps to underline the need for flexibility in defining core genes, not only in compensation for sequencing and annotation artifacts, and the odd rarely lost gene, but also for some genes' tendency to form multidomain proteins.
Table 5.
Prevalent genes computed from cross-phylum analysis
| A: ubiquitous genes (total 34): | ||||||||
| argS | infB | pheS | rplN | secY | ||||
| dnaG | ksgA | proS | rpsB | serS | ||||
| dnaX | leuS | rplA | rpsC | thrS | ||||
| fusA | lysS | rplC | rpsD | trpS | ||||
| gcp | metG | rplE | rpsG | valS | ||||
| gltX | nusA | rplE | rpsH | ychF | ||||
| hisS | nusG | rplK | rpsM | |||||
| B: present in at least 90% of members from each phylum; add 26: | ||||||||
| alaS | map | recA | rpoC | rpsL | ||||
| atpD | pkg | rplB | rpsE | rpsN | ||||
| eno | pheT | rplM | rpsI | rpsS | ||||
| ftsH | pyrG | rplX | rpsJ | topA | ||||
| groEL | pyrH | rpoB | rpsK | trxB | ||||
| ileS | ||||||||
| C: present in at least 80% of members from each phylum; add 11: | ||||||||
| cysS | guaA | nth | rplV | uppS | ||||
| efp | lpdA | pepP | rpmC | yggV | ||||
| glyA | ||||||||
Listed are consensus gene names (CGN) present in genomes from each of the 14 available phyla (12 bacterial, 2 archaeal) of 170 prokaryotes. (A) Present in all genomes from all phyla. Contrast with Table 3 (computed from 147 prokaryotes), where dnaX and rpsM are not included and where rpoB and tufA have dropped out. (B) Present in at least 90% of members from each phylum. Note that rpoB reappears in this list, but not tufA (due to a known annotation inconsistency). (C) Present in at least 80% of members from each phylum. (Note: lysS is a homonym for both class I and class II lysyl-tRNA synthetase genes ([Ibba et al. 1999], and thus represents a false positive entry.)
Gene loss is a known factor in genome evolution, but since ancestral genomes cannot have been much larger than present-day genomes, gene genesis (largely by duplication and divergence) is necessary in order to compensate (Snel et al. 2002; Kunin and Ouzounis 2003; Mirkin et al. 2003). Gene genesis creates paralogs, and gene loss deletes both paralogs and orthologs; the whole process thus inevitably results in a shrinking of an orthologously defined core. Genes remaining in a universal core should thus not only be critically necessary and maintained in all taxa, but should also be resistant to the usage of alternative forms.
The most inclusive core defined in this way contains 71 genes (Table 5). We suspect that its size may increase as more or larger genomes for some of the sparsely sampled phyla— especially Aquificales, Chlorobi, Planctomycetes, and Thermotogales—become available. Although the potentially distorting effect of individually aberrant genomes that are among the few representatives of their respective phyla cannot be ignored, its importance will diminish as more genome sequences appear (Two examples are as follows: ffh appears to be absent from Nanoarchaeum equitans [acceptable to PBC], but also from both strains of Leptospira interrogans and thus, from 40% of our Spirochaetes; ftsY appears to be absent from N. equitans [acceptable], but also from Sulfolobus tokodaii and thus, from 25% of our Crenarchaeota). The decision to choose genes present in 90%, 80%, or some other fraction of genomes in each phylum remains arbitrary. But, the requirement imposed by the PBC approach for presence at such a level in all phyla guarantees a more representative biological sampling, whereas a core defined as including genes present in 80% of all genomes regardless of phyletic distribution would be a bacterial core.
Discussion
Although universal prokaryotic cores as described here and in other recent literature are often suggestively similar in size, this apparent convergence lacks biological significance. When each core gene is required to be in every genome, cores will inevitably be artifactually small. Some genes will be missing because of sequencing, assembly, and annotation errors, and some genuine orthologs will have diverged beyond detectability. Because of such errors alone, the size of ubiquity-requiring genomic cores should continue to decline slowly, as more genome sequences appear. The impact of errors like this might be reduced by relaxing the requirement for ubiquity (to <100% of all genomes). However, the set of almost ubiquitous genes rises almost continuously in number as percent representation is relaxed, and includes more and more genes whose nonubiquity is not artifactual. There is no obvious place to draw the line, other than that at which we can discount all Archaea (about 80%). To do this would be to abandon the claim for (prokaryotic) universality. Defining cores as genes present in some fraction of genomes less than 100% is thus not only arbitrary, but gives disproportionate weight to taxa favored, for whatever reason, by sequencers.
The PBC approach we describe will address the problem of errors and the problem of disproportionate weighting of popular phyla when ubiquity is not required. It also favors universality (within a phylogenetic context), but it still does not tell us where to draw the line. We do find it encouraging that the core defined in this way increases less than 20% in size, when prevalence required within taxa is dropped from 90% to 80%. If, indeed, there is a conserved set of genes that might be considered the basic heritage for all prokaryotes, but which defies precise definition because of occasional loss or orthologous replacement in scattered lineages, the PBC might provide a good approximation.
Although a nonarbitrary delimitation of core size may be impossible, core composition is not random. Cores are generally dominated by genes of the translational apparatus. Why should this be? The widely accepted explanation is that genes of this important informational process are intrinsically less exchangeable than genes of operational processes, like metabolism (Woese 1987, 2002; Jain et al. 2002). There are so many complex coevolved interactions with other cellular informational constituents, this argument holds, that any replacement of one of these genes by a distant homolog or by an analog would be disadvantageous. Although one might object that there are individual instances in which such components (not only ribosomal proteins and translation factors, but ribosomal RNAs themselves) have been transferred, this complexity hypothesis (or annealing hypothesis) remains appealing and popular.
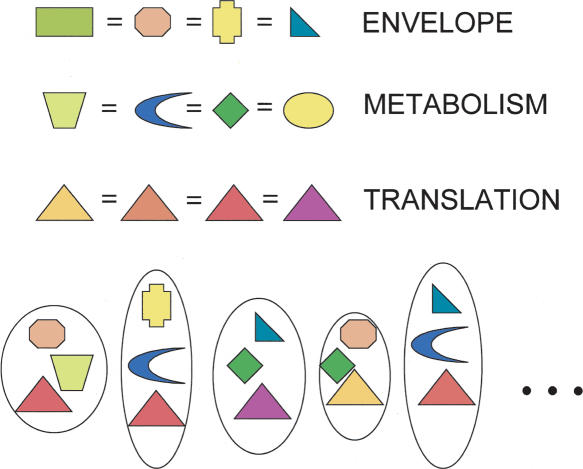
But there is an alternative explanation, which we will sketch out here, and have represented by a cartoon in Figure 4. All cells need genes of each of several functional categories, such as (put very crudely) translation, metabolism, and cell envelope formation. Lake et al. (1999) and Jain et al. (2002) describe genes for replication, transcription, and translation as informational, and other classes as operational, but the distinction is not hard and fast. Imagine that evolution has assembled cells in a mix-and-match fashion, choosing to satisfy each of the several functional categories with genes available from a global repertoire of determinants, whether homologous, analogous, or in the case of a function superfluous in a current niche, null. Evolution no longer makes cells from scratch, so in general, what happens is that one gene or suite of cofunctional genes (a pathway) replaces another, a general process that includes both replacement by close or distant homologs and by analogs, which Koonin and his collaborators call nonorthologous displacement (Koonin et al. 1996). Where there are many analogous (nonhomologous) types of genes (or suites of genes) that can perform the same general function (e.g., energy production or cell envelope formation), the living world will collectively exhibit much variability, and there will be no ubiquitous sets of genes that appear as part of any universal core. Where choices are more limited, most genes performing the needed function (some step in translation for instance) being homologous, there will appear to be little variability. Our analysis and all similar studies of the core will not generally detect orthologous replacements, in which a resident gene is replaced by an ortholog from another species. Such genes will appear as part of a universal core, but this need not mean that they are exchanged any less frequently than genes in other categories (as implied in the complexity hypothesis). Against the objection that there would be no selective advantage to such cryptic orthologous replacements of generally essential genes, we note that antimicrobials are often targeted against the products of such genes, and here the advantages would be obvious and large.
Figure 4.
A mix-and-match model for prokaryotic genome evolution. Every cell needs genes for multiple functions, and new genomic lineages arise in evolution through mixing and matching of genes performing these different functions, by processes of replacement, including nonorthologous displacement (Koonin et al. 1996). The simplest hypothesis would be that all functions are equally subject to such exchange processes. For many functions, available genes include nonhomologs and even null entries (gene and function loss), indicated here by different shapes. Thus, for these functions, no genes or even gene families will likely appear to be shared among all genomes. For some informational functions especially (such as translation), displacement most often involves genes that, although evolutionarily distinct (as indicated by colors), are homologous (as shown by shape). Such genes will appear among those of the ubiquitous core.
Exchanges of this nature need not be cryptic in phylogenetic analyses. Informational genes of the universal prokaryotic core should not produce congruent phylogenetic trees if cells have exchanged them for foreign (but still orthologous) versions as often as they seem to have traded nonorthologous (indeed, nonhomologous) operational genes. Whether or not genes of the universal core do show congruent phylogenies is, however, still a matter of legitimate debate; at this (phylum) depth few individual genes have reliable phylogenetic signal. Several years ago, Teichmann and Mitchison (1999) observed that only three of 32 protein families shared between selected bacterial, archaeal, and eukaryotic genomes showed significant phylogenetic signal, and concluded that this signal was actually due to recent LGT events. Several recent studies that have used concatenated sequences of core genes to construct universal trees have argued that the robustness of these trees reflects an underlying phylogenetic coherence (Brown et al. 2001; Brochier et al. 2002; Matte-Tailliez et al. 2002). But, it was the failure of the genes individually to produce resolved trees that motivated concatenation in the first place, and in any event, all such reports conclude that there is significant divergent signal among core translational genes. Harris et al. (2003) note, from preliminary phylogenetic analyses, that 30 of their 80 universal core genes do not maintain domain monophyly, showing clades in which Bacteria, Archaea, and Eukarya are mixed. Brown et al. (2001) were reduced to a core of only 14 genes they considered congruent phylogenetically. Overall, it might be safe to say that informational genes support the Bacterial/Archaeal dichotomy more often than do operational genes— and perhaps, more often than not. But, a consistent and extensive pattern of congruence among informational genes in branching patterns at the level of bacterial and archaeal phyla has not been established.
Thus, objections to our alternative explanation (Fig. 4) must rest on one or both of two lines of argument as follows: (1) that genes for such components are intrinsically less transferable for some reason related to their function (the complexity hypothesis) or (2) that the general support for the three-domain Tree of Life among informational genes is best explained by their relative nonexchangeability. The first is as yet unproven, and the presence among ubiquitous genes of some presumably noncomplexing proteins outside of the informational class argues against it, and the second is—given that domains have themselves been defined primarily with reference to their translational components—dangerously circular.
Methods
Determining the size and composition of a core of genes ubiquitous among a set of genomes requires a method with which to compute orthologous relationships in bulk. For practical reasons, we can assume that a pair of reciprocally best (or near-best) matching genes are likely to be orthologs, and can thus automate the process of comparative genomic analysis (Charlebois et al. 2003). The BLASTP bit score (Altschul et al. 1997) serves as a convenient measure of sequence similarity, although we concede that it can only represent an approximation to the true similarity between genes, and may therefore generate false positive matches and false negative failures of matching, especially near the match threshold that must be imposed (Ragan and Charlebois 2002).
Using BLASTP scores, we can distinguish between ordinary matches (including many which may be paralogous) and reciprocal best matches (RBMs), which are more often orthologous. Additionally, we can make use of information present in genomic annotations, permitting genes with standardized gene names to find their orthologs despite BLASTP matches that may fall slightly below threshold. Where annotations are reasonably consistent, this permits the union of overlapping sets of genes where BLASTP matches alone might miss members outside of the sets' intersection. Matches are still based on BLASTP, but some-what disconnected outliers can then link through the bridge of a common consensus gene name (CGN). These are computed as follows: For each ORF in a genome, its RBM, if any, is found in each of the other genomes, and the RBM's annotated name is appended to a list. The dominant name in this list (e.g., ftsA) becomes the query ORF's CGN. Lists of prevalent names, found in most or all members of a set of genomes, are generated by the “List named ubiquitous genes” query within NGIBWS (http://www.neurogadgets.com/bws.php) (Charlebois et al. 2003).
A strict definition of a core of genes has those genes present in every member of the set of genomes under consideration. We performed such an analysis on clades of genomes (defined according to a genomic phylogeny [Gophna et al. 2004]), using the “Find ubiquitous genes” query within NGIBWS (Charlebois et al. 2003). Using each of the genomes from the clade in turn, ORFs are found that have an RBM (with allowance for near ties) better than the specified BLASTP threshold, in each of the other genomes in the clade.
Both RBM and CGN approaches present some limitations. False negatives can arise in the RBM approach when BLASTP matches fall below threshold; false positives can arise when match thresholds are set so low as to permit spurious matches, and when paralogs match for want of lost orthologs. The use of CGNs as bridges for weak matches overcomes some of the problems inherent in the pure RBM approach, but introduces new problems relating to inconsistent annotation. Where alternative names for a gene are popular, an orthologous cluster may break up into name cliques, where a gene is truly ubiquitous, but fails to appear as a ubiquitous CGN. False positives are theoretically possible with the CGN approach, but only if homonymous names are used in annotation, which should be rare among core or otherwise prevalent genes, lysyl-tRNA synthetase (lysS) not-withstanding (Ibba et al. 1999).
Both RBM and CGN methods of finding ubiquitous genes are exquisitely sensitive to missed annotation (where a gene is present in the sequence, but is not annotated as an ORF), and to sequencing artifacts (during cloning or sequence assembly). We assessed the extent of the former problem by repeating the work of Lecompte et al. (2002) on ribosomal proteins, where several expected proteins turned up missing in our larger set (147 vs. 59) of prokaryotic genomes. All of our analyses, except that illustrated in Table 5, are based on all complete sequences of Bacterial (130) and Archaeal (17) genomes available in January, 2004.
Acknowledgments
This work was supported by Genome Atlantic (Genome Canada) and the Canadian Institutes for Health Research, and the Canada Research Chairs Program.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3024704.
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher, Y., Douady, C.J., Papke, R.T., Walsh, D.A., Boudreau, M.E., Nesbø, C.L., Case, R.J., and Doolittle, W.F. 2003. Lateral gene transfer and the origins of prokaryotic groups. Annu. Rev. Genet. 37: 283-328. [DOI] [PubMed] [Google Scholar]
- Brochier, C., Bapteste, E., Moreira, D., and Philippe, H. 2002. Eubacterial phylogeny based on translational apparatus proteins. Trends Genet. 18: 1-5. [DOI] [PubMed] [Google Scholar]
- Brown, J.R., Douady, C.J., Italia, M.J., Marshall, W.E., and Stanhope, M.J. 2001. Universal trees based on large combined protein sequence data sets. Nat. Genet. 28: 281-285. [DOI] [PubMed] [Google Scholar]
- Charlebois, R.L., Clarke, G.D.P., Beiko, R.G., and St. Jean, A. 2003. Characterization of species-specific genes using a flexible, web-based querying system. FEMS Microbiol. Lett. 225: 213-220. [DOI] [PubMed] [Google Scholar]
- Charlebois, R.L., Beiko, R.G., and Ragan, M.A. 2004. Genome phylogenies. In Organelles, genomes and eukaryote phylogeny: An evolutionary synthesis in the age of genomics (eds. R.P. Hirt and D.S. Horne), pp. 189-206. CRC Press, Boca Raton, FL.
- Clarke, G.D.P., Beiko, R.G., Ragan, M.A., and Charlebois, R.L. 2002. Inferring genome trees using a filter to eliminate phylogenetically discordant sequences and a distance matrix based on normalized BLASTP scores. J. Bacteriol. 184: 2072-2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubin, V., Gouy, M., and Perrière, G. 2002. A phylogenomic approach to bacterial phylogeny: Evidence of a core of genes sharing a common history. Genome Res. 12: 1080-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubin, V., Moran, N.A., and Ochman, H. 2003. Phylogenetics and the cohesion of bacterial genomes. Science 301: 829-832. [DOI] [PubMed] [Google Scholar]
- Gophna, U., Doolittle, W.F., and Charlebois, R.L. 2004. Weighted genome trees: Refinements and applications. J. Bacteriol. (in press). [DOI] [PMC free article] [PubMed]
- Harris, J.K., Kelley, S.T., Spiegelman, G.B., and Pace, N.R. 2003. The genetic core of the universal ancestor. Genome Res. 13: 407-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibba, M., Losey, H.C., Kawarabayasi, Y., Kikuchi, H., Bunjun, S., and Söll, D. 1999. Substrate recognition by class I lysyl-tRNA synthetases: A molecular basis for gene displacement. Proc. Natl. Acad. Sci. 96: 418-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain, R., Rivera, M.C., Moore, J.E., and Lake, J.A. 2002. Horizontal gene transfer in microbial genome evolution. Theor. Popul. Biol. 61: 489-495. [DOI] [PubMed] [Google Scholar]
- Koonin, E.V. 2003. Comparative genomics, minimal gene-sets and the last universal common ancestor. Nat. Rev. Microbiol. 1: 127-136. [DOI] [PubMed] [Google Scholar]
- Koonin, E.V., Mushegian, A.R., and Bork, P. 1996. Non-orthologous gene displacement. Trends Genet. 12: 334-336. [PubMed] [Google Scholar]
- Kunin, V. and Ouzounis, C.A. 2003. The balance of driving forces during genome evolution in prokaryotes. Genome Res. 13: 1589-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krylov, D.M., Wolf, Y.I., Rogozin, I.B., and Koonin, E.V. 2003. Gene loss, protein sequence divergence, gene dispensability, expression level, and interactivity are correlated in eukaryotic evolution. Genome Res. 13: 2229-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake, J.A., Jain, R., and Rivera, M.C. 1999. Mix and match in the tree of life. Science 283: 2027-2028. [DOI] [PubMed] [Google Scholar]
- Lecompte, O., Ripp, R., Thierry, J.C., Moras, D., and Poch, O. 2002. Comparative analysis of ribosomal proteins in complete genomes: An example of reductive evolution at the domain scale. Nucleic Acids Res. 30: 5382-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerat, E., Daubin, V., and Moran, N.A. 2003. From gene trees to organismal phylogeny in prokaryotes: The case of the γ-Proteobacteria. PLoS Biol. 1: E19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova, K.S. and Koonin, E.V. 2003. Comparative genomics of archaea: How much have we learned in six years, and what's next? Genome Biol. 4: 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova, K.S., Aravind, L., Galperin, M.Y., Grishin, N.V., Tatusov, R.L., Wolf, Y.I., and Koonin, E.V. 1999. Comparative genomics of the Archaea (Euryarchaeota): Evolution of conserved protein families, the stable core, and the variable shell. Genome Res. 9: 608-628. [PubMed] [Google Scholar]
- Matte-Tailliez, O., Brochier, C., Forterre, P., and Phillippe, H. 2002. Archaeal phylogeny based on ribosomal proteins. Mol. Biol. Evol. 19: 631-639. [DOI] [PubMed] [Google Scholar]
- Mirkin, B.G., Fenner, T.I., Galperin, M.Y., and Koonin, E.V. 2003. Algorithms for computing parsimonious evolutionary scenarios for genome evolution, the last universal common ancestor and dominance of horizontal gene transfer in the evolution of prokaryotes. BMC Evol. Biol. 3: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mushegian, A.R. and Koonin, E.V. 1996. A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc. Natl. Acad. Sci. 93: 10268-10273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesbø, C.L., Boucher, Y., and Doolittle, W.F. 2001. Defining the core of nontransferable prokaryotic genes: The euryarchaeal core. J. Mol. Evol. 53: 340-350. [DOI] [PubMed] [Google Scholar]
- Ragan, M.A. and Charlebois, R.L. 2002. Distributional profiles of homologous open reading frames among bacterial phyla: Implications for vertical and lateral transmission. Int. J. Syst. Evol. Microbiol. 52: 777-787. [DOI] [PubMed] [Google Scholar]
- Snel, B., Bork, P., and Huynen, M.A. 2002. Genomes in flux: The evolution of archaeal and proteobacterial gene content. Genome Res. 12: 17-25. [DOI] [PubMed] [Google Scholar]
- Tatusov, R.L., Koonin, E.V., and Lipman, D.J. 1997. A genomic perspective on protein families. Science 278: 631-637. [DOI] [PubMed] [Google Scholar]
- Teichmann, S.A. and Mitchison, G. 1999. Is there a phylogenetic signal in prokaryote proteins? J. Mol. Evol. 49: 98-107. [DOI] [PubMed] [Google Scholar]
- Waters, E., Hohn, M.J., Ahel, I., Graham, D.E., Adams, M.D., Barnstead, M., Beeson, K.Y., Bibbs, L., Bolanos, R., Keller, M., et al. 2003. The genome of Nanoarchaeum equitans: Insights into early archaeal evolution and derived parasitism. Proc. Natl. Acad. Sci. 100: 12984-12988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese, C. 1998. The universal ancestor. Proc. Natl. Acad. Sci. 95: 6854-6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese, C.R. 1987. Bacterial evolution. Microbiol. Rev. 51: 221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2000. Interpreting the universal phylogenetic tree. Proc. Natl. Acad. Sci. 97: 8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2002. On the evolution of cells. Proc. Natl. Acad. Sci. 99: 8742-8747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer, C. 2003. Genomics. Tinker, tailor: Can Venter stitch together a genome from scratch? Science 299: 1006-1007. [DOI] [PubMed] [Google Scholar]
Web site references
- http://www.neurogadgets.com/bws.php; The NeuroGadgets Inc. Bioinformatics Web Service (NGIBWS).