Abstract
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) analysis of base-specific cleavage products is an efficient, highly accurate tool for the detection of single base sequence variations. We describe the first application of this comparative sequencing strategy for automated high-throughput mutation detection in microbial genomes. The method was applied to identify DNA sequence changes that occurred in Escherichia coli K-12 MG1655 during laboratory adaptive evolution to new optimal growth phenotypes. Experiments were based on a genome-scale in silico model of E. coli metabolism and growth. This model computes several phenotypic functions and predicts optimal growth rates. To identify mutations underlying a 40-d adaptive laboratory evolution on glycerol, we resequenced 4.4% of the E. coli-K12 MG1655 genome in several clones picked at the end of the evolutionary process. The 1.54-Mb screen was completed in 13.5 h. This resequencing study is the largest reported by MALDI-TOF mass spectrometry to date. Ten mutations in 40 clones and three deviations from the reference sequence were detected. Mutations were predominantly found within the glycerol kinase gene. Functional characterization of the most prominent mutation shows its metabolic impact on the process of adaptive evolution. All sequence changes were independently confirmed by genotyping and Sanger-sequencing. We demonstrate that comparative sequencing by base-specific cleavage and MALDI-TOF mass spectrometry is an automated, fast, and highly accurate alternative to capillary sequencing.
Adaptive evolution imposes fixed environmental constraints on microorganisms that result in genotypes, proteins, and pathways that are “functionally tuned” in the context of the environment (Elena and Lenski 2003). The genotype holds instructions used and interpreted by the cellular machinery to adapt to environments and display a “fit” phenotype. Delineating sequence variations during the course of adaptive evolution is essential to understanding of the architecture and evolution of genetic circuits. Determining evolutionary changes at the DNA sequence level thus provides information about the plasticity of the genome, the interaction between the lower (molecular or cellular) and higher (whole organism) levels of organization, as well as the “evolvability” of phenotype—the ability of an individual or a population to generate fit variants.
We present resequencing by base-specific cleavage and MALDI-TOF mass spectrometry (MS) to identify DNA sequence changes underlying adaptive evolution of Escherichia coli-K12 MG1655 on nonoptimal carbon substrates (Ibarra et al. 2002). This resequencing effort is the first microbial and the largest by MALDI-TOF MS reported to date. It is interesting to contrast the MALDI-TOF technology with most common mutation screening technologies like denaturing gradient HPLC, which reveals information on the presence or absence of a sequence change without exact localization and characterization, and applications like hybridization to oligonucleotide arrays and capillary sequencing, which locate the exact nature of sequence variations, but sometimes suffer from base-calling errors and variable quality threshold definitions. Resequencing by base-specific cleavage and MALDI-TOF MS on average facilitates the detection of >99.0% of all homozygous base-pair changes in 500-bp target regions. The technology has proven to be successful in the discovery of single nucleotide polymorphisms (SNPs) in human disease candidate genes (Stanssens et al. 2004) and signature sequence-based bacterial identification of pathogens (von Wintzingerode et al. 2002; Lefmann et al. 2004). Compared to earlier approaches of mass spectrometry based Sanger-sequencing (Koester et al. 1996), this new method increases the screened target region length between fivefold and 10-fold and thus represents a major advance in establishing MALDI-TOF MS as a resequencing tool.
Results and Discussion
Study design
A genome-scale in silico model of E. coli metabolism (Edwards et al. 2000; Reed et al. 2003) and growth has been constructed that can compute several phenotypic functions of E. coli correctly (Edwards and Palsson 2000), and can be used to interpret and predict the outcome of adaptive evolutionary experiments (Ibarra et al. 2002; Fong et al. 2003). E. coli-K12 MG1655 evolved reproducibly on glycerol as the sole carbon source for 700 generations from a suboptimal to an optimal growth rate.
These adaptive evolutions—generated at 30°C in 2% glycerol (G)—provided eight closely related clones (G1-A to G1-H) from a heterogeneous population (G1) of E. coli (Ibarra et al. 2002), which were compared to the wild type E. coli-K12 MG1655 reference sequence (Blattner et al. 1997). In an investigation of post-evolutionary genotypic properties, mutations identified in the eight initial clones (G1-A–G1-H) were additionally screened in clones of six populations evolved either prior to or subsequently on the following suboptimal carbon sources: pyruvate (P), malate (M), and α-ketoglutarate (A). Clones are named: GP, GPA, MG, MGA, PG, PGA, where, for example, GP refers to an initial evolution on glycerol and then pyruvate. The rest of these clones are named accordingly.
The extent to which identical genotypic evolutionary changes appear in parallel evolutions was further studied by resequencing seven phenotypically distinct mixed populations evolved on glycerol (G1, G2, GlyA, GlyB, GlyC, GlyD, GlyE).
The study encompassed 202 kb (4.4%) of the E. coli-K12 MG1655 genome containing a total of 125 genes including those related to glycerol metabolism, transcription factors, mutator genes, and σ factors. Regions were chosen using a candidate gene approach, which included information from in silico modeling (Edwards et al. 2001; Reed et al. 2003) and expression profiling (Fong 2004). Segments of 200 bp upstream and downstream of the coding regions were included. Genomic regions were subdivided into 362 amplicons of 500–800 bp in length.
All but 1% of the designed amplicons could be converted into functional assays, which corresponds to a PCR failure rate of three PCR designs (three PCR primer pairs) out of the total of 362 designed assays.
Base-specific cleavage and MALDI-TOF MS
Resequencing by base-specific cleavage and MALDI-TOF MS couples a PCR amplification of the target region of interest with a labeling-free homogeneous in vitro transcription/RNAse system and MALDI-TOF MS analysis. No purification of the PCR product or any subsequent reaction product is required. Reactions are designed for automated liquid handling in 384 microtiter plate formats with a throughput of 16 plates or ∼768 kb (1536 × 500 bp amplicons) per day and operator.
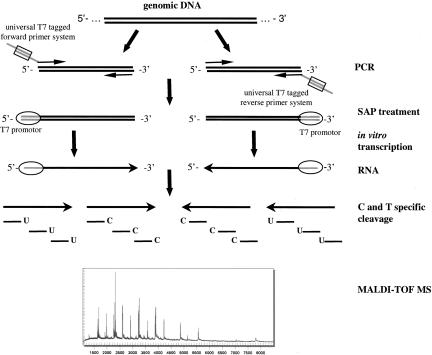
A novel universal primer system for PCR amplification streamlines the previously described amplification system of four tagged gene-specific oligonucleotides (Rodi et al. 2002), reduces primer costs by half, and facilitates the usage of a single polymerase. The methodology is outlined in Figure 1.
Figure 1.
The universal primer system: Scheme of base-specific cleavage and MALDI-TOF MS based resequencing. PCR amplification of the DNA region of interest is followed by T7-mediated in vitro transcription and base-specific cleavage. RNA transcripts of the target regions are cleaved in four reactions corresponding to virtually each of the four bases. This corresponds to an RNAse-specific cleavage at U and C of the forward as well as of the reverse RNA transcript. MALDI-TOF spectra of all four cleavage reactions are subject to subsequent mutation analysis.
The universal primer system is a combination of a universal T7-promoter-tagged forward and a universal T7-promoter-tagged reverse primer with two 18-bp-tagged gene-specific oligonucleotides. In a forward PCR reaction the universal forward primer facilitates the introduction of the T7-promoter sequence at the 5′-end of the forward strand. The universal reverse primer introduces the T7-promoter sequence at the 5′-end of the reverse complement strand in a second parallel PCR reaction.
Base-specific cleavage products were analyzed on matrix-coated chips by real-time quality-controlled MALDI-TOF MS as previously described (Stanssens et al. 2004).
An average data acquisition speed of 18 sec per sample resulted in a total acquisition time of 13.5 h for the whole study. This is among the fastest turnover rates of currently available resequencing technologies (cf. 2.8 Mb/d for a 384-capillary instrument) (Kling 2003). Recent technological improvements such as 200 Hz lasers promise further increase of the throughput by about fivefold.
Simulation
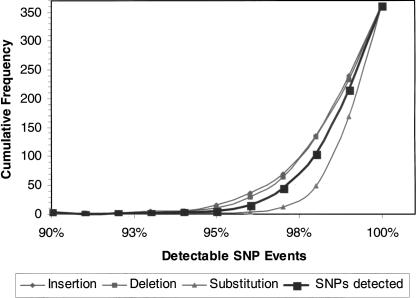
A computational simulation analysis of all 362 amplicons determined the theoretical success rate of the detection of all potential single-base-pair insertions, deletions, and SNPs. All possible single base sequence variations were systematically introduced in all of the amplicons and the cumulative frequency of detectable insertions, deletions, and SNPs were plotted against their rate of detection (Fig. 2). Signals between 1100 Da and 8000 Da were considered with a resolution of (m/Δm) 600. The results show that 97%–100% of all possible deletions can be detected in a total of 319 amplicons, 97%–100% of all possible insertions are detectable in 314 amplicons and 97%–100% of all single nucleotide substitutions are detectable in 355 amplicons. The average detection rate for the total of all single base sequence variations over all amplicons is 99% with a minimum detection rate of 94% in only three out of the total of 362 amplicons.
Figure 2.
Simulated cumulative frequencies of all detectable single base sequence variation events in the 362 amplicons of the E. coli-K-12 reference set. 97%–100% of all possible deletions are detectable in a total of 319 amplicons, 97%–100% of all possible insertions are detectable in 314 amplicons, and 97%–100% of all single nucleotide substitutions can be detected in 355 amplicons. (Black) Total single base variation events; (gray) SNPs, insertions, and deletions.
Resequencing results of E. coli-K12 evolutionary variants
A total of 13 mutations including three sequence deviations from the E. coli-K-12 wild-type reference sequence were detected (Tables 1 and 2).
Table 1.
Identified mutations in E. coli-K12 MG1655 clones underlying adaptive evolution on glycerol
| Gene (protein) | Position | Mutation | Affected amino acid | Mutant clones |
|---|---|---|---|---|
| glpK (glycerol kinase) | 113 | A → C | Q38P | GlyE |
| 160 | T → G | W54G | MG, MGA | |
| 184 | G → T | V62L | GlyC | |
| 218 | A → C | D73A | GlyA, GlyB | |
| 218 | A → T | D73V | PG | |
| 692 | G → A | G231D | G1, GP, GPA | |
| 705 | Ins. AAAGGCGGC | Ins. 235GGK | G2 | |
| aldB (aldehyde dehydrogenase B) | 36 | C → A | P12P | G1, GP, GPA |
| tdcR (threonine dehydratase activator) | +4 | C → A | (L + 2I) | G1-A |
| rpoS (σ factor 38) | 739 | G → T | E247stop | G1, GP, GPA |
Table 2.
Identified sequence deviations in E. coli-K12 MG1655 (ATCC# 47076) in reference to the published sequence of E. coli-K12 MG1655
| Gene (protein) | Position | Sequence deviation | Affected amino acid | |
|---|---|---|---|---|
| gppA1 (pppGpp phosphohydrolase) | 413 | G → C | T86R | Wild type |
| tktA4 (transketolase) | 145 | G → C | G470G | Wild type |
| glcF1 (glycolate oxidase) | 64 | — → C | – 111 | Wild type |
Time-efficient algorithms calculated deviations of detected mass signal pattern from reference sequence based in silico cleavage pattern (Boecker 2003). Single base variations were automatically listed and ranked by score. This score is determined by supporting mass signal events and weighted by spectra quality. Scores are derived from the number of confirmatory signals (missing and additional) and signal intensity changes in four cleavage reactions.
Of all single base sequence variations identified in this study, 100% were detected and autoselected by the software. The detection of all of these single base sequence variations is based on a redundancy of ≥4 confirming mass signal changes. To support the validity of the results, all single base sequence variations were confirmed by genotyping (Storm et al. 2003). Positional uncertainties were resolved, where present.
A duplication of a 9-bp sequence stretch in the glycerol kinase gene (glpK) was detected by a missing signal of the corresponding 21-bp DNA fragment in the T cleavage reaction of the forward transcript. As a result of the exact duplication of the sequence stretch, the mass spectra showed no further deviations from the reference and the amplicon was subject to Sanger-sequencing for unambiguous identification.
In addition, three detected sequence changes—deviations from the published E. coli-K12 MG1655 reference sequence (Blattner et al. 1997)—were identified and verified by Sanger-sequencing (Table 2). These variations were identified within the wild-type E. coli-K12 strain (ATCC #47076) as well as all evolved clones and populations.
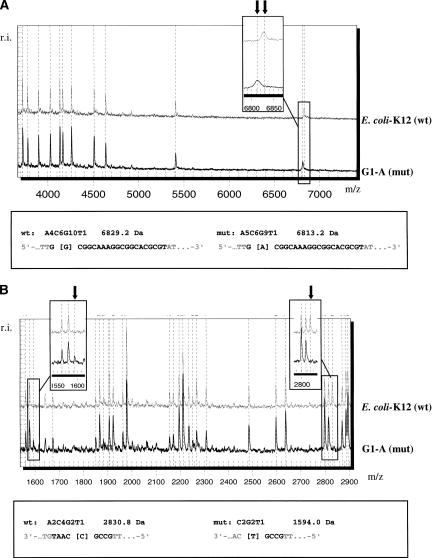
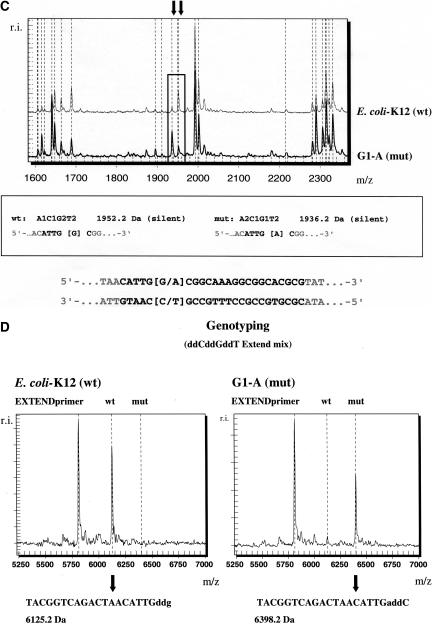
Figure 3 exemplifies the detection and localization of the G→A mutation at position 356 in a 748-bp amplicon of the glycerol kinase gene. The mutation was detected in the E. coli clone G1-A and verified by genotyping. Multiple observations in three out of the four cleavage reactions allow for the exact detection and localization of the mutation. The T-specific cleavage of the forward RNA transcript (Fig. 3A) reveals a missing signal at 6829.2 Da in the G1-A clone as opposed to the wild-type E. coli K-12 strain, where the signal is in concordance with a RNA fragment of defined composition and location. A new signal appears in the G1-A clone at 6813.2 Da. This observation can be explained by the replacement of one of the G residues on the 6829.2-Da wild-type fragment by an A in clone G1-A, reducing the mass of the fragment by 16 Da. The T-specific cleavage reaction of the reverse RNA transcript (Fig. 3B) confirms the observation of a G→A mutation of clone G1-A in comparison to the expected cleavage pattern of the wild type and localizes the mutation to position 356 in the amplicon corresponding to position 692 in the glpK gene. The mutation introduces an additional cleavage site and creates an additional mass signal at 1594.0 Da as well as at 1272.8 Da (data not shown). The wild-type signal at 2830.8 Da is missing. The C-cleavage forward reaction (Fig. 3C) shows confirmatory information based on signal intensity changes. Both the missing as well as the additional signal are generated by two fragments of the exact same composition, but an intensity increase of the signal at 1936.2 Da in the mutant as well as an signal intensity decrease at 1952.2 Da confirm the single base sequence variation event. The C-reverse reaction does not contain any supporting information as all signals are composed of dimers and trimers. They are out of the mass range of detection. The mass range below 1000 Da is excluded from the detection as it can be affected by background noise of matrix signals and is loaded by dimers and trimers, which are generally represented multiple times.
Figure 3.
Base-specific cleavage and MALDI-TOF based discovery of the mutation G→A at position 356 in a 748-bp amplicon of the glycerol kinase gene (glpK). Mutation-specific signal changes as described in the text are highlighted and enlarged. (A) Overlay of mass spectra resulting from the T-specific cleavage reaction of the forward RNA transcript of the wild-type E. coli-K12 strain and the mutant isolate G1-A. Mutation-specific signal changes at 6813.2 Da and 6829.2 Da. (B) Overlay of mass spectra resulting from the T-specific cleavage reaction of the reverse RNA transcript of the wild-type E. coli-K12 strain and the mutant isolate G1-A. Mutation-specific signal changes at 1594.0 Da and 2830.8 Da. (C) Overlay of mass spectra resulting from the C-specific cleavage reaction of the forward RNA transcript of the wild-type E. coli-K12 strain and a mutant isolate G1-A. Intensity changes of mutation-specific mass signals at 1936.2 Da and 1952.2 Da. (D) Verification of the mutation by MALDI-TOF-based genotyping. A G-allele-specific extend product is detected in the wild-type E. coli-K12 strain at 6125.2 Da. The corresponding mutant C allele is detected at 6398.2 Da in clone G1-A.
The detected and localized mutation G→A at position 692 was verified by genotyping (Fig. 3D). A corresponding extend product was unambiguously detected at 6398.2 Da.
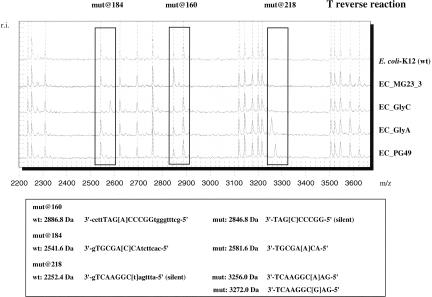
Substitutions T160G, G184T, A218C, and A218T in glpK can be unambiguously identified and differentiated using a single cleavage reaction. The spectral overlay of four genotypically different clones in Figure 4 presents the base-specific cleavage results and corresponding masses of the T-specific reverse reaction. Unique mutation identifiers in comparison to the wild-type spectrum are signal events at 2886 Da for T160G, at 2541.6 Da and 2581.6 Da for G184T, at 3256.0 Da for A218T, and at 3272.0 Da for A218C.
Figure 4.
Mutation detection in the glycerol kinase gene (glpK) by base-specific cleavage and MALDI-TOF MS. Discriminating mass signals in the T-cleavage reaction of the reverse RNA transcript (highlighted in boxes) at 2541.6 Da, 2581.6 Da, 2886.8 Da, 3256.0 Da, and 3272.0 Da allow for unambiguous identification of the substitution T→G at position 160, G→T at position 184, A→C at position 218, and A→T at position 218.
Characterization of identified mutations
Of the 10 single base sequence variations detected in the glycerol-evolved E. coli isolates, eight are nonsynonymous, two are synonymous, and one introduces a premature stop codon. With these mutations identified, one can study their expected biochemical and physiological impact. For a complete list of all mutations refer to Table 1.
Half a dozen distinct single point mutations and a 9-bp insertion were identified in the structural gene for glycerol kinase (glpK). Interestingly, we observed that each of these amino acid substitutions could affect the binding of the allosteric regulator Fructose-1,6-bisphosphate (FBP). Ongoing studies to connect genotype to phenotype include the cloning and biochemical characterization of these mutants. Initial experiments and results are discussed in the next section. Interestingly, different clones derived from the same parental ancestor exhibited the same optimal growth phenotype (Ibarra et al. 2002), although these strains were found to be genotypically different, which clearly demonstrates the heterogeneity of the end population (G1).
Mutation G739T in σ factor 38 introduces a premature stop codon. σ factor 38 is a general stress response factor in E. coli controlling a regulon of at least 30 genes, which are expressed during starvation and at transition into stationary phase (Bjedov et al. 2003; Wei et al. 2000).
A synonymous substitution in the gene aldB changes the codon for proline from ccc to cca. The codon ccc is classified as a rare codon for proline in E. coli, but the abundances of tRNA species cognate to these codons are similar. Proline residues have been implicated in limiting the rate of protein folding due to energy barriers in cis/trans isomerization (Eisenmesser et al. 2002). However, such effects have yet to be determined in the context of codon change and adaptive evolution.
Another mutation was identified in the fourth base pair after the stop codon for threonine dehydratase activator, tdcR, and its effects are unknown.
Mutator genes that can potentially confer an increased rate of mutation in evolving populations were found to be identical to the wild type. This confirms that the observed mutations are a consequence of adaptation rather than an elevated mutation rate due to compromised fidelity in replication and DNA repair.
Functional analysis of mutation G692A in glycerol kinase
In order to assess the metabolic impact of the discovered mutations (Table 1) on the process of adaptive evolution, we focused on one of the mutations as an example for functional analysis. Among all resequenced strains, mutations were most frequently detected in the glycerol kinase gene. The most prevalent mutation present in almost every resequenced strain was the nonsynonymous substitution G692A resulting in replacement of a glycine with an aspartate, G232D. This transition mutation was detected in a walker loop (AA 230–236; motif: GxxGxGKT/S) (Ormo et al. 1998), a conserved motif involved in Fructose-1,6-bisphosphate (FBP) binding in ATP-binding proteins.
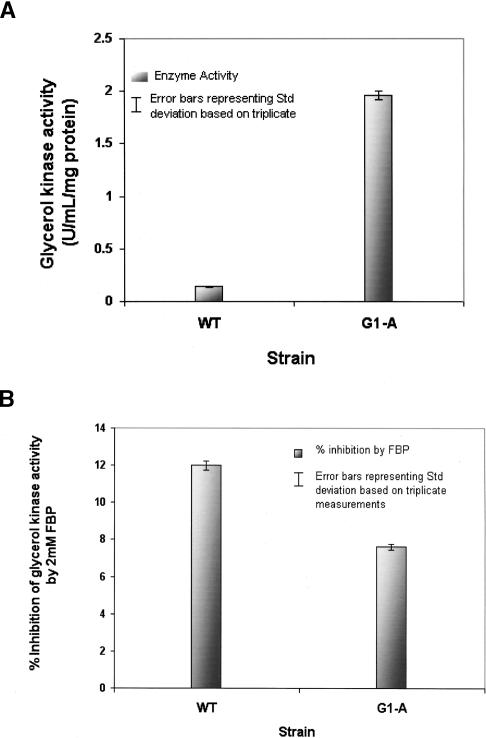
The wild-type glycerol kinase and its variant (G692A) were cloned and kinetically characterized. Kinetic assays for the corresponding G282D amino acid variant of the protein show an increase in glycerol kinase activity (Fig. 5A) and a simultaneous increase in tolerance toward the inhibitor FBP (Fig. 5B).
Figure 5.
Enzyme kinetics of the glycerol kinase variant (G692A, Gly232Asp) of E. coli G1-A in comparison to the wild-type enzyme (wt). Enzyme activities were determined as a decrease in absorbance at 340 nm monitoring ADP release by coupling pyruvate kinase and lactate dehydrogenase activity as detailed in Methods. (A) A 12-fold increase in enzyme activity in the recombinant mutant clone as opposed to the wild type. (B) Enzyme inhibition by an allosteric inhibitor Fructose-1,6-bisphosphate (FBP) was determined by addition of FBP to a final concentration of 2 mM. Inhibitory effects decreased by ∼33% in the enzyme variant when compared to the wild-type enzyme.
For assessment of enzyme activity, we used an assay format that couples the conversion of glycerol to glycerolphosphate with a pyruvate kinase/lactate dehydrogenase system. In this assay, one unit of the enzyme is defined as the amount that will convert 1.0 μmol of glycerol and ATP to L-α-glycerophosphate and ADP per minute at pH 9.8 at 25°C in a coupled system with pyruvate kinase/lactate dehydrogenase. The activity of the enzyme increased from 0.14 U/mL per mg protein for wild type to 1.9 U/mL per mg protein for the G282D recombinant enzyme. This calculates to a 12-fold increase in enzyme activity.
FBP is a known allosteric inhibitor of glycerol kinase. Since the intracellular concentration of FBP in E. coli has been estimated to be ∼1.7 mM, we tested the inhibitory activity of FBP close to the physiological concentration at 2 mM. 2 mM FBP decreased the activity of the wild-type enzyme by 12%. However, the decrease was only 7.6% in the case of the mutant enzyme. This corresponds to a 33% decrease in inhibition by FBP compared to the wild-type enzyme. Figure 5 summarizes enzyme activities in the presence and absence of FBP of the wild-type and mutant (G232D) recombinant glycerol kinase.
The activity of glycerol kinase is known to be a rate-limiting step in E. coli glycerol metabolism. It has been found earlier that increased enzyme activity, generated, for example, through FBP resistance, leads to increased glyceraldehydes-3-phosphate (G-3-P) formation, which subsequently can lead to the death of the cells (Zwaig et al. 1970). For our model of adaptive evolution, this indicates that the improved growth rates on glycerol are achieved while maintaining stringent control of the intracellular levels of G-3-P below those that affect growth and glycerol metabolism. Several further control mechanisms might contribute to this. FBP can prevent glycerol utilization at the level of enzyme catalysis and also at the level of enzyme synthesis (through interference with formation of the inducer G-3-P). A mutant strain less susceptible to the inhibition by FBP would thus grow faster on glycerol as the sole source of carbon and energy than the wild-type parent. Along with the increased activity of the enzyme, this reduced inhibition by FBP is probably an important factor that also contributes to increased growth rates (from 0.22 h-1 to 0.55 h-1) (Ibarra et al. 2002) in the E. coli strains evolved on glycerol. Site-directed mutagenesis and reverse mutagenesis will have to be performed to provide more insights into the effect of these mutations on the overall phenotype.
Additional identified mutations in the glycerol kinase gene (Table 1) are subject to further analysis in order to determine their impact on protein function and phenotype.
Conclusion
Physiological consequences of sequence changes and the elucidation of genotype–phenotype relationships are fundamental to biology. Adaptive evolution has been used to systematically generate changes in phenotype (Elena and Lenski 2003), and we now need to collect the corresponding genotypic data sets. Mutations in the glycerol kinase gene, a key enzyme in glycerol metabolism, suggest their implication in causing the observed phenotype. A future challenge will be the final interpretation of all the mutation data, to determine which of the identified mutations contribute to the selected phenotype and which represent neutral or deleterious mutations that hitchhike with adaptive sequence variations. However, this data set might also contribute further insight into the structure–function relationship of glycerol kinase.
Sanger-sequencing delivers direct DNA sequence information, while the MALDI-TOF based large-scale systematic comparison of mass spectral cleavage pattern to the in silico pattern of a reference sequence focuses on the detection of sequence deviations. The analysis relies on the explanation of combinations of missing and additional signal pairs rather than the more time-consuming and computationally intensive process of sequence alignments as required in conventional Sanger-sequencing. The result is an automated output of mutations and their localization.
The statistical significance of parallel screening of multiple samples and the accuracy of mutation detection, both offered by mass spectrometry, resulted in reliable data sets of sequence variations occurring during adaptive evolution that provide potential insights into the genetic basis of microbial systems. Base-specific cleavage and mass spectrometry offers a viable solution for rapid large-scale resequencing efforts and is ideal for parallel analysis of a multitude of samples, a necessity in genome comparison and epidemiology. Similar studies on pathogens have the potential to provide a mechanistic basis for pathogenesis and management of disease.
Methods
Strains and media
The E. coli-K12 MG1655 annotated genome sequence (NC_000913) and biochemical literature were used to construct the in silico E. coli strain (Edwards and Palsson 2000). Growth and maintenance requirements were imposed in an in silico model as described previously (Ibarra et al. 2002; Fong et al. 2003). E. coli MG1655 (ATCC# 47076, genotype: F-, λ-) was used as the wild type reference strain for all experiments. Briefly, adaptation experiments were carried out at 30°C in M9 minimal medium with the addition of 2 g/L glycerol through serial passaging at midex-ponential phase. For resequencing genomic DNA was extracted using the Qiagen DNeasy Tissue kit (Qiagen).
Primer design
The E. coli-K12 MG1655 reference sequence (NC_000913) was used for primer design. Robust, specific PCR assays were designed with the primer design software GeneTool (Biotools Inc.). Sequence analysis software, NetPrimer (http://www.premierbiosoft.com/netprimer) (Premier Biosoft International) was used to check primer properties. NetPrimer combines the latest primer design algorithms with a Web-based interface. All primers were analyzed for melting temperature (Tm) using the nearest-neighbor thermodynamic theory. Primers were analyzed for all secondary structures including hairpins, self-dimers, and cross-dimers in all primer pairs. The average length of the gene specific primer part was 18–20 bp with a Tm of 60°C and an appropriate 3′-end stability of ΔG ≤ -10 kcal/mol.
A list of all resequenced genes is available in Supplemental Table 1.
To facilitate in vitro transcription primers were extended by an 18-bp forward (5′-CACTATAGGGAAGGAACT-gene-specific primer part-3′) and an 18-bp reverse tag (5′-CACTATAGG GAGAAGGTC-gene-specific primer part-3′).
Sample processing for mutation detection
Target regions of interest were amplified in a total volume of 5 μL. Amplification mixtures contained 1× PCR buffer [Tris-HCl, KCl, (NH4)2SO4, MgCl2 at pH 8.7; final concentration of 1.5 mM], 200 μM of each deoxynucleoside triphosphate, 0.1 U of HotStar Taq polymerase (Qiagen GmbH), 5 ng of DNA and primer.
A concentration of 40 nM of the forward gene-specific primer as well as a concentration of 200 nM of the forward universal primer (5′-CGTAATACGACTCACTATAGGGAAGGAACT-3′) and the gene-specific reverse primer were used in the forward reaction. The reverse reaction contained 40 nM of the reverse gene specific primer as well as 200 nM of the universal reverse primer (5′-CGTAATACGACTCACTATAGGGAGAAGGTC-3′) and the forward gene-specific primer. Following an initial step of HotStarTaq activation (15 min at 95°C) a touchdown thermo cycling program combined 20 cycles of denaturation at 95°C for 20 sec, annealing at 62°C for 30 sec, and extension at 72°C for 1 min with 25 cycles of denaturation at 95°C for 20 sec, annealing at 50°C for 30 sec, and extension at 72°C for 1 min.
Unincorporated dNTPs were dephosphorylated prior to T7-polymerase mediated in vitro transcription by incubation with 0.3 units of shrimp alkaline phosphatase (SAP) at 37°C for 20 min. The enzyme was inactivated at 85°C for 5 min. To mediate a C- as well as a T-specific cleavage reaction two in vitro transcription reactions were carried out in a total volume of 4 μL each. For each of the reactions, 2 μL of the SAP-treated PCR product were incubated with 0.22 μL of either a C- or a T-specific transcription mix (Sequenom Inc.), 5 mM DTT and 20 units of T7 RNA&DNA polymerase (Epicenter) at 37°C for 2 h. Transcription products were cleaved by addition of 2.5 units of RNAseA and incubation at 37°C for 1 h.
In summary, target regions were cleaved in four reactions at positions corresponding to each of the four bases. The RNA transcript of the forward strand was cleaved by RNAse A at U in a dCMP-containing transcript and at C in a dTMP-containing transcript. An A- and G-specific cleavage of the template sequence was facilitated by U or C cleavage of the corresponding reverse RNA transcripts.
Samples were diluted with 21 μL of H2O and conditioned by 6 mg of SpectroCLEAN (Sequenom Inc.) prior to MALDI-TOF analysis. MALDI-TOF MS acquisition of spectra of each of the cleavage reactions was followed by comparative analysis to reference sequence derived in silico cleavage pattern.
Genotyping by MassEXTEND
Candidate single base sequence variations identified by base-specific cleavage were verified by means of primer extension reactions using an extend primer adjacent to the sequence variation of interest and an appropriate dNTP/ddNTP mix. Extend products were analyzed by MALDI-TOF MS as described (Little et al. 1997; Rodi et al. 2002).
MALDI-TOF MS analysis
Aliquots of 15 nL of analyte were robotically dispensed onto matrix loaded silicon chips (SpectroCHIP, Sequenom Inc.). Mass spectra were acquired using a MassARRAY mass spectrometer (Bruker-Sequenom Inc.). Positive ions were analyzed in linear TOF mode using delayed ion extraction and a total acceleration voltage of 20 kV. Analysis of all mass spectra and automated data interpretation were preformed with the Discovery-RT software Version 1.2.2 and Typer software Version 3.1 (Sequenom Inc.).
Cloning of the glycerol kinase gene
The open reading frame (ORF) of the glycerol kinase gene (glpK, b3926) was amplified by PCR. Genomic DNA of both the wild-type E. coli-K12 strain and the evolved strain G1-A of E. coli-K12 served as templates. PCR fragments of the ORFs of interest were agarose gel purified and ligated into the expression vector pCR T7/NT-TOPO according to the manufacturer's instructions (Invitrogen). Characterization and propagation of the construct is done in E. coli TOP10F′ cells. Clones were checked for the presence of insert and correct orientation by restriction digestion. Sequence identity was confirmed by Sanger-sequencing. The recombinant expression vectors were transformed into E. coli BL21(DE3) cells for protein expression. Gene expression in pCR T7/NT TOPO is controlled by the phage T7 promoter, which is recognized by T7 RNA polymerase.
Kinetics of E. coli glycerol kinase
Recombinant glycerol kinase expression was induced by IPTG according to the manufacturer's instructions (Invitrogen). Cells were harvested by centrifugation and resuspended in Tris buffer. Cellular extracts were obtained in subsequent freeze/thaw cycles using published protocols (Pettigrew et al. 1988; Brendan et al. 1998). The protein content of the cellular extract was determined using the BCA (bicinchonic acid) protein assay (Pierce). Enzyme activity assays used were as outlined in Cleland and Janson (Janson and Cleland 1974). The activity of glycerol kinase was determined in an ADP-coupled assay containing 40 IU of pyruvate kinase, 57 IU lactate dehydrogenase, 2.8 mM phosphoenolpyruvate, 0.22 mM NADH, 2 mM MgCl2, 3.4 mM glycerol, and 1.5 mM ATP in glycine buffer (pH 9.8) and 25°C.
The basal activity of the cellular E. coli extract nonspecific to glycerol kinase was estimated in a clone with reverted orientation of the insert, not capable of recombinant expression. Negligible background expression and basal activity of the expression strain was observed and corrected for in all the enzyme activity measurements. All measurements were done in triplicate and the standard deviations are represented as error bars in Figure 5.
All enzyme standards and chemicals were purchased from Sigma Chemicals to perform enzyme kinetics and obtain calibration curves.
Acknowledgments
We thank Nadine Vater for excellent technical assistance and sample processing.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2977704.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Bjedov, I., Tenaillon, O., Gérard, B., Souza, V., Denamur, E., Radman, M., Taddei, F., and Matic, I. 2003. Stress-induced mutagenesis in bacteria. Science 300: 1404-1409. [DOI] [PubMed] [Google Scholar]
- Blattner, F.R., Plunket III, G., Bloch, C.A., Perna, T., Burland, V., Riley, M., Collado-Vides, J., Glasner, J.D., Rode, C.K., Mayhew, G.F., et al. 1997. The complete sequence of Escherichia coli K-12. Science 277: 1453-1462. [DOI] [PubMed] [Google Scholar]
- Boecker, S. 2003. SNP and mutation discovery using base-specific cleavage and MALDI-TOF mass spectrometry. Bioinformatics 19: 44-53. [DOI] [PubMed] [Google Scholar]
- Brendan, B., Mendz, G., and Hazell, S. 1998. Methods for the measurement of a bacterial enzyme activity in cell lysates and extracts. Biol. Proced. Online 1: 17-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, J.S. and Palsson, B.Ø. 2000. The Escherichia coli MG1655 in silico metabolic genotype: Its definition, characteristics, and capabilities. Proc. Natl. Acad. Sci. 97: 5528-5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, J.S., Ibarra, R.U., and Palsson, B.Ø. 2001. In silico predictions of Escherichia coli metabolic capabilities are consistent with experimental data. Nat. Biotechnol. 19: 125-130. [DOI] [PubMed] [Google Scholar]
- Eisenmesser, E.Z., Bosco, D.A., Akke, M., and Kern, D. 2002. Enzyme dynamics during catalysis. Science 295: 1520-1523. [DOI] [PubMed] [Google Scholar]
- Elena, S.F. and Lenski, R.E. 2003. Evolution experiments with microorganisms: The dynamics and genetic bases of adaption. Nat. Rev. Genet. 390: 395-398. [DOI] [PubMed] [Google Scholar]
- Fong, S.S. 2004. “Adaptive evolution of Escherichia coli: Systems biology and a genome-scale metabolic model.” Ph.D. thesis, University of California–San Diego, La Jolla, CA.
- Fong, S.S., Marciniak, J.Y., and Palsson, B.Ø. 2003. Description and interpretation of adaptive evolution of Escherichia coli K-12 MG 1655 using a genome-scale metabolic model. J. Bacteriol. 185: 6400-6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra, R.U., Edwards, J.S., and Palsson, B.Ø. 2002. Escherichia coli K-12 undergoes adaptive evolution to achieve in silico predicted optimal growth. Nature 420: 186-189. [DOI] [PubMed] [Google Scholar]
- Janson, C. and Cleland, W. 1974. The kinetic mechanism of glycerol kinase. J. Biol. Chem. 249: 2562. [PubMed] [Google Scholar]
- Kling, J. 2003. Ultrafast DNA sequencing. Nat. Biotech. 21: 1425-1427. [DOI] [PubMed] [Google Scholar]
- Koester, H., Tang, K., Fu, D.J., Braun, A., van den Boom, D., Smith, C.L., Cotter, R.J., and Cantor, C.R. 1996. A strategy for rapid and efficient DNA sequencing by mass spectrometry. Nat. Biotechnol. 14: 1123-1128. [DOI] [PubMed] [Google Scholar]
- Lefmann, M., Honisch, C., Boecker, S., Storm, N., von Wintzingerode, F., Schloetelburg, C., Moter, A., van den Boom, D., and Goebel, U.B. 2004. Novel mass spectrometry-based tool for genotypic identification of mycobacteria. J. Clin. Microbiol. 42: 339-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little, D.P., Braun, A., O'Donnell, M.J., and Koester, H. 1997. Mass spectrometry from miniaturized arrays for full comparative DNA analysis. Nat. Med. 3: 1413-1416. [DOI] [PubMed] [Google Scholar]
- Ormo, M., Bystrom, C.E., and Remington, S.J. 1998. Crystal structure of a complex of Escherichia coli glycerol kinase and an allosteric effector fructose 1,6-bisphosphate. Biochemistry 37: 16565-16572. [DOI] [PubMed] [Google Scholar]
- Pettigrew, D.W., Ma, D.P., Conrad, C.A., and Johnson, J.R. 1988. Escherichia coli glycerol kinase. Cloning and sequencing of the glpK gene and the primary structure of the enzyme. J. Biol. Chem. 263: 135-139. [PubMed] [Google Scholar]
- Reed, J.L., Vo, T.D., Schilling, C.H., and Palsson, B.Ø. 2003. An expanded genome-scale model of Escherichia coli K-12 (iJR904 GSM/GPR). Genome Biol. 4: R54.1-R54.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodi, C.P., Darnhofer-Patel, B., Stanssens, P., Zabeau, M., and van den Boom, D. 2002. A strategy for the rapid discovery of disease markers using the MassARRAY system. BioTechniques 32: S62-S69. [PubMed] [Google Scholar]
- Stanssens, P., Zabeau, M., Meersseman, G., Remes, G., Gansemans, Y., Storm, N., Hartmer, R., Honisch, C., Rodi, C. Boecker, S., et al. 2004. High-throughput MALDI-TOF discovery of genomic sequence polymorphisms. Genome Res. 14: 126-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storm, N., Darnhofer-Demar, B., van den Boom, D., and Rodi, C.R. 2003. MALDI-TOF mass spectrometry-based SNP genotyping. Methods Mol. Biol. 212: 241-262. [DOI] [PubMed] [Google Scholar]
- von Wintzingerode, F., Boecker, S., Schloetelburg, C., Chiu, N.H., Storm, N., Jurinke, C., Cantor, C.R., Goebel, U.B., and van den Boom, D. 2002. Base-specific fragmentation of amplified 16S rRNA genes analyzed by mass spectrometry: A tool for rapid bacterial identification. Proc. Natl. Acad. Sci. 99: 7039-7044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, B., Shin, S., LaPorte, D., Wolfe, A.J., and Romeo T. 2000. Global regulatory mutations in csrA and rpoS cause severe central carbon stress in Escherichia coli in the presence of acetate. J. Bacteriol. 182: 1632-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaig, N., Kistler, W.S., and Lin, E.C. 1970. Glycerol kinase, the pacemaker for the dissimilation of glycerol in Escherichia coli. J. Bacteriol. 102: 753-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
Web site references
- http://www.premierbiosoft.com/netprimer/; NetPrimer.