Abstract
The sequence of any genome becomes most useful for biological experimentation when a complete and accurate gene set is available. Gene prediction programs offer an efficient way to generate an automated gene set. Manual annotation, when performed by experienced annotators, is more accurate and complete than automated annotation. However, it is a laborious and expensive process, and by its nature, introduces a degree of variability not found with automated annotation. EAnnot (Electronic Annotation) is a program originally developed for manually annotating the human genome. It combines the latest bioinformatics tools to extract and analyze a wide range of publicly available data in order to achieve fast and reliable automatic gene prediction and annotation. EAnnot builds gene models based on mRNA, EST, and protein alignments to genomic sequence, attaches supporting evidence to the corresponding genes, identifies pseudogenes, and locates poly(A) sites and signals. Here, we compare manual annotation of human chromosome 6 with annotation performed by EAnnot in order to assess the latter's accuracy. EAnnot can readily be applied to manual annotation of other eukaryotic genomes and can be used to rapidly obtain an automated gene set.
With any draft or finished genome sequence, a complete and accurate annotated gene set is the first step in transforming the raw sequence into meaningful biological knowledge. Gene sets can be generated either entirely computationally or by a combination of computational and manual annotation. While the former is useful in generating preliminary gene sets, manual annotation is still necessary for ensuring accuracy and completeness.
Recent work in gene prediction has focused on generating complete gene sets directly from genomic sequence. Some ab initio programs, such as Genscan (Burge and Karlin 1997) and Fgenesh (Salamov and Solovyev 2000) are based on intrinsic characteristics of coding sequence (e.g., codon usage, consensus splice sites, etc.) and require training on known genes from the organism. Others, like Twinscan (Korf et al. 2001) and SGP2 (Parra et al. 2003), use sequences conserved between closely related species. Although both of these approaches are becoming more accurate and sensitive, they suffer from certain shortcomings, particularly when dealing with vertebrate genes where exons are small and introns are large (Volfovsky et al. 2003). These programs often overpredict exons and genes, fuse neighboring genes, split genes, and miss exons or entire genes. Other features present problematic challenges as well, in particular, overlapping genes (Zhou and Blumberg 2003; Veeramachaneni et al. 2004), nested genes (Monani and Burghes 1996; Legare et al. 2000; Ponting et al. 2001), tandemly duplicated genes (Ferrier and Minguillon 2003), noncanonical splice sites (Shaw et al. 2003; Tschan et al. 2003), pseudogenes (Torrents et al. 2003), and alternative splicing (Zavolan et al. 2003). Some computational tools, like Ensembl (Birney et al. 2004), have incorporated experimental data into their gene-finding strategy. Although the Ensembl gene-building system has been refined and improved over several years, manual annotation has demonstrated that the accuracy and coverage of genes built by this system can be improved upon, especially with respect to alternative transcripts (Clamp et al. 2003).
Vertebrate genes are rather complex and experimental data appear to be the best source for their accurate identification. In response, the National Institutes of Health launched the Mammalian Gene Collection (MGC) Program in an effort to identify and sequence cDNA clones containing the complete coding sequence for each human and mouse gene (Strausberg et al. 1999). Similar projects have been established internationally (Bannasch et al. 2004; Ota et al. 2004). Large-scale EST sequencing projects have also been undertaken with the goal of representing a substantial fraction of all genes as EST sequences. As a result, a large number of transcribed sequences are publicly available and can be used to confirm and correct predicted genes and to identify genes omitted by gene-prediction programs. While mRNA sequences have been used as markers on the chromosome to locate known genes, EST sequences are useful in detecting novel genes and alternatively spliced forms of known genes.
To take advantage of the rapid growth of experimental data, we developed a computer program called EAnnot to build gene models based on mRNA, EST, and protein alignments, to annotate the supporting evidence, to locate poly(A) addition sites and signals, and to identify potential pseudogenes. EAnnot improves the accuracy of gene predictions by evaluating splice sites, adjusting gene models using protein evidence, making use of clone-linked EST reads, and locating missing exons via local alignments. EAnnot can be used not only to support manual annotation, but also as a computational gene-prediction tool for eukaryotic genomes.
Results
To evaluate the performance of EAnnot, we compared its predicted gene set with the manually annotated gene set of human chromosome 6 (Mungall et al. 2003). The human chromosome 6 gene set was chosen as a test case because it contains not only known genes, but also novel genes, splice variants, and poly(A) signals and sites. This enabled us to evaluate several aspects of EAnnot gene predictions. We also compared EAnnot predictions with several well-known gene-prediction programs using manual annotation as the standard.
Evaluation of EAnnot's gene prediction
We compared the gene set from the manual annotation of human chromosome 6 (build 31) (see Supplemental data) to EAnnot's gene set (see Supplemental data) and found a significant intersection of the two data sets from a comparison of gene boundaries (genomic coordinates of the ends of the genes) (Table 1). Manually annotated and EAnnot predicted gene sets can be viewed at http://gmod.wustl.edu/cgi-bin/gbrowse/human_chr6. Specifically, 87.6% of the manually annotated genes (not including pseudogenes) overlap 80.3% of the EAnnot-predicted genes, while 92.7% of the manually annotated transcripts overlap 91.9% of the EAnnot-predicted transcripts. Among the 193 manually annotated genes not predicted by EAnnot, 169 are novel genes and 24 are known genes, including nine histone genes. This is likely due to clustered repeats of histone genes. The majority of genes not annotated by EAnnot (70.3%) are single or two-exon genes. Similarly, among the 339 EAnnot genes that do not overlap any manually annotated genes (not including pseudogenes), 81.4% are single or two-exon genes. By examining the overlap between the EAnnot novel genes and the manually annotated pseudogenes, we found that 66 could be eliminated as potential pseudogenes. Although this indicates that the EAnnot novel genes must be evaluated individually, our poly(A) data (see below) suggests that some of these genes are likely novel genes identified only by EAnnot.
Table 1.
Comparison of EAnnot gene predictions with manual annotation of human chromosome 6
| Number of genes | Number of overlapping genes | Number of transcripts | Number of overlapping transcripts | |
|---|---|---|---|---|
| Manual | 1557 | 1364 (87.6%) | 3271 | 3033 (92.7%) |
| EAnnot | 1724 | 1385 (80.3%) | 5266 | 4842 (91.9%) |
Analysis does not include pseudogenes unless specified.
To determine the quality of the predicted genes, we compared splice sites in EAnnot-predicted and manually annotated genes. Predicted and manually annotated genes were considered identical if they had the same splice site coordinates, while the predicted gene was considered longer if it included all splice sites of the manually annotated gene but had additional 5′ or 3′ splice sites. We found that 68.2% of the manually annotated multiple exon genes have at least one identical or longer EAnnot prediction. Among the 20,724 manually annotated splice sites, 17,874 (86.2%) splice sites were present in the set of 20,901 splice sites found by EAnnot (Table 2).
Table 2.
Comparison of the accuracy of splice site prediction between different gene prediction programs using manual annotation as a standard
|
Number of identical splice sites
|
||
|---|---|---|
| Sensitivity | Specificity | |
| EAnnot | 86.2% (17874/20724) | 85.5% (17874/20901) |
| Ensembl | 84.4% (17489/20724) | 82.3% (17489/21258) |
| Fgenesh | 66.1% (13698/20724) | 35.6% (13698/38484) |
| Genscan | 62.1% (12870/20724) | 30.3% (12870/42538) |
(Sensitivity) Number predicted correctly over total number annotated manually.
(Specificity) Number predicted correctly over total number predicted.
To further evaluate the performance of EAnnot, we compared EAnnot predictions with Ensembl, Genscan, and Fgenesh predictions using manual annotation as a standard. While Genscan and Fgenesh are ab initio programs, Ensembl takes into account experimental data, a feature shared with EAnnot. Ensembl predicted 1037 known genes with 1798 transcripts and 1457 EST genes with 2308 transcripts for chromosome 6 (build 31), while Fgenesh and Genscan predicted 6230 and 6225 genes, respectively. We evaluated the performance of each program with respect to splice sites, transcripts, and genes across all of chromosome 6.
First, we examined how many splice sites were identical among each set of predictions and manual annotation. We took the complete set of manually annotated splice sites and searched for individual matches among the predicted gene sets (Table 2). EAnnot and Ensembl correctly predicted 86.2% and 84.4% of the splice sites with a specificity of 85.5% and 82.3% respectively, while Fgenesh and Genscan had lower sensitivity and appreciably lower specificity as compared with EAnnot and Ensembl (Table 2).
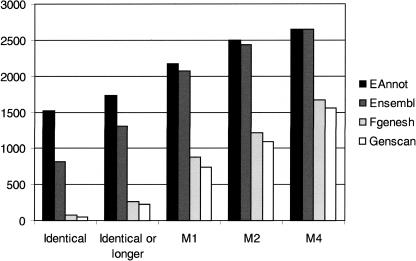
In order to evaluate predictions at the transcript level, we took 3092 manually annotated spliced transcripts and searched for identical or longer transcripts among the predicted gene sets. Since EAnnot uses clone-linking information (see Methods) to build gene models, some EAnnot models contain additional 5′ or 3′ exons compared with manually annotated transcripts. These will be counted as longer transcripts. EAnnot predicted 1524 (49%) identical transcripts and 205 longer transcripts when compared with manual annotation, while Ensembl (build 31) predicted 817 (27%) identical transcripts and 488 longer transcripts compared with manual annotation (Fig. 1). Among the 6230 genes predicted by Fgenesh, 72 have the identical splicing pattern as manually annotated transcripts and 197 are longer. Genscan predicted 47 identical transcripts and 185 identical, but longer transcripts. When we allow transcripts with missing maximum of four splice sites, EAnnot, Ensemble, Fgenesh, and Genscan predicted 85.8%, 85.5%, 54.1%, and 50.3% respectively (Fig. 1).
Figure 1.
Comparison of transcripts predicted by EAnnot, Ensembl, Fgenesh, and Genscan with manual annotation. (Identical) Predictions with identical splice sites (coordinates, order, and number) compared with manual annotation; (Identical or longer) predictions with or including identical splice sites (coordinates, order, and number) compared with manual annotation; (M1) predictions missing a maximum of 1 splice site; (M2) prediction missing a maximum of 2 splice sites; (M4) predictions missing a maximum of 4 splice sites. Missing splice site can be due to incorrect splice site present or a predicted form being shorter than manually annotated form.
To evaluate the accuracy of predictions at the gene level, we examined the number of manually annotated genes with at least one identical transcript identified by each prediction program (see Supplemental Table 1). Of the 1411 multiple exon genes on chromosome 6, EAnnot identified at least one identical transcript for 64.7% of the genes, while Ensembl identified at least one identical transcript for 48.1% of the genes. Fgenesh and Genscan identified at least one identical transcript for only 5.1% and 3.8% of the genes, respectively. When we examined genes with transcripts missing a maximum of four splice sites, EAnnot, Ensemble, Fgenesh, and Genscan predicted 82.1%, 78.6%, 55.6%, and 52.6% such genes, respectively.
Analysis of EAnnot's splice variant prediction
EAnnot is designed to specifically detect alternative splicing events using experimental data. The average number of transcripts per gene on chromosome 6 predicted by EAnnot is 3.05 compared with 2.1 transcripts per gene in the manually annotated data set. EAnnot identified 20901 splice sites, only 177 more than identified by manual annotation, but produced almost one more splice form per gene on average. This appears to be because most novel splice variants do not use new splice sites, but instead use different combinations of splice sites (exons).
Since EAnnot predicts, on average, one more splice form per gene compared with manual annotation, we manually evaluated the quality of the splice variants in a subset of 15 genes and 101 transcripts from chromosome 6. Each of the randomly selected genes included between four and 16 EAnnot-predicted transcripts (Table 3). Transcripts were classified as correct or incorrect on the basis of the manual evaluation of the alternatively spliced junctions using the underlying evidence. EAnnot identified 82.1% of the splice variants correctly. An additional 13.8% of the gene models were intron-retension variants, whose biological significance is questionable (Kan et al. 2002).
Table 3.
Manual evaluation of 101 predicted transcripts from 15 genes
|
Number of correct predictions
|
||||
|---|---|---|---|---|
| Gene name | Number of EAnnot predictions | Standard variants | Intron retension variants | Number of incorrect predictions |
| QRSL1 | 5 | 4 | 0 | 1 |
| RTN4IP1 | 7 | 5 | 2 | 0 |
| BXDC1 | 5 | 5 | 0 | 0 |
| CD164 | 7 | 6 | 1 | 0 |
| FYN | 12 | 8 | 4 | 0 |
| SNX3 | 5 | 5 | 0 | 0 |
| TUBE1 | 5 | 2 | 2 | 1 |
| PKIB | 7 | 5 | 2 | 0 |
| RWDD1 | 6 | 5 | 1 | 0 |
| NICAL | 16 | 13 | 3 | 0 |
| SSR1 | 8 | 6 | 0 | 2 |
| CAGE1 | 5 | 5 | 0 | 0 |
| TXNDC5 | 4 | 4 | 0 | 0 |
| PECI | 7 | 6 | 1 | 0 |
| CDYL | 4 | 4 | 0 | 0 |
| Total | 101 | 83 | 14 | 4 |
| 100.00% | 82.18% | 13.86% | 3.96% | |
Analysis of EAnnot's poly(A) prediction
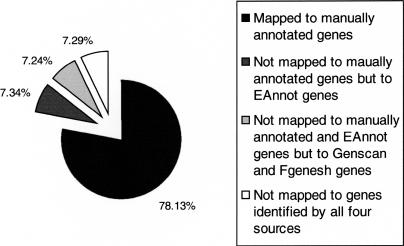
Annotation of a poly(A) site defines the 3′ boundary of the gene. EAnnot uses ESTs and mRNAs with terminal poly(A) runs to locate poly(A) sites and further maps them onto genomic sequence for annotation purpose. EAnnot's poly(A) module identified 2181 unique poly(A) sites and 2512 associated poly(A) signals as compared with the 1303 poly(A) sites and 1211 poly(A) signals annotated manually on chromosome 6. The difference between the number of EAnnot-predicted poly(A) sites and signals is due to more than one poly(A) signal being associated with some of the poly(A) sites, while in the manual annotation, some of the genes have poly(A) sites without annotation of the corresponding poly(A) signals. Among the poly(A) sites identified by EAnnot, 1704 (78.1%) map within 5 kb downstream from the 3′ end of 923 manually annotated genes. This result demonstrates that poly(A) signals and sites predicted by EAnnot are useful in anchoring the majority of manually annotated genes. Of 477 poly(A) sites not mapped to manually annotated genes, 160 mapped within 5 kb of the 3′ end of EAnnot annotated genes. As a result, 119 EAnnot genes that were not included in manual annotation or were longer at their 3′ end were supported with one or more poly(A) sites. Among the 317 poly(A) sites not associated with either EAnnot or manually annotated genes, 158 were associated with Fgenesh or Genscan genes (Fig. 2). This indicates that ab initio gene-prediction programs will continue to be helpful in locating potential novel genes when expression data are absent or rare. We found 69 poly(A) sites that are supported by multiple ESTs, but that are not associated with either manually annotated genes or any of the gene-prediction programs used in this study. We speculate these may come from the 3′ end of unidentified genes. This group of poly(A) signals and sites may serve as anchors for novel genes yet to be discovered. Experimentally, they can be also used as tags to perform 5′ RACE (Rapid Amplification of cDNA ends) to determine the coding sequence of such genes.
Figure 2.
Poly(A) signals and sites identified by EAnnot serve as anchors for genes. A total of 1704 (78.1%) poly(A) sites identified by EAnnot are mapped within 5 kb from the 3′ end of manually annotated chromosome 6 genes. Among the ones not mapped to manually annotated genes, 7.3% are mapped to EAnnot genes. Among those mapped to neither manually annotated nor EAnnot genes, 7.2% are mapped to Fgenesh or Genscan genes; 7.3% are not mapped to any genes identified by the above four gene prediction methods.
Many of the manually annotated genes (440, 28.2%) on chromosome 6 have multiple poly(A) signal/site pairs identified by EAnnot (Table 4). The known genes, LAMA4, PTPRK, QKI, SH3BGRL2, and STX7, all have seven or more poly(A) signal and site pairs. In many cases, different poly(A) pair sets are associated with different alternatively spliced forms. Two splice variants of the QKI gene having terminal exons about 6 kb apart clearly show distinct poly(A) signals and sites. This suggests that differential addition of the poly(A) tail might affect mRNA splicing of the QKI gene, leading to the creation of different splice forms of the QKI gene.
Table 4.
Number of poly-A-signals and sites for manually annotated genes on human chromosome 6
| Number of polyA signals/sites | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
| Number of genes | 483 | 225 | 109 | 56 | 29 | 10 | 6 | 5 |
Analysis of EAnnot gene model annotation
The coding sequence (CDS) was assigned to 3513 predicted chromosome 6 transcripts (66.7% of 5266 transcripts). Of these assignments, 1212 were based on protein alignment of underlying cDNA translation from the GenBank by EAnnot, while 2301 were based on GeneMark calling the longest open reading frame (ORF) in EAnnot models and nonredundant protein database screening. EAnnot first annotates the coding sequence according to predictions using protein evidence. If there is a discrepancy between an EAnnot gene model and the available protein evidence, EAnnot will adjust the gene model as suggested by the supporting protein evidence. If the difference cannot be resolved, EAnnot reports the nature and the position of the difference. For 146 gene models on chromosome 6, EAnnot reported differences between the gene models and the protein evidence. This information is useful to annotators who might be able to resolve the discrepancy and is important for identifying potential EAnnot errors, GenBank mRNA or EST sequence errors, and genomic sequence errors.
In order to estimate how many of the CDS are called correctly, we used BLASTP (W. Gish [1996–2004], http://blast.wustl.edu) to compare our results to the manually annotated chromosome 6 protein database and a nonredundant protein database (NR). For the 1212 proteins predicted by EAnnot based on protein evidence, 725 (60%) have an identical match (100% identity and 100% coverage) to proteins from manually annotated genes, while 871 (72%) have partial matches (>98% identity and >50% coverage). If the same data set is searched against NR, 914 (75%) proteins have identical matches, while 1115 (92%) have partial matches. Some of the forms that didn't have an identical or partial match against the manually annotated protein database are still valid, as shown by a search against the NR database.
EAnnot attaches the supporting evidence used to build the gene model. This includes EST, cDNA, and protein evidence. This allows users to review the supporting evidence, make their own evaluation, and obtain additional information, for example, EST libraries and tissue sources. The number of ESTs and cDNAs supporting a gene model is also an indication of the expression level of individual transcripts. On chromosome 6, 51.6% of gene models (in the EAnnot gene prediction dataset) are supported by multiple pieces of evidence. The fact that ∼48.4% of models are only supported by a single piece of evidence suggests that EAnnot is very sensitive in detecting rare transcripts. Beside attaching supporting evidence, EAnnot assigns predicted genes with a symbol and remark from LocusLink (Pruitt and Maglott 2001) if available. Both the symbol and remark are assigned to all splice variants from the same locus.
For chromosome 6, 101 models were extended based on EST clone-linking information (see Methods). There are several benefits to using clone-linking information. The strand assignment of some nonspliced EST reads can be determined based on the strand assignment of the corresponding clone-linked spliced read. Sometimes an expanded gene boundary can be created using nonoverlapping reads based on their clone origins. When ESTs from the same clone do overlap, a 5′ or 3′ extended gene model can be created using the mapping of ESTs from the same clone.
EAnnot also annotates both processed and unprocessed pseudogenes. Three sets of pseudogenes were created by EAnnot using distinct sets of parameters. EAnnot predicted 267 pseudogenes using parameters listed in Table 5. Among the 267 pseudogenes EAnnot identified, 197 of them have truncated coding sequence due to frameshifts or early termination codons. When we lowered the mRNA identity threshold to 80% and allowed the accumulative gap to be 50 bp, EAnnot predicted 418 pseudogenes. Lowering the threshold even further, mRNA identity to 75% and accumulative gap to 100 bp, EAnnot predicted 514 pseudogenes. This indicates that we could use our empirically established parameters (Table 5) to create an initial gene set and a set of lowered thresholds to capture more pseudogenes.
Table 5.
Percent identity and gap size thresholds for ESTs and mRNAs
| EST | EST | EST | EST | mRNA | mRNA | |
|---|---|---|---|---|---|---|
| Percent identity | 100 | 100 > id ≥ 97 | 97 > id ≥ 90 | 90 > id ≥ 85 | ≥92 | 92 > id ≥ 85 |
| Cumulative gap size (bp) | ≤20 | <15 | <5 | ≤2 | <200 | <10 |
| Local repeatitiveness (bp) | ≤200 | ≤200 | ≤200 | ≤200 | ≤400 | ≤400 |
Discussion
High-quality annotation is essential in deciphering the growing number of sequenced genomes. Automated annotation greatly facilitates the speed of annotation and reduces variability. There are various good gene-prediction programs currently in use. However, what is lacking is an annotation system with integrated functionality including predicting genes and splice variants, capturing partial genes, identifying poly(A) signals and sites, and annotating supporting evidence. To meet this need, we developed EAnnot. This system was originally designed for automating human genome annotation according to HAWK (http://www.sanger.ac.uk/HGP/havana/hawk.shtml) standards. We have since expanded its functionality to annotate other eukaryotic genomes.
To efficiently and accurately identify genes using all available experimental data, EAnnot program defines a gene boundary based on the collective evidence of strand assignment, sequence overlapping of EST, mRNA, and protein and clone-linking of EST. This feature is unique among existing gene prediction and annotation programs and provides the foundation for improved accuracy of gene boundary prediction. Following gene boundary determination, EAnnot clusters sequences within the defined gene boundary to identify different splicing events. The clustering combines all mRNAs and ESTs in one process to accurately predict the number of unique variants per gene. Manual inspections suggest that EAnnot's alternative splice form prediction is sensitive and accurate (Table 3).
To provide further biological information, EAnnot assigns supporting evidence in the form of proteins, mRNAs, and ESTs. This feature enables independent evaluation and analysis. In addition, it allows straightforward tracking of EST tissue origin via their GenBank entries. This feature should benefit researchers interested in tissue distribution, developmental expression profiles, and the pathological changes in the expression of a gene and its splice forms.
Another unique aspect of EAnnot is its use of spliced and nonspliced ESTs and mRNAs with poly(A) tails to identify poly(A) sites and search for corresponding signals in genomic sequence. This not only locates the 3′ end of the gene, but also tags potential novel genes supported solely by poly(A) sequences identified by EAnnot. The discovery of a substantial number of genes with multiple poly(A) signals and sites sets the stage for future studies on the regulation of RNA poly(A) adenylation.
Using manual annotation as the standard, we have demonstrated that EAnnot predicts gene models with high confidence and accuracy compared with several existing gene-prediction programs. In addition, EAnnot is able to discover splice variants of genes, which ab initio gene-prediction programs cannot. Even though both the Ensembl system and EAnnot use experimental evidence to build gene models, EAnnot considers all mRNAs and ESTs as a whole in building a common set of gene models, while Ensembl separates mRNA and EST data to build separate sets of models. Consequently, EAnnot is able to build nonredundant gene models, predict splice variants for each gene, and predict the total number of genes in a given genome. Conversely, some Ensembl transcripts and EST transcripts are redundant. Furthermore, EAnnot is able to make more complete models using the combination of underlying mRNA and EST evidence. Beside using mRNA and EST evidence, EAnnot also uses available protein evidence to fine-tune gene models and improve accuracy.
EAnnot is designed to be versatile. By adjusting the parameters, we have been successfully using EAnnot to annotate other vertebrate and invertebrate genomes. For genomes with minimum experimental data, we rely extensively on the use of cross-species alignments with adjusted thresholds. In addition, we set an option of incorporating protein evidence into EAnnot's gene boundary determination and transcript clustering processes, which further improves the accuracy of gene boundaries and gene models for certain genomes. Due to the abundance of vertebrate mRNA data, we intentionally omitted protein evidence in determining gene boundaries and clustering transcripts for the human annotation effort. However, we did use protein evidence to capture single exon repeat gene families, such as the histone family. To annotate genomes with rare splice variants, we recently implemented evidence merging in order to build the most representative and complete model for each gene. In this process, the predominant splice pattern among evidence at any given splice site within a gene boundary is selected to construct the merged gene model. These adjustments broaden EAnnot's applicability to virtually any sequenced genome, vertebrate, and invertebrate, with or without abundant experimental evidence.
We have demonstrated the effectiveness and the advantages of the EAnnot system in the annotation of the human genome. EAnnot will continue to be used in this capacity, but its use will also be expanded for annotating other organisms. We envision that by implementing an ab initio gene-prediction algorithm into EAnnot, we will be able to extend partial, evidence-based genes into full-length genes and uncover novel genes not revealed by existing evidence.
Methods
Data used by EAnnot
The genomic sequence can be clone, contig, or chromosome based. EAnnot can assemble the sequence and alignments from clones into contigs or chromosomes using an AGP file, facilitating the annotation of genes spanning more than one clone. Gene prediction and annotation are performed at the contig or chromosome level and can be transformed to clone-based coordinates to facilitate viewing. The primary EAnnot input is mRNA, EST, and protein alignments to genomic sequence, in the ACEDB “ace” database format, generated by BLASTN, BLASTX (Altschul et al. 1997) (W. Gish [1996–2004], http://blast.wustl.edu), est2genome (Mott 1997), or other alignment tools.
EAnnot uses clone-linked EST read information (EST reads from the same clone) to help establish gene boundaries (see below). ESTs were downloaded from NCBI (ftp://ftp.ncbi.nih.gov/genbank), and a clone-linking table was generated for multiple reads in which the organism of origin and clone IDs were identical. All reads from clones with more than five clone-linked reads were disregarded. Also, clone IDs containing less than three characters or only numbers were not considered valid IDs and were not used to link reads. These conservative criteria were used to avoid false linking. The clone-linking table generated from data downloaded from NCBI on July 7, 2003 contained 3,615,915 ESTs from 1,770,607 clones or ∼21% of the ESTs in dbEST.
EAnnot uses information from LocusLink to assign gene symbols and to help identify pseudogenes. Gene symbols, descriptions, and chromosomal locations are extracted from the LocusLink LL_tmpl file (ftp://ftp/ncbi.nih.gov/refseq/LocusLink/) into the locuslink.sel table used by EAnnot.
Alignments
EAnnot begins by examining the quality of all alignments and decides which will be used, modified, or disregarded based on a set of adjustable, empirically established parameters, that is, percent identity, continuity, and local repetitiveness (Table 5). Percent identity is the percent of bases or amino acids in the mRNA, EST, or protein alignment that identically match the genomic sequence, or its translation. Continuity refers to the percentage of total bases or amino acids included in the alignments. Local repetitiveness refers to whether any part of the mRNA, EST, or protein aligns to more than one location in the contig or chromosome. EAnnot attempts to correct gaps in alignments by doing local BLASTN searches and looking for nearby splice sites using SplicePredictor (Usuka et al. 2000), evaluating intron size, and determining whether there are other alignments in the area. Repetitive alignments are evaluated based on percent identity, sequence order and orientation, and the use of consensus splice sites. While evaluating each alignment, EAnnot also assigns each EST and mRNA alignment to the appropriate strand based on read orientation and splice-site sequences. Every splice site is examined to see whether the best consensus splice site resides on the plus or minus strand. If a strand assignment cannot be made, EAnnot will omit those ESTs and mRNAs from further analysis. Nonspliced ESTs without a strand assignment are not used for gene prediction, but are used for the identification of poly(A) signals and sites (see below).
Gene boundaries
After evaluating and modifying alignments, EAnnot sets the boundaries it will use to predict genes and alternatively spliced forms. Gene boundaries are set at the ends of alignments and extended using overlapping alignments on the same strand. Boundaries are also extended with nonoverlapping, but clone-linked reads in the clone-linking table described above.
Clusters and gene models
Within each gene boundary, EAnnot clusters mRNAs to create unique transcripts, each representing an alternatively spliced form. It only uses spliced mRNAs (those that span an intron), unless more than one GenBank entry indicates no splicing. It compares each spliced EST mapped within the gene boundary with each mRNA cluster, and either clusters the EST with an mRNA cluster or creates novel EST clusters. If no mRNA alignments map within a gene boundary, then ESTs are clustered. Nonredundant mRNA clusters and novel EST clusters are then used to generate gene models by merging all of the members of a cluster into the longest form. EAnnot only creates gene models based on EST and mRNA sequences from each cluster. It does not attempt to merge clusters within the same gene boundary to avoid building chimeric transcripts for vertebrate genomes. Each mRNA and EST in a cluster is attached to the resulting gene model as supporting evidence (see below).
EAnnot makes three types of gene models; transcripts with a CDS, transcripts in which a CDS could not be determined, and pseudogenes. Each gene can have one or more gene models. Each gene model is translated and aligned with the protein corresponding to the mRNA in the cluster that defined the gene model using FASTX (Pearson et al. 1997). The alignment is used to determine the initiator methionine and stop codon. Any mismatches, frame shifts, deletions, or insertions are identified. Mismatches are allowed due to polymorphisms and differences between species. Frame shifts, deletions, and insertions are further analyzed. EAnnot will adjust the gene model if a nearby consensus splice can be found that removes the frame shift, deletion, or insertion. All other gene models within the gene boundary will be adjusted to use this consensus splice site. GeneMark (Lukashin and Borodovsky 1998) is used to find ORFs for gene models without CDS assignments. Only ORFs >100 nucleotides and having the best hit with the P value <1e-15 against nonredundant protein database (NR) are kept. Genes are annotated as pseudogenes if the chromosomal location is incorrect (obtained from LocusLink). The information regarding frame shifts and incomplete coding sequence is attached to pseudogenes.
To predict additional single exon genes that could have been missed during the clustering process, EAnnot re-examines all protein alignments outside of the identified gene boundaries and makes additional single exon gene models accordingly.
Annotation
EAnnot uses information generated during the prediction process, as well as information from LocusLink and GenBank entries to annotate genes. All of the mRNAs and ESTs used in clustering are attached as supporting evidence. Proteins are attached if their corresponding mRNAs are used as supporting evidence. Each full-length mRNA can only be attached to one gene model. Partial mRNAs and ESTs are attached as supporting evidence to all gene models with a splicing pattern that matches the gene model. Locus information (locus name and description) from LocusLink is also added. If one gene model within a gene boundary is assigned locus information, then all the gene models within that same boundary will adopt the same symbol and description. Information about poly(A) sites and signals is also annotated (see below).
Poly(A) sites and signals
EAnnot has a native module for identifying poly(A) signals and sites. All ESTs and mRNAs (spliced and nonspliced) are examined for runs of As at the 3′ end of the sequence, or runs of Ts at the 5′ end of the sequence in case of reverse reads. The presence of at least eight As or Ts among the last or first 10 bases of the sequence is required before further analysis is initiated. EAnnot requires at least five As or Ts at either end of the sequence not mapped to genomic sequence. Once poly(A) site is determined, poly(A) signals (AATAAA or ATTAAA) will be searched within 50 bp upstream from the poly(A) adenylation site (Beaudoing et al. 2000). If the poly(A) signal is found, the poly(A) sites and signals will be assigned in pairs.
Comparison with human chromosome 6 manual annotation
To eliminate potential discrepancies due to differences in the underlying data (mRNA, EST, and protein libraries) and differences in alignments, EAnnot predictions were based on the same alignments used by the Sanger Institute when doing manual annotation of human chromosome 6 (Mungall et al. 2003). The alignments were accessed from a local ACEDB database (Walsh et al. 1998). The manually annotated gene models from chromosome 6 were obtained from the Sanger Institute in the ACEDB “ace” file format, and were converted from clonal to chromosomal coordinates based on human build 31 AGP file (Supplemental data). We divided chromosome 6 sequence into 20 supercontigs according to the same AGP file. The average size of the supercontig was 11,435,641 bp. The largest supercontig was 19,934,087 bp.
Software and Ensembl data
Genscan and Fgenesh (v.2.0) were installed locally. Ensembl genes and transcripts and Ensembl EST genes and transcripts (build 31) were downloaded from http://atlas.cnio.es/.
Acknowledgments
We thank Michael Wendl for his helpful discussions and comments on this work. We are grateful to scientists of the Wellcome Trust Sanger Institute for providing the mapping of the vertebrate data and the manual annotation of human chromosome 6, especially Jennifer Ashurst, Tim Hubbard, James Gilbert, and Stephen Keenan. This work was supported by a grant from the National Human Genome Research Institute (HG002042, principal investigator, R.K.W.).
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3152604.
Footnotes
[Supplemental material available online at www.genome.org and http://genome.wustl.edu/analysis/EAnnot.]
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannasch, D., Mehrle, A., Glatting, K.H., Pepperkok, R., Poustka, A., and Wiemann, S. 2004. LIFEdb: A database for functional genomics experiments integrating information from external sources, and serving as a sample tracking system. Nucleic Acids Res. 32: D505-D508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoing, E., Freier, S., Wyatt, J.R., Claverie, J.M., and Gautheret, D. 2000. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 10: 1001-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney, E., Andrews, T.D., Bevan, P., Caccamo, M., Chen, Y., Clarke, L., Coates, G., Cuff, J., Curwen, V., Cutts, T., et al. 2004. An overview of Ensembl. Genome Res. 14: 925-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge, C. and Karlin, S. 1997. Prediction of complete gene structure in human genomic DNA. J. Mol. Biol. 268: 78-94. [DOI] [PubMed] [Google Scholar]
- Clamp, M., Andrews, D., Barker, D., Bevan, P., Cameron, G., Chen, Y., Clark, L., Cox, T., Cuff, J., Curwen, V., et al. 2003. Ensembl 2002: Accommodating comparative genomics. Nucleic Acids Res. 31: 38-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrier, D.E. and Minguillon, C. 2003. Evolution of the Hox/ParaHox gene clusters. Int. J. Dev. Biol. 47: 605-611. [PubMed] [Google Scholar]
- Kan, Z., States, D., and Gish, W. 2002. Selecting for functional alternative splices in ESTs. Genome Res. 12: 1837-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf, I., Flicek, P., Duan, D., and Brent, M.R. 2001. Integrating genomic homology into gene structure prediction. Bioinformatics 17: S140-S148. [DOI] [PubMed] [Google Scholar]
- Legare, M.E., Bartlett II, F.S., and Frankel, W.N. 2000. A major effect QTL determined by multiple genes in epileptic EL mice. Genome Res. 10: 42-48. [PMC free article] [PubMed] [Google Scholar]
- Lukashin, A.V. and Borodovsky, M. 1998. GeneMark.hmm: New solutions for gene finding. Nucleic Acids Res. 26: 1107-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monani, U. and Burghes, A.H. 1996. Structure of the human α 2 subunit gene of the glycine receptor—Use of vectorette and Alu-exon PCR. Genome Res. 6: 1200-1206. [DOI] [PubMed] [Google Scholar]
- Mott, R. 1997. EST_GENOME: A program to align spliced DNA sequences to unspliced genomic DNA. Comput. Appl. Biosci. 13: 477-478. [DOI] [PubMed] [Google Scholar]
- Mungall, A.J., Palmer, S.A., Sims, S.K., Edwards, C.A., Ashurst, J.L., Wilming, L., Jones, M.C., Horton, R., Hunt, S.E., Scott, C.E., et al. 2003. The DNA sequence and analysis of human chromosome 6. Nature 425: 805-811. [DOI] [PubMed] [Google Scholar]
- Ota, T., Suzuki, Y., Nishikawa, T., Otsuki, T., Sugiyama, T., Irie, R., Wakamatsu, A., Hayashi, K., Sato, H., Nagai, K., et al. 2004. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat. Genet. 36: 40-45. [DOI] [PubMed] [Google Scholar]
- Parra, G., Agarwal, P., Abril, J.F., Wiehe, T., Fickett, J.W., and Guigo, R. 2003. Comparative gene prediction in human and mouse. Genome Res. 13: 108-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson, W.R., Wood, T., Zhang, Z., and Miller, W. 1997. Comparison of DNA sequences with protein sequences. Genomics 46: 24-36. [DOI] [PubMed] [Google Scholar]
- Ponting, C.P., Mott, R., Bork, P., and Copley, R.R. 2001. Novel protein domains and repeats in Drosophila melanogaster: Insights into structure, function, and evolution. Genome Res. 11: 1996-2008. [DOI] [PubMed] [Google Scholar]
- Pruitt, K.D. and Maglott, D.R. 2001. RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res. 29: 137-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salamov, A.A. and Solovyev, V.V. 2000. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 10: 516-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw, M.A., Brunetti-Pierri, N., Kadasi, L., Kovacova, V., Van Maldergem, L., De Brasi, D., Salerno, M., and Gecz, J. 2003. Identification of three novel SEDL mutations, including mutation in the rare, non-canonical splice site of exon 4. Clin. Genet. 64: 235-242. [DOI] [PubMed] [Google Scholar]
- Strausberg, R.L., Feingold, E.A., Klausner, R.D., and Collins, F.S. 1999. The mammalian gene collection. Science 286: 455-457. [DOI] [PubMed] [Google Scholar]
- Torrents, D., Suyama, M., Zdobnov, E., and Bork, P. 2003. A genome-wide survey of human pseudogenes. Genome Res. 13: 2559-2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschan, M.P., Fischer, K.M., Fung, V.S., Pirnia, F., Borner, M.M., Fey, M.F., Tobler, A., and Torbett, B.E. 2003. Alternative splicing of the human cyclin D-binding Myb-like protein (hDMP1) yields a truncated protein isoform that alters macrophage differentiation patterns. J. Biol. Chem. 278: 42750-42760. [DOI] [PubMed] [Google Scholar]
- Usuka, J., Zhu, W., and Brendel, V. 2000. Optimal spliced alignment of homologous cDNA to a genomic DNA template. Bioinformatics 16: 203-211. [DOI] [PubMed] [Google Scholar]
- Veeramachaneni, V., Makalowski, W., Galdzicki, M., Sood, R., and Makalowska, I. 2004. Mammalian overlapping genes: The comparative perspective. Genome Res. 14: 280-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volfovsky, N., Haas, B.J., and Salzberg, S.L. 2003. Computational discovery of internal micro-exons. Genome Res. 13: 1216-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh, S., Anderson, M., and Cartinhour, S.W. 1998. ACEDB: A database for genome information. Meth. Biochem. Anal. 39: 299-318. [DOI] [PubMed] [Google Scholar]
- Zavolan, M., Kondo, S., Schonbach, C., Adachi, J., Hume, D.A., Hayashizaki, Y., and Gaasterland, T. 2003. Impact of alternative initiation, splicing, and termination on the diversity of the mRNA transcripts encoded by the mouse transcriptome. Genome Res. 13: 1290-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, C. and Blumberg, B. 2003. Overlapping gene structure of human VLCAD and DLG4. Gene 305: 161-166. [DOI] [PubMed] [Google Scholar]
Web site references
- http://gmod.wustl.edu/cgi-bin/gbrowse/human_chr6; Gbrowse view of manual and EAnnot gene sets.
- http://blast.wustl.edu; BLAST
- http://atlas.cnio.es/; Ensembl mirror site.
- http://www.sanger.ac.uk/HGP/havana/hawk.shtml; HAWK.
- http://genome.wustl.edu/analysis/EAnnot; EAnnot program and associated files.
- ftp://ftp.ncbi.nih.gov/refseg/LocusLink; LocusLink.
- ftp://ftp.ncbi.nih.gov/genbank/; major resource for download of ESTS from NCBI.