Summary
Background
Gastric cancer (GC) is one of the most common cancers in the world; however, chemoresistance greatly decreases the efficacy of therapy in gastric cancer. Long noncoding RNAs (IncRNAs) participate in a variety of biological processes, and we hypothesize that lncRNA HULC regulates the multidrug resistance in GC treatment.
Methods
We obtained GC tissue samples from 42 GC patients and detected the expression level of HULC in the plasma and tissues via qRT-PCR. The relationship between HULC expression and survival rate was confirmed by Kaplan-Meier survival analysis. We verified the expression of HULC in GC cell lines via qRT-PCR, and the function of HULC was detected via flow cytometry assay and CCK-8 assay.
Results
HULC was highly expressed in the plasma and tissues of the GC patients compared with controls, with HULC high expression indicating lower survival rate. HULC knockdown enhanced cisplatin-induced apoptosis in GC cells.
Conclusions
Our results suggest that silencing lncRNA HULC could enhance chemotherapy induced apoptosis in GC cells, which could provide a novel approach for therapeutic strategies.
Keywords: LncRNA, HULC, gastric cancer, multidrug resistance
Kratak sadržaj
Uvod
Karcinom želuca jedan je od najčešćih kancera na svetu. Međutim, rezistencija na hemoterapiju umnogome smanjuje efikasnost lečenja karcinoma želuca. Dugi neko-dirajući RNK (lncRNK) učestvuju u mnogim biološkim pro-cesima i naša je hipoteza da lncRNK HULC reguliše rezistenciju na lekove u terapiji za karcinom želuca.
Metode
Uzeli smo uzorke tkiva karcinoma želuca od 42 obolelih i detektovali nivo ekspresije HULC u plazmi i tkivima putem qRT-PCR. Odnos između ekspresije HULC i stope preživljavanja potvrđen je pomoću Kaplan-Majerove analize preživljavanja. Ekspresiju HULC u čelijskim linijama karcinoma želuca verifikovali smo pomoću qRT-PCR, dok je funkcija HUMC detektovana protočnom citometrijom i testom CCK-8.
Rezultati
HULC je bio veoma izražen u plazmi i tkivima obolelih od karcinoma želuca u poređenju s kontrolom, dok je visoka ekspresija HULC ukazivala na nižu stopu preživljavanja. Pad vrednosti HULC uticao je na pojačanu apoptozu izazvanu cisplatinom u ćelijama karcinoma želuca.
Zaključak
Naši rezultati pokazuju da bi prigušivanje lncRNK HULC moglo podstaći apoptozu izazvanu hemoterapijom u ćelijama karcinoma želuca, što bi moglo otvoriti nov pristup za terapijske strategije.
Introduction
Gastric cancer is one most common cancers in the world (1), and chemotherapy is an important strategy against gastric cancer (2). Because multiple chemotherapeutic drugs are used to treat gastric cancer, the sensitivity of anticancer drugs to gastric cancer gradually decreases allowing resistance to many different drugs with various chemical structures and mechanisms of action to occur. This type of chemoresistance has remarkably decreased the efficacy of therapy in gastric cancer. Chemoresistance is becoming a major problem for the successful treatment of gastric cancer (3).
LncRNAs, abundantly transcribed by the mammalian genome, range in size from 200 to 10,000 nucleotides (4, 5). LncRNAs are misregulated in a wide range of human diseases and disorders, including various types of cancer. For example, increased expression of the lncRNA MALAT-1 indicates poorer prognosis in patients with lung cancer (6). LncRNAs PCGEM (7) and DD3 (8) are overexpressed in prostate cancer tissues and both lncRNAs are involved in prostate tumorigenesis (9). BC200RNA overexpression is correlated with the progression of breast tumors and proposed as a potential novel biomarker for breast cancer (10). Taken together, these observations provide evidence and support for the potential roles of lncRNAs in tumor development and progression.
Highly upregulated in liver cancer (HULC) was first confirmed from a hepatocellular carcinoma (HCC)-specific gene library as a novel lncRNA. HULC is aberrantly expressed (11, 12) and associated with the molecular pathogenesis of HCC (13), and its potential as a diagnostic biomarker has been proposed (14, 15). However, the role of HULC expression in patients with gastric cancer (GC) has not been examined. In addition, the function of HULC in GC MDR remains unclear.
In this study, we confirmed that lncRNA HULC is present in the plasma of GC patients and the plasma HULC level is correlated with the overall 5-year survival rate in GC patients. The expression level of HULC was greatly elevated in GC tumor tissues compared with the control group, and the expression level of HULC was associated with the degree of malignancy in GC cell lines. The significance of our study is to reveal that silencing of lncRNA HULC enhances chemotherapy sensitivity in GC cells, and this novel finding would shed light on developing new therapy strategies for GC cancer.
Materials and Methods
Patients and clinical specimens
This study was approved by the institutional review board of Qingdao University Medical College. GC tissue samples from 42 GC patients were obtained from Department of Thyroid Surgery in Gastrointestinal Surgery, the Affiliated Yantai Yuhu-angding Hospital of Qingdao University Medical College. Informed consents were signed by all subjects enrolled in this study. Subjects were recruited between Sep. 2006 and Sep. 2009, and patients with other known tumors and those receiving medical therapy and/or previous cancer therapies were excluded from the study.
Plasma samples were obtained from 42 GC patients and 25 volunteers and stored in Eppendorf tubes. Plasma was collected and centrifuged using FitAmp Plasma/Serum DNA Isolation Kit (Epigentek Group Inc., USA) according to the manufacturer′s instructions and stored at -80 °C.
Cell culture
The normal gastric epithelial cells GES-1, and human gastric adenocarcinoma MGC-803 and MKN-45 cells were purchased from Beijing Zhongyuan Ltd, China. All cells were cultured in high glucose DMEM medium supplemented with 10% FBS and antibiotics (100 U/mL penicillin and streptomycin) with 5% CO2 in a humidified incubator at 37 °C.
Drugs
Cisdiamminedichloroplatinum (CDDP) (Qilu Pharmo Co. Ltd, China) was resuspended in phosphate-buffered saline (PBS) (1 mg/mL) and stored at -20 °C. Adriamycin (ADR) (Sigma-Aldrich, USA) was diluted in PBS (2 mg/mL). 5-fluorouracil (5-FU) (Sigma-Aldrich, USA) was added to the solution (25 mg/mL) and stored at room temperature. The GC cells were treated with 2, 4, 6, 8, 10 μg/mL of CDDP or 0.2, 0.4, 0.6, 0.8, 1 mg/mL of ADR, or 3, 6, 9, 12, 15 μg/mL of 5-FU for 48 hours then applied to designed analysis.
RNA Extraction and qRT-PCR
Total RNA from 1 mL plasma was extracted using Trizol (Invitrogen) according to the manufacturer′s instructions. Reverse transcription was performed using random primers (AUGCT Biotech, Beijing, China). Real-time PCR was carried out in an Applied Biosystems 7300 system using Power SYBR Green PCR master mix (Applied Biosystems). Relative levels of gene expression were determined using 18S rRNA as the control. The DCt value was defined as DCt=CtHUCL - Ct18S, and a lower DCt value implied a higher expression. The primers for 18S rRNA are forward, 5′- AGGATCCATTGGAGGGCAAGT -3′; reverse, 5′- TCCAACTACGAGCTTTTTAACTGCA -3′. The primers for HULC are forward, 5′- TCATGATGGAATTGGAGCCTT -3′; reverse, 5′- CTCTTCCTGGCTTGCAGATTG -3′.
siRNA transfection
The siRNA specially targeting HULC (siHULC) and the scramble siRNA (siCon) were both purchased from RiboBio (Ribo, China), which were chemically synthesized fragments. Transfection was carried out according to the manufacturer′s protocol with Lipofectamine 2000 (Life Science, NY, USA). The sequence of siHULC was as follows: S, 5′-CCGGAAUAUUCUUUGUUUAUU-3′; AS, 5′-UAAACAAAGAAUAUUCCGGUU-3′; the sequence of siCon was as follows: 5′-CCUUAUAUGUUCUGGAAUUUU-3′; AS, 5′-UAAAACGAAUGGAAUUCACUU-3′.
Flow cytometry of apoptosis
After treatment, the cells were harvested by trypsin-EDTA solution to produce single cell suspension. Annexin-V and PI double staining kit (Biosea Biotech, Beijing, China) was used to assess apoptosis. The cells were analyzed by flow cytometry. Apoptotic cells were localized in the lower right quadrant and upper right quadrant of a dot-plot graph using annexin-V-fluorescein and PI.
Chemotherapeutic drugs sensitivity analysis
MGC-803 and MKN-45 cells (2 x 103 cells/ well) were seeded in 96-well plates and treated with different drugs including CDDP, ADR and 5-FU in different concentrations for 48 h. Cell Counting Kit-8 (CCK-8, Dojindo, Japan) was added into each well. After 4 h incubation at 37 °C, the coloring reaction was quantified by an automatic plate reader (BIOTEK, USA). Growth inhibitory effects were calculated and the chemotherapeutic drugs sensitivity was evaluated by IC50 parameter (inhibitory concentration of 50% cells).
Statistical analysis
All data are expressed as the mean ± standard deviation. SPSS software version 18.0 (SPSS, Inc., Chicago, IL, USA) was used for statistical analyses. The differences between groups were evaluated using the Student-Newman-Keuls test. P<0.05 was considered to indicate a statistically significant difference.
Results
The expression levels of plasma HULC were increased in GC patients
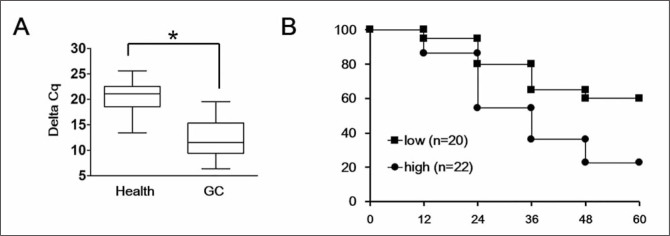
To examine the correlation between the expression levels of plasma HULC and clinicopathologic factors in GC, 42 patient cases were statistically analyzed. We observed that the expression of HULC increased significantly in GC patients compared with healthy volunteers (p<0.05) by qRT-PCR (Figure 1A). Among them, we observed a stratification of the patient samples - the high expression group, 22 samples, showed more than two-fold up-regulation of HULC compared with that in plasma of healthy volunteers, while the remaining 20 samples were classified as the low expression group. Kaplan-Meier survival analysis of the stratified GC patient samples showed the patient survival time in HULC high expression group (n = 22) was significantly shorter than in the low expression group (n = 20) (p< 0.05) (Figure 1B), indicating the potential for HULC to be used as a biomarker in patients with GC.
Figure 1.
The expression levels of plasma HULC were increased in GC patients.
(A) The expression levels of plasma HULC in 42 GC cases and 25 healthy volunteers were detected by qRT-PCR assay as indicated by the box plot ranges. The line indicates the average, whereas the whiskers are the standard deviations. (B) Kaplan-Meier survival analysis confirmed that higher plasma HULC levels result in a decreased overall 5-year survival rate.
Each experiment was repeated at least three times using independent biological replicates. *, p<0.05 compared with control.
The expression levels of HULC were elevated in GC tissues and GC cell lines
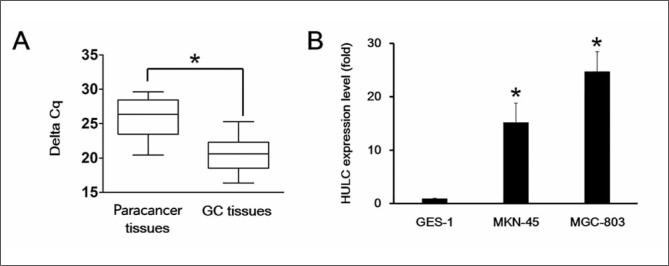
To explore the function of HULC in GC tissues, we first analyzed the expression level of HULC via qRT-PCR. We observed that the expression of HULC was remarkably elevated in GC tissue samples compared with the matched paracancer tissues (p< 0.05) (Figure 2A). Furthermore, we compared HULC expression in human gastric adenocarcinoma MGC-803 and MKN-45 cell lines to normal gastric epithelial cells, GES-1, using qRT-PCR. We observed significant upregulation of HULC in MGC-803 and MKN-45 cells compared with GES-1 cells, which suggested that HULC might exert its function in GC cells (Figure 2B).
Figure 2.
The expression levels of HULC were elevated in GC tissues and GC cell lines.
(A) The expression level of HULC was verified in paracancer tissues and GC tissues via qRT-PCR as indicated by the box plot ranges. The line indicates the average, whereas the whiskers are the standard deviations. (B) The expression level of HULC was elevated in human gastric cancer cell lines, MGC-803 and MKN-45, compared to wild-type gastric cell line, GES-1, by qRT-PCR.
Each experiment was repeated at least three times using independent biological replicates. *, p<0.05 compared with control.
HULC knockdown suppressed apoptosis in GC cells
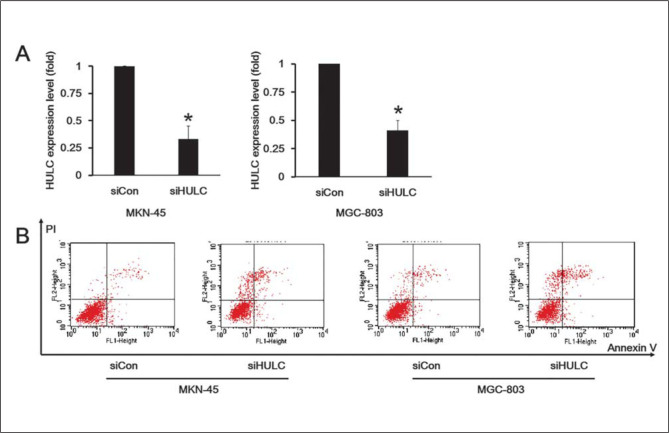
To further determine the function of HULC in GC cells, MGC-803 and MKN-45 cells were transfected with HULC siRNA. The efficiency of HULC knockdown was confirmed via qRT-PCR assay in both cell lines (Figure 3A). To examine whether HULC knockdown in MGC-803 and MKN-45 cells induced apoptosis, we double-stained the cells with Annexin V and PI. Additionally, MGC-803 and MKN-45 cells with HULC knockdown were treated with a low dose of CDDP (2 μg/mL) for 6 h. The results demonstrated that knockdown of HULC increased the percentage of MGC-803 and MKN-45 cells undergoing apoptosis respectively (Figure 3B).
Figure 3.
HULC knockdown suppressed GC cells apoptosis.
(A) The efficiency of HULC knockdown was confirmed via qRT-PCR in two independent human gastric cancer cell lines, MGC-803 and MKN-45. (B) Flow cytometry detected the apoptosis rate using double-staining with Annexin V and PI in MGC-803 and MKN-45 cells exposed to CDDP.
Each experiment was repeated at least three times using independent biological replicates. *, p<0.05 compared with control.
HULC knockdown enhanced the sensitivity of GC cells to chemotherapeutic drugs
The pro-apoptotic effect of HULC knockdown on GC cells prompted us to speculate that it may also contribute to the drug resistance of GC cells. To test this hypothesis, we assayed chemosensitivity to three common chemotherapy drugs: CDDP, ADR, and 5-FU. To analyze the influence of HULC on chemoresistance, we compared HULC knockdown cells (siHULC) to the non-targeting control group (siCon), and the untransfected cells (Empty). We observed that HULC knockdown in both MGC-803 and MKN-45 cells significantly decreased the IC50 values of CDDP, ADR, and 5-FU (Table I). Therefore, we concluded that HULC knockdown contributes to the sensitivity of GC cells to chemotherapeutic drugs.
Table I.
TIC50 values in MGC-803 and MKN-45 cells to different drugs.
| Cell line | Group | CDDP (mg/mL) | ADR (mg/mL) | 5-FU (mg/mL) |
|---|---|---|---|---|
| MGC-803 | Empty | 4.56±0.41 | 0.44±0.05 | 7.84±0.94 |
| siCon | 4.75±0.48 | 0.41±0.04 | 7.69±0.88 | |
| siHULC | 2.75±0.31** | 0.25±0.03* | 3.56±0.59** | |
| MKN-45 | Empty | 3.52±0.18 | 0.69±0.08 | 8.43±1.21 |
| siCon | 3.94±0.21 | 0.74±0.09 | 8.13±0.95 | |
| siHULC | 1.94±0.11** | 0.41±0.05** | 6.63±0.77* |
Note: siCon group and siHULC group were compared with untransfected cells.
Each experiment was repeated in triplicate at least three times.
p<0.05 compared with control;
p<0.01 compared with control.
Discussion
Gastric carcinoma is one of the most commonly occurring malignancies in humans, with a high incidence in China. While surgical resection remains the main treatment approach, chemotherapy is occasionally useful in patients with advanced gastric carcinoma. However, the effectiveness of chemotherapy is often hindered by simultaneous resistance of tumor cells to a variety of cytotoxic drugs, known as chemoresistance. Until now, chemoresistance remains a major obstacle to successful gastric cancer chemotherapy in the clinic (16). The exact molecular mechanisms behind chemoresistance remain unclear.
To improve the efficiency of GC treatment, the reversal of chemoresistance and enhancing the sensitivity of tumor cells to chemotherapeutic agents are very important aspects of GC therapy. The molecular basis of resistance to cancer therapeutics is generally complex, involving multiple processes such as drug metabolism and transport, apoptosis, and DNA repair (17–19). However, other mechanisms are also involved in the development of MDR in tumor cells, and exhibit chemotherapy-related effects, including the activation of detoxifying systems, the change in drug targets, alterations in cell cycle control and the interruption of signaling pathways (20). However, whether other lncRNAs are related to chemoresistance remains to be investigated.
Only 2% of RNAs encode proteins in human cells. Since the large majority is not translated, noncoding RNAs (ncRNAs) still play important roles in regulating transcriptional and non-transcriptional processes (21). ncRNAs consist of microRNAs, small interfering RNAs, and long noncoding RNAs (lncRNAs). LncRNAs are eukaryotic RNAs longer than 200 nucleotides, with no coding capacity. Altered lncRNA expression has been associated with the development of cancer and other diseases (22). Furthermore, several lncRNAs have shown promise as cancer biomarkers and potential therapeutic targets in several cancer subtypes (23, 24). Wang et al. (25) reported that a lncRNA MRUL (MDR-related and upregulated lncRNA) was significantly upregulated in two multidrug-resistant GC cell sublines, SGC7901/ADR and SGC7901/VCR. Their findings indicated that MRUL promotes ABCB1 (ATP-binding cassette, subfamily B, member 1) expression and is a potential target to reverse the multidrug resistance phenotype of GC cell sublines. LncRNA HULC was first found in 2007 (13) and is reported to modulate abnormal lipid metabolism in hepatoma cells through a miR-9-mediated RXRA signaling pathway (26). Additionally, the elevation of HULC by hepatitis B virus X protein promotes hepatoma cell proliferation via down-regulating p18 (27) and HULC is a novel biomarker of poor prognosis in patients with pancreatic cancer (28). An up-to-date study by Zhao et al. (29) discovered that HULC may play an important role in the growth and tumorigenesis of human GC by activating autophagy, and silencing of HULC effectively reversed the epithelial-to-mesenchymal transition (EMT) phenotype. All these evidence point to lncRNA HULC as an important molecule in gastric tumorigenesis and survival.
In the current paper, we identified that HULC is differentially expressed in the tissues and plasma of GC patients compared with those of controls. Knockdown of HULC using specific siRNA enhances the apoptosis rate in GC cells treated with CDDP, which indicates that HULC is potentially implicated in GC chemoresistance including CDDP, ADR, and 5′-FU. Taken together, our results provide new evidence for lncRNA-mediated regulation of chemoresistance in GC, which has potential for novel therapeutic strategies.
Conflict of interest statement
The authors stated that they have no conflicts of interest regarding the publication of this article.
References
- 1.Parkin DM, Pisani P, Ferlay J. Estimates of the worldwide incidence of 25 major cancers in 1990. International journal of cancer Journal international du cancer. 1999;80(6):827–41. doi: 10.1002/(sici)1097-0215(19990315)80:6<827::aid-ijc6>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 2.Ajani JA. Chemotherapy for gastric carcinoma: new and old options. Oncology (Williston Park) 1998;12(10 Suppl 7):44–7. [PubMed] [Google Scholar]
- 3.Borst P, Elferink RO. Mammalian ABC transporters in health and disease. Annual review of biochemistry. 2002;71:537–92. doi: 10.1146/annurev.biochem.71.102301.093055. [DOI] [PubMed] [Google Scholar]
- 4.Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X. et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306(5705):2242–6. doi: 10.1126/science.1103388. 24. [DOI] [PubMed] [Google Scholar]
- 5.Kapranov P, Drenkow J, Cheng J, Long J, Helt G, Dike S. et al. Examples of the complex architecture of the human transcriptome revealed by RACE and high-density tiling arrays. Genome research. 2005;15(7):987–97. doi: 10.1101/gr.3455305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM. et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22(39):8031–41. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 7.Srikantan V, Zou Z, Petrovics G, Xu L, Augustus M, Davis L. et al. PCGEM1, a prostate-specific gene, is overexpressed in prostate cancer. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(22):12216–21. doi: 10.1073/pnas.97.22.12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bussemakers MJ, van Bokhoven A, Verhaegh GW, Smit FP, Karthaus HF, Schalken JA. et al. DD3: a new prostate-specific gene, highly overexpressed in prostate cancer. Cancer research. 1999;59(23):5975–9. [PubMed] [Google Scholar]
- 9.Petrovics G, Zhang W, Makarem M, Street JP, Connelly R, Sun L. et al. Elevated expression of PCGEM1, a prostate-specific gene with cell growth-promoting function, is associated with high-risk prostate cancer patients. Oncogene. 2004;23(2):605–11. doi: 10.1038/sj.onc.1207069. [DOI] [PubMed] [Google Scholar]
- 10.Iacoangeli A, Lin Y, Morley EJ, Muslimov IA, Bianchi R, Reilly J. et al. BC200 RNA in invasive and preinvasive breast cancer. Carcinogenesis. 2004;25(11):2125–33. doi: 10.1093/carcin/bgh228. [DOI] [PubMed] [Google Scholar]
- 11.Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP, Wei Wl. Mature miR-184 as Potential Oncogenic microRNA of Squamous Cell Carcinoma of Tongue. Clinical cancer research: an official journal of the American Association for Cancer Research. 2008;14(9):2588–92. doi: 10.1158/1078-0432.CCR-07-0666. [DOI] [PubMed] [Google Scholar]
- 12.Park NJ, Zhou H, Elashoff D, Henson BS, Kastratovic DA, Abemayor E. et al. Salivary microRNA: discovery, characterization, and clinical utility for oral cancer detection. Clinical cancer research: an official journal of the American Association for Cancer Research. 2009;15(17):5473–7. doi: 10.1158/1078-0432.CCR-09-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Panzitt K, Tschernatsch MM, Guelly C, Moustafa T, Stradner M, Strohmaier HM. et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132(1):330–42. doi: 10.1053/j.gastro.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 14.Liu CJ, Kao SY, Tu HF, Tsai MM, Chang KW, Lin SC. Increase of microRNA miR-31 level in plasma could be a potential marker of oral cancer. Oral diseases. 2010;16(4):360–4. doi: 10.1111/j.1601-0825.2009.01646.x. [DOI] [PubMed] [Google Scholar]
- 15.Gorenchtein M, Poh CF, Saini R, Garnis C. MicroRNAs in an oral cancer context - from basic biology to clinical utility. Journal of dental research. 2012;91(5):440–6. doi: 10.1177/0022034511431261. [DOI] [PubMed] [Google Scholar]
- 16.Chuman Y, Sumizawa T, Takebayashi Y, Niwa K, Yamada K, Haraguchi M. et al. Expression of the multidrug-resistance-associated protein (MRP) gene in human colorectal, gastric and non-small-cell lung carcinomas. International journal of cancer Journal international du cancer. 1996;66(2):274–9. doi: 10.1002/(SICI)1097-0215(19960410)66:2<274::AID-IJC23>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 17.Novaković I, Maksimović N. Pavlović A, Žrković M, Rovčanin B, Mirković D, Pekmezović T, Cvetković D. Introduction to molecular genetic diagnostics. J Med Biochem. 2014;33:3–7. [Google Scholar]
- 18.Pavlović S, Zukić B, Stojiljković Petrović M. Molecular genetic markers as a basis for personalized medicine. J Med Biochem. 2014;33:8–21. [Google Scholar]
- 19.Dai Z, Huang Y, Sadee W. Growth factor signaling and resistance to cancer chemotherapy. Current topics in medicinal chemistry. 2004;4(13):1347–56. doi: 10.2174/1568026043387746. [DOI] [PubMed] [Google Scholar]
- 20.Fetisova EK, Avetisyan AV, Izyumov DS, Korotetskaya MV, Chernyak BV, Skulachev VP. Mitochondria-targeted antioxidant SkQR1 selectively protects MDR (Pgp 170)-negative cells against oxidative stress. FEBS letters. 2010;584(3):562–6. doi: 10.1016/j.febslet.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 21.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes & development. 2009;23(13):1494–504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends in cell biology. 2011;21(6):354–61. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 23.Crea F, Watahiki A, Quagliata L, Xue H, Pikor L, Parolia A. et al. Identification of a long non-coding RNA as a novel biomarker and potential therapeutic target for metastatic prostate cancer. Oncotarget. 2014;5(3):764–74. doi: 10.18632/oncotarget.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi Y, Lu J, Zhou J, Tan X, He Y, Ding J. et al. Long noncoding RNA Loc554202 regulates proliferation and migration in breast cancer cells. Biochemical and biophysical research communications. 2014;446(2):448–53. doi: 10.1016/j.bbrc.2014.02.144. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, Zhang D, Wu K, Zhao Q, Nie Y, Fan D. Long noncoding RNA MRUL promotes ABCB1 expression in multidrug-resistant gastric cancer cell sublines. Mol Cell Biol. 2014;34(17):3182–93. doi: 10.1128/MCB.01580-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cui M, Xiao Z, Wang Y, Zheng M, Song T, Cai X. et al. Long noncoding RNA HULC modulates abnormal lipid metabolism in hepatoma cells through an miR-9-mediated RXRA signaling pathway. Cancer Res. 2015;75(5):846–57. doi: 10.1158/0008-5472.CAN-14-1192. [DOI] [PubMed] [Google Scholar]
- 27.Du Y, Kong G, You X, Zhang S, Zhang T, Gao Y. et al. Elevation of highly up-regulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via down-regulating p18. J Biol Chem. 2012;287(31):26302–11. doi: 10.1074/jbc.M112.342113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peng W, Gao W, Feng J. Long noncoding RNA HULC is a novel biomarker of poor prognosis in patients with pancreatic cancer. Med Oncol. 2014;31(12):346. doi: 10.1007/s12032-014-0346-4. [DOI] [PubMed] [Google Scholar]
- 29.Zhao Y, Guo Q, Chen J, Hu J, Wang S, Sun Y. Role of long non-coding RNA HULC in cell proliferation, apoptosis and tumor metastasis of gastric cancer: a clinical and in vitro investigation. Oncol Rep. 2014;31(1):358–64. doi: 10.3892/or.2013.2850. [DOI] [PubMed] [Google Scholar]