Abstract
Background: Studies of thyroid stem/progenitor cells have been hampered due to the small organ size and lack of tissue, which limits the yield of these cells. A continuous source that allows the study and characterization of thyroid stem/progenitor cells is desired to push the field forward.
Method: A cell line was established from Hoechst-resistant side population cells derived from mouse thyroid that were previously shown to contain stem/progenitor-like cells. Characterization of these cells were carried out by using in vitro two- and three-dimensional cultures and in vivo reconstitution of mice after orthotopic or intravenous injection, in conjunction with quantitative reverse transcription polymerase chain reaction, Western blotting, immunohisto(cyto)chemistry/immunofluorescence, and RNA seq analysis.
Results: These cells were named SPTL (side population cell-derived thyroid cell line). Under low serum culturing conditions, SPTL cells expressed the thyroid differentiation marker NKX2-1, a transcription factor critical for thyroid differentiation and function, while no expression of other thyroid differentiation marker genes were observed. SPTL cells formed follicle-like structures in Matrigel® cultures, which did not express thyroid differentiation marker genes. In mouse models of orthotopic and intravenous injection, the latter following partial thyroidectomy, a few SPTL cells were found in part of the follicles, most of which expressed NKX2-1. SPTL cells highly express genes involved in epithelial–mesenchymal transition, as demonstrated by RNA seq analysis, and exhibit a gene-expression pattern similar to anaplastic thyroid carcinoma.
Conclusion: These results demonstrate that SPTL cells have the capacity to differentiate into thyroid to a limited degree. SPTL cells may provide an excellent tool to study stem cells, including cancer stem cells of the thyroid.
Keywords: : thyroid stem cells, thyroid cancer, RNA seq analysis, thyroid side population, epithelial-to-mesenchymal transition
Introduction
Adult stem cells are undifferentiated cells that include hematopoietic stem cells, mesenchymal stem cells, and endothelial stem cells that are rich in bone marrow, and tissue resident progenitor cells. They can divide to replenish dying cells and regenerate damaged tissues, and are considered a powerful tool in regenerative medicine (1–3). Adult stem cells are thought to be found throughout the body. However, it remains unclear whether adult stem cells are present in all organs (4).
The thyroid gland is a dormant organ with very slow turnover, while it retains the capacity to grow through cell hypertrophy and proliferation in response to stimuli, suggesting the presence of adult stem/progenitor cells (5). The stimuli are those that perturb the pituitary–thyroid axis, including xenobiotics, physiological alterations, alterations in iodine intake, and direct damage to the thyroid gland such as radiation exposure and partial thyroidectomy (6–8). In recent years, the presence of adult stem/progenitor cells in the thyroid has been described by using thyroid tissues, monolayer cultures, and/or sphere cultures of thyroid tissues or side population (SP) cells from mice and humans (9–12). However, the origin of these stem/progenitor cells is yet to be determined.
It has previously been demonstrated that the SP cells of the mouse thyroid, ranging from 0.3% to 1.4% of the total population of cells, exhibit stem/progenitor cell–like characteristics (9). SP cells were originally identified as a putative stem-cell population, based on their ability to efflux Hoechst 33342 dye in an ATP-binding cassette transporter-dependent manner (13). Mouse thyroid SP cells are composed of two groups of cells characterized as CD45(–)/CD117(c-kit)(–)/SCA1(+) and CD45(–)/CD117(c-kit)(–)/SCA1(–), each representing approximately half of SP cells. Thyroid SP cells highly express genes involved in cancer and/or cancer invasion/metastasis, as determined by microarray analysis, compared with non-SP cells, suggesting that thyroid SP cells may contain cancer stem cells of the thyroid (14). Further, partial thyroidectomy was established as a model for studying thyroid regeneration (15). In the partial thyroidectomized thyroid, many clear cells due to scantiness of the cytoplasm, indicative of immature cells, were observed that have characteristics of either C cells or follicular cells. It was suggested that these immature cells may participate in the repair and/or regeneration of the thyroid gland (15). The formation of clear cells was considered as one of the pathways for thyroid regeneration (16). By using the β-galactosidase reporter mouse in conjunction with partial thyroidectomy and bromodeoxyuridine (BrdU) long label–retaining cell analysis, it has been demonstrated that stem cell antigen 1 (SCA1)-expressing non-follicular mesenchymal cells are the origin of follicular cells during thyroid regeneration, although it remains unclear where these cells originate from (17). The SCA1-positive cells, initially found in non-follicular areas, were found 35 days after partial thyroidectomy in part of follicles that co-expressed NKX2-1, a critical transcription factor for thyroid development and function (18,19). The SCA1-positive cells accounted for only ≤0.1% of intra-follicular thyroid cells (17). This pathway was suggested to be an alternative for thyroid regeneration (16).
Even though studies on thyroid stem/progenitor cells are slowly emerging, one of the biggest challenges is the availability of a tool, which is currently very limited, partly due to the small size of the mouse thyroid. The study described herein was initiated to obtain a stable continuous cell line that allows the study and characterization of thyroid stem/progenitor cells. A cell line was generated from mouse SP cells that partially exhibited stem/progenitor characteristics based on the expression of various marker genes for stem cells and thyroid differentiation, the formation of follicle-like structure in vitro, and some of these cells being found in part of the follicles in vivo when orthotopically or intravenously injected into mice. RNA seq analysis demonstrated that SP-derived cells are enriched in the expression of genes involved in epithelial–mesenchymal transition (EMT), and have a gene-expression pattern similar to those found in anaplastic thyroid carcinoma (ATC). The SP-derived cell line may provide an excellent tool to study stem/progenitor cells, including cancer stem cells of the thyroid.
Materials and Methods
Cell culture
Thyroid glands of 30–40 C57BL/6NCr mice were used to prepare thyroid cell suspensions and Hoechst 33342 staining, followed by subjection to fluorescence-activated cell sorting (FACS) to obtain thyroid SP cells, as previously described (14), except that fumitremorgin C (Sigma–Aldrich, St. Louis, MO) was used instead of verapamil as an ABC transporter inhibitor at a final concentration of 10 μM. SP cells were continuously cultured in Dulbecco's modified Eagle's medium (DMEM)/F12 50/50 mix medium containing 10% fetal bovine serum (FBS) in a 37°C CO2 incubator, which resulted in the establishment of SPTL (side population cell-derived thyroid cell line) cells. For the in vitro experiments, SPTL cells (4.2 × 105 cells/10 cm culture dish) were seeded in DMEM/F12 50/50 mix medium containing 10% FBS in a 37°C CO2 incubator. The serum concentration in the medium was switched from 10% to 2% one day after seeding (day 0). SPTL cells were cultured in the medium with 2% FBS and collected at various time points for gene expression analysis. SPTL cells between passages 36 and 40 were used in all in vitro and in vivo experiments.
Three-dimensional Matrigel® culture
After two-dimensional culture for six days with DMEM/F12 50/50 mix medium containing 2% FBS, 1.6 × 105 SPTL cells in 100 μL of Matrigel® were laid onto cover glass, which was then put into 24-well culture plates. The medium containing 2% FBS and 1 mIU/mL of bovine thyrotropin (TSH; Sigma–Aldrich) was added onto the Matrigel®. SPTL cells together with Matrigel® were subjected to hematoxylin and eosin (H&E) staining after 16 days of three-dimensional culture.
Animal procedures
SPTL cells were either directly injected to one of the thyroid lobes or by intravenous injection through the tail vein of non-obese diabetic severe combined immunodeficiency (NOD/SCID) mice. For direct injection, 5 μL of 5 × 104 SPTL cells/μL were injected into one of the thyroid lobes using a Hamilton syringe (Hamilton Company, Reno, NV). For tail-vein injection of SPTL cells, mice were first subjected to partial thyroidectomy where the caudal third of both thyroid lobes were resected. One day later, the SPTL cells (100 μL of 5 × 105 cells/μL) were injected through the tail vein. All animal studies were performed in accordance with the Using Animals in Intramural Research Guidelines (National Institutes of Health Animal Research Advisory Committee, National Institutes of Health, Bethesda, MD) and after approval of the institutional Animal Care and Use Committee.
Immunofluorescence analyses
Immunofluorescence analysis of mouse thyroid tissue was carried out using OCT-embedded (Sakura Finetek USA, Inc., Torrance, CA) thyroid tissue cryo specimens sectioned at 8 μm using a CM1950 cryostat (Leica, Buffalo Grove, IL), followed by brief fixation in 4% paraformaldehyde (PFA) for 10 min at room temperature. The sections were washed three times with Tris-buffered saline (TBS) after each step. Non-specific binding sites were blocked using 1% bovine serum albumin in TBS for 1 h at room temperature before incubation with an antibody. For double labeling using two primary antibodies from the same species, the sections were first incubated with rabbit polyclonal anti-green fluorescent protein (GFP; 598; 1:250; MBL International Corp, Woburn, MA) at 4°C overnight, which was then treated with the secondary antibody, labeled goat anti-rabbit IgG (Dylight 650; 1:200; Abcam, Cambridge, United Kingdom) for 1 h at room temperature. The sections were then incubated with 5% rabbit serum (the same host species) for 1 h at room temperature, followed by incubation with an unconjugated Fab antibody derived from the same host species as the primary antibody (AffiniPure Fab Fragment Goat Anti-Rabbit IgG; Jackson ImmunoResearch Laboratories, West Grove, PA) for 1 h at room temperature. The sections were finally incubated with the second primary antibody, rabbit anti-NKX2-1 (H-190; 1:200; Santa Cruz Biotechnology, Dallas, TX), followed by incubation with a labeled donkey anti-rabbit IgG secondary antibody. DAPI dye (D1306; Molecular Probes/Life Technologies, Eugene, OR) was used to stain the nuclei of cells, and the sections were mounted using Fluoromount-G (SouthernBiotech, Birmingham, AL). Confocal images were obtained with a Zeiss 780 LSM (Carl Zeiss, Oberkochen, Germany).
Sample preparation for RNA seq analysis
Total RNAs were prepared from SP and MP (main population, non-SP) cells immediately after sorting, and SPTL cells using RNAlater (Life Technologies) and TRIzol reagent (Life Technologies), followed by RNeasy Mini Kit (Qiagen, Valencia, CA). Two biological replicates were used for each experimental group. Total RNA was converted to cDNA and amplified using Ovation RNA seq System V2 (NuGEN, San Carlos, CA). Following amplification, 1 μg of cDNA was fragmented using a Covaris S1 ultrasonicator (Covaris, Woburn, MA). The sheared cDNAs (100 ng) were used for library construction using Ovation SP Ultraflow Library System (NuGEN). Library droplets were collected and subjected to library enrichment by low-cycle polymerase chain reaction (PCR). The 250–450 bp gel fragments were size selected by 2% agarose gel electrophoresis and then purified with a Zymo column.
RNA seq
The RNA seq libraries (5 ng) were subjected to sequencing on a Hiseq 2000 (Illumina, San Diego, CA). BWA (Burrows-Wheeler Aligner) software (20) was used to map RNA seq reads to the mm9 mouse reference genome (http://genome.ucsc.edu) with default parameters. Only uniquely mapped reads were retained for the following downstream analysis. Transcript expression levels were quantified by using the rpkmForGene python program (21). EdgeR (22) was used to identify differentially expressed transcripts. Fold changes were calculated using ratios of the arithmetic mean of the normalized read counts for each pair of replicates. Statistical significance of differential gene expression in RNA seq data was determined using EdgeR. Differentially expressed genes were selected by requiring both fold changes ≥2 and a false discovery rate (FDR) of ≤0.01. The gene ontology (GO) analysis of the differentially expressed genes was carried out using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (23).
Quantitative reverse transcription PCR
Approximately 2 μg of RNA was treated with DNase I (Life Technologies) and then reverse transcribed into cDNA using Superscript III (Life Technologies) and Random Primers (Life Technologies). ES cell cDNA was kindly provided by Dr. Tao Deng (NCI) (24). Real-time PCR was performed in triplicate using Power SYBR Green PCR Master Mix (Life Technologies) and a 7900 HT Fast Real-Time PCR system (Life Technologies). The primers used were:
Nkx2-1 (forward) 5′-ACA GCC AAG CAA ATT CAA CC-3′, (reverse) 5′-GGG TGC ATC CAC AGA AAA GT-3′; Pax8 (forward) 5′-GCC ACC TGT TTG GTC TTC AT-3′, (reverse) 5′-AGC CAC TGG TTG AGA GGC TA-3′; Foxe1 (forward) 5′-GGC GGC ATC TAC AAG TTC AT-3′, (reverse) 5′-GGA TCT TGA GGA AGC AGT CG-3; Tshr (forward) 5′-CAA GGA GCT CCA CCG AAT C-3′, (reverse) 5′-ATT GGG GAG ACT CGA AAA TG-3′; Nis/Scl5a5 (forward) 5′-AGC TGC CAA CAC TTC CAG AG-3′, (reverse) 5′-GAT GAG AGC ACC ACA AAG CA-3′; Tg (forward) 5′-GTC CAA TGC CAA AAT GAT GGT C-3′, (reverse) 5′-GAG AGC ATC GGT GCT GTT AAT-3′; Tpo (forward) 5′-ACA GTC ACA GTT CTC CAC GGA TG-3′, (reverse) 5′-ATC TCT ATT GTT GCA CGC CCC-3′; Gata4 (forward) 5′-CCC TAC CCA GCC TAC ATG G-3′, (reverse) 5′-ACA TAT CGA GAT TGG GGT GTC T3′; Gata6 (forward) 5′-TTG CTC CGG TAA CAG CAGT G-3′, (reverse) 5′-GTG GTC GCT TGT GTA GAA GGA-3′; Sca1/Ly6a (forward) 5′-CAA TTA CCT GCC CCT ACC CT-3′, (reverse) 5′-GGC AGA TGG GTA AGC AAA GA-3′; Oct4/Pou5f1 (forward) 5′-ATC GGA CCA GGC TCA GAG GTA TTG-3′, (reverse) 5′-TTC TCC AAC TTC ACG GCA TTG-3′; Nanog (forward) 5′-AGC CCT GAT TCT TCT ACC AGT CCC-3′, (reverse) 5′-GCT TCT GAA ACC TGT CCT TGA GTG-3′; Rex1/Zfp42 (forward) 5′-CCC TCG ACA GAC TGA CCC TAA-3′, (reverse) 5′-TCG GGG CTA ATC TCA CTT TCA T-3′; Tgfb2 (forward) 5′-CTT CGA CGT GAC AGA CGC T-3′, (reverse) 5′-GCA GGG GCA GTGT AAA CTT ATT-3′; Tgfb3 (forward) 5′-CCT GGC CCT GCT GAA CTT G-3′, (reverse) 5′-TTG ATG TGG CCG AAG TCC AAC-3′; Snai1 (forward) 5′-CAC ACG CTG CCT TGT GTC T-3′, (reverse) 5′-GGT CAG CAA AAG CAC GGT T-3′; Snai2 (forward) 5′-TGG TCA AGA AAC ATT TCA ACG CC-3′, (reverse) 5′-GGT GAG GAT CTC TGG TTT TGG TA-3′; Cdh1 (forward) 5′-GAG GTC TAC ACC TTC CCG GT-3′, (reverse) 5′-AAA AGA AGG CTG TCC TTG GC-3′; Vim (forward) 5′-AGA GAG AGG AAG CCG AAA GC-3′, (reverse) 5′-TCC ACT TTC CGT TCA AGG TC-3′, and Ppia (forward) 5′-GTG TTC TTC GAC ATC ACG GC-3, (reverse) 5′-CAG TGC TCA GAG CTC GAA AGT-3′. The PCR reaction conditions used were: 95°C for 2 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 45 sec. Relative levels of gene expression were calculated using the ΔΔCq method with normalization to peptidylprolyl isomerase A (PPIA, cyclophilin A).
Results
Generation of a mouse thyroid SP cell-derived cell line
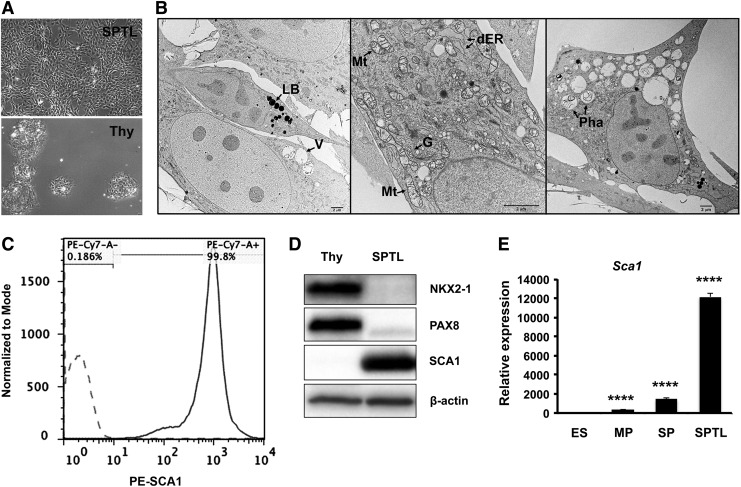
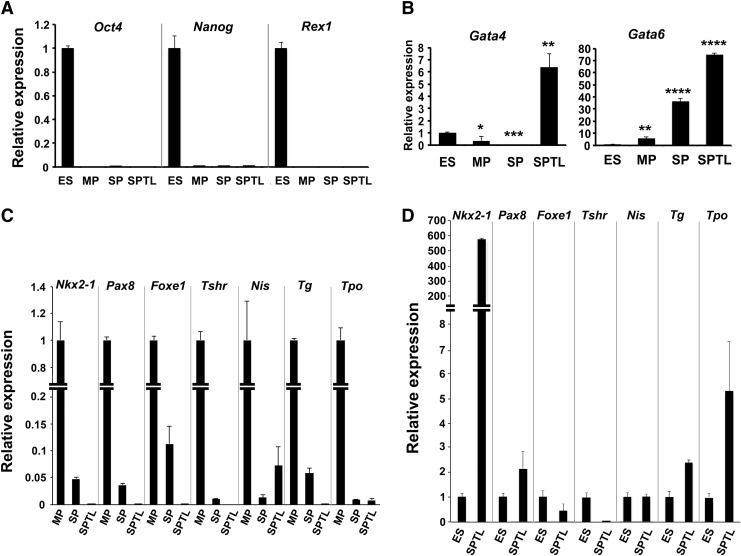
Mouse thyroid SP cells were subjected to culture right after FACS. The majority of cells died after several passages as expected, while a few cells remained viable, which eventually started growing and forming an established cell line. They were named SPTL cells. SPTL cells were passaged over 36 generations. SPTL cells were more stellate, elongated, and mesenchymal in shape compared with primary thyroid cells, which were round and tended to cluster (Fig. 1A). The doubling time of SPTL cells was approximately 35 hours. Electron microscopy demonstrated that SPTL cells were generally small and round, and some were elongated with three- or four-pointed star-shaped cells having heterogeneous cytoplasm containing many mitochondria, Golgi apparatus, a dilated endoplasmic reticulum (ER), medium to large vacuoles, phagosomes, and lipid bodies (Fig. 1B). Most mitochondrial matrixes were not enriched, and the cytoplasm, especially in small cells, was slightly granular and less contrasted, suggesting that SPTL cells were not fully differentiated. These characteristics were in contrast to those found in rat thyroid-derived FRTL-424 and FRTL-5 cells, and rat follicular cells, where many enriched mitochondria and well-developed ER were found (25,26). In a previous study, half of the thyroid SP cells were positive for SCA1, a candidate stem-cell marker for various tissues (27–29). SPTL cells showed high expression of SCA1, as determined by FACS (Fig. 1C), Western blotting (Fig. 1D), and quantitative reverse transcription PCR (qRT-PCR), compared with primary thyroid cells, and/or embryonic stem (ES) cells, and MP (main population, non-SP cells) and SP cells (Fig. 1E). This suggests that SCA1 expression might confer an advantage for the survival of SPTL cells. The SPTL cells as well as the original SP and MP cells did not express the stem cell markers Oct4(Pou5f1), Nanog, and Rex1(Zfp42) mRNAs (Fig. 2A), while the endoderm specification markers Gata4 and Gata6 mRNAs were highly expressed in SPTL cells (Fig. 2B). SPTL cells did not express or expressed very low levels of mRNAs of thyroid-specific genes compared with MP cells (Figs. 2C and 1D). Nkx2-1 was the only gene highly expressed compared with ES cells (Fig. 2D). The relative expression levels of Nkx2-1 mRNA in MP, SP, SPTL, and ES cells using SPTL as 1 were estimated to be MP (∼4000) > SP (∼200) > SPTL (1) > ES (∼0.001). These results suggest that unlike ES cells, SPTL cells are not pluripotent or differentiated thyroid cells, but rather they may be characterized as partially thyroid-specified progenitor-like cells.
FIG. 1.
Characteristics of side population cell-derived thyroid cell line (SPTL) cells. (A) Monolayer culture of SPTL cells (upper: SPTL) and primary thyroid cells (lower: Thy). (B) Representative electron microscopic images. Cells are small, round, elongated, or three- or four-pointed star-shaped (most right panel). LB, lipid body; V, vacuole; Mt, mitochondria; dER, dilated endoplasmic reticulum; G, Golgi apparatus; Pha, Phagosome. The matrix of most mitochondria is not enriched. (C) Fluorescence-activated cell sorting (FACS) analysis for SCA1. Dotted line and straight line show cells treated without and with PE-conjugated SCA1 antibody, respectively. (D) Western blot analysis of SPTL cells for the expression of SCA1, as well as thyroid differentiation markers, NKX2-1 and PAX8, compared with mouse thyroid primary cells (Thy). SPTL cells were those cultured under 10% serum. β-Actin was used as a loading control (15 μg/lane). (E) Quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis of Sca1 mRNA expression in SPTL, original side population (SP) and main population (MP), and embryonic stem (ES) cells, cultured under 10% serum. Relative expression is shown based on the expression level of ES cells as 1. Mean ± standard deviation (SD) from representative experiment in triplicate are shown. Note that qRT-PCR was carried out at least twice using samples prepared at different times. Similar results to those shown were obtained. ****p < 0.0001 for ES vs. MP, SP, and SPTL by unpaired t-test.
FIG. 2.
qRT-PCR analyses of various genes from ES, MP, SP, and SPTL cells. (A) Expression of stem cell marker gene mRNAs, Oct4, Nanog, and Rex1. Relative expression is shown based on the expression level of ES cells as 1. p < 0.0001 for ES versus MP, SP, or SPTL for all genes. (B) Expression of Gata4 and Gata 6 mRNAs. Relative expression is shown based on the expression level of ES cells as 1. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001 for ES vs. MP, SP, or SPTL. (C) Expression of various mRNAs encoded by thyroid differentiation marker genes as indicated. Relative expression is shown based on the expression level of MP cells as 1. p < 0.0001 for MP versus SP or SPTL for all genes. (D) Expression of mRNAs encoded by various thyroid differentiation marker genes in SPTL cells compared with ES cells. Relative expression is based on the expression level of ES cells as 1. p < 0.0001 for ES versus SPTL for all genes. All cells used in these experiments were cultured in the presence of 10% serum. For each mRNA, mean ± SD from representative experiment in triplicate are shown. Note that qRT-PCR was carried out at least twice using samples prepared at different times. Similar results to those shown were obtained. Statistical analysis by unpaired t-test.
Expression of thyroid-specific genes in SPTL cells under low serum conditions
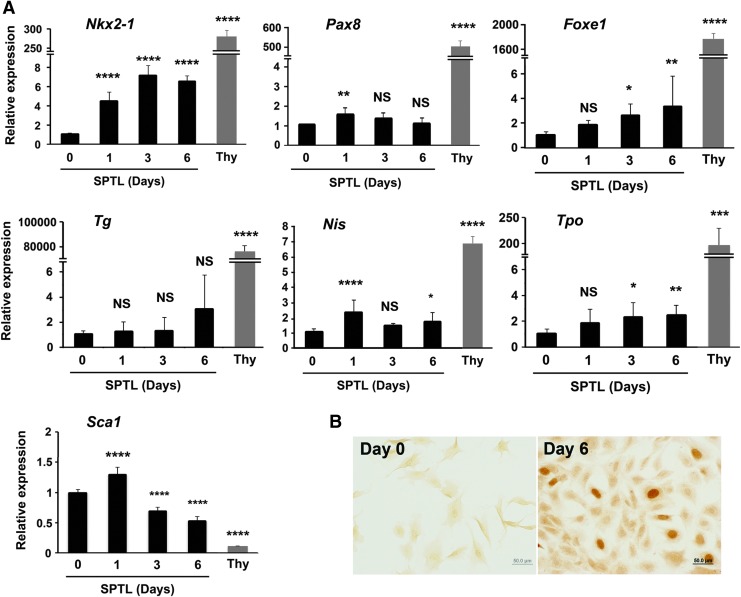
SPTL cells were maintained in media containing 10% FBS. Under this serum condition, SPTL cells did not express three transcription factors critical for thyroid development: NKX2-1, PAX8, and FOXE1 (Fig. 1D and see below). When the serum concentration was changed to 2%, which switches cells from proliferation to differentiation (30,31), the expression of mRNA encoding SCA1 decreased, while mRNAs encoding NKX2-1 increased during 6 days of 2% FBS culture (Fig. 3A). The expression of Foxe1, sodium iodide symporter (Nis;Scl5a5), and thyroid peroxidase (Tpo) mRNAs appeared to have slightly increased, while Pax8 and thyroglobulin (Tg) mRNA levels were not changed. The levels of TSH receptor (Tshr) mRNA at both day 0 and day 6 were too low to be detected. Even though Nkx2-1 expression increased as demonstrated by qRT-PCR, the level was significantly lower compared with that of thyroid primary cells. Immunocytochemistry analysis revealed that some cells clearly expressed NXK2-1 in their nuclei by day 6, but not PAX8 (Fig. 3B, data not shown for PAX8). The NKX2-1 positive cell ratio was calculated as 1.3%, averaged from nine microscopic fields. The addition of other growth factors such as FGFs, EGF, and activin A with and without TSH (32,33) did not enhance expression of Nkx2-1, nor did it induce the expression of other genes. These results demonstrate that under low serum conditions, SPTL cells may start differentiating into thyroid cells, albeit at a limited degree.
FIG. 3.
Induction of thyroid differentiation markers in monolayer culture of SPTL cells. (A) SPTL cells in monolayer culture were collected at days 0, 1, 3, and 6 after serum concentration was changed to 2%. Day 0 is equivalent to 10% culture condition. The expression levels of thyroid primary culture cells (Thy) are shown as comparison. Relative expression is based on the expression level of SPTL day 0 cultured cells as 1. Mean ± SD from three experiments, determined in triplicate are shown. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; NS, not significant for day 0 vs. days 1, 3, and 6 of SPTL cultures by one-way analysis of variance. ****p < 0.0001 for day 0 SPTL cells versus Thy (thyroid primary culture cells) by unpaired t-test. (B) Immunocytochemistry for NKX2-1 using SPTL cells collected at days 0 and 6 under 2% serum conditions. Experiments were carried out more than three times, and each time, similar results were obtained. Representative results are shown.
Three-dimensional culture of SPTL cells
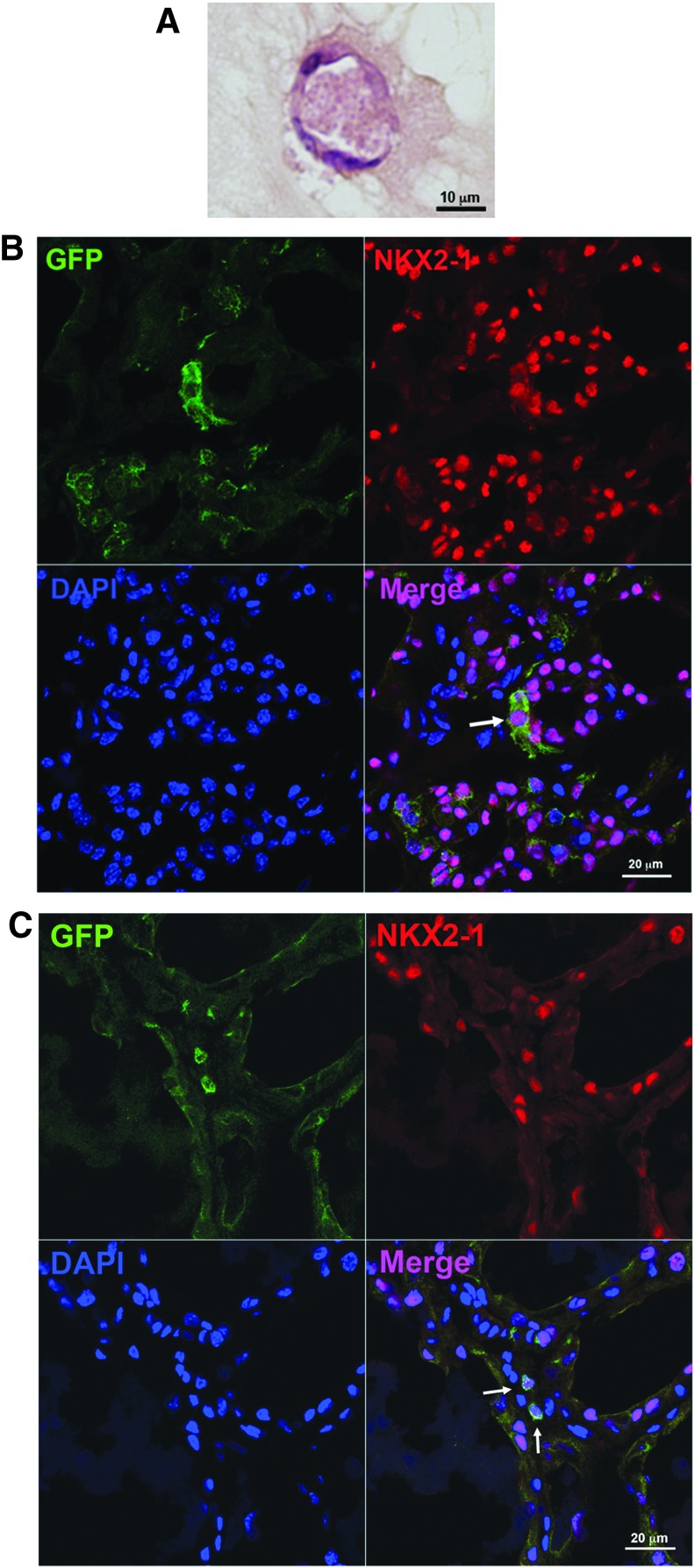
After six days of monolayer culture under low serum conditions, SPTL cells were embedded into Matrigel and further cultured over two weeks. On day 16, the presence of follicle-like structures was noted in H&E-stained sections (Fig. 4A). Immunofluorescence analysis did not show any expression of NKX2-1, PAX8, or thyroglobulin (data not shown). The results suggest that SPTL cells may not differentiate into functional thyroid follicles in vitro.
FIG. 4.
In vitro and in vivo analysis of SPTL cells. (A) Matrigel® culture of SPTL cells. After culturing for six days in 2% serum, SPTL cells were subjected to Matrigel® culture for 16 days, which were then used for hematoxylin and eosin (H&E) staining. (B) Co-immunofluorescence analysis of orthotopically injected SPTL cells for GFP, NKX2-1, and DAPI. The analysis was carried out 14 days after injection of cells. (C) Co-immunofluorescence analysis of intravenously injected SPTL cells for GFP, NKX2-1, and DAPI. The analysis was carried out 28 days after injection of cells. Representative GFP and NKX2-1 co-expressing cells are indicated by arrows in the Merge panel.
Presence of SPTL cells in part of the follicles in vivo
In order to examine how SPTL cells behave in vivo, two methods were used: (i) GFP-expressing SPTL cells were directly injected into one lobe of the thyroid, and (ii) GFP-expressing SPTL cells were injected through the tail vein after mice underwent partial thyroidectomy. For the latter, it was previously demonstrated that partial thyroidectomy can be used as a model for thyroid regeneration (15). When SPTL cells were orthotopically injected into the thyroid, many GFP-positive cells were found outside as well as inside the thyroid (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/thy). In the latter, most GFP-positive cells resided in the interfollicular space. However, a few cells were found, albeit in low numbers, in part of the follicles (Fig. 4B and Supplementary Fig. S1). When these thyroids were subjected to immunofluorescence analysis, strong GFP signals that overlapped with NKX2-1 signals were found. GFP-expressing SPTL cells were also injected through the tail vein following partial thyroidectomy (Fig. 4C). This was based on the assumption that SPTL cells might home to the damaged thyroid trying to participate in organ repair, as described for other organs (34). The GFP-expressing cells, which weakly expressed NKX2-1, were occasionally found in part of the follicles at least up to 42 days following partial thyroidectomy (day 28 results for Fig. 4C, day 42 results for Supplementary Fig. S2). However, they did not express PAX8 (data not shown). These results demonstrate that SPTL cells have a capacity to be integrated as part of the follicle structure in vivo. Orthotopically injected SPTL cells remained unchanged for at least six months with no evidence of tumor development.
RNA seq analysis of SP and SPTL cells
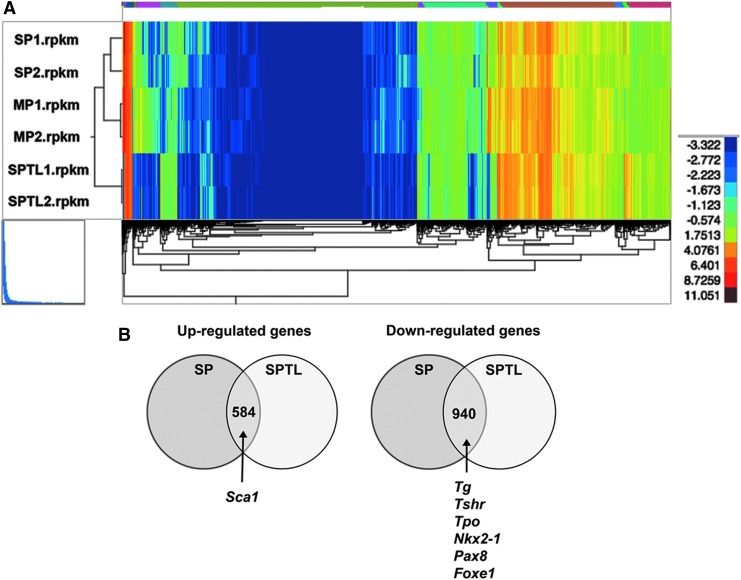
To understand the nature of SPTL cells compared with SP cells in terms of gene-expression patterns, SPTL cells and freshly FACS-sorted SP cells and MP cells in biological duplicates were subjected to RNA seq analysis. A total of 22,194 genes were compared between SPTL, SP, and MP cells (each with two replicates). Reads per kilobase per million mapped reads were used to quantify gene expression levels. SPTL cells had 4269 and 2591 up- and downregulated genes, respectively, compared with MP cells, while SP cells had 1388 and 1020 up- and downregulated genes, respectively, compared with MP cells, with fold changes ≥2 (log2FC ≥1) and a FDR ≤0.01. Hierarchical clustering of gene expression demonstrated very little difference between the three groups of samples (Fig. 5A). In addition, pair-wise correlation analysis showed that the expression in SPTL correlates better with the expression in SP than in MP cells (Supplementary Fig. S3). Among genes altered in SPTL and SP cells, 584 and 940 genes were commonly up- and downregulated respectively (Supplementary Tables S1 and S2). The commonly upregulated genes included Sca1, while commonly downregulated genes included Tg, Tshr, Tpo, Nkx2-1, Pax8, and Foxe1, the genes encoding critical transcription factors and/or proteins for development, homeostasis, and function of the thyroid, and/or thyroid hormone synthesis (Fig. 5B) (18,19). The expression of Tg, Tshr, Tpo, Nkx2-1, Pax8, and Foxe1 when examined by qRT-PCR was indeed downregulated in SP as well as SPTL cells compared with MP cells (Fig. 2C).
FIG. 5.
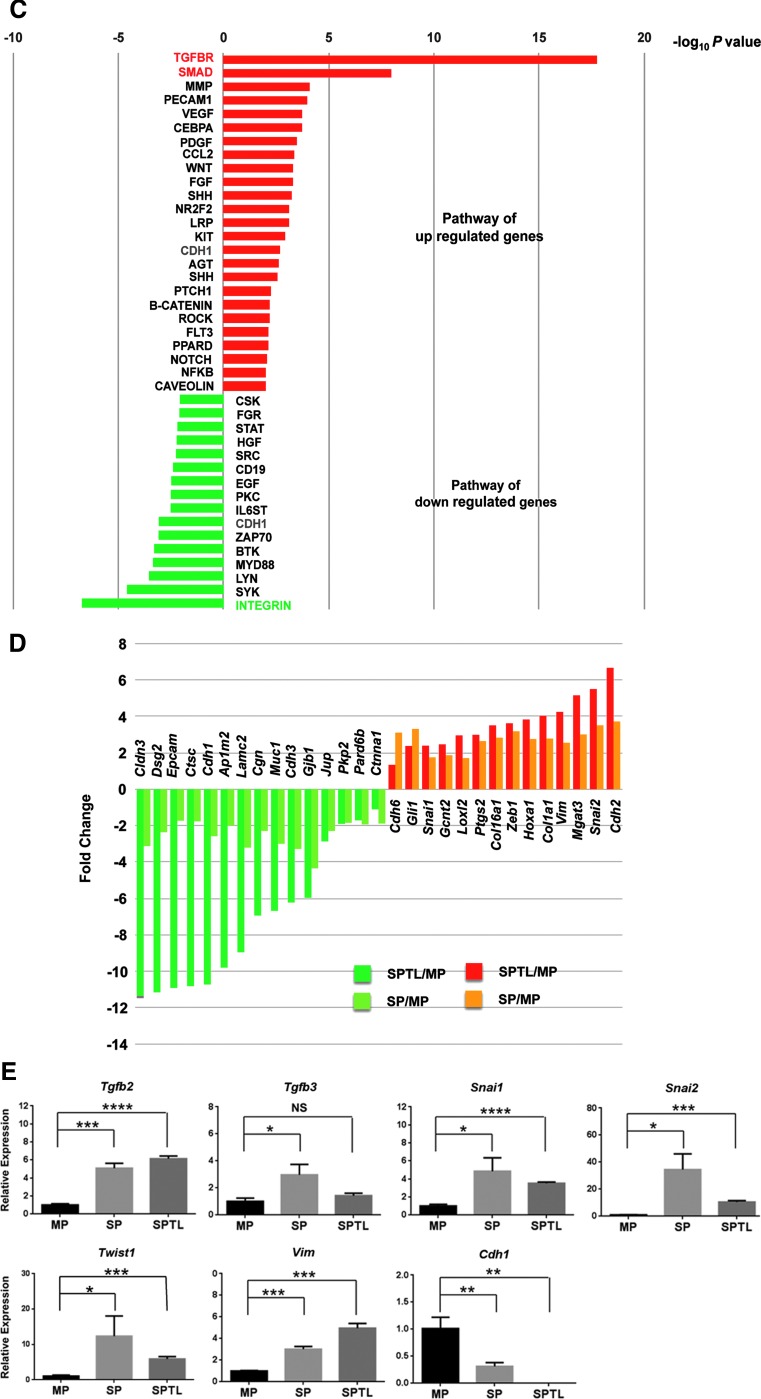
RNA seq analysis. (A) Dendrogram using total reads per kilobase million from SP1, SP2, MP1, MP2, and SPTL1, and SPTL2. (B) Venn diagram of commonly up- and downregulated genes between SP and SPTL cells compared with MP cells. Commonly upregulated genes include Sca1, while commonly downregulated genes include Tg, Tshr, Tpo, Nkx2-1, Pax8, and Foxe1 mRNAs. (C) Signal transduction pathways altered in SP and SPTL cells compared with MP cells were plotted based on –log10 p-values. Red bars are upregulated pathways, while green bars are downregulated pathways. The TGF-β and SMAD pathways (highlighted in red) are the two most upregulated pathways, while the integrin pathway (highlighted in green) is the most downregulated pathway. The cadherin pathway (CDH1, highlighted in blue) is both up- and downregulated. (D) Several genes in the cadherin pathways are plotted based on fold changes for SP/SPTL cells versus MP cells. (E) qRT-PCR analysis of several commonly up- and downregulated mRNAs using RNA isolated from MP, SP, and SPTL cells. The values obtained with the MP was set as 1. Mean ± SD in triplicate experiments are shown. Statistical significance between MP versus SP, or MP versus SPTL by unpaired t-test. *p < 0.05; **p < 0.005; ***p < 0.0005; ****p < 0.0001; NS, not significant.
GO analysis
GO analysis for “biological processes” demonstrated that the commonly upregulated genes compared between SP and SPTL cells were involved in morphogenesis and development, while the downregulated genes were involved in adhesion, junction, localization, and transport (Table 1). Various biological and/or cellular processes and their regulation were found in both up- and downregulated genes. GO analysis for “molecular functions” identified genes involved in protein binding, ion binding, and receptor binding for both up- and downregulated genes (Table 2). The list of observed genes in the GO “biological processes” and “molecular functions” share many transcription factors such as Gata2/4, various Hox genes, Meox2, and Tbx1, among others—transcription factors that play critical roles in development. Furthermore, pathway analysis of the commonly up- and downregulated genes for signal transduction pathway association determined using ingenuity pathway analysis revealed that most of the up- and downregulated signal transduction pathways were those related to EMT (Fig. 5C and Table 3). In particular, transforming growth factor beta (TGF-β; p = 0.00000000000000000169) and SMAD pathways (p = 0.0000000106), the two representative pathways of EMT, were the top two upregulated pathways with extremely low p-values, while the third top pathway of MMP had a much lower p value (0.0000790). On the other hand, the integrin pathway was the most downregulated pathway, and the cadherin 1 (E-cadherin) pathway was found in both up- and downregulated pathways. Several genes involved in the E-cadherin pathway were plotted based on fold changes relative to MP cells (Fig. 5D). SP and SPTL cells showed the same up- or downregulation for all the genes listed. The fold changes were in general larger for SPTL cells than SP cells, particularly for the downregulated genes, suggesting that the expression of the listed genes may have been enriched in SPTL cells. The qRT-PCR analysis showed that the expression of EMT-related genes such as Tgfb2, Tgfb3, Snai1, Snai2, Twist1, and Vim was higher in SP and/or SPTL cells compared with MP cells, while the expression of Cdh1, the epithelial marker, was lower in SP and SPTL cells (Fig. 5E). The results demonstrate that the gene-expression patterns obtained by RNA seq correlate well with the actual gene-expression patterns. These results altogether demonstrate that SPTL and SP cells share the common feature representative of EMT.
Table 1.
GO Term Analysis for “Biological Processes”
| GO term | GO term ID | p-Value |
|---|---|---|
| Upregulated genes | ||
| Anatomical structure morphogenesis | GO:0009653 | 1.89E-40 |
| Developmental process | GO:0032502 | 2.00E-38 |
| System development | GO:0048731 | 3.86E-38 |
| Organ morphogenesis | GO:0009887 | 7.70E-38 |
| Multicellular organismal development | GO:0007275 | 7.97E-36 |
| Cell adhesion | GO:0007155 | 3.34E-35 |
| Biological adhesion | GO:0022610 | 6.34E-35 |
| Anatomical structure development | GO:0048856 | 8.20E-35 |
| Single-organism developmental process | GO:0044767 | 8.88E-35 |
| Positive regulation of biological process | GO:0048518 | 1.27E-32 |
| Regulation of multicellular organismal process | GO:0051239 | 3.40E-32 |
| Single-organism process | GO:0044699 | 1.19E-31 |
| Negative regulation of biological process | GO:0048519 | 2.23E-31 |
| Organ development | GO:0048513 | 4.68E-31 |
| Negative regulation of cellular process | GO:0048523 | 1.05E-29 |
| Skeletal system morphogenesis | GO:0048705 | 8.98E-29 |
| Positive regulation of cellular process | GO:0048522 | 4.27E-28 |
| Locomotion | GO:0040011 | 2.41E-27 |
| Regulation of developmental process | GO:0050793 | 3.03E-27 |
| Skeletal system development | GO:0001501 | 2.58E-26 |
| Downregulated genes | ||
| Biological adhesion | GO:0022610 | 9.19E-24 |
| Cell adhesion | GO:0007155 | 6.91E-23 |
| Localization | GO:0051179 | 1.50E-20 |
| Single-organism process | GO:0044699 | 8.10E-18 |
| Immune response | GO:0006955 | 3.70E-16 |
| Establishment of localization | GO:0051234 | 4.76E-16 |
| Cell–cell adhesion | GO:0016337 | 5.47E-16 |
| Single-organism cellular process | GO:0044763 | 2.08E-15 |
| Single-organism transport | GO:0044765 | 4.31E-15 |
| Transport | GO:0006810 | 8.75E-15 |
| Regulation of response to stimulus | GO:0048583 | 1.00E-14 |
| Immune system process | GO:0002376 | 6.14E-14 |
| Cellular process | GO:0009987 | 7.46E-14 |
| Response to wounding | GO:0009611 | 3.72E-13 |
| Inflammatory response | GO:0006954 | 8.22E-13 |
| Positive regulation of biological process | GO:0048518 | 1.21E-12 |
| Regulation of localization | GO:0032879 | 1.27E-12 |
| Cell junction organization | GO:0034330 | 5.31E-12 |
| Ion transport | GO:0006811 | 7.26E-12 |
| System development | GO:0048731 | 1.83E-11 |
GO, gene ontology.
Table 2.
GO Term Analysis for “Molecular Functions”
| GO term | GO term ID | p-Value |
|---|---|---|
| Upregulated genes | ||
| Binding | GO:0005488 | 4.94E-37 |
| Protein binding | GO:0005515 | 6.38E-24 |
| Nucleic acid binding transcription factor activity | GO:0001071 | 3.98E-18 |
| Sequence-specific DNA binding transcription factor activity | GO:0003700 | 1.45E-17 |
| Ion binding | GO:0043167 | 7.25E-17 |
| Metal ion binding | GO:0046872 | 1.79E-16 |
| Cation binding | GO:0043169 | 2.25E-16 |
| Sequence-specific DNA binding | GO:0043565 | 3.90E-16 |
| Carbohydrate derivative binding | GO:0097367 | 8.51E-12 |
| Calcium ion binding | GO:0005509 | 1.55E-11 |
| Glycosaminoglycan binding | GO:0005539 | 3.91E-11 |
| DNA binding | GO:0003677 | 1.25E-10 |
| Sulfur compound binding | GO:1901681 | 1.30E-09 |
| Identical protein binding | GO:0042802 | 1.16E-08 |
| Chromatin binding | GO:0003682 | 2.18E-08 |
| Growth factor binding | GO:0019838 | 2.32E-08 |
| Heparin binding | GO:0008201 | 5.35E-08 |
| Receptor binding | GO:0005102 | 6.70E-08 |
| Extracellular matrix structural constituent | GO:0005201 | 1.13E-07 |
| Extracellular matrix binding | GO:0050840 | 1.74E-07 |
| Downregulated genes | ||
| Protein binding | GO:0005515 | 2.08E-26 |
| Binding | GO:0005488 | 2.14E-26 |
| Calcium ion binding | GO:0005509 | 1.31E-16 |
| Actin binding | GO:0003779 | 1.34E-10 |
| Cytoskeletal protein binding | GO:0008092 | 2.79E-09 |
| Ion binding | GO:0043167 | 1.73E-07 |
| Receptor binding | GO:0005102 | 3.08E-07 |
| Phospholipid binding | GO:0005543 | 6.96E-07 |
| Lipid binding | GO:0008289 | 8.23E-07 |
| Structural molecule activity | GO:0005198 | 1.07E-06 |
| S100 protein binding | GO:0044548 | 3.42E-06 |
| Anion binding | GO:0043168 | 8.43E-06 |
| Transporter activity | GO:0005215 | 1.41E-05 |
| Identical protein binding | GO:042802 | 2.99E-05 |
| Chemokine receptor binding | GO:0042379 | 4.62E-05 |
| Chemokine activity | GO:0008009 | 4.89E-05 |
| Actin filament binding | GO:0051015 | 6.15E-05 |
| Immunoglobulin binding | GO:0019865 | 6.73E-05 |
| Transmembrane transporter activity | GO:0022857 | 8.18E-05 |
| Protein complex binding | GO:0032403 | 9.45E-05 |
Table 3.
Signal Transduction Pathway Associations
| Pathway | Pathway ID | p-Value |
|---|---|---|
| Upregulated | ||
| TGF-β | PW_TGFBR_MUS_MUSCULUS | 1.69E-18 |
| Mothers against DPP homolog | PW_SMAD_MUS_MUSCULUS | 1.06E-08 |
| Matrix metalloproteinase | PW_MMP_MUS_MUSCULUS | 7.90E-05 |
| Platelet/endothelial cell adhesion molecule 1 (CD31) | PW_PECAM1_MUS_MUSCULUS | 1.08E-04 |
| Vascular endothelial growth factor | PW_VEGF_MUS_MUSCULUS | 1.90E-04 |
| CCAAT/enhancer binding protein (C/EBP), Alpha | PW_CEBPA_MUS_MUSCULUS | 1.94E-04 |
| Platelet-derived growth factor | PW_PDGF_MUS_MUSCULUS | 3.20E-04 |
| Chemokine (C C Motif) ligand 2 | PW_CCL2_MUS_MUSCULUS | 4.71E-04 |
| Wingless type | PW_WNT_MUS_MUSCULUS | 5.05E-04 |
| Fibroblast growth factor | PW_FGF_MUS_MUSCULUS | 5.26E-04 |
| Indian hedgehog | PW_SHH_MUS_MUSCULUS | 6.22E-04 |
| Nuclear receptor subfamily 2, group F, member 2 | PW_NR2F2_MUS_MUSCULUS | 7.42E-04 |
| Low-density lipoprotein receptor related protein | PW_LRP_MUS_MUSCULUS | 8.34E-04 |
| C kit | PW_C KIT_MUS_MUSCULUS | 1.26E-03 |
| Cadherin 1, type 1, E cadherin (Epithelial) | PW_CDH1_MUS_MUSCULUS | 2.15E-03 |
| Angiotensin | PW_AGT_MUS_MUSCULUS | 2.57E-03 |
| Hedgehog | PW_HH_MUS_MUSCULUS | 2.70E-03 |
| Patched homolog 1 (drosophila) | PW_PTCH1_MUS_MUSCULUS | 5.87E-03 |
| Beta catenin | PW_BETA CATENIN_MUS_MUSCULUS | 6.45E-03 |
| Rho associated, coiled coil containing protein kinase | PW_ROCK_MUS_MUSCULUS | 6.65E-03 |
| FMS-like receptor tyrosine kinase 3 | PW_FLT3_MUS_MUSCULUS | 7.73E-03 |
| Peroxisome proliferator activated receptor delta | PW_PPARD_MUS_MUSCULUS | 8.07E-03 |
| Notch | PW_NOTCH_MUS_MUSCULUS | 8.98E-03 |
| NF kappa B | PW_NFKB_MUS_MUSCULUS | 9.51E-03 |
| Caveolin 1 | PW_CAVEOLIN_MUS_MUSCULUS | 9.80E-03 |
| Downregulated | ||
| Integrin | PW_INTEGRIN_MUS_MUSCULUS | 1.87E-07 |
| Spleen tyrosine kinase | PW_SYK_MUS_MUSCULUS | 2.58E-05 |
| V yes1 Yamaguchi sarcoma viral related oncogene Homolog; Lck/yes-related novel protein tyrosine kinase | PW_LYN_MUS_MUSCULUS | 2.84E-04 |
| Myeloid differentiation primary response gene (88) | PW_MYD88_MUS_MUSCULUS | 4.42E-04 |
| Bruton's tyrosine kinase | PW_BTK_MUS_MUSCULUS | 5.06E-04 |
| Zeta chain (TCR) associated protein kinase 70 kDa | PW_ZAP_70_MUS_MUSCULUS | 8.22E-04 |
| Cadherin 1, type 1, E cadherin (epithelial) | PW_CDH1_MUS_MUSCULUS | 8.34E-04 |
| Interleukin 6 signal transducer (GP130, oncostatin M receptor) | PW_IL6ST_MUS_MUSCULUS | 3.04E-03 |
| Protein kinase C | PW_PKC_MUS_MUSCULUS | 3.13E-03 |
| Epidermal growth factor | PW_EGF_MUS_MUSCULUS | 3.28E-03 |
| CD19 | PW_CD19_MUS_MUSCULUS | 3.96E-03 |
| Tyrosine protein kinase SRC | PW_SRC_MUS_MUSCULUS | 5.53E-03 |
| Hepatocyte growth factor receptor | PW_HGF_MUS_MUSCULUS | 5.96E-03 |
| Signal transducer and activator of transcription | PW_STAT_MUS_MUSCULUS | 6.44E-03 |
| Gardner Rasheed feline sarcoma viral (V FGR) Oncogene homolog | PW_FGR_MUS_MUSCULUS | 7.97E-03 |
| C terminal SRC kinase | PW_CSK_MUS_MUSCULUS | 8.38E-03 |
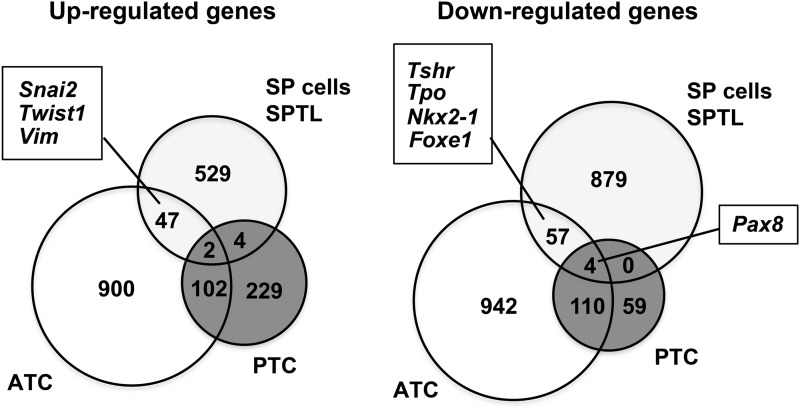
Gene-expression patterns of SP/SPTL cells were compared with mRNA profiles of 59 thyroid tumors (11 ATC and 48 papillary thyroid carcinoma [PTC]) obtained by microarray analysis, as described in (35). A Venn diagram was drawn after combining all altered genes together (Fig. 6 and Supplementary Table S3). The SP/SPTL cells and ATC shared 49 commonly up- and 61 commonly downregulated genes, while SP/SPTL cells and PTC shared only six commonly up- and four downregulated genes. The commonly upregulated genes in SP/SPTL cells and ATC included Snai2, Twist1, and Vim, while the commonly downregulated genes included Tshr, Tpo, Nkx2-1, and Foxe1. Pax 8 was found in commonly downregulated genes among all three groups: SP/SPTL, ATC, and PTC cells. Thus, SPTL/SP cells and ATC share enriched EMT markers.
FIG. 6.
Venn diagram showing the similarity of SP/SPTL cells to anaplastic thyroid carcinoma (ATC) in gene-expression patterns. Data on ATC gene-expression patterns are from Hebrant et al. (35). Commonly up- and downregulated genes between SP/SPTL cells, ATC, and/or papillary thyroid carcinoma are shown.
Discussion
The establishment of a mouse thyroid cell line SPTL, derived from mouse thyroid SP cells that partially exhibit stem/progenitor characteristics, is herein described. This cell line offers a continuous source that can be obtained in large quantities for the study of adult thyroid stem/progeny cells. Previously, it has been demonstrated that approximately half of the thyroid SP cells express SCA1 (9). The newly established SPTL cell line expresses SCA1 at 100%, while SCA1 expression decreases once the SPTL cells start differentiating, thus suggesting that SCA1 could be used as a marker for thyroid stem/progenitor cells. Electron microscopy revealed that SPTL cells are not fully differentiated compared with rat thyroid-derived FRTL-424 and FRTL-5 cells and rat follicular cells (25,26). It is believed that this cell line is not the result of contaminated fibroblasts that overgrew during passages because (i) an increased expression of Nkx2-1 was not observed when NIH3T3 cells as representative mouse fibroblast cells were cultured in low serum; (ii) no fibers were detected in the electron microscope images, which quite often can be seen if the cells are fibroblast; and (iii) the expression of the thyroid-specific transcription factor NKX2-1 was increased when cells were cultured in low serum (see below).
SPTL cells did not express typical stemness genes such as Oct4, Nanog, and Rex1. Instead, they expressed Gata transcription factors, known to be endoderm specification genes (36,37), suggesting that SPTL cells may have already been committed to form endoderm lineage cells. This seems reasonable, given that SPTL cells were established from thyroid SP cells. The stem/progenitor-like characteristics of SPTL cells were clearly demonstrated by their ability to grow continuously and by the induction of transcription factors critical for thyroid differentiation such as NKX2-1 in low serum culture in vitro, and the incorporation into follicles in vivo. It is known that various growth factors contained in serum play a role in the proliferation of cells. Cells continuously proliferate under high serum concentrations (10%), while low serum concentrations (1%) permit their terminal differentiation (30,31,38). Under low serum culturing of SPTL cells, the induction of thyroid differentiation marker genes other than Nkx2-1 was not observed, although the expression of Foxe1, Nis (Scl5a5), and Tpo mRNAs appeared to have slightly increased toward the end of six days in culture. It is felt that this increase may not be biologically meaningful due to the high qRT-PCR Cq values (for Foxe1 and Tpo >32), absent expression of PAX8 (particularly on day 6), and the extremely low relative expression levels of Pax8 and Foxe1 mRNAs compared with primary thyroid cells. PAX8 and FOXE1, in addition to NKX2-1, are the critical transcription factors for the expression of most thyroid differentiation marker genes including Nis and Tpo (39–41). The addition of other growth factors to the media such as FGFs, EGF, and activin A, with and without TSH (32,33), did not enhance Nkx2-1 expression, nor did it induce the expression of any other thyroid differentiation marker genes. Thus, SPTL cells appear to start differentiating into thyroid-like cells under low serum conditions, but they cannot fully differentiate.
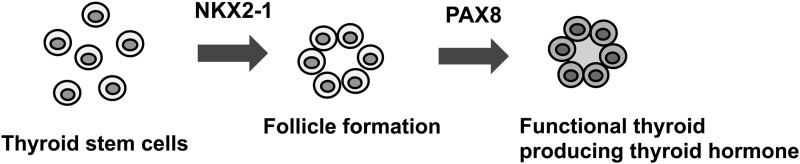
Induced expression of NKX2-1 was found in only 1.3% of SPTL cells, in which no PAX8 expression was observed. Others found that high expression of Nkx2-1 and Pax8 was required and sufficient to induce the expression of all thyroid differentiation marker genes and to form functional follicles from ES cells (42). In follicle-like structures obtained in 3D cultures in vitro, no expression of NKX2-1, PAX8, or thyroglobulin as thyroid differentiation markers were found. Results for the expression of TPO, TSHR, and NIS could not be obtained due to the lack of availability of appropriate antibodies that work in mouse tissues. A number of different techniques/maneuvers, including those to produce spheres before subjection to 3D cultures, and the use of different cell/sphere numbers and culture conditions were tried to obtain a condition that better induces differentiation of SPTL cells in 3D culture. Unfortunately, it was not possible to produce follicles that are positive for the aforementioned thyroid differentiation markers. The low number of NKX2-1-positive cells with no PAX8 expression may be one of the reasons that SPTL cells did not form functional follicles in vitro. It is possible that many differentiated cells, or cells having the capacity to differentiate fully, may be required to produce functional follicles. On the other hand, in the mouse thyroid in vivo, the majority of cells are differentiated follicular cells, which may result in a few GFP-positive SPTL cells with weak expression of NKX2-1 to become part of the follicles through a currently unknown mechanism. Alternatively, folliculogenesis may consist of two steps: NKX2-1 is required for the formation of follicles, followed by the expression of PAX8 that is required for the development of functional follicles, a stage where many thyroid differentiation marker genes become expressed (Fig. 7). It is currently not known whether the NKX2-1-positive cells that were found in mice eventually express PAX8 and differentiate into functional follicular cells. It is also worth noting that SPTL cells when injected through the tail vein, homed to the thyroid, the tissue of their origin. Homing of stem cells to a target tissue has been described for many years, mainly for bone marrow and other source-derived mesenchymal stem cells, as well as other stem cells such as epidermal neural crest stem cells and cardiac progenitor cells (34,43,44). To the authors' knowledge, this is the first report showing that thyroid stem/progenitor-like cells home to the thyroid.
FIG. 7.
Two-step process of thyroid folliculogenesis. Thyroid follicle formation requires NKX2-1 expression, while the expression of PAX8 is required for the follicle to become functional, resulting in the expression of many thyroid differentiation marker genes and eventual thyroid hormone synthesis.
One of the interesting aspects of the current study is that SPTL and SP cells share common features, representative of EMT compared with MP cells, and that SPTL cells appear to be enriched in the expression of EMT-related genes. EMT is known to play a role in the maintenance of stemness, development, and diseases, particularly cancer (45–48). In a comparative study of gene-expression patterns between ATC versus PTC and FTC, the TGF-β signaling pathway was among the top identified pathways (49). Further, using human thyroid tissues, the expression of SNAI1, SNAI2, TWIST1, and/or ZEB1 was found only in ATC or ATC-derived cells, but not normal cells or other thyroid tumor tissues or cell lines, whereas CDH1 was expressed only in the latter thyroid tissues/cells, but not in ATC tissues/cells, as examined by immunohistochemistry and/or qRT-PCR (50–52). Based on these results, it is suggested that SPTL cells have more similarity to ATC than they do to PTC in gene expression profiles. However, it should be noted that there is a possibility that SPTL cells express genes representative of EMT, as found in ATC, simply because they are already in a fibroblastoid mode, as seen in the two-dimensional culture. It was recently demonstrated that mouse thyroid SP cells highly express genes involved in cancer and/or cancer metastasis/invasion compared with non-SP cells by microarray analysis (14). However, the fact that SPTL cells did not develop tumors for at least six months following injection suggests that an EMT-rich condition does not always predispose to carcinogenesis, although there remains the possibility that it may take a longer latency time.
In conclusion, SPTL cells were established from mouse thyroid SP cells that can partially differentiate into thyroid-like cells, and be incorporated as part of follicles in in vivo mouse injection models. SPTL cells have gene-expression patterns enriched in EMT, the main player involved in stemness and cancer. Thus, SPTL cells may provide an excellent tool to study thyroid adult stem/progenitor cells, including thyroid cancer stem cells, and to understand their roles and the mechanisms of thyroid carcinogenesis.
Supplementary Material
Acknowledgments
We would like to thank Subhadra Banerjee and Karen M. Wolcott (Flow Cytometry Core Facility, Laboratory of Genome Integrity, CCR, NCI) for their help in FACS analysis; Poonam Mannan, Langston Lim, and Susan H. Garfield (CCR Confocal Microscopy Core Facility, Laboratory of Cancer Biology and Genetics, NCI) for their help in confocal microscopy; Jerrold Ward (VetPathology, Montgomery Village, MD) for his advice on electron microscope analysis; and Tao Deng (NCI) for providing us with ES cells cDNA for qRT-PCR analysis.
This work was supported by an Intramural Research Program of National Cancer Institute, Center for Cancer Research ZIABC005522 (to S.K.), National Heart Lung and Blood Institute 1ZICHL006058 (to J.Z.), Federal funds from the National Cancer Institute, National Institutes of Health under contract HHSN26120080001E (to K.N.), and Japan Society for the Promotion of Science Research Fellowship for Japanese Biomedical and Behavioral Researchers at NIH (to T.M.).
Author Disclosure Statement
The authors have nothing to disclose.
References
- 1.Zhang Y, Xu L, Wang S, Cai C, Yan L. 2014. Concise review: differentiation of human adult stem cells into hepatocyte-like cells in vitro. Int J Stem Cells 7:49–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aggarwal S, Moggio A, Bussolati B. 2013. Concise review: stem/progenitor cells for renal tissue repair: current knowledge and perspectives. Stem Cells Transl Med 2:1011–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liao SY, Tse HF. 2013. Multipotent (adult) and pluripotent stem cells for heart regeneration: what are the pros and cons? Stem Cell Res Ther 4:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouwens L, Houbracken I, Mfopou JK. 2013. The use of stem cells for pancreatic regeneration in diabetes mellitus. Nat Rev Endocrinol 9:598–606 [DOI] [PubMed] [Google Scholar]
- 5.Dumont JE, Lamy F, Roger P, Maenhaut C. 1992. Physiological and pathological regulation of thyroid cell proliferation and differentiation by thyrotropin and other factors. Physiol Rev 72:667–697 [DOI] [PubMed] [Google Scholar]
- 6.Capen CC, Martin SL. 1989. The effects of xenobiotics on the structure and function of thyroid follicular and C-cells. Toxicol Pathol 17:266–293 [DOI] [PubMed] [Google Scholar]
- 7.Capen CC. 1997. Mechanistic data and risk assessment of selected toxic end points of the thyroid gland. Toxicol Pathol 25:39–48 [DOI] [PubMed] [Google Scholar]
- 8.Hancock SL, McDougall IR, Constine LS. 1995. Thyroid abnormalities after therapeutic external radiation. Int J Radiat Oncol Biol Phys 31:1165–1170 [DOI] [PubMed] [Google Scholar]
- 9.Hoshi N, Kusakabe T, Taylor BJ, Kimura S. 2007. Side population cells in the mouse thyroid exhibit stem/progenitor cell-like characteristics. Endocrinology 148:4251–4258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lan L, Cui D, Nowka K, Derwahl M. 2007. Stem cells derived from goiters in adults form spheres in response to intense growth stimulation and require thyrotropin for differentiation into thyrocytes. J Clin Endocrinol Metab 92:3681–3688 [DOI] [PubMed] [Google Scholar]
- 11.Fierabracci A, Puglisi MA, Giuliani L, Mattarocci S, Gallinella-Muzi M. 2008. Identification of an adult stem/progenitor cell-like population in the human thyroid. J Endocrinol 198:471–487 [DOI] [PubMed] [Google Scholar]
- 12.Thomas T, Nowka K, Lan L, Derwahl M. 2006. Expression of endoderm stem cell markers: evidence for the presence of adult stem cells in human thyroid glands. Thyroid 16:537–544 [DOI] [PubMed] [Google Scholar]
- 13.Goodell MA, Brose K, Paradis G, Conner AS, Mulligan RC. 1996. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J Exp Med 183:1797–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayase S, Sasaki Y, Matsubara T, Seo D, Miyakoshi M, Murata T, Ozaki T, Kakudo K, Kumamoto K, Ylaya K, Cheng SY, Thorgeirsson SS, Hewitt SM, Ward JM, Kimura S. 2015. Expression of stanniocalcin 1 in thyroid side population cells and thyroid cancer cells. Thyroid 25:425–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ozaki T, Matsubara T, Seo D, Okamoto M, Nagashima K, Sasaki Y, Hayase S, Murata T, Liao XH, Hanson J, Rodriguez-Canales J, Thorgeirsson SS, Kakudo K, Refetoff S, Kimura S. 2012. Thyroid regeneration: characterization of clear cells after partial thyroidectomy. Endocrinology 153:2514–2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kimura S.2014Thyroid regeneration: how stem cells play a role? Front Endocrinol (Lausanne) 5:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okamoto M, Hayase S, Miyakoshi M, Murata T, Kimura S. 2013. Stem cell antigen 1-positive mesenchymal cells are the origin of follicular cells during thyroid regeneration. PLoS One 8:e80801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kimura S, Hara Y, Pineau T, Fernandez-Salguero P, Fox CH, Ward JM, Gonzalez FJ. 1996. The T/ebp null mouse: thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain, and pituitary. Genes Dev 10:60–69 [DOI] [PubMed] [Google Scholar]
- 19.De Felice M, Di Lauro R. 2004. Thyroid development and its disorders: genetics and molecular mechanisms. Endocr Rev 25:722–746 [DOI] [PubMed] [Google Scholar]
- 20.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25:1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramskold D, Wang ET, Burge CB, Sandberg R. 2009. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput Biol 5:e1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang da W, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57 [DOI] [PubMed] [Google Scholar]
- 24.Deng T, Zhu ZI, Zhang S, Leng F, Cherukuri S, Hansen L, Marino-Ramirez L, Meshorer E, Landsman D, Bustin M. 2013. HMGN1 modulates nucleosome occupancy and DNase I hypersensitivity at the CpG island promoters of embryonic stem cells. Mol Cell Biol 33:3377–3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Avvedimento VE, Monticelli A, Tramontano D, Polistina C, Nitsch L, Di Lauro R. 1985. Differential expression of thyroglobulin gene in normal and transformed thyroid cells. Eur J Biochem 149:467–472 [DOI] [PubMed] [Google Scholar]
- 26.De Felice M, Di Lauro R. 2015. Anatomy and development of the thyroid. Available at: http://clinicalgate.com/anatomy-and-development-of-the-thyroid/ (accessed February10, 2017)
- 27.Welm BE, Tepera SB, Venezia T, Graubert TA, Rosen JM, Goodell MA. 2002. Sca-1(pos) cells in the mouse mammary gland represent an enriched progenitor cell population. Dev Biol 245:42–56 [DOI] [PubMed] [Google Scholar]
- 28.Burger PE, Xiong X, Coetzee S, Salm SN, Moscatelli D, Goto K, Wilson EL. 2005. Sca-1 expression identifies stem cells in the proximal region of prostatic ducts with high capacity to reconstitute prostatic tissue. Proc Natl Acad Sci U S A 102:7180–7185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oh H, Bradfute SB, Gallardo TD, Nakamura T, Gaussin V, Mishina Y, Pocius J, Michael LH, Behringer RR, Garry DJ, Entman ML, Schneider MD. 2003. Cardiac progenitor cells from adult myocardium: homing, differentiation, and fusion after infarction. Proc Natl Acad Sci U S A 100:12313–12318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ram-Liebig G, Meye A, Hakenberg OW, Haase M, Baretton G, Wirth MP. 2004. Induction of proliferation and differentiation of cultured urothelial cells on acellular biomaterials. BJU Int 94:922–927 [DOI] [PubMed] [Google Scholar]
- 31.Yoshiko Y, Hirao K, Sakabe K, Seiki K, Takezawa J, Maeda N. 1996. Autonomous control of expression of genes for insulin-like growth factors during the proliferation and differentiation of C2C12 mouse myoblasts in serum-free culture. Life Sci 59:1961–1968 [DOI] [PubMed] [Google Scholar]
- 32.Ma R, Latif R, Davies TF. 2009. Thyrotropin-independent induction of thyroid endoderm from embryonic stem cells by activin A. Endocrinology 150:1970–1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Longmire TA, Ikonomou L, Hawkins F, Christodoulou C, Cao Y, Jean JC, Kwok LW, Mou H, Rajagopal J, Shen SS, Dowton AA, Serra M, Weiss DJ, Green MD, Snoeck HW, Ramirez MI, Kotton DN. 2012. Efficient derivation of purified lung and thyroid progenitors from embryonic stem cells. Cell Stem Cell 10:398–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jackson JS, Golding JP, Chapon C, Jones WA, Bhakoo KK. 2010. Homing of stem cells to sites of inflammatory brain injury after intracerebral and intravenous administration: a longitudinal imaging study. Stem Cell Res Ther 1:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hebrant A, Dom G, Dewaele M, Andry G, Tresallet C, Leteurtre E, Dumont JE, Maenhaut C. 2012. mRNA expression in papillary and anaplastic thyroid carcinoma: molecular anatomy of a killing switch. PLoS One 7:e37807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fujikura J, Yamato E, Yonemura S, Hosoda K, Masui S, Nakao K, Miyazaki Ji J, Niwa H. 2002. Differentiation of embryonic stem cells is induced by GATA factors. Genes Dev 16:784–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rojas A, Schachterle W, Xu SM, Black BL. 2009. An endoderm-specific transcriptional enhancer from the mouse Gata4 gene requires GATA and homeodomain protein-binding sites for function in vivo. Dev Dyn 238:2588–2598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kniss DA, Burry RW. 1988. Serum and fibroblast growth factor stimulate quiescent astrocytes to re-enter the cell cycle. Brain Res 439:281–288 [DOI] [PubMed] [Google Scholar]
- 39.Ohno M, Zannini M, Levy O, Carrasco N, di Lauro R. 1999. The paired-domain transcription factor Pax8 binds to the upstream enhancer of the rat sodium/iodide symporter gene and participates in both thyroid-specific and cyclic-AMP-dependent transcription. Mol Cell Biol 19:2051–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zannini M, Francis-Lang H, Plachov D, Di Lauro R. 1992. Pax-8, a paired domain-containing protein, binds to a sequence overlapping the recognition site of a homeodomain and activates transcription from two thyroid-specific promoters. Mol Cell Biol 12:4230–4241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fernandez LP, Lopez-Marquez A, Martinez AM, Gomez-Lopez G, Santisteban P. 2013. New insights into FoxE1 functions: identification of direct FoxE1 targets in thyroid cells. PLoS One 8:e62849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Antonica F, Kasprzyk DF, Opitz R, Iacovino M, Liao XH, Dumitrescu AM, Refetoff S, Peremans K, Manto M, Kyba M, Costagliola S. 2012. Generation of functional thyroid from embryonic stem cells. Nature 491:66–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bussolati B, Hauser PV, Carvalhosa R, Camussi G. 2009. Contribution of stem cells to kidney repair. Curr Stem Cell Res Ther 4:2–8 [DOI] [PubMed] [Google Scholar]
- 44.Sharif F, Bartunek J, Vanderheyden M. 2011. Adult stem cells in the treatment of acute myocardial infarction. Catheter Cardiovasc Interv 77:72–83 [DOI] [PubMed] [Google Scholar]
- 45.Thiery JP, Acloque H, Huang RY, Nieto MA. 2009. Epithelial–mesenchymal transitions in development and disease. Cell 139:871–890 [DOI] [PubMed] [Google Scholar]
- 46.Gomes LR, Terra LF, Sogayar MC, Labriola L. 2011. Epithelial–mesenchymal transition: implications in cancer progression and metastasis. Curr Pharm Biotechnol 12:1881–1890 [DOI] [PubMed] [Google Scholar]
- 47.Lamouille S, Xu J, Derynck R. 2014. Molecular mechanisms of epithelial–mesenchymal transition. Nat Rev Mol Cell Biol 15:178–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sakaki-Yumoto M, Katsuno Y, Derynck R. 2013. TGF-beta family signaling in stem cells. Biochim Biophys Acta 1830:2280–2296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Montero-Conde C, Martin-Campos JM, Lerma E, Gimenez G, Martinez-Guitarte JL, Combalia N, Montaner D, Matias-Guiu X, Dopazo J, de Leiva A, Robledo M, Mauricio D. 2008. Molecular profiling related to poor prognosis in thyroid carcinoma. Combining gene expression data and biological information. Oncogene 27:1554–1561 [DOI] [PubMed] [Google Scholar]
- 50.Buehler D, Hardin H, Shan W, Montemayor-Garcia C, Rush PS, Asioli S, Chen H, Lloyd RV. 2013. Expression of epithelial–mesenchymal transition regulators SNAI2 and TWIST1 in thyroid carcinomas. Mod Pathol 26:54–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hardy RG, Vicente-Duenas C, Gonzalez-Herrero I, Anderson C, Flores T, Hughes S, Tselepis C, Ross JA, Sanchez-Garcia I. 2007. Snail family transcription factors are implicated in thyroid carcinogenesis. Am J Pathol 171:1037–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salerno P, Garcia-Rostan G, Piccinin S, Bencivenga TC, Di Maro G, Doglioni C, Basolo F, Maestro R, Fusco A, Santoro M, Salvatore G. 2011. TWIST1 plays a pleiotropic role in determining the anaplastic thyroid cancer phenotype. J Clin Endocrinol Metab 96:E772–781 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.