Abstract
In comparison to genome-wide association studies (GWAS), there has been poor replication of gene expression studies in chronic obstructive pulmonary disease (COPD). We performed microarray gene expression profiling on a large sample of resected lung tissues from subjects with severe COPD. Comparing 111 COPD cases and 40 control smokers, 204 genes were differentially expressed; none were at significant GWAS loci. The top differentially expressed gene was HMGB1, which interacts with AGER, a known COPD GWAS gene. Differentially expressed genes showed enrichment for putative interactors of the first three identified COPD GWAS genes IREB2, HHIP, and FAM13A, based on gene sets derived from protein and RNA binding studies, RNA-interference, a murine smoking model, and expression quantitative trait locus analyses. The gene module most highly associated for COPD in Weighted Gene Co-Expression Network Analysis (WGCNA) was enriched for B cell pathways, and shared seventeen genes with a mouse smoking model and twenty genes with previous emphysema studies. As in other common diseases, genes at COPD GWAS loci were not differentially expressed; however, using a combination of network methods, experimental studies and careful phenotype definition, we found differential expression of putative interactors of these genes, and we replicated previous human and mouse microarray results.
Chronic obstructive pulmonary disease (COPD) is characterized by progressive airflow obstruction accompanied by chronic inflammation. It is the third leading cause of morbidity and mortality worldwide1. Although cigarette smoking is the major risk factor, multiple studies have demonstrated a genetic component to COPD susceptibility2,3,4,5. Genome-wide association studies (GWAS) have identified multiple genetic loci associated with COPD susceptibility, with replication of the results across different populations6,7,8.
Many published studies have performed microarray gene expression profiling in COPD lungs9,10,11,12,13,14,15,16,17. In contrast to the GWAS, there has been minimal overlap between the differentially expressed genes identified in each microarray study18; greater overlap has been found for differentially expressed pathways. These microarray studies have been limited by variable definitions of COPD, incomplete consideration of past and current smoking status, failure to consider quantitative traits and COPD heterogeneity (such as emphysema and airway disease), and until recently, small sample sizes19,20. The COPD microarray studies have not found robust differential expression of the genes at GWAS loci, which has been shown in other complex diseases and traits, including coronary heart disease, height, type 2 diabetes, and autism21,22,23,24. Although most GWAS single nucleotide polymorphisms (SNPs) are likely regulatory variants and perhaps eQTLs25,26,27,28, the magnitude of effect of these SNPs on gene expression may be too subtle to capture with genome-wide expression studies in disease tissue.
We hypothesized that the genes identified at COPD GWAS loci (GWAS genes) may point to pathways and interacting gene networks important for COPD pathogenesis, and that genes in these pathways may be differentially expressed in lung tissues. However, the pathways relevant for many COPD GWAS genes are largely unknown. Therefore, we focused on the first three identified COPD GWAS genes, which have the strongest association signals: IREB2 (iron responsive element binding protein 2), HHIP (hedgehog interacting protein) and FAM13A (family with sequence similarity 13 member A)6, for which multiple omics datasets have been previously generated. Other COPD GWAS genes lacked the breadth of omics data required for a similarly thorough study. We used these available data, from in vitro and in vivo models29,30,31,32 and in silico analyses, to identify gene sets functionally related to these genes. We merged these results with gene expression profiling from human COPD lung tissues and former smoker controls to demonstrate that the genes related to the GWAS genes, but not the GWAS genes themselves, were differentially expressed in the disease-relevant tissue.
A Weighted Gene Co-expression Network Analysis (WGCNA) was performed to identify gene modules associated with COPD status, as network medicine approaches33,34 provide insight into the understanding of complex disease and have been applied to the study of COPD32,35,36, heart disease21, diabetes23 and autism spectrum disorder24. By leveraging these methods in lung tissue, and examining interacting gene set enrichment, co-expression networks and pathways instead of individual genetic determinants, this study sought to identify key genes and gene modules involved in the molecular pathogenesis and etiology of COPD.
Results
Gene expression association with phenotype variables
We performed microarray gene expression profiling on lung tissue samples from 111 COPD cases and 40 control smokers with normal lung function (Supplemental Table S1). The differential expression analysis using R and the package limma (see Methods) identified 214 probes (mapped to 204 genes) associated with COPD at a 5% FDR; the top results are provided in Table 1 and all significant results may be found in Supplemental Table S2. The top differentially expressed probe is annotated to HMGB1. This probe was also the top gene expression association with lung function measures forced expiratory volume in 1 sec (FEV1)% predicted and the ratio of FEV1 to forced vital capacity (FVC). None of the previously identified genome-wide significant COPD GWAS genes (p-value < 5e-8) from the NHGRI-EBI Catalog (www.ebi.ac.uk/gwas/) were significantly differentially expressed (Table 2). A summary of the results for the other COPD-related phenotypes is provided in Supplemental Table S3. There were 1,556 differentially expressed probes (mapped to 1,366 genes) at a 5% FDR for FEV1%predicted and 1,689 differentially expressed probes (mapped to 1,429 genes) for the ratio of FEV1 to FVC. In lung function analyses in COPD cases only, the numbers of differentially expressed probes were lower and the overlap with the results for the full cohort was modest (Supplemental Figure S1). However, nearly half of the case-only results are recapitulated in the full-cohort analyses.
Table 1. Top 20 genes differentially expressed in COPD vs. control.
| Probe name | Log Fold Difference* | p-value | FDR q-value | Gene symbol |
|---|---|---|---|---|
| ILMN_1676938 | −0.9164 | 5.40E-010 | 1.77E-005 | HMGB1 |
| ILMN_3187508 | −0.6609 | 1.70E-009 | 2.79E-005 | FLJ40504 |
| ILMN_2050921 | −0.6510 | 3.16E-009 | 3.46E-005 | C3orf78 |
| ILMN_2398474 | −0.7957 | 2.11E-008 | 1.73E-004 | RAP1B |
| ILMN_3245015 | −0.6185 | 3.18E-008 | 2.09E-004 | LOC440563 |
| ILMN_1665333 | −0.7186 | 6.52E-008 | 3.57E-004 | SUMO2 |
| ILMN_1705330 | −0.7185 | 8.61E-008 | 4.04E-004 | CDC42 |
| ILMN_1656898 | −0.7223 | 1.31E-007 | 5.37E-004 | PTCD1 |
| ILMN_3266294 | −0.6043 | 1.66E-007 | 6.05E-004 | SREK1IP1 |
| ILMN_1696031 | −0.5292 | 1.99E-007 | 6.22E-004 | C15orf21 |
| ILMN_1691485 | −0.6776 | 2.08E-007 | 6.22E-004 | GTF2H2 |
| ILMN_3191922 | −0.8825 | 2.28E-007 | 6.24E-004 | FLJ46111 |
| ILMN_2068122 | −0.6563 | 3.42E-007 | 8.63E-004 | TMEM65 |
| ILMN_3239871 | −0.5563 | 4.85E-007 | 1.07E-003 | ARPP19 |
| ILMN_3219808 | −0.5897 | 4.90E-007 | 1.07E-003 | HNRNPA1 |
| ILMN_1716246 | 0.8208 | 5.94E-007 | 1.22E-003 | FRZB |
| ILMN_3183789 | −0.3577 | 7.60E-007 | 1.47E-003 | UQCRFS1 |
| ILMN_1799837 | −0.6066 | 1.07E-006 | 1.95E-003 | POTEM |
| ILMN_2141398 | −0.6715 | 1.32E-006 | 2.24E-003 | VEZT |
| ILMN_1674399 | 0.3467 | 1.40E-006 | 2.24E-003 | ZNF143 |
*Reference group is controls: positive log fold difference corresponds to higher expression in cases.
Table 2. Differential expression results for putative genes at previously identified genome-wide significant COPD and emphysema GWAS loci (probe with highest ranking result shown).
| Gene | Probe | P-value | FDR q-value | Log fold difference* |
|---|---|---|---|---|
| MMP12 | ILMN_2073758 | 0.02 | 0.30 | 0.91 |
| HHIP | ILMN_1675453 | 0.04 | 0.42 | 0.41 |
| AGER | ILMN_1729777 | 0.11 | 0.59 | −0.31 |
| IREB2 | ILMN_1726554 | 0.12 | 0.60 | 0.21 |
| DLC1 | ILMN_1698020 | 0.16 | 0.66 | −0.24 |
| CHRNA5 | ILMN_1770044 | 0.19 | 0.67 | 0.12 |
| RAB4B | ILMN_1761896 | 0.33 | 0.80 | 0.096 |
| FAM13A | ILMN_1800267 | 0.34 | 0.80 | −0.09 |
| CHRNA3 | ILMN_2154157 | 0.53 | 0.89 | 0.13 |
| RIN3 | ILMN_1731736 | 0.64 | 0.93 | 0.07 |
| TGFB2 | ILMN_1812526 | 0.89 | 0.98 | −0.03 |
*Reference group is controls: positive log fold difference corresponds to higher expression in cases.
Pathway enrichment tests were performed using the gene symbol annotations for the 214 probes (204 unique genes) differentially expressed by COPD case/control status (Supplemental Table S4). The two nominally significant Reactome results (FDR ≤ 0.1) were Regulation of actin dynamics for phagocytic cup formation::2029482 and Fcgamma receptor (FCGR) dependent phagocytosis::2029480. The higher false discovery rate threshold was chosen to highlight pathways or processes that may be of interest in future studies. The GO results identified a broad group of pathways, with processes related to protein modification and apoptosis being dominant.
Interactor enrichment in differentially expressed genes
While we did not observe differential expression of genes at GWAS loci, we noted that the top differentially expressed gene, HMGB1 (high mobility group box 1), has been identified as an important interacting partner of AGER (advanced glycosylation end product specific receptor)37, a gene implicated by GWAS for emphysema susceptibility38,39,40. This led us to test whether interacting partners of other COPD GWAS genes were also differentially expressed. The available in vitro, in vivo, and in silico datasets, described in the Methods section, defined sets of genes that may interact with COPD GWAS genes HHIP, FAM13A, and IREB2. These were tested for enrichment within the 204 COPD differentially-expressed genes. Table 3 shows that the genes differentially expressed in COPD lungs are enriched for interacting partners of IREB2, HHIP and FAM13A. There was similar enrichment in the 1,366 genes with expression associated with FEV1% predicted and the 1,429 genes associated with FEV1/FVC ratio. In the UpSet41 plot of the intersecting genes across the enrichment results (Supplemental Figure S2), the rows correspond to the rows of Table 3, and the gene sets for each row are the genes that overlap with the 204 differentially expressed genes (column three of Table 3). The intersecting genes for each column are listed in Supplemental Table S5. HMGB1 and CD79A were present in the intersection of IREB2 RNA immunoprecipitation sequencing (RIP-seq) and the FAM13A trans-eQTL overlaps, and SERPINE2 was found in the overlap for IREB2 RIP-seq (Supplemental Table S5 and Supplemental Figure S2). POU2AF1 and BCL11A were present in overlaps involving two different Hhip+/− mouse smoking models. BCL2 was found in the intersection of gene list overlaps for an Hhip+/− mouse model, IREB2 RIP-seq and FAM13A trans-eQTL.
Table 3. Enrichment of functional interactors of COPD GWAS genes in the gene expression results (differentially expressed genes with FDR < 0.05).
| Experiment | Number of genes or proteins* | Overlap with 204 COPD genes | Enrichment p-value for COPD genes | Overlap with 1,366 FEV1% predicted genes | Enrichment p-value for FEV1% predicted genes | Overlap with 1,429 FEV1/FVC genes | Enrichment p-value for FEV1/FVC genes |
|---|---|---|---|---|---|---|---|
| IREB2 RIP-seq | 4008 | 64 | 2.4e-5 | 359 | 3.6e-11 | 400 | <1.0e-12 |
| HHIP affinity purification | 216 | 5 | 0.062 | 34 | 1.8e-6 | 45 | 1.6e-11 |
| FAM13A affinity purification | 97 | 0 | 1 | 8 | 0.31 | 8 | 0.35 |
| Hhip+/+ vs. Hhip+/− mouse, 6 mo smoke | 492 | 15 | 0.00011 | 60 | 2.7e-6 | 61 | 5.4e-6 |
| Hhip mouse, genotype x smoke interaction | 549 | 12 | 0.0082 | 46 | 0.054 | 46 | 0.095 |
| IREB2 Irp2+/+ vs. Irp2−/− mouse knockout | 31 | 0 | 1 | 3 | 0.33 | 3 | 0.36 |
| HHIP shRNA knockdown | 266 | 3 | 0.49 | 20 | 0.30 | 19 | 0.46 |
| FAM13A siRNA knockdown | 598 | 13 | 0.0063 | 54 | 0.011 | 61 | 0.0013 |
| IREB2 trans-eQTLs at p < 0.05 | 1612 | 31 | 0.00023 | 171 | 1.4e-10 | 159 | 1.7e-6 |
| HHIP trans-eQTLs at p < 0.05 | 1560 | 21 | 0.087 | 143 | 2.4e-05 | 156 | 9.4e-7 |
| FAM13A trans-eQTL at p < 0.05 | 1753 | 38 | 2.7e-06 | 209 | <1.0e-12 | 194 | 8.7e-12 |
*Unique gene annotations found in the final expression data.
In a COPD GWAS dataset8, nominal association with case-control status was observed for SNPs annotated to BCL2 (rs1481031, p = 0.0003), BCL11A (rs191541310, p = 0.007), POU2AF1 (rs2282637, p = 0.008), HMGB1 (rs117938771, p = 0.01), and SERPINE2 (rs6721140, p = 0.03). No association was observed for CD79A (rs112580282, p = 0.2).
Cis-eQTL analysis
For the HHIP (rs11724319, p = 0.03) and FAM13A (rs2609255, p = 0.001) eQTLs, the risk allele dosage corresponds to increased expression, while for the IREB2 eQTL (rs2009746, p < 1.0e-10) the risk allele dosage corresponds to decreased expression8.
Network analysis
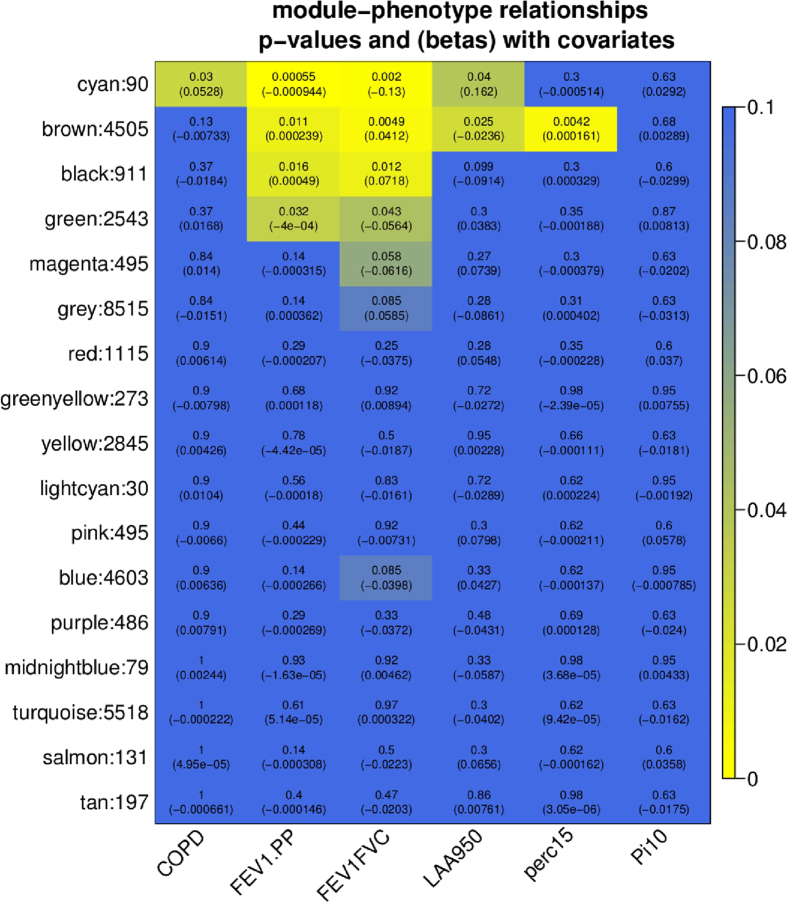
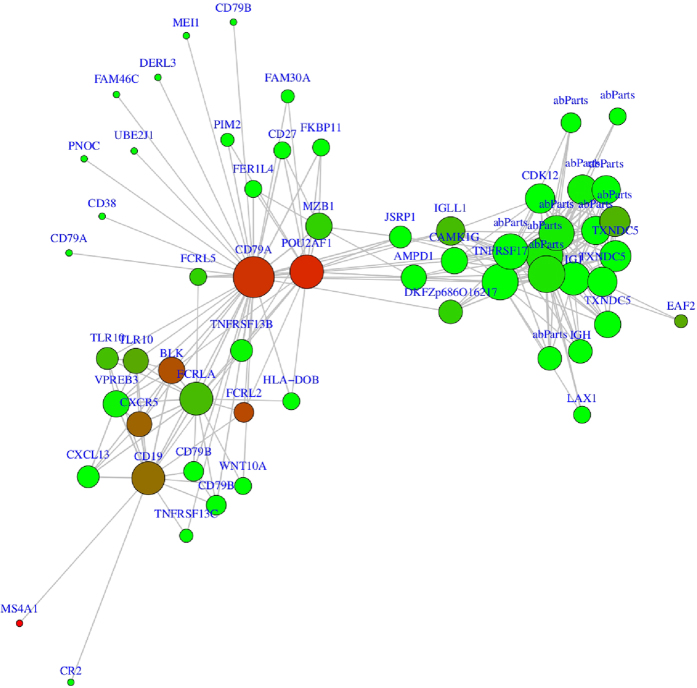
A weighted gene co-expression network was constructed to group together co-expressed genes prior to pathway analysis42,43. The final network consisted of 17 modules, ranging in size from 30 to 5,518 probes (Supplemental Table S6). There was high correlation within the modules as expected, with the mean module membership (MM), or mean correlation of gene expression with the module eigengene, varying from 0.53 to 0.79 across all modules. The grey module is a grouping of probes with outlying gene expression profiles and was not considered further. Tests of association between phenotype variables of interest and the module eigengenes were performed for each model (see Methods), and the results were summarized in a heatmap (Fig. 1). Only the cyan module (number of probes = 90) was significantly associated with COPD case-control status (FDR < 0.05); however, the cyan module and brown module (number of probes = 4505) were associated with multiple COPD-related phenotypes, including case-control status, two measures of lung function – FEV1% predicted and FEV1/FVC ratio – and emphysema, measured as low attenuation areas <−950 HU on chest computed tomography (CT) scans. The probes within the cyan module and the top probes from the brown module after sorting by gene significance and module membership values, are listed in Supplemental Tables S7 and S8; putative driver genes for the two modules are highlighted at the top. We define driver genes by gene significance (GS) FDR <0.05 in COPD differential expression analysis and higher module membership (MM) correlations. These include CD79A (GS = 0.03, MM = 0.93) and POU2AF1 (GS = 0.03, MM = 0.91) for cyan, and HMGB1 (GS = 1.77e-5, MM = 0.84) for the brown module. The probes annotated to AGER were found in the grey module, and therefore co-expression with other genes, including HMGB1, was not observed (Pearson r = 0.09 for HMGB1:AGER). The gene network in the cyan module is shown in Fig. 2. In this module, CD79A and POU2AF1 have high degree (high number of edges) and therefore appear as hubs. Seventeen genes in this module overlap with the mouse Hhip+/+ vs. Hhip+/− 6 month smoking model gene set (Supplemental Table S7, enrichment p-value 6.5e-14), demonstrating that pathways associated with lung damage and B cell aggregate formation32 are shared across species. This module includes 9 genes (15 probes) associated with emphysema in a previous lung tissue microarray expression study10 and 14 genes (17 probes) found to be differentially expressed between emphysema and bronchiolitis44. The brown module was relatively large and did not demonstrate sub-structures like those observed in the cyan module; sub-structures could have informed a network pruning step. In the COPD case-control analysis, the top probes annotated to HHIP, FAM13A and IREB2 were found in the grey, grey and blue modules, respectively.
Figure 1. Heatmap of module association with phenotype variables (color scale for adjusted p-value).
The top number in each cell corresponds to the FDR q-value and the bottom number is the beta coefficient from the linear regression model. COPD = COPD case-control status, FEV1.PP = forced expiratory volume in 1 sec, percent predicted; FEV1FVC = ratio of FEV1 to forced vital capacity (FVC); LAA950 = Low attenuation areas at −950 HU on chest computed tomography (CT) scans; perc15 = 15th percentile of the lung density histogram on chest CT scans; SRWA-Pi10 = square root wall area of a hypothetical airway with 10mm internal perimeter. The phenotype variables FEV1, FEV1FVC and perc15 decrease with COPD, while LAA950 and SRWA-Pi10 increase with disease.
Figure 2. Module that was significantly associated with COPD case-control status (cyan module sub-network).
Node size is proportional to the node degree and the color is related to the p-value in the differential expression analysis for COPD status (red lower p-value and green higher p-value).
Pathway enrichment tests were performed for the genes in the cyan and brown modules (Supplemental Tables S9 and S10). In contrast to the pathway enrichment results for the top COPD associated genes, the pathways enriched in the cyan module are more specific, demonstrating enrichments for B cell related processes. For the brown module, the results are mixed and broad, highlighted by RNA processing, apoptosis, immune system, and protein modification pathways.
Discussion
In a microarray study of surgically-resected human lung tissue from ex-smoking severe COPD subjects and controls, we discovered 214 differentially expressed probes, corresponding to 204 unique genes. We found that none of the significant genes from previous COPD GWAS in the NHGRI-EBI Catalog or emphysema GWAS39 were differentially expressed. As the top differentially expressed gene was HMGB1, a known interactor of the COPD and emphysema gene AGER, we sought to determine whether there was differential expression of the putative interactors for the first three identified COPD GWAS genes: IREB2, HHIP and FAM13A. Using a combination of experimental datasets and network analyses, we observed strong enrichment for putative interactors of IREB2, HHIP, and FAM13A. We identified a gene co-expression module strongly associated with COPD, and enriched for B cell functions. This module contained seventeen overlapping genes compared to the Hhip+/− smoking mouse model32, highlighting a cross-species signal of B cell related pathogenesis in COPD and emphysema. Using the strict phenotype definition of severe COPD in former smokers, we were able to replicate significant gene and B cell pathway findings from a previous emphysema gene expression study10, in contrast to the poor replication in many previous COPD microarray studies18.
Our top differentially expressed gene was HMGB1. The protein encoded by HMGB1 is an interacting partner of AGER (which encodes the emphysema biomarker sRAGE), a replicated lung function and emphysema associated gene38,39,45. Plasma HMGB1, along with sRAGE, has been found to be higher during acute exacerbations in COPD patients and lower during convalescence46. Increased HMGB1 has been observed in blood and lung of smokers with COPD relative to healthy smokers47. There is also evidence of elevated HMGB1 protein levels in the sputum48 and bronchoalveolar lavage (BAL)37 of COPD patients. The nature of HMGB1 involvement in COPD is unclear49, and the reduced expression of the HMGB1 gene in our COPD lung tissue study, in contrast to the increased protein levels in sputum and BAL in these previous studies, and in a study of plasma levels in COPD patients50, requires further investigation; differences in smoking status, disease severity and tissue of origin may explain the differing results. HMGB1 was a driver gene in the brown module, which was associated with COPD, lung function and emphysema.
Our analysis also identified interacting genes that may play a role in COPD pathogenesis. SERPINE2 (sepin peptidase inhibitor clade E member 2) has been suggested as a COPD and asthma susceptibility gene51,52, and CD79A (CD79a molecule, immunoglobulin-associated alpha) was identified as a putative driver gene for the cyan module. Another putative driver for the cyan module, POU2AF1 (POU class 2 associating factor 1), was present in the intersection with the Hhip+/− mouse smoking model. BCL2 (Bcell CLL/lymphoma 2) has been implicated in COPD via regulation of apoptosis through mitochondrial maintenance functions53,54,55.
Construction of co-expression networks is a method to group similarly expressed probes into correlated network modules, with potentially common functions. Using WGCNA, we identified modules associated with COPD, lung function and quantitative CT emphysema. The cyan module was most significantly associated with COPD status, and the pathway enrichment analyses provided insight into the functions of the module genes, namely B cell activation, proliferation, aggregation and signaling. This is consistent with the histologic findings of lymphoid aggregates in severe COPD patients and the importance of immune pathways in COPD56,57,58,59,60,61. In addition, lymphoid aggregates were observed in the lungs of the Hhip+/− smoking mouse32; the overlap of 17 genes in the cyan module with the mouse Hhip+/+ vs. Hhip+/− 6 month smoke gene set provides further evidence of a cross-species signature of B cell involvement in COPD.
In a previous study restricted to former smokers, B cell receptor signaling pathways were enriched in genes whose expression was associated with emphysema severity10. Nine of the 127 genes that were differentially expressed in the Campbell et al. study are found in our cyan module, demonstrating replication of the B cell signature. A key member of this overlap set is CD79A, which is a hub gene in the cyan module. The gene BCL11A (B-cell CLL/lymphoma 11A) was also replicated. BCL11A is a transcription factor critical to B cell development62,63. In contrast to previous COPD gene expression studies20, our study and the Campbell et al. study both included subjects with severe COPD and eliminated the effects of current smoking on gene expression by limiting to former smokers64,65, allowing for replication of results at both the gene and pathway level. The second putative driver gene for the cyan module POU2AF1, was also identified in a study of emphysema by Faner et al.44 and is a B-lymphocyte specific transcription factor66. In this recent study, also of former smokers44, we observed replication of our results at the gene level.
In COPD and other complex diseases, there is often little overlap between genes at GWAS loci and differentially expressed genes in the target tissue20,21,22,23,24, despite the fact that most GWAS associations are likely due to regulatory variants. We found that genes that potentially interact with COPD GWAS genes were differentially expressed. These GWAS genes identified relevant pathways, but they themselves were not differentially expressed. It has been shown that GWAS genes tend to be located on the periphery of gene interaction networks and are not hub genes67; GWAS genes were not found in the hubs of our co-expression modules. Genetic variants affecting hub genes may be too deleterious to cells to be maintained in the population. Using network and pathway methods, we are able to capture the overall genetic perturbation and relate it to the gene expression analysis.
We demonstrated significant differential gene expression related to COPD in this study; however increased sample sizes in future studies will improve the power for gene identification. Inclusion of subjects with mild to moderate COPD will be necessary to determine whether B cells and other pathways are relevant earlier in the disease course, as infections are a common component of more severe COPD. Additional CT scan data in future studies should allow us to dissect COPD phenotypes of emphysema and airway disease. Data from specific cell types would provide more specificity than homogenized lung tissue. In addition, the variation in significance seen across our interactor enrichment results was not surprising given the different potential mechanisms of interaction and the heterogeneity with respect to species and cell types. Future studies may involve other COPD GWAS genes beyond the top three in the current study, as additional omics datasets are generated for those genes. Given the presence of samples from patients undergoing lung nodule resection surgery, we note that no enrichment for cancer related gene sets was observed in our REACTOME pathway analyses, which argues against bias by cancer diagnosis. With respect to GWAS integration, a more complete set of loci and integration across multiple putative functional variants, along with more uniform interaction measurement methods would provide a deeper understanding of the subtle effects of genetic variants. Although candidate eQTL analyses were included in this study, a future direction would be a genome-wide eQTL analysis28,68 to examine the direct association between COPD GWAS loci and the expression of genes of interest, including the genes identified in these interactor and network analyses.
By incorporating gene expression, experimental interaction, and network methods, we found significant differences in expression for genes interacting with genes at COPD GWAS loci. Our methodology could be applied to expression data from target tissues in other common diseases. Network analyses highlighted the importance of B cell pathways in COPD, which was recapitulated in the human lung tissue and a mouse smoking model. Replication of previous gene expression results was made possible by careful attention to subject enrollment, including former smokers with severe COPD. Consistent phenotype definitions are likely to be important for other diseases as well.
Methods
Study Population
Lung tissue samples were collected from subjects undergoing thoracic surgery for lung transplantation, lung volume reduction surgery or lung nodule resection at three medical centers: Brigham and Women’s Hospital (Boston, MA), St. Elizabeth’s Hospital (Boston, MA) and Temple University Hospital (Philadelphia, PA). Subjects provided written informed consent for use of excess lung tissue for research. IRB approval was obtained at the three centers. All subjects were former smokers, who quit smoking at least one month prior to surgery. Distant normal tissue was used from nodule resection samples. Phenotypes extracted from the medical records included demographics, anthropometrics, smoking history, and spirometry to measure lung function. COPD cases had severe or very severe airflow obstruction (GOLD grades 3-4)1. Control smokers had normal lung function. When available, chest computed tomography (CT) scans were retrieved for quantitative image analysis using 3-D Slicer software (www.slicer.org). Emphysema was assessed by the fraction of lung voxels with attenuation less than −950HU (LAA-950) and by the 15th percentile of the lung density histogram (Perc15). Airway disease was quantified by the square root wall area of a hypothetical 10mm internal perimeter airway (SRWA-Pi10)69.
Lung tissue gene expression profiling
Lung tissue samples were snap frozen and stored at −80 °C. RNA and DNA were simultaneously extracted from the homogenized lung tissue using the AllPrep kit (Qiagen, Valenica, CA). RNA quality was assessed on a BioAnalyzer (Agilent, Santa Clara, CA). Gene expression profiling was performed using HumanHT-12 BeadChips (Illumina, San Diego, CA). Quality control was performed using quantile, signal-to-noise, correlation matrix, MA, and principal component analysis (PCA) plots using R statistical software (v 3.2.0) to identify outliers and samples with questionable or low-quality levels, distributions, or associations. In addition, information from other omics data for this cohort was used to cross-check for sample issues during the data cleaning process. This process yielded 151 samples for analysis. There were 32,831 probes retained, after filtering for low variance and percentage of high detection p-values, which denote probes mapped to low expressed genes. This set of probes contains 20,794 unique gene symbol annotations. These data were background corrected, log2 transformed and quantile normalized using the R Bioconductor package lumi70. Microarray data has been deposited in Gene Expression Omnibus (GEO accession GSE76925).
RNA-interference
To find genes that could be targets of FAM13A, three FAM13A siRNA (Ambion, S19751, S19752 and S19753) were transfected into 16HBE cells. The experiment was controlled using Ambion Silencer Negative Control #1 siRNA (Ambion, 4390843). This control siRNA has no significant sequence similarity to human gene sequences. Cells were collected 48 hours after transfection for RNA extraction, and microarray expression analyses were performed using Illumina HT12 microarrays. Comparisons were made using 2-way ANOVA to determine differentially expressed probes between siRNA-treated and control cells. The intersection of differentially expressed probes from all three comparisons became the final gene list for FAM13A siRNA experiments.
External Datasets
As previously reported, microarray gene expression profiling was performed in human bronchial epithelial cells (Beas-2B) stably infected with four different lentivirus-based shRNAs against HHIP and one non-targeting shRNA (control), using the Illumina HT12 gene expression platform31. To identify differentially expressed probes after HHIP silencing, two linear regression models were used. The set of significantly differentially expressed genes found using both models became the analysis gene list.
Affinity purification followed by mass spectrometry (AP-MS) was performed to identify proteins that bind to either HHIP71 or FAM13A30. Briefly, HEK 293 cells were transfected with FLAG-tagged HHIP or control vector. Seventy-two hours after transfection, cells were lysed, and protein complexes were immunoprecipitated with an anti-FLAG antibody. HHIP-bound proteins were identified by mass spectrometry. Identified proteins were mapped to genes and the control results were subtracted from the experimental results to produce a final gene list for comparison with COPD expression results.
Because the IREB2 protein binds mRNA, RNA immunoprecipitation sequencing (RIP-Seq) was used to identify IREB2 targets29. IREB2-RNA complexes were immunoprecipitated from the Beas2B cells and whole transcriptome sequencing was performed. Two replicates in stimulated cells, with deferoxamine (DFO) and DFO-free controls (CTL), were included. In all cases, IREB2 was precipitated, along with IgG (background). The software application HOMER72 was used to call peaks accounting for the background using an hg19 read alignment, and four comparisons were made: CTL/IgG, CTL/DFO, DFO/IgG, and DFO/CTL. Peaks common between CTL/IgG and CTL/DFO, CTL/IgG and DFO/IgG, or DFO/IgG and DFO/CTL were mapped to genes and defined the IREB2 target gene list.
Homozygous deletion of Hhip in mice is embryonic lethal73. Therefore, Hhip+/− heterozygous mice (C57BL/6 background) and wild-type littermates were exposed to cigarette smoke or filtered air for six months32. RNA was extracted from mouse lungs and profiled in Illumina MouseRef-8 arrays. The differentially expressed genes (FDR < 0.10) were mapped to human orthologs for comparisons to the human lung tissue.
The GeneChip Mouse Gene 1.0 ST Array (Affymetrix) was used to assess gene expression in the Irp2 (IREB2 gene in humans) knockout and WT mice29. The expression difference between the Irp2 knockout and WT mice was quantified using an unpaired two-tailed t-test. A set of genes was created using the human orthologs from these results.
Differential gene expression and pathway analyses
Microarray batch effects were identified using MDS plots, and these along with latent effects were addressed by using surrogate variables, obtained via the R/Bioconductor package SVA74, as covariates in the linear models. For each expression probe or module eigengene, we fitted a linear regression model to detect association with variables of interest using R statistical software (v 3.2.0). In the microarray data analysis, an empirical Bayes shrinkage method was used to obtain a moderated t-test statistic and its p-value in limma75; the log fold change (logFC) values from limma are reported here as fold difference for improved clarity. Adjustment for multiple testing controlled for false discovery rate (FDR). The regression models for each clinical phenotype, including covariates, are provided in Supplemental Table S11. We found no significant association with time since smoking cessation, so this variable was not included as a covariate in subsequent analyses. Using the hypergeometric test in the GeneAnswers package76, we examined enrichment of curated gene sets from the Reactome database and from the biological process (BP) category in the gene ontology (GO) database. Hypergenometric tests were performed using the phyper function in the stats R package to observe enrichment of interactors in the differentially expressed gene sets with respect to the background (genes represented in the expression data following QC).
Weighted Gene Co-expression Analysis (WGCNA)
The WGCNA method was used to identify groups of probes that have similar expression characteristics in the sample population, using the R package WGCNA43. Signed networks were built using biweight midcorrelation as the correlation function, and a soft thresholding power of 12. WGCNA produces a set of modules (labeled by color), each containing a set of unique probes. The module eigengenes were used in the regression models35 from Supplemental Table S11. Driver or hub genes within each module were identified using previously described methods35, and through observation of the degree for each node.
Genotyping
Genomewide genotyping of extracted lung DNA was performed using the HumanOmni2.5Exome-8 V1.0 BeadChip (Illumina, Inc; San Diego, CA) and quality control assessment was performed using Python and R scripts and PLINK 277. Subject data were excluded after examining missingness, relatedness, sex, and inbreeding. Relatedness was examined using rgGRR and KING78, and sex assignment was based on X homozygosity estimates; discordant samples were removed. Principal components were generated for the lung tissue cohort and HapMap3 Public Release #3 panel using EIGENSTRAT79, and two additional subjects were subsequently excluded based on genetic ancestry that differed from reported. Manufacturer-identified poorly performing markers, duplicate markers, indels and markers exhibiting ambiguous or failed mapping were removed. Markers with missingness >1%, Hardy-Weinberg equilibrium deviation (p-value < 1e-7) or minor allele frequency <5% were excluded. To account for population stratification, two principal components (based on the Tracy-Widom statistic) for the Caucasian population were retained for use in the statistical analysis.
Candidate gene expression quantitative trait locus (eQTL) analysis
The eQTL analyses were performed using the 117 Caucasian subjects for whom both expression and genotyping data were available. The R/Bioconductor package Matrix eQTL80 was used to perform eQTL analysis to identify associations between genotype and gene expression levels, adjusted for the covariates age, sex, pack-years of smoking and two ancestry principal components. In addition, an iterative method was used to determine the number of expression matrix principal components (PC) to add as covariates to the model to mitigate batch effects. This procedure involves finding the number of PC covariates that produces a maximum number of significant (FDR < 0.05) cis-eQTLs (window of 1 Mb) using genotyping data from chromosome 21 and 22. The maximum was achieved with 13 PCs, and these were included as covariates in the eQTL analyses. Candidate cis-eQTL and trans-eQTL analyses were performed using all probes in the microarray analysis that passed QC and the 108 markers (p-value < 1.0e-5) from a recent GWAS for COPD case-control status8 that were available in our genotyping data and annotated by proximity to the three candidate genes of interest. All nominally significant trans-eQTL results (p-value < 0.05) for the three top cis-eQTLs located in each of the three loci (IREB2 rs2009746, HHIP rs11724319, FAM13A rs2609255) were used to produce each gene list.
GWAS results for interactors
The SNPs annotated by proximity to the interactor genes BCL2, BCL11A, POU2AF1, HMGB1, SERPINE2 and CD79A were downloaded from dbSNP. Association p-values for these SNPs were extracted from a published COPD case-control GWAS8.
Ethics Statement
Subjects provided written informed consent for use of excess lung tissue for research. IRB approval was obtained at Partners Healthcare (parent company of Brigham and Women’s Hospital), Temple University and St. Elizabeth’s Hospital. The methods for lung tissue research were carried out in accordance with the relevant guidelines. The mouse protocols were approved by the Institutional Animal Care and Use Committee, Harvard Medical School, and the methods were carried out in accordance with the relevant guidelines, as detailed in previous publications29,32,71.
Additional Information
How to cite this article: Morrow, J. D. et al. Functional interactors of three genome-wide association study genes are differentially expressed in severe chronic obstructive pulmonary disease lung tissue. Sci. Rep. 7, 44232; doi: 10.1038/srep44232 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We thank Drs. Amund Gulsvik, Per Bakke, Augusto Litonjua, Pantel Vokonas, Ruth Tal-Singer, and the GenKOLS, NETT/NAS, ECLIPSE, and COPDGene studies for use of their GWAS meta-analysis data.The COPDGene® study (R01 HL089856 and R01 HL089897). (NCT00608764) was funded by the National Institutes of Health and is also supported by the COPD Foundation through contributions made to an Industry Advisory Board comprised of AstraZeneca, Boehringer Ingelheim, Novartis, Pfizer, Siemens, GSK and Sunovion. The National Emphysema Treatment Trial was supported by the NHLBI N01HR76101, N01HR76102, N01HR76103, N01HR76104, N01HR76105, N01HR76106, N01HR76107, N01HR76108, N01HR76109, N01HR76110, N01HR76111, N01HR76112, N01HR76113, N01HR76114, N01HR76115, N01HR76116, N01HR76118 and N01HR76119, the Centers for Medicare and Medicaid Services and the Agency for Healthcare Research and Quality. The Normative Aging Study is supported by the Cooperative Studies Program/ERIC of the US Department of Veterans Affairs and is a component of the Massachusetts Veterans Epidemiology Research and Information Center (MAVERIC). The Norway GenKOLS study (Genetics of Chronic Obstructive Lung Disease, GSK code RES11080), and the ECLIPSE study (NCT00292552; GSK code SCO104960) were funded by GlaxoSmithKline. This work was supported by National Institutes of Health [P01HL105339 to EKS, R01HL111759 to JQ, R01HL094635 to CPH, R01HL130512 to CPH, R01HL125583 to CPH, R01HL113264 to MHC, R01HL127200 to XZ].
Footnotes
Drs. Morrow, Zhou, Lao, Jiang, DeMeo, Cho, Qiu, Cloonan, Pinto-Plata, Celli, Marchetti, Criner, Bueno, Glass, Quackenbush, Choi report no competing interests related to this manuscript. Dr. Washko has been a consultant for GSK, Genentech, Emmes, PulmonX. Dr. Silverman has received honoraria and consulting fees from Merck, grant support and consulting fees from GlaxoSmithKline, and honoraria from Novartis. Dr. Hersh has been a consultant for Mylan, Concert Pharmaceuticals and AstraZeneca.
Author Contributions J.M.: concept and design, analysis and interpretation of data, manuscript preparation, approval of final manuscript. X.Z.: acquisition of data, analysis and interpretation of data, manuscript preparation, approval of final manuscript. T.L.: acquisition of data, analysis and interpretation of data, approval of final manuscript. Z.J.: acquisition of data, analysis and interpretation of data, approval of final manuscript. D.D.: manuscript preparation, and approval of the final manuscript. M.C.: acquisition of data, analysis and interpretation of data, manuscript preparation, approval of final manuscript. W.Q.: analysis and interpretation of data, statistical support, approval of the final manuscript. S.C.: acquisition of data, analysis and interpretation of data, review and approval of final manuscript. V.P.P.: acquisition of data, approval of final manuscript. B.C.: acquisition of data, approval of final manuscript. N.M.: acquisition of data, analysis and interpretation of data, approval of final manuscript. G.C.: acquisition of data, approval of final manuscript. R.B.: acquisition of data, approval of final manuscript. G.W.: acquisition of data, analysis and interpretation of data, approval of final manuscript. K.G.: analysis and interpretation of data, statistical support, manuscript preparation, approval of final manuscript. J.Q.: statistical support, approval of final manuscript. A.C.: concept and design, acquisition of data, analysis and interpretation of data, review and approval of final manuscript. E.S.: concept and design, acquisition of data, manuscript preparation, approval of final manuscript. C.H.: concept and design, acquisition of data, analysis and interpretation of data, manuscript preparation, approval of final manuscript.
References
- Vestbo J. et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am. J. Respir. Crit. Care Med. 187, 347–65 (2013). [DOI] [PubMed] [Google Scholar]
- Hersh C. P. et al. Family history is a risk factor for COPD. Chest 140, 343–350 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCloskey S. et al. Siblings of patients with severe chronic obstructive pulmonary disease have a significant risk of airflow obstruction. Am J Respir Crit Care Med 164, 1419–1424 (2001). [DOI] [PubMed] [Google Scholar]
- Silverman E. K. In Chronic Obstructive Pulmonary Disease: Pathogenesis to Treatment (eds. Organizer D. C. & Goode J. A.) 45–64 (John Wiley & Sons, Ltd, 2000). [Google Scholar]
- Silverman E. K. et al. Genetic epidemiology of severe, early-onset chronic obstructive pulmonary disease. Risk to relatives for airflow obstruction and chronic bronchitis. Am. J. Respir. Crit. Care Med. 157, 1770–8 (1998). [DOI] [PubMed] [Google Scholar]
- Cho M. H. et al. Variants in FAM13A are associated with chronic obstructive pulmonary disease. Nat. Genet. 42, 200–202 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho M. H. et al. A genome-wide association study of COPD identifies a susceptibility locus on chromosome 19q13. Hum. Mol. Genet. 21, 947–57 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho M. H. et al. Risk loci for chronic obstructive pulmonary disease: a genome-wide association study and meta-analysis. Lancet Respir. Med. 2, 214–25 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya S. et al. Molecular Biomarkers for Quantitative and Discrete COPD Phenotypes. Am. J. Respir. Cell Mol. Biol. 40, 359–367 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell J. D. et al. A gene expression signature of emphysema-related lung destruction and its reversal by the tripeptide GHK. Genome Med. 4, 67 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezzie M. E. et al. Gene expression networks in COPD: microRNA and mRNA regulation. Thorax 67, 122–131 (2012). [DOI] [PubMed] [Google Scholar]
- Francis S. M. et al. Expression profiling identifies genes involved in emphysema severity. Respir. Res. 10, 81 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golpon H. A. et al. Emphysema Lung Tissue Gene Expression Profiling. Am. J. Respir. Cell Mol. Biol. 31, 595–600 (2004). [DOI] [PubMed] [Google Scholar]
- Ning W. et al. Comprehensive gene expression profiles reveal pathways related to the pathogenesis of chronic obstructive pulmonary disease. Proc. Natl. Acad. Sci. USA 101, 14895–14900 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savarimuthu Francis S. M. et al. Genes and Gene Ontologies Common to Airflow Obstruction and Emphysema in the Lungs of Patients with COPD. PLoS ONE 6, e17442 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spira A. et al. Gene Expression Profiling of Human Lung Tissue from Smokers with Severe Emphysema. Am. J. Respir. Cell Mol. Biol. 31, 601–610 (2004). [DOI] [PubMed] [Google Scholar]
- Wang I.-M. et al. Gene Expression Profiling in Patients with Chronic Obstructive Pulmonary Disease and Lung Cancer. Am. J. Respir. Crit. Care Med. 177, 402–411 (2008). [DOI] [PubMed] [Google Scholar]
- Zeskind J. E., Lenburg M. E. & Spira A. Translating the COPD Transcriptome. Proc. Am. Thorac. Soc. 5, 834–841 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.-H., Kim H. P., Ryter S. W. & Choi A. M. Identifying targets for COPD treatment through gene expression analyses. Int. J. Chron. Obstruct. Pulmon. Dis. 3, 359 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs B. D. & Hersh C. P. Integrative genomics of chronic obstructive pulmonary disease. Biochem. Biophys. Res. Commun. 452, 276–286 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huan T. et al. A systems biology framework identifies molecular underpinnings of coronary heart disease. Arterioscler. Thromb. Vasc. Biol. 33, 1427–1434 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui J. C. et al. Synthesizing genome-wide association studies and expression microarray reveals novel genes that act in the human growth plate to modulate height. Hum. Mol. Genet. 21, 5193–5201 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneera J. et al. A Systems Genetics Approach Identifies Genes and Pathways for Type 2 Diabetes in Human Islets. Cell Metab. 16, 122–134 (2012). [DOI] [PubMed] [Google Scholar]
- Voineagu I. et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature 474, 380–384 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurano M. T. et al. Systematic Localization of Common Disease-Associated Variation in Regulatory DNA. Science 337, 1190–1195 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy A. et al. Mapping of numerous disease-associated expression polymorphisms in primary peripheral blood CD4+ lymphocytes. Hum. Mol. Genet. 19, 4745–4757 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolae D. L. et al. Trait-Associated SNPs Are More Likely to Be eQTLs: Annotation to Enhance Discovery from GWAS. PLoS Genet 6, e1000888 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obeidat M. et al. Molecular mechanisms underlying variations in lung function: a systems genetics analysis. Lancet Respir. Med. 3, 782–795 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloonan S. et al. Mitochondrial iron as a therapeutic target for IRP2-regulated cigarette smoke-induced bronchitis and emphysema. Nat. Med. 22, 163–74 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z. et al. A Chronic Obstructive Pulmonary Disease Susceptibility Gene, FAM13A, Regulates Protein Stability of β-catenin. Am. J. Respir. Crit. Care Med 194, 185–97 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X. et al. Gene expression analysis uncovers novel hedgehog interacting protein (HHIP) effects in human bronchial epithelial cells. Genomics 101, 263–272 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao T. et al. Haploinsufficiency of Hedgehog interacting protein causes increased emphysema induced by cigarette smoke through network rewiring. Genome Med. 7, 12 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B. E. et al. Adipose co-expression networks across Finns and Mexicans identify novel triglyceride-associated genes. BMC Med. Genomics 5 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman E. & Loscalzo J. Network Medicine Approaches to the Genetics of Complex Diseases. Discov. Med. 14, 143–152 (2012). [PMC free article] [PubMed] [Google Scholar]
- Morrow J. D. et al. Identifying a gene expression signature of frequent COPD exacerbations in peripheral blood using network methods. BMC Med. Genomics 8, 1 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue J. et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity 40, 274–88 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferhani N. et al. Expression of High-Mobility Group Box 1 and of Receptor for Advanced Glycation End Products in Chronic Obstructive Pulmonary Disease. Am. J. Respir. Crit. Care Med. 181, 917–927 (2010). [DOI] [PubMed] [Google Scholar]
- Cheng D. T. et al. Systemic Soluble Receptor for Advanced Glycation Endproducts Is a Biomarker of Emphysema and Associated with AGER Genetic Variants in Patients with Chronic Obstructive Pulmonary Disease. Am. J. Respir. Crit. Care Med. 188, 948–957 (2013). [DOI] [PubMed] [Google Scholar]
- Cho M. H. et al. A Genome-Wide Association Study of Emphysema and Airway Quantitative Imaging Phenotypes. Am. J. Respir. Crit. Care Med. 192, 559–569 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichaikul A. et al. Genome-Wide Study of Percent Emphysema on Computed Tomography in the General Population. The Multi-Ethnic Study of Atherosclerosis Lung/SNP Health Association Resource Study. Am. J. Respir. Crit. Care Med. 189, 408–418 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lex A., Gehlenborg N., Strobelt H., Vuillemot R. & Pfister H. UpSet: Visualization of Intersecting Sets. IEEE Trans. Vis. Comput. Graph. 20, 1983–1992 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S. Weighted Network Analysis. (Springer New York, 2011). [Google Scholar]
- Langfelder P. & Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faner R. et al. Network Analysis of Lung Transcriptomics Reveals a Distinct B Cell Signature in Emphysema. Am. J. Respir. Crit. Care Med. 193, 1242–53 (2016). [DOI] [PubMed] [Google Scholar]
- Repapi E. et al. Genome-wide association study identifies five loci associated with lung function. Nat. Genet. 42, 36–44 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. et al. Changes of HMGB1 and sRAGE during the recovery of COPD exacerbation. J. Thorac. Dis. 6, 734 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko H.-K. et al. High expression of high-mobility group box 1 in the blood and lungs is associated with the development of chronic obstructive pulmonary disease in smokers. Respirology 19, 253–261 (2014). [DOI] [PubMed] [Google Scholar]
- Hou C. et al. High mobility group protein B1 (HMGB1) in Asthma: comparison of patients with chronic obstructive pulmonary disease and healthy controls. Mol. Med. 17, 807 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouwels S. D. et al. DAMPs activating innate and adaptive immune responses in COPD. Mucosal Immunol. 7, 215–226 (2014). [DOI] [PubMed] [Google Scholar]
- Mizumura K. et al. Mitophagy-dependent necroptosis contributes to the pathogenesis of COPD. J. Clin. Invest. 124, 3987–4003 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMeo D. L. et al. The SERPINE2 Gene Is Associated with Chronic Obstructive Pulmonary Disease. Am. J. Hum. Genet. 78, 253–264 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himes B. E. et al. Association of SERPINE2 with asthma. Chest. 140, 667–674 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge S., Hodge G., Holmes M. & Reynolds P. N. Increased peripheral blood T-cell apoptosis and decreased Bcl-2 in chronic obstructive pulmonary disease. Immunol. Cell Biol. 83, 160–166 (2005). [DOI] [PubMed] [Google Scholar]
- Siganaki M. et al. Deregulation of apoptosis mediators’ p53 and bcl2 in lung tissue of COPD patients. Respir. Res. 11, 46 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H. et al. Apoptosis and Bcl-2 family proteins, taken to chronic obstructive pulmonary disease. Eur Rev Med Pharmacol Sci 16, 711–727 (2012). [PubMed] [Google Scholar]
- Baraldo S. et al. Immune Activation in α 1 -Antitrypsin-Deficiency Emphysema. Beyond the Protease–Antiprotease Paradigm. Am. J. Respir. Crit. Care Med. 191, 402–409 (2015). [DOI] [PubMed] [Google Scholar]
- Brusselle G. G., Demoor T., Bracke K. R., Brandsma C.-A. & Timens W. Lymphoid follicles in (very) severe COPD: beneficial or harmful? Eur. Respir. J. 34, 219–230 (2009). [DOI] [PubMed] [Google Scholar]
- Faner R., Cruz T. & Agusti A. Immune response in chronic obstructive pulmonary disease. Expert Rev. Clin. Immunol. 9, 821–833 (2013). [DOI] [PubMed] [Google Scholar]
- Hogg J. C. et al. The nature of small-airway obstruction in chronic obstructive pulmonary disease. N. Engl. J. Med. 350, 2645–2653 (2004). [DOI] [PubMed] [Google Scholar]
- Polverino F. et al. B Cell–Activating Factor. An Orchestrator of Lymphoid Follicles in Severe Chronic Obstructive Pulmonary Disease. Am. J. Respir. Crit. Care Med. 192, 695–705 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Strate B. W. A. et al. Cigarette Smoke–induced Emphysema: A Role for the B Cell? Am. J. Respir. Crit. Care Med. 173, 751–758 (2006). [DOI] [PubMed] [Google Scholar]
- Liu P. et al. Bcl11a is essential for normal lymphoid development. Nat. Immunol. 4, 525–532 (2003). [DOI] [PubMed] [Google Scholar]
- Singh H., Medina K. L. & Pongubala J. M. R. Contingent gene regulatory networks and B cell fate specification. Proc. Natl. Acad. Sci. USA. 102, 4949–4953 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durham A. L. et al. Regulation of Wnt4 in chronic obstructive pulmonary disease. FASEB J. 27, 2367–2381 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilley A. E. et al. Biologic Phenotyping of the Human Small Airway Epithelial Response to Cigarette Smoking. PLoS ONE 6, e22798 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teitell M. A. OCA-B regulation of B-cell development and function. Trends Immunol. 24, 546–553 (2003). [DOI] [PubMed] [Google Scholar]
- Goh K.-I. et al. The human disease network. Proc. Natl. Acad. Sci. 104, 8685–8690 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao K. et al. Lung eQTLs to Help Reveal the Molecular Underpinnings of Asthma. PLOS Genet 8, e1003029 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano Y. et al. The Prediction of Small Airway Dimensions Using Computed Tomography. Am. J. Respir. Crit. Care Med. 171, 142–146 (2005). [DOI] [PubMed] [Google Scholar]
- Du P., Kibbe W. A. & Lin S. M. lumi: a pipeline for processing Illumina microarray. Bioinformatics 24, 1547–1548 (2008). [DOI] [PubMed] [Google Scholar]
- Lao T. et al. Hhip haploinsufficiency sensitizes mice to age-related emphysema. Proc. Natl. Acad. Sci. 113, E4681–E4687 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S. et al. Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang P.-T., Kawcak T. & McMahon A. P. Feedback control of mammalian Hedgehog signaling by the Hedgehog-binding protein, Hip1, modulates Fgf signaling during branching morphogenesis of the lung. Genes Dev. 17, 342–347 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek J. T. & Storey J. D. Capturing Heterogeneity in Gene Expression Studies by Surrogate Variable Analysis. PLoS Genet 3, e161 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G. K. In Bioinformatics and Computational Biology Solutions Using {R} and Bioconductor (eds Gentleman R., Carey V., Dudoit S., Irizarry R. & Huber W.) 397–420 (Springer, 2005). [Google Scholar]
- Feng G. et al. A collection of bioconductor methods to visualize gene-list annotations. BMC Res. Notes 3, 10–10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience 4, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichaikul A. et al. Robust relationship inference in genome-wide association studies. Bioinformatics 26, 2867–2873 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A. L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006). [DOI] [PubMed] [Google Scholar]
- Shabalin A. A. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics 28, 1353–1358 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.