In the crystal, the 5FC+ cation interacts with the PA− anion through three-centre N—H⋯O hydrogen bonds, forming two conjoined rings having  (6) and
(6) and  (6) motifs, and is extended by N—H⋯O hydrogen bonds and C—H⋯O interactions into a two-dimensional sheet structure lying parallel to (001). Also present in the crystal structure are weak C—F⋯π interactions.
(6) motifs, and is extended by N—H⋯O hydrogen bonds and C—H⋯O interactions into a two-dimensional sheet structure lying parallel to (001). Also present in the crystal structure are weak C—F⋯π interactions.
Keywords: crystal structure, 5-fluorocytosine, picrate, hydrogen bonding, bifurcated interactions
Abstract
In the crystal structure of the title compound, 5-fluorocytosinium picrate, C4H5FN3O+·C6H2N3O7 −, one N heteroatom of the 5-fluorocytosine (5FC) ring is protonated. The 5FC ring forms a dihedral angle of 19.97 (11)° with the ring of the picrate (PA−) anion. In the crystal, the 5FC+ cation interacts with the PA− anion through three-centre N—H⋯O hydrogen bonds, forming two conjoined rings having R 2 1(6) and R 1 2(6) motifs, and is extended by N—H⋯O hydrogen bonds and C—H⋯O interactions into a two-dimensional sheet structure lying parallel to (001). Also present in the crystal structure are weak C—F⋯π interactions.
Chemical context
Crystal engineering is defined as the rational design of crystalline solids through control of intermolecular interactions (hydrogen bonding, hydrophobic forces, van der Waals forces, π–π interactions and electrostatic forces). New solid forms of pharmaceuticals are designed using the crystal engineering approach. These engineered solids have technological and legal importance. Among the intermolecular interactions, hydrogen bonding is the master key for molecular recognition in biological systems because of its strength and directionality (Almarsson & Zaworoko, 2004 ▸; Desiraju, 1995 ▸). It plays a dominant role in molecular aggregates (Samuel, 1997 ▸; Tutughamiarso & Egert, 2012 ▸) and three-dimensional structure, stability and function of biomacromolecules (Gould, 1986 ▸). In particular, pyrimidine derivatives are used in the treatment of antiviral, antifungal, antitumor and cardiovascular diseases. 5-fluorocytosine (5FC) is a synthetic antimycotic compound, first synthesized in 1957 and widely used as an antitumor agent it is also active against fungal infection (Heidelberger et al., 1957 ▸; Portalone & Colapietro, 2007 ▸; Vermes et al., 2000 ▸). It becomes active by deamination of 5FC into 5-fluorouracil by the enzyme cytosine deaminase (CD) and inhibits RNA and DNA synthesis (Morschhäuser, 2003 ▸). Picric acid forms charge-transfer complexes with many organic compounds. It functions not only as an acceptor to form π-stacking complexes with aromatic biomolecules, but also as an acidic ligand to form salts with polar biomolecules through specific electrostatic hydrogen-bonding interactions (In et al., 1997 ▸). The present work is focused on the understanding of supramolecular hydrogen-bonding patterns exhibited by the interaction of 5FC and picric acid, giving the (1:1) title salt, C4H5FN3O+·C6H2N3O7
− whose structure and hydrogen-bonding patterns are reported on herein.
Structural commentary
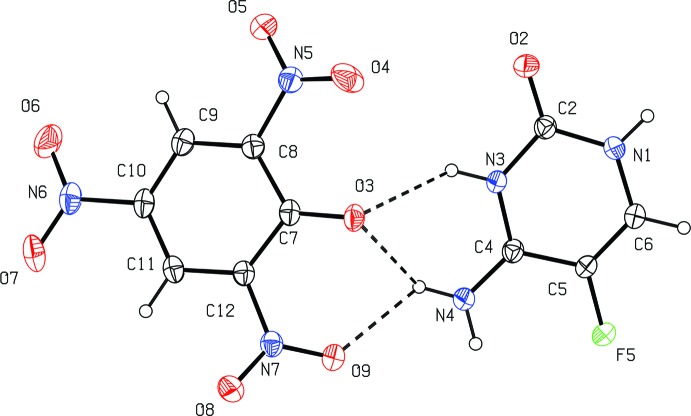
The asymmetric unit contains one 5-fluorocytosinium cation (5FC+) and one picrate anion (PA−) (Fig. 1 ▸). The 5-fluorocytosine cation is protonated at the N3 atom, as is evident from the widening of the corresponding internal angle from 120.8 (5)° to 125.37 (17)° compared to neutral 5FC (Louis et al., 1982 ▸). The dihedral angle between the planes of the rigs in the cation and anion is 19.97 (11)°. In the picrate (PA−) anion, the nitro groups lie variously out of the parent benzene ring, with torsion angles C9—C8—N5—O4, C9—C10—N6—O7 and C11—C12—N7—O9 of 166.2 (2), −171.7 (2) and 147.2 (2)°, respectively.
Figure 1.
The naming scheme for the 5FC+ cation and the PA− anion in the title compound, showing 30% probability level displacement ellipsoids. Dashed lines represent hydrogen bonds.
Supramolecular features
In this crystal structure, the N4-amino group and protonated N3 atom of the 5FC+ cation interact with atoms O3 and O9 of the picrate anion through three-centre N—H⋯O hydrogen bonds, forming two fused-ring motifs with graph-sets  (6) and
(6) and  (6) (Fig. 1 ▸). One of the N4-amino hydrogen atoms of the 5FC+ cation acts as a three-centre donor and the O3 atom of the PA− anion acts as a three-centre acceptor. This type of interaction has also been reported in the crystal structures of 2-amino-4,6-dimethylpyrimidinium picrate (Subashini et al., 2006 ▸) and 2-amino-4,6- dimethoxypyrimidinium picrate, pyrimethaminium picrate dimethyl sulfoxide (Thanigaimani et al., 2009 ▸). Similarly, the other hetero nitrogen atom (N1) of the cation and both the phenolate O3i and a nitro O4i atom of a PA− anion form an
(6) (Fig. 1 ▸). One of the N4-amino hydrogen atoms of the 5FC+ cation acts as a three-centre donor and the O3 atom of the PA− anion acts as a three-centre acceptor. This type of interaction has also been reported in the crystal structures of 2-amino-4,6-dimethylpyrimidinium picrate (Subashini et al., 2006 ▸) and 2-amino-4,6- dimethoxypyrimidinium picrate, pyrimethaminium picrate dimethyl sulfoxide (Thanigaimani et al., 2009 ▸). Similarly, the other hetero nitrogen atom (N1) of the cation and both the phenolate O3i and a nitro O4i atom of a PA− anion form an  (6) ring motif through N—H⋯O hydrogen bonds with a second C—H⋯O4i interaction, closing an
(6) ring motif through N—H⋯O hydrogen bonds with a second C—H⋯O4i interaction, closing an  (5) ring (Table 1 ▸). A similar type of interaction has also been observed in the crystal structure of cytosinium hydrogen chloroanilate monohydrate (Gotoh et al., 2006 ▸).
(5) ring (Table 1 ▸). A similar type of interaction has also been observed in the crystal structure of cytosinium hydrogen chloroanilate monohydrate (Gotoh et al., 2006 ▸).
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1⋯O3i | 0.88 | 1.92 | 2.794 (3) | 175 |
| N1—H1⋯O4i | 0.88 | 2.56 | 3.021 (3) | 114 |
| N3—H3⋯O3 | 0.88 | 2.22 | 2.915 (2) | 136 |
| N4—H4A⋯O3 | 0.88 | 2.10 | 2.828 (2) | 139 |
| N4—H4A⋯O9 | 0.88 | 2.18 | 2.782 (3) | 125 |
| N4—H4B⋯O2ii | 0.88 | 1.96 | 2.832 (3) | 171 |
| C6—H6⋯O4i | 0.95 | 2.51 | 3.003 (3) | 113 |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
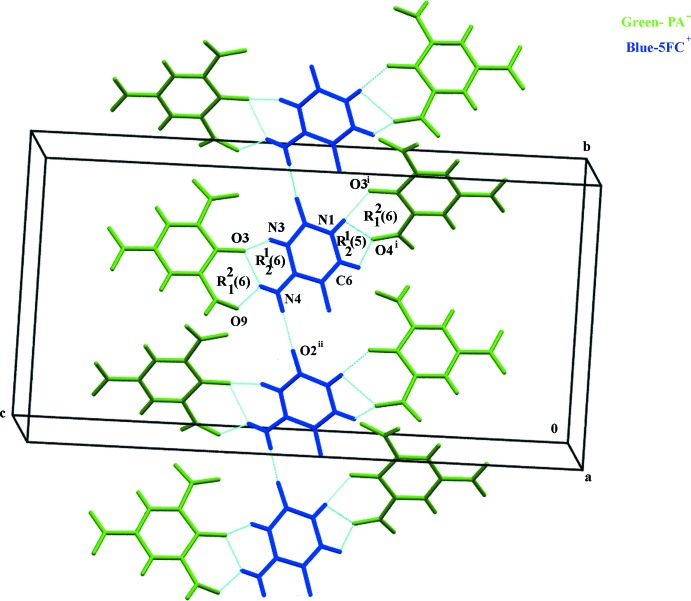
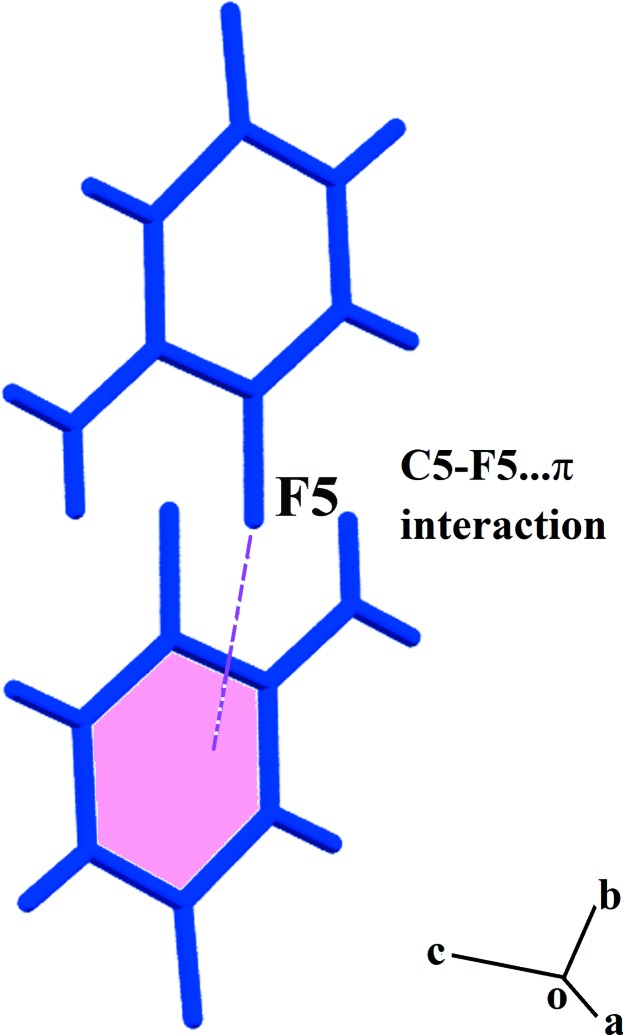
Further, the symmetry-related O2ii atom and the amino group of the 5FC+ cation are connected through an N—H⋯O hydrogen bond, forming a two-dimensional supramolecular network lying parallel to (001) (Fig. 2 ▸). Also present in the crystal structure is a weak C5—F5⋯π interaction (Fig. 3 ▸) between 5FC+ cations [C5⋯Cg iv = 3.4002 (19) Å; C—F⋯Cg = 88.34 (12)°, where Cg is the centroid of the N1–C6 ring; symmetry code: (iv) −x, −y, −z + 1]. A similar angle [90.5 (2)°] has been reported for a C—F⋯Cg interaction in an acridinium trifluoromethane sulfonate compound (Sikorski et al., 2005 ▸).
Figure 2.
A view of the supramolecular network formed via N—H⋯O and C—H⋯O interactions. Dashed lines represent hydrogen bonds. For symmetry codes, see Table 1 ▸.
Figure 3.
A view of the C5—F5⋯π interaction between 5FC+ cations.
Database survey
The crystal structures of 5-fluorocytosine monohydrates (Louis et al., 1982 ▸; Portalone & Colapietro, 2006 ▸; Portalone & Colapietro, 2007 ▸; Portalone, 2011 ▸), polymorphs (Hulme & Tocher, 2006 ▸; Tutughamiarso & Egert, 2012 ▸), salts (Perumalla et al., 2013 ▸) and co-crystals (Tutughamiarso et al., 2012 ▸; da Silva et al., 2014 ▸) have been reported in the literature. From our laboratory, 5-fluorocytosinium salicylate (Prabakaran et al., 2001 ▸), 5-fluorocytosinium 3-hydroxypicolinate (Karthikeyan et al., 2014 ▸) and 5-fluorocytosine melamine (Mohana et al., 2016 ▸) have been reported. Various salts and co-crystals of picric acid have also been reported in the literature (Subashini et al., 2006 ▸; Thanigaimani et al., 2009 ▸; Nagata et al., 1995 ▸; Smith et al., 2004 ▸; Gotoh et al., 2004 ▸).
Synthesis and crystallization
A hot aqueous solution of 5-fluorocytosine (32 mg) and picric acid (57 mg) were mixed in a 1:1 molar ratio. The resulting solution was warmed to 353 K wrong symmetry description - inversion centre in central benzene ring over a water bath for half an hour and kept for slow evaporation. After a week, colourless prismatic crystals were obtained.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. All hydrogen atoms were positioned geometrically (C—H = 0.95 Å and N—H = 0.88 Å) and were refined using a riding model with U iso(H) = 1.2U eq(parent atom).
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C4H5FN3O+·C6H2N3O7 − |
| M r | 358.22 |
| Crystal system, space group | Orthorhombic, P b c a |
| Temperature (K) | 200 |
| a, b, c (Å) | 7.7463 (15), 13.235 (3), 25.642 (5) |
| V (Å3) | 2628.9 (9) |
| Z | 8 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.17 |
| Crystal size (mm) | 0.65 × 0.58 × 0.20 |
| Data collection | |
| Diffractometer | Rigaku AFC-8S |
| Absorption correction | Multi-scan multi-scan |
| T min, T max | 0.899, 0.967 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 21815, 2779, 2367 |
| R int | 0.041 |
| (sin θ/λ)max (Å−1) | 0.633 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.058, 0.174, 1.09 |
| No. of reflections | 2779 |
| No. of parameters | 226 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.28, −0.36 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S205698901700216X/zs2375sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S205698901700216X/zs2375Isup2.hkl
Supporting information file. DOI: 10.1107/S205698901700216X/zs2375Isup3.cml
CCDC reference: 1531927
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
MM thanks UGC-BSR, India, for the award of an RFSMS. PTM thanks the UGC for a one-time BSR–faculty grant.
supplementary crystallographic information
Crystal data
| C4H5FN3O+·C6H2N3O7− | Dx = 1.810 Mg m−3 |
| Mr = 358.22 | Mo Kα radiation, λ = 0.71073 Å |
| Orthorhombic, Pbca | Cell parameters from 2779 reflections |
| a = 7.7463 (15) Å | θ = 3.1–26.7° |
| b = 13.235 (3) Å | µ = 0.17 mm−1 |
| c = 25.642 (5) Å | T = 200 K |
| V = 2628.9 (9) Å3 | Prism, colorless |
| Z = 8 | 0.65 × 0.58 × 0.20 mm |
| F(000) = 1456 |
Data collection
| Rigaku AFC-8S diffractometer | 2779 independent reflections |
| Radiation source: fine focus sealed tube | 2367 reflections with I > 2σ(I) |
| Detector resolution: 14.6199 pixels mm-1 | Rint = 0.041 |
| ω scans | θmax = 26.7°, θmin = 3.1° |
| Absorption correction: multi-scan multi-scan | h = −9→7 |
| Tmin = 0.899, Tmax = 0.967 | k = −16→16 |
| 21815 measured reflections | l = −32→32 |
Refinement
| Refinement on F2 | 0 restraints |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.058 | H-atom parameters constrained |
| wR(F2) = 0.174 | w = 1/[σ2(Fo2) + (0.1025P)2 + 1.2366P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.09 | (Δ/σ)max < 0.001 |
| 2779 reflections | Δρmax = 0.28 e Å−3 |
| 226 parameters | Δρmin = −0.36 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O3 | 0.4311 (2) | 0.65661 (11) | 0.61738 (6) | 0.0381 (4) | |
| O9 | 0.5493 (2) | 0.46460 (13) | 0.62650 (7) | 0.0440 (4) | |
| O7 | 0.5646 (3) | 0.58196 (18) | 0.85486 (7) | 0.0566 (5) | |
| O8 | 0.7432 (2) | 0.45799 (14) | 0.68664 (7) | 0.0469 (4) | |
| C7 | 0.4418 (3) | 0.65628 (15) | 0.66650 (8) | 0.0317 (5) | |
| N7 | 0.6116 (2) | 0.49512 (14) | 0.66734 (7) | 0.0353 (4) | |
| C12 | 0.5282 (3) | 0.57726 (16) | 0.69555 (8) | 0.0317 (4) | |
| C8 | 0.3714 (3) | 0.73340 (16) | 0.70030 (8) | 0.0338 (5) | |
| C10 | 0.4631 (3) | 0.65060 (18) | 0.77790 (8) | 0.0365 (5) | |
| O4 | 0.2970 (3) | 0.83845 (18) | 0.63172 (8) | 0.0664 (7) | |
| N5 | 0.2811 (3) | 0.81933 (15) | 0.67800 (8) | 0.0390 (4) | |
| O5 | 0.1900 (3) | 0.87035 (16) | 0.70710 (7) | 0.0595 (6) | |
| C11 | 0.5410 (3) | 0.57451 (16) | 0.74889 (8) | 0.0344 (5) | |
| H11 | 0.601989 | 0.521474 | 0.765720 | 0.041* | |
| O6 | 0.3831 (4) | 0.70675 (18) | 0.85946 (8) | 0.0733 (7) | |
| N6 | 0.4708 (3) | 0.64684 (16) | 0.83447 (8) | 0.0448 (5) | |
| C9 | 0.3783 (3) | 0.72956 (17) | 0.75414 (9) | 0.0366 (5) | |
| H9 | 0.325057 | 0.780799 | 0.774507 | 0.044* | |
| F5 | 0.2136 (2) | 0.44284 (10) | 0.45073 (6) | 0.0484 (4) | |
| O2 | 0.0618 (2) | 0.81443 (11) | 0.51702 (7) | 0.0412 (4) | |
| N3 | 0.1841 (2) | 0.66094 (13) | 0.53164 (6) | 0.0320 (4) | |
| H3 | 0.209477 | 0.679151 | 0.563747 | 0.038* | |
| N1 | 0.0672 (3) | 0.69947 (14) | 0.45092 (7) | 0.0371 (4) | |
| H1 | 0.019456 | 0.742373 | 0.429023 | 0.044* | |
| N4 | 0.3114 (3) | 0.50588 (14) | 0.54809 (7) | 0.0383 (4) | |
| H4A | 0.338817 | 0.526201 | 0.579703 | 0.046* | |
| H4B | 0.339680 | 0.444694 | 0.537710 | 0.046* | |
| C2 | 0.1011 (3) | 0.73142 (15) | 0.50045 (8) | 0.0325 (5) | |
| C4 | 0.2293 (3) | 0.56620 (15) | 0.51674 (8) | 0.0316 (4) | |
| C5 | 0.1785 (3) | 0.53785 (16) | 0.46564 (8) | 0.0348 (5) | |
| C6 | 0.1035 (3) | 0.60450 (18) | 0.43367 (9) | 0.0392 (5) | |
| H6 | 0.075454 | 0.585605 | 0.398941 | 0.047* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O3 | 0.0529 (10) | 0.0359 (8) | 0.0256 (7) | −0.0065 (7) | −0.0044 (6) | 0.0023 (6) |
| O9 | 0.0528 (10) | 0.0445 (9) | 0.0347 (8) | 0.0036 (7) | −0.0071 (7) | −0.0055 (7) |
| O7 | 0.0523 (11) | 0.0861 (15) | 0.0315 (9) | −0.0037 (10) | −0.0075 (8) | 0.0120 (9) |
| O8 | 0.0448 (9) | 0.0477 (10) | 0.0483 (10) | 0.0065 (7) | −0.0101 (8) | 0.0000 (8) |
| C7 | 0.0347 (10) | 0.0337 (10) | 0.0268 (10) | −0.0097 (8) | −0.0011 (8) | 0.0030 (7) |
| N7 | 0.0386 (10) | 0.0369 (9) | 0.0305 (9) | −0.0042 (7) | −0.0001 (7) | 0.0035 (7) |
| C12 | 0.0335 (10) | 0.0331 (10) | 0.0286 (10) | −0.0057 (8) | −0.0010 (8) | 0.0025 (8) |
| C8 | 0.0373 (11) | 0.0315 (10) | 0.0327 (10) | −0.0057 (8) | −0.0022 (8) | 0.0014 (8) |
| C10 | 0.0408 (12) | 0.0434 (12) | 0.0252 (10) | −0.0119 (9) | −0.0015 (8) | 0.0014 (8) |
| O4 | 0.0806 (15) | 0.0708 (14) | 0.0478 (11) | 0.0296 (11) | 0.0199 (10) | 0.0262 (10) |
| N5 | 0.0424 (10) | 0.0379 (10) | 0.0368 (10) | −0.0034 (8) | 0.0003 (8) | 0.0009 (8) |
| O5 | 0.0814 (15) | 0.0547 (12) | 0.0424 (10) | 0.0228 (10) | −0.0041 (9) | −0.0079 (8) |
| C11 | 0.0357 (11) | 0.0380 (10) | 0.0296 (10) | −0.0076 (8) | −0.0047 (8) | 0.0058 (8) |
| O6 | 0.122 (2) | 0.0654 (13) | 0.0320 (9) | 0.0087 (13) | 0.0120 (11) | −0.0020 (9) |
| N6 | 0.0538 (12) | 0.0518 (12) | 0.0287 (10) | −0.0135 (10) | −0.0011 (9) | 0.0027 (8) |
| C9 | 0.0396 (12) | 0.0386 (11) | 0.0317 (11) | −0.0097 (9) | 0.0015 (8) | −0.0019 (8) |
| F5 | 0.0673 (10) | 0.0373 (7) | 0.0404 (8) | 0.0123 (6) | −0.0140 (7) | −0.0128 (6) |
| O2 | 0.0538 (10) | 0.0292 (8) | 0.0406 (9) | 0.0029 (7) | −0.0023 (7) | −0.0004 (6) |
| N3 | 0.0404 (10) | 0.0302 (9) | 0.0252 (8) | 0.0019 (7) | −0.0033 (7) | −0.0028 (6) |
| N1 | 0.0491 (11) | 0.0347 (9) | 0.0274 (9) | 0.0077 (8) | −0.0028 (8) | 0.0032 (7) |
| N4 | 0.0514 (11) | 0.0310 (9) | 0.0324 (9) | 0.0053 (8) | −0.0094 (8) | −0.0031 (7) |
| C2 | 0.0380 (11) | 0.0308 (11) | 0.0288 (10) | −0.0023 (8) | 0.0005 (8) | 0.0012 (8) |
| C4 | 0.0360 (10) | 0.0303 (10) | 0.0285 (10) | −0.0039 (8) | −0.0006 (8) | 0.0000 (8) |
| C5 | 0.0439 (11) | 0.0312 (10) | 0.0294 (10) | 0.0031 (8) | −0.0032 (9) | −0.0057 (8) |
| C6 | 0.0473 (12) | 0.0418 (12) | 0.0284 (10) | 0.0055 (10) | −0.0053 (9) | −0.0049 (8) |
Geometric parameters (Å, º)
| O3—C7 | 1.262 (3) | O6—N6 | 1.225 (3) |
| O9—N7 | 1.222 (3) | C9—H9 | 0.9500 |
| O7—N6 | 1.240 (3) | F5—C5 | 1.342 (2) |
| O8—N7 | 1.235 (3) | O2—C2 | 1.217 (3) |
| C7—C8 | 1.446 (3) | N3—C4 | 1.357 (3) |
| C7—C12 | 1.448 (3) | N3—C2 | 1.387 (3) |
| N7—C12 | 1.457 (3) | N3—H3 | 0.8800 |
| C12—C11 | 1.372 (3) | N1—C6 | 1.362 (3) |
| C8—C9 | 1.383 (3) | N1—C2 | 1.364 (3) |
| C8—N5 | 1.452 (3) | N1—H1 | 0.8800 |
| C10—C9 | 1.377 (3) | N4—C4 | 1.299 (3) |
| C10—C11 | 1.389 (3) | N4—H4A | 0.8800 |
| C10—N6 | 1.453 (3) | N4—H4B | 0.8800 |
| O4—N5 | 1.219 (3) | C4—C5 | 1.419 (3) |
| N5—O5 | 1.229 (3) | C5—C6 | 1.337 (3) |
| C11—H11 | 0.9500 | C6—H6 | 0.9500 |
| O3—C7—C8 | 124.81 (19) | C10—C9—C8 | 119.2 (2) |
| O3—C7—C12 | 123.1 (2) | C10—C9—H9 | 120.4 |
| C8—C7—C12 | 112.08 (18) | C8—C9—H9 | 120.4 |
| O9—N7—O8 | 122.5 (2) | C4—N3—C2 | 125.37 (17) |
| O9—N7—C12 | 119.83 (18) | C4—N3—H3 | 117.3 |
| O8—N7—C12 | 117.63 (18) | C2—N3—H3 | 117.3 |
| C11—C12—C7 | 124.4 (2) | C6—N1—C2 | 123.29 (18) |
| C11—C12—N7 | 116.32 (18) | C6—N1—H1 | 118.4 |
| C7—C12—N7 | 119.24 (18) | C2—N1—H1 | 118.4 |
| C9—C8—C7 | 123.9 (2) | C4—N4—H4A | 120.0 |
| C9—C8—N5 | 116.14 (19) | C4—N4—H4B | 120.0 |
| C7—C8—N5 | 119.89 (18) | H4A—N4—H4B | 120.0 |
| C9—C10—C11 | 121.4 (2) | O2—C2—N1 | 123.8 (2) |
| C9—C10—N6 | 119.2 (2) | O2—C2—N3 | 121.46 (19) |
| C11—C10—N6 | 119.5 (2) | N1—C2—N3 | 114.70 (18) |
| O4—N5—O5 | 122.3 (2) | N4—C4—N3 | 121.33 (19) |
| O4—N5—C8 | 119.8 (2) | N4—C4—C5 | 123.0 (2) |
| O5—N5—C8 | 117.87 (19) | N3—C4—C5 | 115.65 (19) |
| C12—C11—C10 | 118.9 (2) | C6—C5—F5 | 122.10 (19) |
| C12—C11—H11 | 120.6 | C6—C5—C4 | 120.8 (2) |
| C10—C11—H11 | 120.6 | F5—C5—C4 | 117.05 (19) |
| O6—N6—O7 | 123.5 (2) | C5—C6—N1 | 120.0 (2) |
| O6—N6—C10 | 118.5 (2) | C5—C6—H6 | 120.0 |
| O7—N6—C10 | 117.9 (2) | N1—C6—H6 | 120.0 |
| O4—N5—C8—C7 | −16.2 (3) | O3—C7—C8—C9 | 177.2 (2) |
| O4—N5—C8—C9 | 166.2 (2) | C12—C7—C8—N5 | 179.5 (2) |
| O5—N5—C8—C7 | 163.6 (2) | C12—C7—C8—C9 | −3.0 (3) |
| O5—N5—C8—C9 | −14.0 (3) | O3—C7—C12—N7 | 1.8 (3) |
| O6—N6—C10—C9 | 9.2 (4) | O3—C7—C12—C11 | −179.6 (2) |
| O6—N6—C10—C11 | −170.7 (2) | C8—C7—C12—N7 | −178.00 (19) |
| O7—N6—C10—C9 | −171.7 (2) | N5—C8—C9—C10 | −179.5 (2) |
| O7—N6—C10—C11 | 8.4 (3) | C7—C8—C9—C10 | 3.0 (4) |
| O8—N7—C12—C7 | 147.0 (2) | C8—C9—C10—N6 | 179.8 (2) |
| O8—N7—C12—C11 | −31.8 (3) | C8—C9—C10—C11 | −0.3 (4) |
| O9—N7—C12—C7 | −34.0 (3) | N6—C10—C11—C12 | 178.0 (2) |
| O9—N7—C12—C11 | 147.2 (2) | C9—C10—C11—C12 | −1.9 (3) |
| C2—N1—C6—C5 | −1.1 (4) | C10—C11—C12—N7 | −179.6 (2) |
| C6—N1—C2—O2 | −176.4 (2) | C10—C11—C12—C7 | 1.7 (4) |
| C6—N1—C2—N3 | 3.1 (3) | N3—C4—C5—F5 | −176.70 (18) |
| C2—N3—C4—C5 | −3.0 (3) | N3—C4—C5—C6 | 5.1 (3) |
| C4—N3—C2—O2 | 178.6 (2) | N4—C4—C5—F5 | 2.2 (3) |
| C4—N3—C2—N1 | −0.9 (3) | N4—C4—C5—C6 | −175.9 (2) |
| C2—N3—C4—N4 | 178.0 (2) | F5—C5—C6—N1 | 178.7 (2) |
| O3—C7—C8—N5 | −0.2 (3) | C4—C5—C6—N1 | −3.3 (4) |
| C8—C7—C12—C11 | 0.7 (3) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···O3i | 0.88 | 1.92 | 2.794 (3) | 175 |
| N1—H1···O4i | 0.88 | 2.56 | 3.021 (3) | 114 |
| N3—H3···O3 | 0.88 | 2.22 | 2.915 (2) | 136 |
| N4—H4A···O3 | 0.88 | 2.10 | 2.828 (2) | 139 |
| N4—H4A···O9 | 0.88 | 2.18 | 2.782 (3) | 125 |
| N4—H4B···O2ii | 0.88 | 1.96 | 2.832 (3) | 171 |
| C6—H6···O4i | 0.95 | 2.51 | 3.003 (3) | 113 |
Symmetry codes: (i) x−1/2, −y+3/2, −z+1; (ii) −x−1/2, y−3/2, z.
References
- Almarsson, Ö. & Zaworotko, M. J. (2004). Chem. Commun. pp. 1889–1896. [DOI] [PubMed]
- Cason, C. J. (2004). POV-RAY for Windows. Persistence of Vision, Raytracer Pvt. Ltd, Victoria, Australia. URL: http://www.povray.org.
- Desiraju, G. R. (1995). Angew. Chem. Int. Ed. Engl. 34, 2311–2327.
- Gotoh, K., Ishikawa, R. & Ishida, H. (2006). Acta Cryst. E62, o4738–o4740.
- Gotoh, M., Kanno, H., Sugaya, E., Osa, Y. & Takayanagi, H. (2004). Anal. Sci. 20, x39–x40.
- Gould, P. J. (1986). Int. J. Pharm. 33, 201–217.
- Heidelberger, C., Chaudhuri, N. K., Danneberg, P., Mooren, D., Griesbach, L., Duschinsky, R., Schnitzer, R. J., Pleven, E. & Scheiner, J. (1957). Nature, 179, 663–666. [DOI] [PubMed]
- Hulme, A. T. & Tocher, D. A. (2006). Cryst. Growth Des. 6, 481–487.
- In, Y., Nagata, H., Doi, M., Ishida, T. & Wakahara, A. (1997). Acta Cryst. C53, 367–369.
- Karthikeyan, A., Thomas Muthiah, P. & Perdih, F. (2014). Acta Cryst. E70, 328–330. [DOI] [PMC free article] [PubMed]
- Louis, T., Low, J. N. & Tollin, P. (1982). Cryst. Struct. Commun. 11, 1059–1064.
- Macrae, C. F., Bruno, I. J., Chisholm, J. A., Edgington, P. R., McCabe, P., Pidcock, E., Rodriguez-Monge, L., Taylor, R., van de Streek, J. & Wood, P. A. (2008). J. Appl. Cryst. 41, 466–470.
- Mohana, M., Muthiah, P. T., Sanjeewa, L. D. & McMillen, C. D. (2016). Acta Cryst. E72, 552–555. [DOI] [PMC free article] [PubMed]
- Morschhäuser, J. (2003). Pharm. Unserer Zeit, 32, 124–128. [DOI] [PubMed]
- Nagata, H., In, Y., Doi, M., Ishida, T. & Wakahara, A. (1995). Acta Cryst. B51, 1051–1058.
- Perumalla, S. R., Pedireddi, V. R. & Sun, C. C. (2013). Mol. Pharm. 10, 2462–2466. [DOI] [PubMed]
- Portalone, G. (2011). Chem. Cent. J. 5, 1–8. [DOI] [PMC free article] [PubMed]
- Portalone, G. & Colapietro, M. (2006). Acta Cryst. E62, o1049–o1051.
- Portalone, G. & Colapietro, M. (2007). J. Chem. Crystallogr. 37, 141–145.
- Prabakaran, P., Murugesan, S., Muthiah, P. T., Bocelli, G. & Righi, L. (2001). Acta Cryst. E57, o933–o936. [DOI] [PubMed]
- Rigaku/MSC (2008). CrystalClear. Rigaku Americas Corporation, The Woodlands, Texas, USA.
- Samuel, H. G. (1997). Chem. Rev. 5, 1231–1232.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
- Sikorski, A., Krzymiński, K., Niziołek, A. & Błażejowski, J. (2005). Acta Cryst. C61, o690–o694. [DOI] [PubMed]
- Silva, C. C. P. da, Pepino, R. de O., de Melo, C. C., Tenorio, J. C. & Ellena, J. (2014). Cryst. Growth Des. 14, 4383–4393.
- Smith, G., Wermuth, U. D. & Healy, P. C. (2004). Acta Cryst. E60, o1800–o1803. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Subashini, A., Muthiah, P. T., Bocelli, G. & Cantoni, A. (2006). Acta Cryst. E62, o3847–o3849.
- Thanigaimani, K., Subashini, A., Muthiah, P. T., Lynch, D. E. & Butcher, R. J. (2009). Acta Cryst. C65, o42–o45. [DOI] [PubMed]
- Tutughamiarso, M., Bolte, M. & Egert, E. (2009). Acta Cryst. C65, o574–o578. [DOI] [PubMed]
- Tutughamiarso, M. & Egert, E. (2012). Acta Cryst. B68, 444–452. [DOI] [PubMed]
- Vermes, A., Guchelaar, H. J. & Dankert, J. J. (2000). J. Antimicrob. Chemother. 46, 171–179. [DOI] [PubMed]
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S205698901700216X/zs2375sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S205698901700216X/zs2375Isup2.hkl
Supporting information file. DOI: 10.1107/S205698901700216X/zs2375Isup3.cml
CCDC reference: 1531927
Additional supporting information: crystallographic information; 3D view; checkCIF report