Abstract
Herbivory has evolved in many groups of vertebrates, but it is rare among both extinct and extant nonavian reptiles. Among squamate reptiles, (lizards, snakes, and their relatives), <2% of the >7,800 species are considered to be herbivorous, and herbivory is restricted to lizards. Here, we show that within a group of South American lizards (Liolaemidae, ≈170 species), herbivory has evolved more frequently than in all other squamates combined and at a rate estimated to be >65 times faster. Furthermore, in contrast to other herbivorous lizards and to existing theory, most herbivorous liolaemids are small bodied and live in cool climates. Herbivory is generally thought to evolve only in reptile species that are large bodied, live in warm climates, and maintain high body temperatures. These three well known “rules” of herbivory are considered to form the bases of physiological constraints that explain the paucity of herbivorous reptile species. We suggest that the recurrent and paradoxical evolution of herbivory in liolaemids is explained by a combination of environmental conditions (promoting independent origins of herbivory in isolated cool-climate regions), ecophysiological constraints (requiring small body size in cool climates, yet high body temperatures for herbivores), and phylogenetic history. More generally, our study demonstrates how integrating information from ecophysiology and phylogeny can help to explain macroevolutionary trends.
Keywords: ecophysiology, macroevolution
Diet is a fundamental aspect of an organism's biology, and the evolution of dietary strategies may have important consequences for both lineages and ecosystems (1–5). Although herbivory is common in some groups of animals, it is rare among both extinct and extant nonavian reptiles (2, 3, 6–8). Among squamate reptiles (lizards, snakes, and their relatives), <2% (9) of the >7,800 currently recognized species (European Molecular Biology Laboratory reptile database, www.embl-heidelberg.de/~uetz/livingreptiles.html) are considered to be herbivorous, and herbivory is confined entirely to lizards (2, 7–9, 10). The paucity of plant-eating reptiles suggests that there are constraints limiting the evolution herbivory in this group.
In this study, we find that herbivory has evolved repeatedly in a clade of South American lizards (Liolaemidae with nearly 170 species), likely more times than are known for all other squamates combined, and at a rate that is >65 times faster. We also find that herbivores in this clade have converged repeatedly on a unique combination of morphological, ecological, and physiological characteristics that are strikingly different from those reported for other Recent herbivorous reptiles. We combine data on phylogeny, diet, ecology, and physiology to document and explain the seemingly paradoxical evolution of herbivory in liolaemid lizards. More generally, our study suggests how the interplay of ecophysiology and phylogenetic history may lead to remarkable shifts in macroevolutionary trends in diet.
Many researchers have noted that herbivorous lizards are generally large bodied, live in warm climates, and maintain high body temperatures (7–14). These widely reported “rules” of herbivory have been proposed as explanations for the paucity of herbivorous reptiles (i.e., herbivory should not evolve in lineages that are small bodied, live in cool climates, or maintain low body temperatures; refs. 2, 7, 8, 11, 13, and 14). These traits may be associated with physiological constraints imposed by herbivory. For example, because most plant tissues are relatively low in digestible energy, large body size is favorable because it affords a low mass-specific rate of energy expenditure (7, 13–16). A large body can also support a voluminous gut that will increase digestive efficiency by increasing the time that food is fermented and assimilated (8, 9, 17, 18). Likewise, high body temperatures may be necessary for microbial fermentation of plant tissues (9, 12, 18, 19), given that herbivorous vertebrates lack endogenous cellulases and must rely on gut endosymbionts (bacteria and protozoa) to release the energy bound in plant cell walls (17, 18, 20). The need to maintain high body temperatures may explain why herbivorous reptiles are largely confined to the tropics or warm deserts (9, 11, 12). Given that these ecophysiological rules may explain the scarcity of herbivory in reptiles, changes in one or more of these three apparent constraints might allow for a dramatic increase in the rate at which herbivory evolves in a given clade.
Liolaemidae consists of three genera, Ctenoblepharys, Liolaemus, and Phymaturus, and 168 species (European Molecular Biology Laboratory reptile database) of small-bodied iguanian lizards that are distributed primarily in southern South America (21–24). The monotypic Ctenoblepharys is insectivorous (23) and is the sister taxon of the clade Liolaemus plus Phymaturus (25). Liolaemus contains 157 species, including insectivores, omnivores, and herbivores (9, 21–24). Phymaturus contains 10 species, all of which are herbivorous (9, 21–24). Although some researchers have suggested or provided evidence that small lizards (including some liolaemids) eat primarily plants (17, 21, 22, 26–32), reviews of herbivory in reptiles have overlooked or dismissed reports of herbivory in liolaemids (2, 3, 7, 8, 18) or were unaware of the extent of herbivory and its unexpected correlates in these lizards (10, 17, 26–32).
Materials and Methods
Definition and Coding of Herbivory. After a review of herbivory in lizards (10), which found that diets are more discrete than previously recognized (figure 2 in ref. 10), we adopted the following definitions: insectivore = 0–10%, omnivore = 11– 50%, and herbivore = 70–100%, where the percentage corresponds to the volumetric proportion of plant matter in the diet (Data Set 1, which is published as supporting information on the PNAS web site). Each taxon was assigned to a diet based on data from the literature or volumetric estimates of gut contents or freshly deposited feces (means of individuals for each taxon).
Phylogeny Reconstruction. Phylogenetic analyses (performed with parsimony and Bayesian methods) were based on combined data matrices consisting of published (23, 33, 34) morphological data (23 characters; Data Set 2, which is published as supporting information on the PNAS web site), reproductive mode (Data Set 1), and published (35, 36) and previously unreported mitochondrial DNA sequences (Data Set 3, which is published as supporting information on the PNAS web site), totaling 1,026 parsimony informative characters (pic). Parsimony analyses were performed by using paup* 4.0B10 (37) and Bayesian analyses were performed by using mrbayes 3.0B4 (38). Sequence data for 61 liolaemid taxa and four outgroup species from a 1.7-kb fragment [referred to as NADH dehydrogenase subunit II (ND2) hereafter, but consisting of eight tRNAs, the ND2 gene, and a portion of NADH dehydrogenase subunit I; 852 pic] were taken from refs. 25, 35, and 36. Two additional Liolaemus sequences were provided by J. A. Schulte (U.S. National Museum, Smithsonian Institution, Washington, D.C.) (personal communication). Sequences from 17 additional taxa for an overlapping 1.4-kb fragment were obtained by using methods described in ref. 36. An additional 685-bp fragment (139 pic) of the 12S (small) ribosomal subunit was obtained for 30 taxa (emphasizing members of the alticolor–bibronii and elongatus–kriegi species groups of Liolaemus) by using methods described in ref. 39. Details of the phylogenetic analyses are provided in Supporting Methods, which is published as supporting information on the PNAS web site.
The majority-rule consensus tree from the first Bayesian analysis of the combined data was used as the preferred topology (Fig. 1), and results from comparative analyses (described below) are based primarily on this tree. However, to address the sensitivity of these results to alternative topologies, comparative analyses were repeated on the two shortest trees from the combined data parsimony analysis, the majority-rule consensus tree from a second Bayesian analysis, and five trees from each Bayesian analysis having the highest posterior probabilities. Results were generally very similar among these topologies. For more information on the Bayesian analyses, see Tables 1 and 2, which are published as supporting information on the PNAS web site.
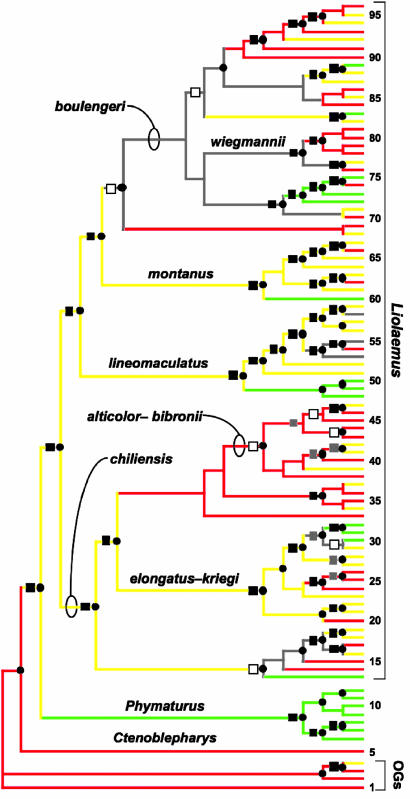
Fig. 1.
Phylogeny of liolaemid lizards showing multiple origins of herbivory. The topology is based on a Bayesian analysis of combined morphological and DNA data. Circles at nodes indicate Bayesian posterior probabilities of ≥95%, and squares indicate concordant nonparametric bootstrap support (≥70%) from a parsimony analysis (black, 90–100%; gray, 80–89%; white, 70–79%). Colored branches indicate diet (red, insectivore; yellow, omnivore; green, herbivore; gray, equivocal). OGs, outgroups. Major species groups within Liolaemus (21–23, 36) are identified and numbered taxa are listed in Data Set 4, which is published as supporting information on the PNAS web site. Although diet reconstructions are ambiguous for some clades, almost all of this uncertainty is associated with transitions between insectivory and omnivory rather than the evolution of herbivory.
Origins of Herbivory. For parsimony reconstructions, each taxon was assigned one of three character states for diet based on categories assigned in Data Set 1. Evolution of diet was reconstructed as an ordered (insectivore → omnivore → herbivore) three-state character by using parsimony with macclade 4.0 (40). We estimated the number of origins of herbivory in liolaemids as the number of transitions from omnivory or insectivory to herbivory. Diet evolution was also analyzed as a continuous character in a likelihood framework by using linear generalized least squares (GLS-linear; ref. 41) with compare 4.5 (http://compare.bio.indiana.edu). We assumed equal branch lengths (1.0) for this analysis because no single molecular data set was available for all of the taxa. We reconstructed the proportion of plant matter in the diet at each node, classified ancestors as herbivores, omnivores, or insectivores based on the categories defined above, and tallied the number of origins of herbivory.
Our phylogenetic analyses included 87 of the 168 currently recognized species of Liolaemidae. To estimate the number of origins of herbivory for virtually all Liolaemidae, we obtained data on diet from 162 taxa of Liolaemidae (described above; Data Set 1) and constructed a phylogenetic supertree for these species. Although we prefer direct character-based analyses to a supertree approach, the only information available on the placement of many liolaemid taxa is taxonomy. Details of supertree construction are described in Supporting Methods. Interestingly, our estimated number of origins of herbivory (18.5) derived from the supertree analysis is similar to an estimate based on simply extrapolating the number of origins of herbivory from the phylogenetic analysis to a similar number of unsampled species (i.e., given there are ≈9 origins of herbivory among 91 species one might expect to see ≈18 origins of herbivory among a sample of species that is roughly twice as large, given many assumptions).
We derived crude estimates of the rates of evolution of herbivory among liolaemids and among nonliolaemid squamates by using the number of estimated transitions to herbivory divided by the total number of branches (terminal plus internal) along which such a change could occur. We considered there to be 168 described species of liolaemids and 7,833 species of squamates overall (European Molecular Biology Laboratory reptile database). For a given tree, we used the median number of reconstructions of herbivory (mean for the supertree analysis). The number of internal and terminal branches for a rooted tree of N species is (N – 1) + N (42). More sophisticated estimates of evolutionary rates would have been difficult given the very large number of squamate taxa and incomplete information on species-level phylogeny and branch lengths.
Body Size of Liolaemids. Maximum snout–vent lengths (SVLs) of liolaemid species were taken from the literature or from museum specimens (references and specimen numbers in Data Set 1). Maximum SVL was used because of the difficulty in establishing minimum adult size for many poorly known species.
Herbivory and Body Size in Nonliolaemid Lizards. The number of independent origins of herbivory in Recent nonliolaemid squamates was reconstructed by using macclade (40) based on reviews of squamate herbivory (9, 10). Monophyly and relationships among squamate families followed revisions and summaries (43, 44); however, our results should be insensitive to uncertainty about interfamilial relationships because herbivory almost always arises within families. We summarize the basic results here by family (number of independent origins of herbivory, herbivorous taxa; ref. 9): Iguanidae (1, all genera); Agamidae (2, Hydrosaurus and Uromastyx); Lacertidae (1, Gallotia simonyi); Scincidae (4, Corucia, Egernia, Macroscincus, and Tiliqua); Gerrhosauridae (1, Angolosaurus); and Teiidae (1–2, Cnemidophorus arubensis, Cnemidophorus murinus, and Cnemidophorus sp.). Note that not all species within each genus are necessarily herbivorous. The genera listed above that are not followed by specific species are conservatively estimated to contain a single origin of herbivory.
Maximum body sizes (SVLs) of Recent herbivorous lizards were assembled from the literature as summarized in ref. 9.
Climate-Based Temperature Indices. Latitudinal and elevational ranges of each species were taken from previous studies, museum specimens, or fieldwork (Data Set 1). Temperature indices were calculated for the latitudinal and elevation midpoints of the range of each species. Temperature indices were constructed from a regression of air temperatures recorded in January (month of peak lizard activity), assuming a starting temperature of 20°C (mean diel temperature in January at the estimated geographic latitudinal and elevational midpoint for liolaemids) and corrected for latitudinal and elevational deviations based on climatic lapse-rate functions (45). The temperature index regression for latitude is y = 25.79 + 0.28263x – 0.017708x2 + 0.000010727x3, where y is the corrected temperature (index) for latitude x (45). The lapse-rate function to correct air temperatures at sea level to those at different elevations (45) is 0.65°C/100 m. The sum of the products of each equation, less the starting temperature (here, 20°C), provide a single index for a given latitude and elevation. Because many factors determine the thermal environment at a given place and time (including cloud cover, day length, and vegetation), this index cannot predict actual environmental temperatures. However, it provides an index of environmental temperatures that can be used for comparing taxa from different thermal environments resulting from differences in latitude and elevation.
Correlations Between Herbivory and Climate. We examined the relationship between evolutionary changes in diet and changes in climate by using independent contrasts (46) as implemented in compare. One set of analyses used branch lengths estimated from the ND2 gene and included 75 taxa. We used the topology in Fig. 1 (pruned to include only these 75 taxa) with branch lengths for the ND2 gene region (reduced to the same 1.4-kb region present in almost all taxa), estimated by using maximum likelihood with the general time reversible (GTR) + I + Γ model (implemented with paup*). Two data points were extreme and obvious outliers in climate (with no corresponding change in diet) and were removed from subsequent regression analyses. These two points had contrasts for climate that were ≈10 times higher than all others (despite all species having relatively similar climatic index values), apparently because of their extremely short branch lengths in the mitochondrial phylogeny. Results from this analysis were highly significant (r2 = 0.23, P < 0.0001; range among alternative trees r2 = 0.21–0.23, P < 0.0001). Another set of analyses assumed equal branch lengths and included 11 additional taxa for which ND2 data were not available. These results were also highly significant (r2 = 0.12, P = 0.0011; range among alternative trees r2 = 0.11–0.14, P = 0.0005–0.0015).
Body Size Reconstructions. The ancestral SVL of Liolaemidae was estimated by using linear generalized least squares (as for diet). Branch-length information from the ND2 gene was available for 78 taxa for which SVLs were also available and was estimated as described above. The reconstructed ancestral SVL is 82.7 ± 37.6 mm (range among alternate trees 82.6–82.7 ± 37.5–37.7 mm). Although the SEs for these and several other reconstructions are high, simulation studies suggest that the estimates of error for ancestral-trait reconstructions may be inappropriately high, even when the reconstructions are accurate (47). An analysis including 11 additional taxa for which ND2 data were not available (assuming equal branch lengths for all taxa) estimates an ancestral SVL of 79.9 ± 11.6 mm (range among alternative trees 79.2–80.0 ± 11.4–11.6 mm).
Body Temperature Reconstructions. Body temperatures of active liolaemids (Data Set 1) were taken from previous studies (18 species) or collected in nature (49 species) by using standard methods described in Supporting Methods. The body temperature of the common ancestor of Liolaemidae (reconstructed value 34.0 ± 5.8°C) was estimated by using linear generalized least squares with branch lengths from the ND2 gene (described above). Almost all nodes of the liolaemid tree have reconstructed values within the range of 33–37°C. The tree in Fig. 1 was pruned to include the 52 taxa for which body temperatures and ND2 sequences were available. Among the parsimony and Bayesian trees, the ancestral body temperature range was 33.9–34.0 ± 5.7–5.8°C. The set of taxa for which body temperatures are available is almost identical to the set of taxa for which both body temperatures and ND2 data are available, so analyses assuming equal branch lengths were not performed.
Ancestral Climate Reconstructions. The temperature indices described above were reconstructed by using linear generalized least squares on the phylogeny (Fig. 1) to estimate the ancestral climatic regime of Liolaemidae. Reconstructions were performed for 75 taxa by using branch lengths estimated for the ND2 data (reconstructed temperature 33.2 ± 37.7°C; range for alternate trees 33.2–33.3 ± 37.6–37.7°C) and for 86 taxa assuming equal branch lengths (37.1 ± 4.9°C; range for alternate trees 36.9–37.2 ± 4.9°C). These analyses concur that the ancestor of Liolaemidae occurred in a relatively hot environment (>33°C) relative to other liolaemids (mean temperature index 22.5°C for these 86 taxa).
Results and Discussion
Our reconstructed phylogeny, based on a combined analysis of DNA sequences and nonmolecular characters, includes 92 liolaemid taxa (plus four outgroup species). Mapping diet onto this phylogeny shows eight or nine independent origins of herbivory among the sampled species (Fig. 1; mean 8.5, range eight to nine origins among reconstructions for all alternative trees). However, we estimate the number of origins of herbivory in liolaemids to be at least 18, based on reconstructions including 161 liolaemid taxa for which data on diet are available (mean 18.5 origins across trees and equally parsimonious reconstructions in the supertree analysis). In contrast, there are only 10 or 11 independent origins of herbivory inferred for all other lizard species combined (9, 10). These origins are scattered widely among major lizard clades, and no family besides Liolaemidae contains more than four origins of herbivory. As in other lizard clades that include herbivores (9, 10), most origins of herbivory in liolaemids appear to represent transitions from omnivory to herbivory rather than direct transitions from insectivory to herbivory (Fig. 1).
Herbivory appears to evolve at least 66 times more rapidly in liolaemids than in nonliolaemid squamates. There are changes to herbivory on ≈4.6% of the branches of the liolaemid tree in Fig. 1 (and on 5.7% of the branches on the liolaemid supertree). In contrast, only 0.07% of the branches on a species-level phylogeny of nonliolaemid squamates would have changes to herbivory. This dramatic difference suggests that, in liolaemids, there has been a release from the constraints that have apparently limited the evolution of herbivory in other squamate reptiles.
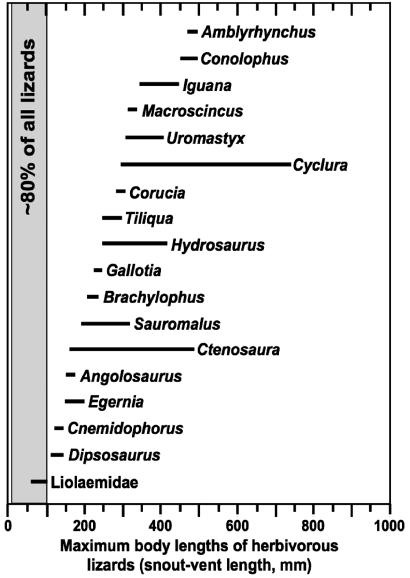
In support of this hypothesis, herbivorous liolaemids differ greatly from other herbivorous lizards in body size and climatic distribution. Liolaemids are smaller than other herbivorous lizards (Fig. 2). Furthermore, most herbivorous liolaemids live in cooler climates than other herbivorous lizards. Thirty-one of 34 known herbivorous liolaemid species live at either high latitude or high elevation. Those living in the Patagonian steppe (13 species) range from ≈37 to 54°S latitude (mean geographic midpoint of the latitudinal ranges of the Patagonian species = 44°S), whereas those distributed in the high Andes (18 species) range from 1,800 to 5,000 m (mean elevational midpoint = 3,580 m). Herbivorous lizards from other clades occur in warm climates at considerably lower latitudes (0–37°) and elevations (0–1,830 m) (9–11).
Fig. 2.
Body sizes of herbivorous Liolaemidae relative to other Recent herbivorous lizards. Bar lengths indicate ranges of maximum body sizes for species within each taxonomic group. Data on body sizes are from ref. 9 and Data Set 1. The gray bar indicates that ≈80% of all lizard species have body masses corresponding to a SVL of ≤100 mm (4, 7).
Given that there are sound ecological and physiological reasons to expect herbivorous lizards to be large and live in warm climates (2, 7, 8, 11, 13, 14), how can the seemingly incongruous traits associated with herbivory in liolaemids be reconciled with the longstanding rules of herbivory in reptiles? We suggest that the repeated and paradoxical evolution of herbivory in liolaemids is explained by a unique combination of climatic conditions, ecophysiological constraints and opportunities, and phylogenetic history.
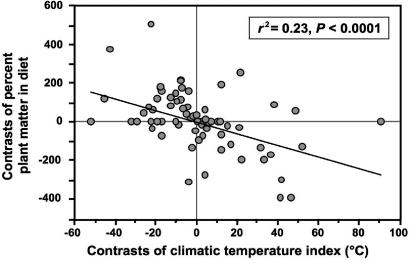
Regression analyses using phylogenetically independent contrasts (46) show that herbivory is correlated with cool climates in Liolaemidae (albeit weakly) (Fig. 3), the opposite of the pattern reported for other herbivorous lizards (9–11). This finding suggests that the invasion of cool climates facilitates the evolution of herbivory in liolaemids. Herbivory is likely favored in cool climates because insects may be a more ephemeral and less abundant food source than are edible plants in high-latitude and high-elevation habitats (26). Most herbivorous lizards live in hot, dry environments or on islands, which are similarly harsh and likely to have correspondingly low arthropod abundance and diversity (8, 10, 48, 49). Although most herbivorous liolaemids live in relatively cool climates, the strength of the correlation between climate and herbivory in the independent contrasts analysis is weakened by the presence of a few herbivorous liolaemids occurring in warm climates (e.g., Liolaemus zapallarensis), and because many nonherbivorous liolaemids live in cool regions (e.g., Liolaemus andinus, Liolaemus sarmientoi, and Liolaemus tari). Thus, herbivory is not a requisite for living in cool climates or harsh deserts, although plant consumption appears to be favored in these environments in several liolaemid lineages.
Fig. 3.
The evolution of herbivory is correlated with cold climates in liolaemid lizards. Results are based on a regression of phylogenetically independent contrasts (46) of the gross proportion of plant matter in the diet of each species and an index of the environmental temperature for each species. Contrasts were calculated by using the phylogeny in Fig. 1 for a sample of 75 species for which data on diet and ND2 sequences (for estimating branch lengths) were available.
The unique association between herbivory and cool climates in liolaemids (Fig. 3; see also ref. 26) may help explain the unusually high frequency with which herbivory has evolved in this group. Because high montane habitats tend to be geographically isolated from each other (and from low-elevation, high-latitude regions), it may be difficult for herbivorous, cold-adapted liolaemids to disperse among these regions. Thus, different lineages of liolaemids may evolve herbivory independently in different high-latitude or high-montane regions, thereby promoting multiple origins of herbivory. In contrast, warm-climate, low-elevation regions are intrinsically more connected than high-elevation regions. For example, the majority of nonliolaemid herbivorous squamates belong to a single clade (Iguanidae, sensu stricto; refs. 2 and 7–10) that is widely distributed in the warm-climate regions of the New World and Oceania. On the mainland, these warm regions are connected geographically, and the range of each continental iguanid genus overlaps the distribution of at least one other (50).
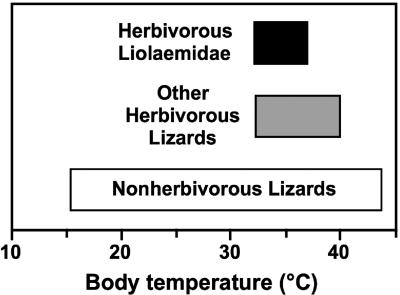
Although most herbivorous liolaemids occur in regions that are much cooler than those inhabited by other herbivorous reptiles, our data indicate that herbivorous liolaemids also seem to require high body temperatures, similar to those recorded for other herbivorous lizards (refs. 9, 11, and 12 and Fig. 4). The need for high body temperatures is likely a general ecophysiological constraint on the evolution of herbivory because the fermentation of plant fiber appears to require high body temperatures (9, 11, 12, 18, 19). Consequently, the absence of herbivores in some reptile lineages may be explained by environmental or evolutionary constraints on achieving high body temperatures (9).
Fig. 4.
Body temperatures of herbivorous Liolaemidae compared with other herbivorous lizards and lizards generally. Results are based on previous studies and this study (ref. 9 and Data Set 1). Herbivorous lizards generally maintain higher body temperatures than nonherbivorous species (9, 11, 12, 19), and body temperatures of herbivorous liolaemids are similar to those of other herbivorous lizards.
Given that herbivorous liolaemids must maintain high body temperatures despite living in cool climates, ecophysiological constraints may explain why they are smaller than other herbivorous lizards. Warm temperatures may occur infrequently and briefly at the high latitudes and high elevations where most herbivorous liolaemids occur (9, 21, 22, 51). Biophysical principles suggest that small size is advantageous in these environments because a small lizard can warm faster than can a large lizard, given the low thermal inertia associated with a small body mass (52). Thus, the small body size of liolaemids can facilitate rapid heating in unpredictable thermal environments, even if the lizards are unlikely to maintain high body temperatures for as long (daily or seasonally) as would a lizard of similar size in the tropics or a hot desert (9).
If ecophysiological constraints imposed by eating plants necessitate high body temperatures and small body size in cool-climate herbivores, then phylogenetic history also may have played a critical role in promoting the repeated evolution of herbivory in liolaemids. Ancestral state reconstructions indicate that the common ancestor of Liolaemidae had a small body size (reconstructed SVL 80.4 ± 11.7 mm for 86 taxa) and a preference for high body temperatures (reconstructed value 33.8 ± 5.8°C for 52 taxa), even though this ancestor was most likely insectivorous (Fig. 1) and lived in a warm climate (reconstructed value 37.0 ± 4.9°C for 86 taxa). Thus, the evidence suggests that the critical ecophysiological traits permitting the repeated evolution of herbivory in cold-climate liolaemids were inherited from an ancestor that evolved in a very different climate and with a very different diet.
In summary, our studies reveal repeated and unexpected origins of herbivory in liolaemid lizards that challenge many preconceptions about the evolution of dietary strategies in vertebrates. The discovery of the repeated evolution of herbivory in small-bodied, cool-climate-dwelling liolaemids also raises many questions for future research, such as how liolaemids overcome the disadvantages of small size for digestive efficiency and why other groups of lizards with similar attributes (i.e., small size, cool-climate distribution, and high body temperatures) have not evolved herbivory (e.g., some lacertids, Phrynocephalus, and Sceloporus). More generally, our results show how characteristics of the environment, ecophysiology, and phylogenetic history can interact in unexpected ways to cause striking changes in macroevolutionary trends in diet.
Supplementary Material
Acknowledgments
We thank J. Navarro (Universidad de Chile, Departamento de Biología Celular y Genetica, Santiago, Chile), S. Kretzschmar, R. Laurent, E. Lavilla, and G. Scrocchi (Instituto Fundación Miguel Lillo, Tucumán, Argentina), A. Resetar and H. Voris (Field Museum of Natural History, Chicago), H. Núñez (Museo Nacional de Historia Natural, Santiago, Chile), B. Stein and D. Wake (University of California, Museum of Vertebrate Zoology, Berkeley), R. Etheridge (San Diego State University, San Diego), F. Lobo (Universidad Nacional de Salta, Salta, Argentina), M. Christie, and J. Valladeres for permitting us to examine specimens or for tissues; C. Abdala, F. Cruz, R. Etheridge, M. Halloy, E. Lavilla, F. Lobo, I. Oliver, G. Perotti, and S. Torres for assistance in Argentina; S. Berman, J. Fetzner, C. Parkinson, T. Reeder, and J. Schulte for assistance in obtaining molecular data; R. Etheridge and H. Núñez for unpublished data and advice; and R. Etheridge, J. Hogue, M. Servedio, D. Wake, and two anonymous reviewers for offering suggestions to improve the manuscript. Our study was funded by a Porter Fellowship from the American Physiological Society, a Rea Fellowship and support from the Carnegie Museum of Natural History, the Graduate School, Biology Department, and Biological Resources Research Center at the University of Nevada, Reno, the Probationary Faculty Support Program at California State University, Northridge, the American Society of Ichthyologists and Herpetologists, the Chicago and Upstate (New York) Herpetological Societies, the Explorers Club, the Fondo para la Investigación Científica y Tecnológica of Argentina, the National Geographic Society, the National Science Foundation (DEB 0331747), and the Society for Comparative and Integrative Biology.
Author contributions: R.E.E., J.J.W., and C.R.T. designed research; R.E.E. and J.J.W. performed research; R.E.E. and J.J.W. analyzed data; and R.E.E., J.J.W., and C.R.T. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ND2, NADH dehydrogenase subunit II; SVL, snout–vent length.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY661892–AY661908 and AY662050–AY662079).
See Commentary on page 16713.
References
- 1.Crawley, M. J. (1983) Herbivory: The Dynamics of Animal–Plant Interactions (Univ. of California Press, Berkeley), Vol. 10.
- 2.King, G. (1996) Reptiles and Herbivory (Chapman & Hall, London).
- 3.Sues, H.-D., ed. (2000) Evolution of Herbivory in Terrestrial Vertebrates: Perspectives from the Fossil Record (Cambridge Univ. Press, Cambridge, U.K.).
- 4.Pough, F. H., Janis, C. M. & Heiser, J. B. (2001) Vertebrate Life (Prentice–Hall, Englewood Cliffs, NJ), 6th Ed.
- 5.Owen-Smith, R. N. (2002) Adaptive Herbivore Ecology: From Resources to Populations in Variable Environments (Cambridge Univ. Press, New York).
- 6.Hotton, N., III, Olson, E. C. & Beerbower, R. (1997) in Amniote Origins: Completing the Transition to Land, eds. Sumida, S. S. & Martin, K. L. M. (Academic, San Diego), pp. 207–264.
- 7.Pough, F. H. (1973) Ecology 54, 837–844. [Google Scholar]
- 8.Iverson, J. B. (1982) in Iguanas of the World: Their Behavior, Ecology, and Conservation, eds. Burghardt, G. M. & Rand, A. S. (Noyes, Park Ridge, NJ), pp. 60–76.
- 9.Espinoza, R. E. (2002) Ph.D. dissertation (Univ. of Nevada, Reno).
- 10.Cooper, W. E., Jr., & Vitt, L. J. (2002) J. Zool. 257, 487–517. [Google Scholar]
- 11.Zimmerman, L. C. & Tracy, C. R. (1989) Physiol. Zool. 62, 374–409. [Google Scholar]
- 12.Espinoza, R. E & Tracy, C. R. (1997) in The Biology, Husbandry, and Health Care of Reptiles, ed. Ackerman, L. J. (T.F.H., Neptune City, NJ), Vol. 1, pp. 159–194. [Google Scholar]
- 13.Wilson, K. J. & Lee, A. K. (1974) Copeia, 338–348.
- 14.Van Devender, R. W. (1982) in Iguanas of the World: Their Behavior, Ecology, and Conservation, eds. Burghardt, G. M. & Rand, A. S. (Noyes, Park Ridge, NJ), pp. 162–183.
- 15.McNab, B. K. (2002) The Physiological Ecology of Vertebrates: A View from Energetics (Cornell Univ. Press, Ithaca, NY).
- 16.Alexander, R. M. (1999) Energy for Animal Life (Oxford Univ. Press, Oxford).
- 17.Troyer, K. (1991) in Biosynthesis and Biodegradation of Cellulose, eds. Haigler, C. H. & Wimer, P. J. (Dekker, New York), pp. 311–325.
- 18.Bjorndal, K. A. (1997) in Gastrointestinal Microbiology. Vol. 1: Gastrointestinal Ecosystems and Fermentations, eds. Mackie, R. I. & White, B. A. (Chapman & Hall, New York), pp. 199–230.
- 19.Schall, J. J. & Dearing, M. D. (1994) J. Herpetol. 28, 526–528. [Google Scholar]
- 20.Stevens, C. E. & Hume, I. D. (1995) Comparative Physiology of the Vertebrate Digestive System (Cambridge Univ. Press, Cambridge, U.K.), 2nd Ed.
- 21.Cei, J. M. (1986) Reptiles del Centro, Centro-oeste y Sur de la Argentina (Mus. Reg. Sci. Nat., Turin, Italy), Monogr. 4.
- 22.Cei, J. M. (1993) Reptiles del Noroeste, Nordeste y Este de la Argentina (Mus. Reg. Sci. Nat., Turin, Italy), Monogr. 14.
- 23.Etheridge, R. (1995) Amer. Mus. Nov. 3142, 1–34. [Google Scholar]
- 24.Etheridge, R. & Espinoza, R. E. (2000) Smithsonian Herpetol. Info. Serv. 126, 1–64. [Google Scholar]
- 25.Schulte, J. A., II, Valladares, J. P. & Larson, A. (2003) Herpetologica 59, 399–419. [Google Scholar]
- 26.Hurtubia, J. & di Castri, F. (1973) in Ecological Studies: Analysis and Synthesis, eds. di Castri, F. & Mooney, H. A. (Springer, Berlin), Vol. 7, pp. 349–360. [Google Scholar]
- 27.Fuentes, E. R. & di Castri, F. (1975) Anal. Mus. Hist. Nat. Valparaíso 8, 66–75. [Google Scholar]
- 28.Jaksic, F. (1978) Anal. Mus. Hist. Nat. Valparaíso 11, 113–116. [Google Scholar]
- 29.Greene, H. W. (1982) in Environmental Adaptation and Evolution, eds. Mossakowsky, D. & Roth, G. (Fischer, Stuttgart), pp. 107–128.
- 30.Valencia, J., Veloso, A. & Sallaberry, M. (1982) in El Hombre y los Ecosistemas de Montaña (Of. Reg. Cien. Tech. UNESCO Am. Latina Caribe, Montevideo, Uruguay), pp. 269–291.
- 31.Jaksic, F. M. & Schwenk, K. (1983) Herpetologica 39, 457–461. [Google Scholar]
- 32.Schwenk, K. (2000) in Feeding: Form, Function, and Evolution in Tetrapod Vertebrates, ed. Schwenk, K. (Academic, San Diego), pp. 175–291.
- 33.Frost, D. R. & Etheridge, R. (1989) Misc. Pap. Mus. Nat. Hist., Univ. Kansas 81, 1–65. [Google Scholar]
- 34.Etheridge, R. (2000) Herpetol. Monogr. 14, 293–352. [Google Scholar]
- 35.Schulte, J. A., II, Macey, J. R., Larson, A. & Papenfuss, T. J. (1998) Mol. Phylogenet. Evol. 8, 367–376. [DOI] [PubMed] [Google Scholar]
- 36.Schulte, J. A., II., Macey, J. R., Espinoza, R. E. & Larson, A. (2000) Biol. J. Linn. Soc. 69, 75–102. [Google Scholar]
- 37.Swofford, D. L. (2001) paup*, Phylogenetic Analysis Using Parsimony (*and Other Methods) (Sinauer, Sunderland, MA), Version 4.
- 38.Huelsenbeck, J. P. & Ronquist, F. (2001) Bioinformatics 17, 754–755. [DOI] [PubMed] [Google Scholar]
- 39.Wiens, J. J., Reeder, T. W. & Nieto Montes de Oca, A. (1999) Evolution 53, 1884–1897. [DOI] [PubMed] [Google Scholar]
- 40.Maddison, W. P. & Maddison, D. R. (2000) macclade: Analysis of Phylogeny and Character Evolution (Sinauer, Sunderland, MA), Version 4.
- 41.Martins, E. P. & Hansen, T. F. (1997) Am. Nat. 149, 646–667. [Google Scholar]
- 42.Felsenstein, J. (1978) Syst. Zool. 27, 27–33. [Google Scholar]
- 43.Frost, D. R., Etheridge, R., Janies, D. & Titus, T. A. (2001) Amer. Mus. Nov. 3343, 1–38. [Google Scholar]
- 44.Pough, F. H., Andrews, R. M., Cadle, J. E., Crump, M. L., Savitzky, A. H. & Wells, K. D. (2001) Herpetology (Prentice–Hall, Englewood Cliffs, NJ), 2nd Ed.
- 45.Conrad, V. & Pollak, L. W. (1950) Methods in Climatology (Harvard Univ. Press, Cambridge, MA).
- 46.Felsenstein, J. (1985) Am. Nat. 125, 1–15. [Google Scholar]
- 47.Martins, E. P. (1999) Syst. Biol. 48, 642–650. [Google Scholar]
- 48.Pietruszka, R. D., Hanrahan, S. A., Mitchell, D. & Seely, M. K. (1986) Oecologia 70, 587–591. [DOI] [PubMed] [Google Scholar]
- 49.Van Damme, R. (1999) J. Herpetol. 33, 663–674. [Google Scholar]
- 50.Hollingsworth, B. D. (2004) in Iguanas: Biology and Conservation, Alberts, A. C., Carter, R. L., Hayes, W. K. & Martins, E. P. (Univ. of California Press, Berkeley), pp. 19–44.
- 51.Pearson, O. P. (1954) Copeia 1954, 111–116. [Google Scholar]
- 52.Tracy, C. R. (1982) in Biology of the Reptilia, eds. Gans, C. & Pough, F. H. (Academic, New York), Vol. 12, pp. 275–321. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.