Abstract
Nonsteroidal antiinflammatory drugs (NSAIDs) form a paradigm for the chemoprevention of cancer, preventing colonic tumor progression in both experimental animals and humans. However, the mechanisms underlying the antineoplastic effects of NSAIDs are currently unclear. We found that the mitochondrial second mitochondrial-derived activator of caspase (SMAC)/direct inhibitor of apoptosis protein-binding protein with low pI (Diablo) protein translocates into the cytosol during NSAID-induced apoptosis in colon cancer cells. When SMAC/Diablo is disrupted by homologous recombination and RNA interference in these cells, the NSAID-induced apoptosis is abrogated. Biochemical markers of apoptosis, such as caspase activation, cytosolic release of cytochrome c and apoptosis-inducing factor, and mitochondrial membrane potential change, are accordingly decreased. These results establish that SMAC/Diablo is essential for the apoptosis induced by NSAIDs in colon cancer cells.
Prevention of human cancers through the use of chemical agents or dietary manipulation is an attractive option to reduce morbidity and mortality from these diseases. Colorectal cancer not only is a prototype for identifying genetic alterations underlying tumorigenesis but also provides a paradigm for developing chemoprevention strategies (1). In particular, the widely used nonsteroidal antiinflammatory drugs (NSAIDs) suppress the formation of colorectal tumors in both humans and rodents (2).
NSAIDs have been shown to inhibit a variety of enzymes, but the most well documented of them is cyclooxygenase-2 (COX-2) (2). Growing evidence has indicated that the antineoplastic effects of NSAIDs are mediated by the induction of apoptosis secondary to the inhibition of COX-2 (3). In mammalian cells, apoptotic cell death is unleashed through a cascade of events, among which a key event is the translocation of at least two apoptogenic proteins from the mitochondria into the cytosol to trigger the activation of caspase proteases (4–7). Cytochrome c is released from the mitochondria into cytosol, where it binds to other proteins and forms an “apoptosome” (8). Formation of this complex leads to activation of the caspase cascade (8). Second mitochondrial-derived activator of caspase (SMAC), also called direct inhibitor of apoptosis protein (IAP)-binding protein with low pI (Diablo), is the second mitochondrial protein found to be released into cytosol during apoptosis (9, 10). SMAC/Diablo (hereafter referred to as SMAC) promotes caspase activation, presumably by antagonizing IAPs such as X-linked IAP (XIAP) (10). Expression of SMAC sensitizes human cancer cells to apoptosis induced by a variety of anticancer agents (11). However, SMAC-knockout (KO) mice were not found to have abnormalities in apoptosis in vivo, raising the question of whether SMAC is required for apoptotic initiation (12). In this study, we investigated the role of SMAC in apoptosis induced by NSAIDs in colon cancer cells using gene targeting and RNA interference (RNAi).
Materials and Methods
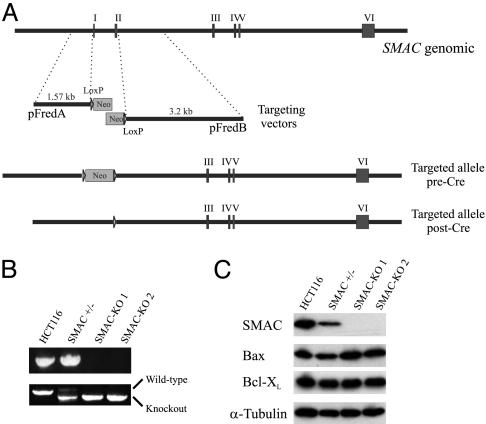
Targeting SMAC in HCT116 Cells. SMAC-KO constructs were generated by cloning a 1.57- and a 3.20-kb SMAC genomic DNA fragment flanking exons 1 and 2 of the SMAC gene, respectively, into the bipartite targeting vectors depicted in Fig. 2A, as described (13). HCT116 colon cancer cells with two intact alleles of SMAC were transfected with the KO constructs by Lipofectamine (Invitrogen) and selected in G418 (0.4 mg/ml) for 3 weeks. Neomycin-resistant clones were pooled and screened for targeting events by PCR by using a SMAC primer (5′-GAAACGAAACTGTTCGCAC-3′) and a neomycin-resistance gene primer (5′-TTGTGCCCAGTCATAGCCG-3′). The targeting events were confirmed by PCR by using different primer pairs (sequences available upon request). To target the second allele of SMAC, the neomycin-resistance gene flanked by two Lox P sites was excised from a heterozygous clone by infection with an adenovirus (Ad) expressing the Cre recombinase (Ad-Cre). The same targeting constructs were used to target the second allele. Targeting at the SMAC locus was verified by PCR and Western blotting.
Fig. 2.
Targeted deletion of SMAC in HCT116 colon cancer cells. (A) SMAC genomic locus and targeting constructs. Each targeting construct consists of a homologous arm (1.57 and 3.20 kb, respectively) and an overlapping fragment of the neomycin-resistance gene (Neo). Homologous recombination resulted in deletion of exons 1 and 2 of SMAC. The same constructs were used in the second round of gene targeting after the Neo was excised from the heterozygous cells by Cre recombinase. (B) SMAC-KO clones identified by genomic PCR. (Upper) PCR products amplified from cells with different SMAC genotypes using primers specific for the deleted region. (Lower) Larger PCR products represent the wild-type SMAC allele, whereas smaller products represent the recombinant allele, which was amplified from an endogenous PCR priming site and a PCR priming site introduced into the genomic locus by the KO construct. (C) The expression of SMAC protein in cells with different SMAC genotypes. Lysates from the cells with the indicated genotypes were analyzed by Western blotting using a SMAC-specific antibody. The same blot was stripped and reprobed for Bax and Bcl-XL. α-Tubulin was used as a loading control.
Cell Culture and Drug Treatment. All cell lines were maintained in McCoy's 5A media (Invitrogen) supplemented with 10% FBS (HyClone), 100 units/ml penicillin, and 100 μg/ml streptomycin at 37°C and 5% CO2. To reconstitute SMAC expression in the SMAC-KO cells, we transfected a SMAC-expressing plasmid (a gift from Chunying Du, Stowers Institute for Medical Research, Kansas City, MO) into the SMAC-KO cells and identified stable clones expressing SMAC by Western blotting.
Cells were plated in 12-well plates at 20–30% density 24 h before treatment with anticancer agents. DMSO stock solutions of anticancer agents were diluted into appropriate concentrations by cell culture medium and added to the cells. The anticancer agents used in this experiment are: ALLN (Sigma), Camptothecin (Sigma), Cisplatin (Sigma), 5-Fluorouracil (5-FU, Sigma), Indomethacin (Sigma), Mitomycin C (Sigma), Staurosporine (Sigma), Sulindac Sulfide (Merck), Taxol (Sigma), tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) (Alexis Biochemicals, San Diego) and Vinblastine (Sigma). In some cases, caspase inhibitors (R & D Systems), including the pan-caspase inhibitor Z-VAD-fmk, caspase-3 inhibitor Z-DEVD-fmk, and caspase-9 inhibitor Z-LEHD-fmk, were used to treat cells before drug treatment.
Apoptosis Assays. After treatment, attached and floating cells were harvested at various time points. The fractions of apoptotic cells were evaluated by nuclear staining and annexin V/propidium iodide (PI) staining. For nuclear staining, cells were fixed in a solution containing final concentrations of 3.7% formaldehyde, 0.5% Nonidet P-40 and 10 μg/ml DAPI in PBS, and assessed for apoptosis through microscopic visualization of condensed chromatin and micronucleation. For each measurement, at least three independent experiments and a minimum of 300 cells were analyzed. For annexin V/PI staining, cells were stained by using the Apo-Alert Annexin V kit (BD Biosciences), following the manufacturer's instruction, and then analyzed by flow cytometry. The long-term survival of cells after drug treatment was evaluated by colony formation assays. In brief, the harvested cells were plated in 25-cm2 flasks at appropriate dilutions. Cells were allowed to grow for 10–14 days before staining with Crystal Violet (Sigma). All experiments were repeated at least three times. For detection of mitochondrial membrane potential change, harvested cells were stained by Mito Tracker Red CMXRos (Molecular Probes) for 15 min at room temperature and then analyzed by flow cytometry on a Beckman Coulter EPICS XL using the FL3 channel according to the manufacturer's instructions.
Western Blotting and Antibodies. Cell lysates were collected at various time points, and immunoblotting was performed as described (13). The antibodies used for Western blotting include rabbit polyclonal antibodies for SMAC (a generous gift from Eileen White, Rutgers University, Piscataway, NJ), caspase 9 (Cell Signaling Technology, Beverly, MA), Bax (N20, Santa Cruz Biotechnology, Santa Cruz, CA), Bcl-XL (BD Biosciences), active caspase 3 (BD Biosciences), and α-tubulin (Oncogene Science).
Cellular Fractionation. Two 75-cm2 flasks of cells were harvested after a PBS wash at 4°C. Whole-cell pellets were resuspended in homogenization buffer (0.25 M sucrose/10 mM Hepes, pH 7.4/1 mM EGTA) and subjected to 40 strokes in a 2-ml Dounce homogenizer at 4°C. The homogenates were subjected to centrifugation at 1,000 × g for 15 min at 4°C to pellet nuclei and unbroken cells. The supernatant was subsequently subjected to centrifugation at 10,000 × g for 15 min at 4°C to obtain cytosolic fractions (supernatant) and mitochondrial fractions (pellet).
RNAi. Nucleotides 204–224 of the SMAC coding sequence (TAGTAGTGAAGCATTGATGAG) were chosen as the target for RNAi. Two 64-base oligonucleotides containing this sequence were designed based on criteria previously described (14). The oligonucleotides were annealed, and the duplex was cloned into the pSUPER vector (Oligoengine, Seattle) to direct the synthesis of small interference RNA (15). RNAi constructs were transfected into DLD1 colon cancer cells by Lipofectamine, and cells were plated in 96-well plates in the presence of puromycin (5 μg/ml, Invitrogen). Puromycin-resistant clones were isolated. Western blotting was used to identify several clones with significant down-regulation of SMAC.
Results
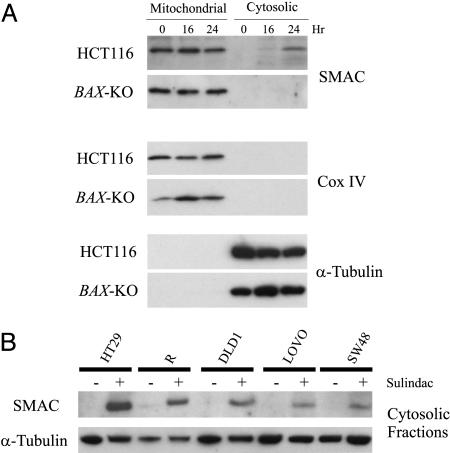
NSAIDs Induce the Release of SMAC in Colon Cancer Cells. In previous studies, we found that NSAIDs induce a fulminant apoptosis in HCT116 colorectal cancer cells (16). The apoptosis in these cells is conventional, as indicated by alterations of Bcl-2 family proteins, disruption of mitochondrial membrane potential, release of apoptogenic mitochondrial proteins, and activation of the caspase cascade (13, 16–18). Moreover, isogenic BAX-KO HCT116 cells are completely resistant to sulindac, an NSAID and prototypic agent used for the prevention of colorectal cancers (19). To determine whether SMAC plays a role in this process, we analyzed HCT116 and BAX-KO cells. We found that SMAC was released into the cytosol of the parental HCT116 cells undergoing apoptosis induced by sulindac (120 μM) (Fig. 1A), whereas SMAC was not found in the cytosol of the BAX-KO cells (Fig. 1A). These results indicate that sulindac induces a Bax-dependent release of SMAC. We then analyzed SMAC release in five other colon cancer cell lines. These cell lines have different genetic backgrounds, as assessed by mutational status of RAS, p53, APC, TGF-β RII, and other genes. It is therefore not surprising that different concentrations of sulindac were required to induce apoptosis in the various lines. However, in each case, SMAC was found to be released into the cytosol after treatment of the cells with sulindac at the appropriate concentration (Fig. 1B). Similar results were obtained when the cells were treated with indomethacin, another NSAID (data not shown). These results suggest a consistent involvement of SMAC in NSAID-induced apoptosis in colorectal cancer cells.
Fig. 1.
Release of SMAC during sulindac-induced apoptosis in colon cancer cells. (A) Bax-dependent release of SMAC induced by sulindac in HCT116 cells. HCT116 and BAX-KO cells were treated with sulindac (120 μM) for the indicated time and analyzed for mitochondrial and cystolic SMAC expression by Western blotting after fractionation. Cytochrome oxidase subunit IV (Cox IV) and α-tubulin were used as controls for the mitochondrial and cytosolic fractionations, respectively. (B) SMAC release induced by sulindac in five other colon cancer cell lines. Colon cancer cell lines were treated with sulindac at different concentrations (HT29, 200 μM; R, 200 μM; DLD1, 180 μM; LOVO, 120 μM; and SW48, 120 μM) for 48 h to induce apoptosis. Cytosolic fractions were isolated and examined for SMAC expression by Western blotting. α-Tubulin, which is exclusively expressed in cytosol, was used as a control for loading and for fractionation. Reduced α-tubulin levels in several cell lines after sulindac treatment reflect decreases in cell number.
Targeted Deletion of SMAC Abrogates NSAID-Induced Apoptosis. Is SMAC release important for NSAID-induced apoptosis or just a coincidental effect? To answer this question, we used homologous recombination to knock out the SMAC genomic locus in HCT116 cells. After two rounds of gene targeting (Fig. 2A), we isolated two SMAC-KO cell lines (SMAC-KO1 and -2) (Fig. 2B). The expression of SMAC protein was significantly decreased in the heterozygous KO cells (Fig. 2C), which is similar to what was reported for heterozygous mice (12). As expected, SMAC protein was completely absent in the homozygous KO cells (Fig. 2C). The cell morphology and growth characteristics of parental, heterozygous, and homozygous cells were indistinguishable under normal growth conditions.
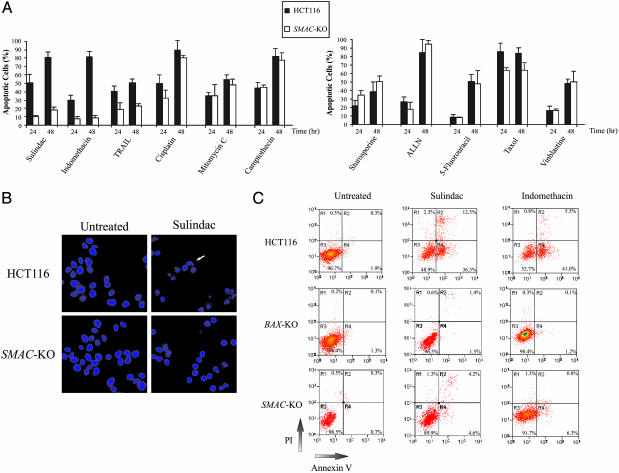
We determined the response of the SMAC-KO and the parental cells to a variety of anticancer agents, including NSAIDs, DNA-damaging agents, microtubule-targeting agents, kinase inhibitors, proteasome inhibitors, antimetabolites, and death receptor ligands. Nuclear morphological analysis revealed a major decrease in NSAID-induced apoptosis and a moderate decrease in TRAIL-induced apoptosis in the SMAC-KO cells compared with parental cells (Fig. 3 A and B). On the other hand, apoptosis induced by the other agents was not significantly affected in the SMAC-KO cells (Fig. 3A). Analysis of apoptosis by annexin V staining and cell cycle analysis confirmed that NSAID-induced apoptosis was abrogated in the SMAC-KO cells (Fig. 3C and data not shown). However, the long-term viability of the SMAC-KO cells as measured by colony formation assays was decreased after sulindac treatment (120 μM for 48 h) and was not significantly different from that of the parental cells after the same treatment.
Fig. 3.
Apoptosis induced by anticancer agents in the SMAC-KO cells. (A) Apoptosis induced by anticancer agents quantified by nuclear staining. Parental HCT116 cells and SMAC-KO cells were treated with the indicated anticancer agents for 24 and 48 h. Cells were fixed and analyzed by fluorescence microscopy after DAPI staining. At least three independent experiments were carried out for each condition, and a minimum of 300 cells were counted for each measurement. The anticancer agents include the NSAIDs sulindac sulfide (120 μM) and indomethacin (500 μM), the death receptor ligand TRAIL (50 ng/ml), the DNA crosslinking agents cisplatin (100 μM) and mitomycin C (30 μM), the topoisomerase inhibitor camptothecin (600 nM), the microtubule-targeting agents taxol (20 nM) and vinblastine (25 nM), the kinase inhibitor staurosporine (200 nM), the proteasome inhibitor ALLN (N-acetyl-leu-leu-norleu-al, 20 μM), and the antimetabolite 5-fluorouracil (375 μM). (B) Nuclear morphology of the cells undergoing apoptosis. Parental and SMAC-KO cells treated with sulindac (120 μM) for 48 h were subject to fluorescence microscopy analysis after staining with DAPI. Cells containing fragmented and condensed nuclei (an example indicated by the arrow) are apoptotic cells. (C) Apoptosis quantified by annexin V/PI staining. Parental, BAX-KO, and SMAC-KO cells were treated with sulindac (120 μM) and indomethacin (500 μM) for 36 h. Cells were stained by annexin V and PI and then analyzed by flow cytometry. Four subpopulations and their quantities are indicated: nonapoptotic dead cells (R1), late apoptotic cells (R2), viable cells (R3), and early apoptotic cells (R4).
The data in Fig. 3A show that isogenic SMAC-KO cells are resistant to apoptosis induced by NSAIDs and TRAIL, whereas previous studies have shown that BAX-KO cells are resistant to a wider spectrum of anticancer agents (16, 18, 20). Because BAX is frequently mutated in mismatch repair-deficient colon tumors, and these mutations can cause resistance to NSAIDs and TRAIL (16, 18), we investigated whether SMAC was mutated in colon tumors. Analysis of the SMAC coding sequences in 36 colon tumors did not reveal any mutations.
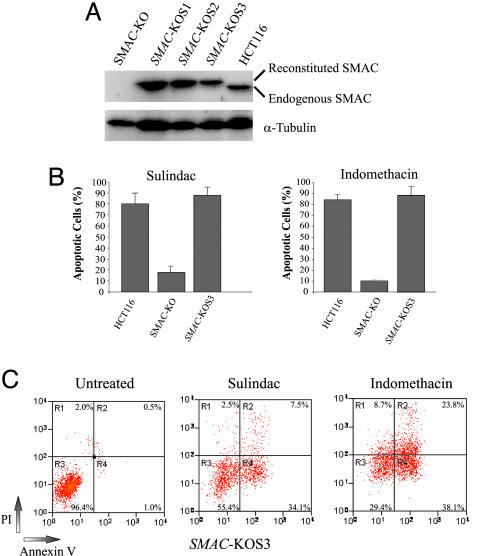
NSAID-Induced Apoptosis Can Be Restored by the Reconstituted SMAC. The apoptotic deficiency in the SMAC-KO cells was not due to an abnormality of Bax, because the genomic sequence of BAX was normal and the Bax protein was expressed in the SMAC-KO cells (Fig. 2C and data not shown). If the deficiency in apoptosis was caused by the absence of SMAC, apoptosis should be restored by introduction of SMAC into the SMAC-KO cells. To test this hypothesis, we expressed SMAC in the SMAC-KO cells by transfecting a SMAC expression plasmid (Fig. 4A). We found that NSAID-induced apoptosis in the SMAC-KO cells was completely restored (Fig. 4 B and C). We also expected that disruption of SMAC in other colon cancer cells would have similar effects on apoptosis. To test this expectation, we used RNAi to knock down expression of SMAC in DLD1 cells, a line in which SMAC is also released during NSAID-induced apoptosis (Fig. 1B). Stable DLD1 cell lines with markedly decreased expression of SMAC were found to be significantly more resistant to sulindac-induced apoptosis (Fig. 5).
Fig. 4.
NSAID-induced apoptosis restored by a reconstituted SMAC. (A) Reconstitution of SMAC expression in the SMAC-KO cells. Expression of SMAC was reconstituted in the SMAC-KO cells by transfection of a Flag-tagged SMAC expression construct. Cell clones resistant to G418 were examined for expression of SMAC by Western blotting. SMAC-KOS1, -2, and -3 are three independent clones with reconstituted SMAC expression. The reconstituted SMAC protein contains a Flag tag and is slightly larger than the endogenous SMAC protein. (B) Restoration of NSAID-induced apoptosis in the SMAC-KO cells by the reconstituted SMAC. SMAC-KOS3 cells, in which the expression level of SMAC is similar to that in the parental HCT116 cells, were analyzed for apoptosis induced by sulindac (120 μM) and indomethacin (500 μM) by nuclear staining. (C) Apoptosis in the cells with the reconstituted SMAC analyzed by annexin V/PI staining. SMAC-KOS3 cells were treated with sulindac (120 μM) or indomethacin (500 μM) for 48 h, stained by annexin V/PI, and analyzed by flow cytometry.
Fig. 5.
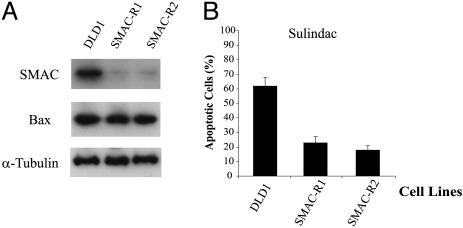
Inhibition of NSAID-induced apoptosis in DLD1 cells by RNAi knockdown of SMAC. (A) RNAi knockdown of SMAC in DLD1 cells. DLD1 cells were transfected with a SMAC RNAi construct. Puromycin-resistant clones with knockdown of SMAC (SMAC-R1 and -R2) were identified by Western blotting. Bax and α-tubulin were used as loading controls. (B) Inhibition of apoptosis in the SMAC-knockdown cells. Parental DLD1 cells, SMAC-R1, and SMAC-R2 cells were treated with sulindac (200 μM) for 48 h to induce apoptosis. Apoptosis were analyzed by nuclear staining.
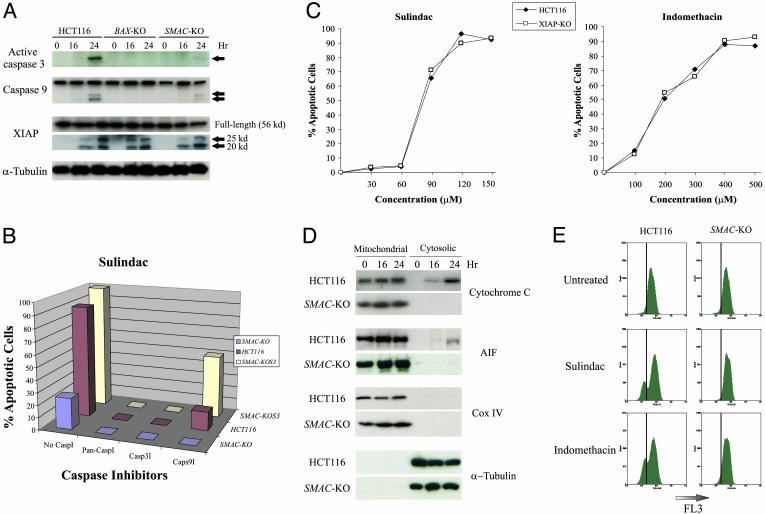
SMAC Deficiency Abrogates Caspase Activation, Cytosolic Release of Cytochrome c and Apoptosis-Inducing Factor (AIF), and Mitochondrial Membrane Potential Change. SMAC has been shown to promote caspase activation after release from the mitochondria (9, 10). We found that caspase activation was deficient in NSAID-treated SMAC-KO cells. The active form of caspase-3, whose primary function is to execute apoptosis, was abundant in parental HCT116 cells undergoing sulindac-induced apoptosis but barely detectable in SMAC-KO cells (Fig. 6A). The blockade of caspase 3 activation could be reverted by the reconstituted SMAC (data not shown). Furthermore, a caspase-3 inhibitor completely blocked NSAID-induced apoptosis (Fig. 6B). On the other hand, activation of caspase-9, which initiates the cascade of caspase activation, was also decreased in SMAC-KO cells, although to a lesser extent (Fig. 6A). This suggested that caspase-9 activation was in part mediated by SMAC. Consistent with these findings, a caspase-9 inhibitor completely blocked apoptosis in the SMAC-KO cells but only partially blocked apoptosis in the parental cells, indicating that SMAC-independent activation of caspase-9 accounts for the residual apoptosis in the SMAC-KO cells (Fig. 6 A and B). Previous in vitro studies have shown that SMAC antagonizes XIAP to promote caspase activation (10). Nonetheless, we did not find any alterations in XIAP, such as changes in its expression level or ability to bind caspases in SMAC-KO cells, despite the observation that XIAP was cleaved in sulindac-treated cells (Fig. 6A). To further investigate the role of XIAP in NSAID-induced apoptosis in colon cancer cells, we examined HCT116 cells in which the XIAP gene had been disrupted through targeted homologous recombination (21). It would be expected that XIAP-KO cells would be more sensitive to NSAIDs if SMAC functioned by inhibiting the antiapoptotic activity of XIAP. However, no differences were detected in the XIAP-KO cells compared with the parental cells with respect to NSAID-induced apoptosis, either with sulindac or indomethacin (Fig. 6C).
Fig. 6.
Mechanisms of NSAID-induced apoptosis mediated by SMAC. (A) Cleavage of caspases and XIAP. Parental HCT116 cells, BAX-KO, and SMAC-KO cells were treated with 120 μM sulindac. Cell lysates collected at the indicated time points were analyzed for active caspase 3, caspase 9, and XIAP by Western blotting. The arrows indicate the cleavage fragments of caspases and XIAP. α-Tubulin was used as a control for loading. (B) Effects of caspase inhibitors on apoptosis. Caspase inhibitors, including a pan-caspase inhibitor (pan-CaspI, Z-VAD-fmk), a caspase-3 inhibitor (Casp3I, Z-DEVD-fmk), and a caspase-9 inhibitor (Casp9I, Z-LEHD-fmk), were diluted to 20 μM with cell culture medium and added to the cells 8 h before sulindac treatment. After cells were treated with sulindac (120 μM) for 48 h, apoptosis was measured by nuclear staining. (C) NSAIDs induced apoptosis in parental and XIAP-KO cells. Cells with the indicated genotypes were treated with sulindac and indomethacin for 48 h, and apoptosis was analyzed by nuclear staining. A minimum of 300 cells were counted for each measurement, and the results are averages of three independent measurements. (D) Blockade of the release of cytochrome c and AIF in the SMAC-KO cells. Mitochondrial and cytosolic fractions from HCT116 and the SMAC-KO cells treated with sulindac (120 μM) for the indicated time were subject to Western analysis for cytochrome c and AIF. Cytochrome oxidase subunit IV (Cox IV) and α-tubulin were used as controls for fractionation. (E) Lack of mitochondrial membrane potential collapse in the SMAC-KO cells. Parental HCT116 cells and SMAC-KO cells were treated with sulindac (120 μM) and indomethacin (500 μM) for 24 h. Cells were harvested, stained by MitoTracker Red CMXRos, and immediately analyzed by flow cytometry. The mitochondrial membrane potential was detected by FL3.
The major mechanism underlying SMAC function has heretofore been thought to be inhibition of XIAP. Our results suggest otherwise. It was not expected that disruption of SMAC alone would have significant effects on apoptosis and caspase activation, because several other mitochondrial apoptogenic proteins, including cytochrome c, endonuclease G, and AIF, are released into the cytosol during apoptosis (22–24), and the release of cytochrome c is thought to be the major caspase activation mechanism (8). To reconcile these observations, we analyzed the release of cytochrome c and AIF in cells treated with NSAIDs. Surprisingly, both cytochrome c and AIF release were reduced in the SMAC-KO cells relative to the parental cells (Fig. 6D). Further analysis showed that mitochondrial membrane potential change was significantly blocked in the SMAC-KO cells compared with the parental cells (Fig. 6E). These results indicate that cytochrome c and AIF cannot be properly released into cytosol in the absence of SMAC, and the effects of SMAC may be mediated by the regulation of mitochondrial integrity.
Discussion
The results described above show that SMAC is essential for the apoptosis induced by NSAIDs in colon cancer cells. A number of biochemical studies have shown that SMAC and its homologues play an important role in programmed cell death in different organisms. Disruption of the SMAC homologues in Drosophila results in expected changes in apoptosis during fly development (25). However, targeted deletion of SMAC in mice did not show obvious alterations in apoptosis (12), raising questions about the importance of SMAC in apoptosis in mammalian cells. These observations indicate that the role of SMAC in apoptosis varies depending on specific organisms, cell types and apoptotic stimuli.
Although cytosolic release of mitochondrial proteins is known to be critical for apoptosis and caspase activation, the mechanism of this process is poorly understood. Previous studies suggested that full caspase activation requires release of both cytochrome c and SMAC from mitochondria to induce assembly of apoptosome and relieve the inhibition of caspase activation by IAPs (26). Cytochrome c release and SMAC release appear to be two independent events that are mediated via different mechanisms (27, 28). However, our results suggest that these two events are related to each other and cytochrome c cannot properly translocate to the cytosol without SMAC. Cytochrome c-mediated caspase activation is therefore affected by the absence of SMAC. This explains why a majority of the apoptosis induced by NSAIDs is inhibited in the SMAC-KO cells with a concomitant decrease in caspase activation.
The concentrations of NSAIDs used in our study are much higher than those achieved in the serum of patients or animals treated with NSAIDs, raising important questions about the physiological relevance of our results. It has been suggested that intestinal epithelial cells are exposed to NSAIDs at much higher concentrations than those measured in serum assays because these drugs are taken orally and are metabolized in a unusual fashion (29). It has also been reported that sulindac, the most effective NSAID in tumor suppression, is metabolized to the active sulfide form by aerobic gut flora on the surface of the colon (30, 31), and that systemic sulindac sulfide concentrations are significantly reduced in patients who have undergone colectomy (32). This unusual mode of metabolism can create locally high concentration of NSAIDs, which has been shown to topically suppress tumor growth in rodents (33), and this may explain why it takes such high concentrations of drugs in vitro to induce apoptosis.
Although SMAC-KO cells are deficient in NSAID-induced apoptosis, the long-term viability of these cells was decreased after sulindac treatment, as measured by colony formation assays, suggesting that mechanisms other than apoptosis are involved in the growth-suppressing effect of NSAIDs. As the apoptotic effects of NSAIDs have been described in a number of in vivo studies using animal models and familial adenomatous polyposis patients (3), our observations should stimulate further in vivo studies to clarify these issues.
Although the role of SMAC in apoptosis in mammalian cells has been less than definitive, substantial evidence has indicated that SMAC is important for the apoptosis induced by anticancer agents. SMAC overexpression and use of SMAC-derived peptides or small molecules can sensitize human cancer cells to apoptosis induced by TRAIL (34, 35). Knockdown of SMAC by RNAi inhibited apoptosis induced by tumor necrosis factor α (36). Our results demonstrate that SMAC is required for apoptosis induced by NSAIDs in colon cancer cells. Elucidating the role of SMAC in NSAID-induced apoptosis may have important implications for cancer prevention. SMAC and SMAC-derived reagents can potentially be used to enhance the antitumor effects of NSAIDs and other anticancer agents (11), and to overcome drug resistance caused by deficiencies in apoptosis due to genetic alterations in cancer cells (37).
Acknowledgments
We thank Dr. Eileen White for providing the SMAC antibody, Dr. Chunying Du for providing the SMAC expression plasmid, Dr. Fred Bunz (Johns Hopkins Oncology Center, Baltimore) for the XIAP-KO cell line, Drs. Robert W. Sobol, Jr. and Federica Marini for help with RNAi, Dr. Miaomiao Wang and Mrs. Sagarika Tiwari for technical assistance, and Drs. Daniel E. Johnson and Chunying Du for critical comments on the manuscript. This work is supported by the Edward Mallinckrodt, Jr., Foundation; the Elsa U. Pardee Foundation; the Clayton Fund; and National Institutes of Health Grants CA 043460 and CA106348. L.Z. is a scholar of the General Motors Cancer Research Foundation and the V Foundation for Cancer Research.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: NSAID, nonsteroidal antiinflammatory drug; SMAC, second mitochondrial-derived activator of caspase; IAP, inhibitor of apoptosis protein; Diablo, direct IAP-binding protein with low pI; XIAP, X-linked IAP; KO, knockout; PI, propidium iodide; RNAi, RNA interference; TRAIL, TNF-related apoptosis-inducing ligand; AIF, apoptosis-inducing factor.
References
- 1.Vogelstein, B. & Kinzler, K. W. (2002) The Genetic Basis of Human Cancer (McGraw–Hill Health Professions Division, New York).
- 2.Gupta, R. A. & Dubois, R. N. (2001) Nat. Rev. Cancer 1, 11-21. [DOI] [PubMed] [Google Scholar]
- 3.Keller, J. J. & Giardiello, F. M. (2003) Cancer Biol. Ther. 2, S140-9. [PubMed] [Google Scholar]
- 4.Cory, S., Huang, D. C. & Adams, J. M. (2003) Oncogene 22, 8590-8607. [DOI] [PubMed] [Google Scholar]
- 5.Danial, N. N. & Korsmeyer, S. J. (2004) Cell 116, 205-219. [DOI] [PubMed] [Google Scholar]
- 6.Green, D. R. & Reed, J. C. (1998) Science 281, 1309-1312. [DOI] [PubMed] [Google Scholar]
- 7.Yu, J. & Zhang, L. (2004) Curr. Opin. Oncol. 16, 19-24. [DOI] [PubMed] [Google Scholar]
- 8.Wang, X. (2001) Genes Dev. 15, 2922-2933. [PubMed] [Google Scholar]
- 9.Du, C., Fang, M., Li, Y., Li, L. & Wang, X. (2000) Cell 102, 33-42. [DOI] [PubMed] [Google Scholar]
- 10.Verhagen, A. M., Ekert, P. G., Pakusch, M., Silke, J., Connolly, L. M., Reid, G. E., Moritz, R. L., Simpson, R. J. & Vaux, D. L. (2000) Cell 102, 43-53. [DOI] [PubMed] [Google Scholar]
- 11.Arnt, C. R. & Kaufmann, S. H. (2003) Cell Death Differ. 10, 1118-1120. [DOI] [PubMed] [Google Scholar]
- 12.Okada, H., Suh, W. K., Jin, J., Woo, M., Du, C., Elia, A., Duncan, G. S., Wakeham, A., Itie, A., Lowe, S. W., et al. (2002) Mol. Cell. Biol. 22, 3509-3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu, J., Wang, Z., Kinzler, K. W., Vogelstein, B. & Zhang, L. (2003) Proc. Natl. Acad. Sci. USA 100, 1931-1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elbashir, S. M., Harborth, J., Weber, K. & Tuschl, T. (2002) Methods 26, 199-213. [DOI] [PubMed] [Google Scholar]
- 15.Brummelkamp, T. R., Bernards, R. & Agami, R. (2002) Science 296, 550-553. [DOI] [PubMed] [Google Scholar]
- 16.Zhang, L., Yu, J., Park, B. H., Kinzler, K. W. & Vogelstein, B. (2000) Science 290, 989-992. [DOI] [PubMed] [Google Scholar]
- 17.Deng, Y., Lin, Y. & Wu, X. (2002) Genes Dev. 16, 33-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LeBlanc, H., Lawrence, D., Varfolomeev, E., Totpal, K., Morlan, J., Schow, P., Fong, S., Schwall, R., Sinicropi, D. & Ashkenazi, A. (2002) Nat. Med. 8, 274-281. [DOI] [PubMed] [Google Scholar]
- 19.Chan, T. A., Morin, P. J., Vogelstein, B. & Kinzler, K. W. (1998) Proc. Natl. Acad. Sci. USA 95, 681-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu, J., Tiwari, S., Steiner, P. & Zhang, L. (2003) Cancer Biol. Ther. 2, 694-699. [PubMed] [Google Scholar]
- 21.Cummins, J. M., Kohli, M., Rago, C., Kinzler, K. W., Vogelstein, B. & Bunz, F. (2004) Cancer Res. 64, 3006-3008. [DOI] [PubMed] [Google Scholar]
- 22.Liu, X., Kim, C. N., Yang, J., Jemmerson, R. & Wang, X. (1996) Cell 86, 147-157. [DOI] [PubMed] [Google Scholar]
- 23.Li, L. Y., Luo, X. & Wang, X. (2001) Nature 412, 95-99. [DOI] [PubMed] [Google Scholar]
- 24.Susin, S. A., Lorenzo, H. K., Zamzami, N., Marzo, I., Snow, B. E., Brothers, G. M., Mangion, J., Jacotot, E., Costantini, P., Loeffler, M., et al. (1999) Nature 397, 441-446. [DOI] [PubMed] [Google Scholar]
- 25.Martin, S. J. (2002) Cell 109, 793-796. [DOI] [PubMed] [Google Scholar]
- 26.Chai, J., Du, C., Wu, J. W., Kyin, S., Wang, X. & Shi, Y. (2000) Nature 406, 855-862. [DOI] [PubMed] [Google Scholar]
- 27.Springs, S. L., Diavolitsis, V. M., Goodhouse, J. & McLendon, G. L. (2002) J. Biol. Chem. 277, 45715-45718. [DOI] [PubMed] [Google Scholar]
- 28.Adrain, C., Creagh, E. M. & Martin, S. J. (2001) EMBO J. 20, 6627-6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moore, B. C. & Simmons, D. L. (2000) Curr. Med. Chem. 7, 1131-1144. [DOI] [PubMed] [Google Scholar]
- 30.Strong, H. A., Renwick, A. G., George, C. F., Liu, Y. F. & Hill, M. J. (1987) Xenobiotica 17, 685-696. [DOI] [PubMed] [Google Scholar]
- 31.Lee, S. C. & Renwick, A. G. (1995) Biochem. Pharmacol. 49, 1567-1576. [DOI] [PubMed] [Google Scholar]
- 32.Strong, H. A., Warner, N. J., Renwick, A. G. & George, C. F. (1985) Clin. Pharmacol. Ther. 38, 387-393. [DOI] [PubMed] [Google Scholar]
- 33.Seed, M. P., Brown, J. R., Freemantle, C. N., Papworth, J. L., Colville-Nash, P. R., Willis, D., Somerville, K. W., Asculai, S. & Willoughby, D. A. (1997) Cancer Res. 57, 1625-1629. [PubMed] [Google Scholar]
- 34.Fulda, S., Wick, W., Weller, M. & Debatin, K. M. (2002) Nat. Med. 8, 808-815. [DOI] [PubMed] [Google Scholar]
- 35.Li, L., Thomas, R. M., Suzuki, H., De Brabander, J. K., Wang, X. & Harran, P. G. (2004) Science 305, 1471-1474. [DOI] [PubMed] [Google Scholar]
- 36.Deng, Y., Ren, X., Yang, L., Lin, Y. & Wu, X. (2003) Cell 115, 61-70. [DOI] [PubMed] [Google Scholar]
- 37.Vogelstein, B. & Kinzler, K. W. (2004) Nat. Med. 10, 789-799. [DOI] [PubMed] [Google Scholar]