Abstract
Microbial communities in the endosphere of Salicaceae plants, poplar (Populus trichocarpa) and willow (Salix sitchensis), have been demonstrated to be important for plant growth promotion, protection from biotic and abiotic stresses, and degradation of toxic compounds. Our study aimed to investigate bio-control activities of Salicaceae endophytes against various soil borne plant pathogens including Rhizoctonia solani AG-8, Fusarium culmorum, Gaeumannomyces graminis var. tritici, and Pythium ultimum. Additionally, different plant growth promoting traits such as biological nitrogen fixation (BNF), indole-3-acetic acid (IAA) biosynthesis, phosphate solubilization, and siderophore production were assessed in all bio-control positive strains. Burkholderia, Rahnella, Pseudomonas, and Curtobacterium were major endophyte genera that showed bio-control activities in the in-vitro assays. The bio-control activities of Burkholderia strains were stronger across all tested plant pathogens as compared to other stains. Genomes of sequenced Burkholderia strains WP40 and WP42 were surveyed to identify the putative genes involved in the bio-control activities. The ocf and hcnABC gene clusters responsible for biosynthesis of the anti-fungal metabolites, occidiofungin and hydrogen cyanide, are present in the genomes of WP40 and WP42. Nearly all endophyte strains showing the bio-control activities produced IAA, solubilized tricalcium phosphate, and synthesized siderophores in the culture medium. Moreover, some strains reduced acetylene into ethylene in the acetylene reduction assay, a common assay used for BNF. Salicaceae endophytes could be useful for bio-control of various plant pathogens, and plant growth promotion possibly through the mechanisms of BNF, IAA production, and nutrient acquisition.
Keywords: bio-control, Salicaceae endophytes, soil borne plant pathogens, Burkholderia
Introduction
Biotic stress, especially due to pathogenic microorganisms, causes major crop losses worldwide which is equivalent to nearly $220 billion lost every year (Chakraborty and Newton, 2011). In response, growers often rely on a variety of chemicals to control these plant pathogens; however, such widespread use comes at both economic and environmental costs, causing undesirable consequences to human health through air, water, and soil pollution. Resistance to chemicals also is commonplace. Alternatively, the use of microbial organisms to manage plant diseases, termed bio-control, offers an environmentally-friendly and more sustainable replacement to the chemical pesticides. Several past studies demonstrated that endophytes have the potential to control many plant diseases caused by different plant pathogens (Ryan et al., 2008; Compant et al., 2010). Endophytic bacteria including Aureobacterium, Bacillus, Paenibacillus, Phyllobacterium, Pseudomonas, and Burkholderia recovered from host plants or seeds showed antagonistic activities against the plant pathogens, Fusarium oxysporum, Rhizoctonia solani, Sclerotium rolfsii, Verticillium dahlia and many other fungi (Chen et al., 1995; Rybakova et al., 2016).
Endophytes are bacterial and fungal communities that colonize the plant interior and contribute to the growth, development, fitness, and adaptation of the host plant (Rodriguez et al., 2008; Hardoim et al., 2015). Endophytes often confer considerable benefits to the host plants they inhabit. The diazotrophic (nitrogen fixing) endophytes can convert dinitrogen gas into nitrogen (N) compounds such as ammonium and nitrate, which are potentially available for N metabolism by the plant (Bhattacharjee et al., 2008; Santi et al., 2013). Furthermore, endophytes can also produce phytohormones and siderophores, and can solubilize inorganic phosphates (Khan et al., 2015; Santoyo et al., 2016). Siderophores are organic compounds that are produced by organisms during iron limiting conditions. Previous studies showed that plants can utilize microbial siderophores for iron acquisition. Iron deficient tomato plants supplemented with microbial siderophores, for example, produced higher crop yields, and had increased chlorophyll and iron content in the leaves (Radzki et al., 2013). In addition, siderophores are considered to be helpful in the biological control of plant pathogens (Verma et al., 2011; Ahmed and Holmström, 2014). Many plant associated rhizo- or endophytic bacteria can solubilize insoluble inorganic phosphates, which is potentially available for uptake by plants. Positive growth response has been reported in different crop plants inoculated with phosphate solubilizing endophytes (Manoel et al., 2015; Oteino et al., 2015; Passari et al., 2015). Endophytes also have the potential to synthesize phytohormones including IAA, gibberellic acid, cytokinin, and abscisic acid (Patten and Glick, 2002; Pirttilä et al., 2004; Feng et al., 2006; Sgroy et al., 2009; Shi et al., 2009; Videira et al., 2012).
Dozens of microbial endophyte strains were isolated from poplar and willow plants that may support the host plant growth in the nutrient limited, cobble-dominated riparian ecosystem in western Washington from which they were isolated (Doty et al., 2005, 2009). Several poplar and willow endophytes are diazotrophs with the ability of producing phytohormones and siderophores, and solubilizing inorganic phosphates (Khan et al., 2015; Doty et al., 2016). A recent study showed that inoculated Salicaceae endophytes in hybrid poplar plants can contribute about 65% of the total N in the leaves and increase plant biomass through BNF (Knoth et al., 2014). Additionally, a significant amount of IAA production has been observed by poplar endophytes in vitro (Xin et al., 2009a,b). Cross inoculation of poplar and willow endophytes in other plant species (rice, maize, tomato, pepper, grasses, and conifer seedlings) showed substantial growth enhancement in nutrient poor conditions (Khan et al., 2012, 2015; Kandel et al., 2015). Furthermore, inoculated maize plants with Salicaceae endophytes showed improvement in photosynthetic capacity (higher CO2 assimilation rate) of leaves and higher biomass, and also resulted in early flowering in tomato and pepper (Khan et al., 2012; Knoth et al., 2013).
Burkholderia, Pseudomonas, Curtobacterium, and Sphingomonas were the most common endophyte genera discovered in poplar and willow plants through culture dependent and independent methods (Doty et al., 2009, 2016). Previous studies have shown that plant associated endophytic or rhizospheric Burkholderia species can degrade toxic compounds, promote plant growth, fix atmospheric N, and inhibit the growth of plant pathogenic fungi or oomycetes (Perin et al., 2006; Suárez-Moreno et al., 2012; Mitter et al., 2013; Bernabeu et al., 2015). Furthermore, bio-control strains of bacteria release different metabolic compounds including antibiotics, and lytic enzymes effective in growth inhibition of phytopathogenic fungi or oomycetes (Compant et al., 2005; Gagne-Bourgue et al., 2013; Pageni et al., 2014). More recent studies suggested the potential application of Burkholderia in agriculture for plant growth promotion, and biological disease control (Govindarajan et al., 2008; Mattos et al., 2008; Paungfoo-Lonhienne et al., 2014; Bernabeu et al., 2015).
The objectives of this study were to explore the capabilities of Salicaceae endophytes to control the in vitro growth of several soil borne plant pathogens including R. solani AG-8, Fusarium culmorum, Gaeumannomyces graminis var. tritici, and Pythium ultimum. These are widespread pathogens of many economically important crops including small grain crops such as wheat and barley, grain legumes, and brassicas worldwide (Paulitz, 2006; Hane et al., 2014). Additionally, we aimed to discover the possible molecular mechanisms used by endophytes to arrest the fungal growth through genomic comparisons with known bacterial strains that produce anti-fungal metabolites. In order to more fully characterize the isolates, we tested the anti-fungal strains for other potential plant growth promoting abilities.
Materials and methods
Endophyte strains
Endophyte strains (except PD1) were isolated from poplar and willow plants that were collected at the Three Forks Natural Area in King County, WA in the riparian zone of the Snoqualmie River (47° 31′ 14.30″ N, 121° 46′ 28.32″ W). The plant samples were surface-sterilized with 10% bleach (10 min) and 1% Iodophor (5 min), and rinsed three times in sterile deionized water. Isolated colonies of individual endophyte strains from surface sterilized plant tissues were flash frozen in 33% (v/v) glycerol, and retained at −70°C for utilization in various studies. Many of the endophyte strains available in our collection, a total of 55 poplar and 4 willow strains (Table 1 and Table S1), were examined for their in vitro antagonistic activities initially against R. solani AG-8. The endophyte strains capable of in vitro growth inhibition of R. solani AG-8 were further tested against other plant pathogens including F. culmorum, G. graminis var. tritici, and P. ultimum. Thirteen out of the total number of strains had shown antagonistic activities against at least one plant pathogen. Two endophyte strains (PD1 and WW7) were characterized in previous studies (Doty et al., 2009; Khan et al., 2014), and the remaining 11 strains were characterized in this study. In addition, endophyte strains having antagonistic activities were further studied for different plant growth promoting traits including phosphate solubilization, siderophore production, BNF, and IAA production.
Table 1.
Endophyte strains used in this experiment.
| Strain | Identity | Origin | Appearance | PGPAa | GenBank accession no. | rRNA sequence size, partial (bp) |
|---|---|---|---|---|---|---|
| WP 4-2-2 | Burkholderia sp. | Poplar root | Buff | IAA, NA | KU495920 | 903 |
| WP 4-3-1 | Rhodotorula graminis | Poplar stem | Pink | CLP, IAA | KU500895 | 554 |
| WP 4-3-2 | Burkholderia sp. | Poplar stem | Buff, mucoid | Sd, NA | KU500893 | 947 |
| WP 4-3-3 | Curtobacterium sp. | Poplar stem | Bright yellow | IAA | KU550576 | 962 |
| WP 4-4-2 | Rahnella sp. | Poplar stem | Buff, mucoid | IAA, Sd | KU500894 | 966 |
| WP 4-4-6 | Pseudomonas sp. | Poplar stem | Pale yellow | IAA, NA | KU500891 | 1,032 |
| WP 4-5-3 | Rahnella sp. | Poplar stem | Buff, mucoid | Sd, IAA | KU500892 | 1,076 |
| WP 4-10-4 | Curtobacterium sp. | Poplar leaf | Bright yellow | NA, IAA | KU550577 | 859 |
| WP40 | Burkholderia sp. | Poplar stem | Buff | ofn, orb, hcn, Sd, NA, IAA | KF597274 | 1,494 |
| WP41 | Burkholderia sp. | Poplar stem | Buff | ofn, orb, Sd, NA, IAA | KF597275 | 1,494 |
| WP42 | Burkholderia sp. | Poplar stem | Buff | ofn, orb, Sd, hcn, NA, IAA | KF597276 | 1,494 |
| WW7 | Curtobacterium sp. | Willow stem | Pale yellow | IAA, Sd, NA | KU523564 | 926 |
| PD1 | Pseudomonas putida | Poplar stem | Buff | IAA | KF443801 | 1,496 |
| Pf 2-79 | P. fluorescens | Soil | Bright yellow | CLP, phz | ||
| Pf Q8r1-96 | P. brassicacearum | Soil | Buff | CLP, dapg, plt, prn |
Plant growth promoting activities: cyclic lipopetide (CLP) determined using the drop collapse assay; indole acetic acid (IAA) determined on YEM medium plus L-trpytophan using Salkowski reagent; gene clusters for the antifungal metabolites occidiofungin (ofn), ornibactin (orb), hydrogen cyanide (hcn), diacetylphloroglucinol (dapg), phenazine (phz), pyoluteorin (plt) and pyrrolnitrin (prn) loci determined using PCR; siderophore (Sd) determined by orange halo on CAS medium, nitrogenase activity (NA) determined by acetylene reduction assay.
Fungal isolates, bacterial controls, and media
Rhizoctonia solani AG-8 isolate C1 (Weller et al., 1986) and F. culmorum isolate 70110023 from oat grain were cultured on potato dextrose (PD) agar. Gaeumannomyces graminis var. tritici (Ggt), isolate ARS-A1 (Kwak et al., 2009) was maintained on 1/5X PD agar. P. ultimum isolate 217, isolated from a chickpea field at Spillman Farm, Pullman, Washington in 2014, was cultured on SY agar (5.0 g sucrose, 0.5 g Difco yeast extract per liter). To optimize growth for inhibition assays, each of the four fungi were transferred to Mannitol Glutamate/Luria (MG/L; Cangelosi et al., 1991), PD, 1/5X PD and SY agar, depending upon the isolate, and grown at 25°C in darkness for 5–10 d. Bacterial controls for inhibition assays were Pseudomonas fluorescens strain 2–79 (Thomashow et al., 1990), P. protegens Pf-5 (Howell and Stipanovic, 1980), and P. brassicacearum strain Q8r1-96 (Raaijmakers et al., 1999). These were cultured in Luria broth (LB) or MG/L at 27°C, 200 rpm for 16–18 h.
Endophyte growth on fungal media
Endophytes were tested to determine whether endophyte growth was altered, especially inhibited, on the media used to culture fungi. Endophyte strains were grown in MG/L broth at 27°C, 200 rpm for 16–18 h. Cultures were diluted to an OD630 of 0.2 using MG/L, PD, 1/5X (1/5th strength) PD or SY broth. Four replicated volumes of 2.5 μL each were transferred to the different agar media. Diameters of the colonies were measured after 22, 40, 54, and 70 h of growth at 27°C.
Dual plate inhibition assays
Each endophyte strain was cultured in either PD or MG/L broth, and diluted to an OD630 of 0.2 as above. Fungal pathogens were transferred to appropriate agar medium and grown at 25°C until the leading edge of the colony was 1–3 cm from the plate perimeter. For the assays, 5-mm agar disks were cored from the leading edge. The “dual plate” assay consisted of the fungus disk in the center of the plate and two 2.5 μL volumes of endophyte culture at opposite sides of the fungal disk, placed 1 cm from the perimeter of the plate. Assays were done on both PD and MG/L agar. Positive controls were Pseudomonas strains 2-79 and Q8r1-96 known to have bio-control activity; and negative bacterial controls were medium without bacteria.
Four plating regimens were developed to accommodate the differential between growth rates of the endophytes and each fungus: (1) Endophytes were plated about 80 h before P. ultimum, and inhibition was assessed 30–34 h later; (2) R. solani was plated 24 h before the endophytes, and inhibition was assessed 4.5 d later; (3) Ggt was plated about 46 h before the endophytes, and assessed for inhibition after 9 d; (4) F. culmorum was plated 3 d before the endophytes, with inhibition assessed after 9 d. Dual plate assays were incubated at 25°C in darkness. Experiments were done two to three times, with four replicates per strain per experiment.
Growth and inhibition measurements were taken when the fungus reached the outer edge of the control plate. Inhibition index was calculated as [y/(x + y)](100), where y is the distance between the center of the bacterial colony and edge of the fungus at the bacterial-fungal interface, and x is distance between the center of the agar plug and the leading edge of the fungus (McSpadden Gardener and Weller, 2001; Mavrodi et al., 2012). Only strains that were positive for antifungal activities were used to test further plant growth promoting properties, and investigated for potential molecular mechanisms through genome analyses in the cases where the genome had been sequenced.
Drop collapse assay
Cyclic lipopeptide (CLP) surfactant production by the endophytes was assessed by transferring 5 μL of a 16–18-h non-diluted broth culture onto a hydrophobic surface, such as Parafilm (de Bruijn and Raaijmakers, 2009). Culture supernatants, obtained by centrifugation at 10,000 × g for 3 min, and 1:10 dilutions of supernatants were also tested. Pseudomonas fluorescens strains 2-79 and Q8r1-96 served as the positive and negative control, respectively. Three independent experiments of 3-5 replicates per strain were done.
Inhibition of R. solani AG-8 by volatile compounds
Petri plates with center partitions (I plates, Carolina Biological Supply Company, Burlington, NC) were used to physically separate the source and target microbes. Assays for strains 4-4-2, 4-4-6, 4-5-3, 4-10-4, WP40, WP41, Pseudomonas brassicacearum Q8r1-96 and P. protegens Pf-5 were done on PDA; those for 4-3-2 and P. fluorescens 2-79 were done on MG/L agar. Strains WP41 and 2-79 were negative controls for the PDA and MG/L sets, respectively. No-endophyte controls (media only) for both PDA and MG/L also were included. An agar plug of R. solani AG-8 was transferred to the perimeter of one half of the I plate and incubated at 25°C for 24 h. Endophytes were cultured in MG/L broth as described previously, and cultures were diluted to OD630 of 0.2. A 2.5-μL volume of diluted culture was applied to the center of the second half of the I plate 24 h after the pathogen. Plates were wrapped with Parafilm and incubated for an additional 3–4 d. When the leading edges of the R. solani colony in the negative controls were close to, but not touching, the center partition, pathogen colony radii were measured in all plates. Each endophyte was tested in triplicate for each of two experiments.
Phosphate solubilization assay
Phosphate solubilizing properties of poplar and willow endophytes were determined using National Botanical Research Institute's Phosphate (NBRIP) agar medium (Nautiyal, 1999). A few colonies of individual endophyte strains were introduced per NBRIP plate in quadruplicate positions using sterile inoculating sticks (Puritan Medical Products Company, Maine). The clear halo area around an endophyte colony was observed after incubating 1 week at 30°C. The phosphate solubilization index (PSI) was calculated in endophyte strains capable of producing a distinct clear halo as a zone of solubilization (Khan et al., 2015). The poplar endophyte WP5 (Doty et al., 2009) was used as a reference strain for comparisons.
Siderophore production assay
M9 minimal medium supplemented with Chrome Azurol S (CAS) was used for the siderophore production assay. Minimal medium (Yun et al., 2000) was prepared, autoclaved, and mixed with filter-sterilized, pre-warmed MgSO4 (2 mM final), CaCl2 (0.1 mM final), sucrose (0.2% final), and Casamino acids (0.9% final) (Loewen, 1984). CAS solution was prepared according to the protocol developed by Schwyn and Neilands (1987), autoclaved, and combined with the minimal medium prior to the assay. The area of color conversion from blue to orange around an endophyte colony was measured after incubating 1 week at 30°C for each endophyte strains per plate.
IAA quantification assay
For IAA quantification, endophytes were grown in triplicate in 25 ml TYC broth (g L−1: 5 tryptone, 3 yeast extract, and 0.872 g CaCl2.2H2O) with 0.1% (w/v) L- tryptophan for 4 days in 125-ml flasks, and pelleted through centrifugation. One ml of the supernatant was incubated with 2 ml of Salkowski reagent (2 ml of 0.5 M FeCl3, and 98 ml of 35% HClO4; Gordon and Weber, 1951) for half an hour, and the optical density at a wavelength of 530 nm was observed (Xin et al., 2009a). A standard curve was developed with known amounts of IAA using Salkowski reagent and TYC broth with tryptophan and without endophytes, and the IAA amount produced by each endophyte strain was calculated using the standard curve.
Acetylene reduction assay (ARA)
Endophyte strains were grown overnight at 30°C in MG/L or N-limited combined carbon medium (NLCCM) on a rotatory shaker. The overnight grown strains in MG/L cultures were microfuged at 4°C at 8,000 rpm for 10 min, combined with NLCCM cultures already in progress, and incubated on a shaker for another night. Cell density was determined through spectrophotometry, and the OD600 was adjusted to 1.0. Sixteen milliliter culture of each strain was transferred into balch tubes, dosed with 500 μl acetylene, and incubated for 24 h. Three tubes per strain were prepared; two were for the dosed cultures and one was for undosed control. After 24 h, 5 ml air from autosampler vials was removed and replaced with 5 ml headspace from the balch tubes. Samples were analyzed in the DeLuca Biogeochemistry Lab at the University of Washington, Seattle.
Analysis of the nitrogenase subunit gene, nifH
The nitrogenase subunit gene, nifH, was amplified through PCR using universal nifH, nifH-b1 (Bürgmann et al., 2004), and nifH (Poly et al., 2001) primers. Colony PCR was performed in the same way as described below.
Identification of endophytes through 16S/26S rRNA sequencing
Overnight grown bacterial colonies were used for colony PCR by transferring a single colony into 20 μl of sterile water, vortexing briefly, and using 1 μl as DNA templates for PCR analysis. The universal primers for 16S rRNA gene, 8F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) were used to amplify the 16S rRNA, which produced 1.5 kb amplicon products. PCR reactions of 25 μl consisted of 1 μl template DNA, 12.5 μl of PCR premix Buffer E (EpiCentre, Madison, WI), 0.8 μl of each primer (at 0.2 μg μl−1), 0.3 μl of Taq DNA polymerase (New England BioLabs, Inc., Ipswich, MA), and 9.6 μl of sterile water. Amplified PCR products were incubated with ExoSAP PCR cleanup reagent (Affymetrix, Inc., Cleveland, OH) at 37°C for 30 min followed by 80°C for 15 min in P100 Thermal Cycler (Bio-Rad, Inc., Hercules, CA). The ExoSAP cleaned 16S rRNA PCR products were sequenced by the Sanger sequencing approach (GENEWIZ, South Plainfield, NJ). For the yeast strain WP 4-3-1, the D1/D2 region of the large subunit (26S) of rRNA was amplified using primers F63 (5′-GCATATCAATAAGCGGAGGAAAAG-3′) and LR3 (5′-CGTCCGTGTTTCAAGACGG-3′), and PCR products were sequenced as mentioned above.
The 16S/26S rRNA sequences of all poplar endophytes were deposited in GenBank (NCBI) database under accession numbers KF597274, KF597275, KF597276, KU495920, KU500894, KU500893, KU550576, KU500895, KU500892, KU500891, and KU550577 for WP40, WP41, WP42, WP 4-2-2, WP 4-3-1, WP 4-3-2, WP 4-3-3, WP 4-4-2, WP 4-5-3, WP 4-4-6, and WP 4-10-4 respectively.
Comparative analysis of genomic regions related to the synthesis of anti-Rhizoctonia metabolites in Burkholderia species
The sequence of the entire genomes of Burkholderia spp. (WP40 and WP42) were compared against phylogenetically related strains where the synthesis of anti-fungal metabolites was functionally assessed through knock-out studies (Gu et al., 2009a). Other gene clusters encoding for putative non-ribosomal peptidase, polyketide synthases, and other enzymes involved in the synthesis of secondary metabolites characterized for a generic anti-microbial and anti-fungal effect were detected using antiSMASH, a bioinformatics tool for automatic genomic identification and analysis of biosynthetic gene clusters (Weber et al., 2015). We set a minimum ClusterFinder probability of 0.5 and searched for gene clusters with a minimum size of 4 open reading frames (orf) characterized by 5 or more biosynthesis PFAM-related domains. The presence of biosynthetic gene clusters for anti-fungal metabolites were assessed through a multi-genome alignment approach using Mauve (Darling et al., 2004). Gene clusters encoding for anti-fungal compounds used in this work as reference, are listed: occidiofungin (ocf) gene cluster from Burkholderia contaminans MS14 (EU938698.5); afc (AFC-BC11) gene cluster from B. cepacia BC11 (AF076477.1); pyrrolnitrin gene cluster prnABCD from B. pyrrocinia CH-67 (AF161186.1); the polyketide synthases (PKSs) genomic island (locus tag: Bamb_5918-5933), and the 4-hydroxy-2-alkylquinolines (HAQs) biosynthetic gene luster hmqABCDEFG (locus tag: Bamb_5763-5769) from B. cepacia AMMD (GCA_000203915.1), hcnABC gene cluster form P. fluorescens PF5 (AF053760); the phenazine-1-carboxylic acid gene cluster phzABCDEFG from P. fluorescens 2-79 (L48616); the 2,4-diacetylphloroglucinol synthetic gene cluster phlABCDEF from P. fluorescens Q2-87 (U41818.1) and pyrrolnitrin gene cluster prnABCD from P. fluorescens (U74493).
Assessing the plant growth promoting properties in Burkholderia spp. (WP40 and WP42) genomes
The draft genomes Burkholderia sp. WP40 and Burkholderia sp. WP42 had been sequenced and annotated at the Joint Genome Institute as a part of the sequencing project “Defining the functional diversity of the Populus root microbiome (Bioproject accession: PRJNA247585 and PRJNA247584).” Genomes are publically available at: https://img.jgi.doe.gov/. The Integrated Microbial Genome platform (Markowitz et al., 2012) was used for data-mining the genome of Burkholderia species WP40 and WP42 for genes with potential beneficial effects on plant fitness.
Statistical analysis
Bartlett's test for homogeneity of the variances (Statistix vers. 8.1, Analytical Software, Tallahassee, FL) was applied to inhibition index values from replicated experiments. Mean values for inhibition index, percent inhibition (split plate assays) and endophyte colony diameter were obtained using ANOVA (Statistix vers. 8.1). Significant differences among means were determined using Fisher's protected least significant difference (LSD) test at P < 0.05.
One way analysis of variance, and Fisher pairwise comparisons were used for ARA and siderophore assays using Minitab 17 (Minitab Inc., State College, PA, USA). The single point measurement was presented in the IAA assay.
Results
Identification of endophytes
Poplar endophytes were identified through 16S/26S rRNA gene sequencing. The 16S/26S rRNA gene for each strain was sequenced and identified using BLAST on the NCBI database from the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/BLAST). As shown in Table 1, the best 16S rRNA matches of poplar endophytes were Burkholderia, Rahnella, Curtobacterium, and Pseudomonas.
Endophyte growth on MG/L and PDA
To determine the optimal agar medium for inhibition assays, the endophytes were grown on MG/L, PDA, 1/5X PDA, and SY agar that were used to culture the fungi. The endophytes grew best on either MG/L or PDA, but not on 1/5X PDA or SY, as indicated by colony size after 70 h of growth (Table 2). Colonies of strain PD1 on MG/L reached the maximum diameter, that is, before undergoing confluent growth, by 54 h. Pseudomonas WP 4-4-6 and all of the Burkholderia spp. grew more rapidly on PDA, whereas Rhodotorula graminis, Rahnella aquatilis, Curtobacterium spp. and P. putida PD1 appeared to prefer MG/L. Strain PD1 also produced a diffusible pale yellow pigment on MG/L that was not observed on PDA (data not shown).
Table 2.
Growth of endophytes on four agar media.
| Strain | Colony diameter (mm)1 | |||
|---|---|---|---|---|
| MG/L | PDA | 1/5X PDA | SY | |
| WP 4-2-2 | 5.5 g | 6.6 cd | 5.0 b | 4.5 g |
| WP 4-3-1 | 7.4 d | 4.9 f | 4.1 c | 5.4 f |
| WP 4-3-2 | 5.9 efg | 6.1 de | 5.2 b | 4.8 fg |
| WP 4-3-3 | 6.9 de | 4.6 fg | 5.0 b | 5.2 fg |
| WP 4-4-2 | 11.2 a | 7.2 bc | 6.0 a | 8.4 b |
| WP 4-4-6 | 5.8 fg | 7.1 bc | 5.9 a | 5.0 fg |
| WP 4-5-3 | 8.6 c | 7.8 b | 4.9 b | 9.2 a |
| WP 4-10-4 | 7.2 d | 4.9 g | 5.9 a | 6.4 e |
| WP40 | 7.5 d | 9.2 a | 6.0 a | 7.5 cd |
| WP41 | 6.8 def | 9.1 a | 5.1 b | 7.8 bc |
| WP42 | 5.2 g | 7.9 b | 5.1 b | 6.9 de |
| WW7 | 9.5 bc | 5.8 e | 5.0 b | 6.4 e |
| PD12 | 10.4 ab2 | 7.8 b | n.g. | 5.0 fg |
Measured after 70 h at 27°C; starting diameter = 4.25–4.75 mm. n.g., no growth. Letters indicate significance (P < 0.05) classes of means within each column, determined by Fisher's LSD.
PD1 on MG/L was measured at 54 h.
Inhibition index on dual plate assays
Dual plate inhibition assays were done using both MG/L and PDA, and significant differences in mean diameter of growth were determined using Fisher's least significant differences (Table 3). Of the 59 strains tested, 13 were positive for anti-fungal activity (Figure 1). The Burkholderia spp. WP40, WP41, WP42 consistently reduced fungal growth of all four pathogens on both media, and were as effective as the Pseudomonas controls 2-79 and Q8r1-96. Burkholderia 4-2-2 and R. aquatilis 4-4-2 displayed inhibition against three of the four pathogens only when grown on PDA. None of the endophytes were active against F. culmorum when grown on MG/L; however eight gave quantifiable inhibition on PDA. In contrast, PD1 showed inhibition against Ggt, P. ultimum and R. solani AG-8 when grown on MG/L but not on PDA. In general, F. culmorum and Ggt were responsive to more of the endophytes than were P. ultimum and R. solani AG-8. While endophyte strains 4-3-1, 4-3-3 and 4-10-4 had no activity against any of the pathogens on full-strength media, in our initial screening on ¼X PDA, all the endophyte strains listed in Table 1 showed some antagonistic activities against R. solani AG-8. Our findings suggest that nutrient composition has a role in the inhibitory activities of the endophytes. In some dual plate inhibition assays, we observed that R. solani AG-8 and P. ultimum did not grow to the perimeter of the agar plate if certain endophytes were present. The leading edge of the fungal hyphae in these cases was shorter than that of control plates (data not shown). The observations suggested the presence of inhibitory volatile compounds (VOCs).
Table 3.
In vitro inhibition of four soilborne fungal pathogens by bacterial strains on potato dextrose agar (PDA) and mannitol/glutamate agar (MG/L).
| Strain | Inhibition index1 | ||||||
|---|---|---|---|---|---|---|---|
| F. culmorum | G. graminis tritici | R. solani AG8 | P. ultimum | ||||
| PDA | PDA | MG/L | PDA | MG/L2 | PDA | MG/L | |
| WP 4-2-2 | 10.0 c | 29.0 a | na | na | na | 8.0 b | na |
| WP 4-3-1 | na | na | na | na3 | na | na | na |
| WP 4-3-2 | 9.4 c | 18.9 b | na | na | na | na | na |
| WP 4-3-3 | na | na | na | na3 | na | na | na |
| WP 4-4-2 | 5.1 d | 18.2 bcd | na | 13.1 b | na | na | na |
| WP 4-4-6 | 8.6 cd | 9.7 c | na | na | na | na | 0.0 e |
| WP 4-5-3 | 9.9 c | 15.0 bcd | na | na | na | na | na |
| WP 4-10-4 | na | na | na | na3 | na | na | na |
| WP40 | 18.3 a | 23.4 ab | 23.6 a | 16.1 b | 8.0 c | 20.3 a | 24.6 ab |
| WP41 | 17.1 ab | 23.5 ab | 24.8 a | 16.1 b | 8.8 bc | 20.8 a | 25.7 a |
| WP42 | 14.0 b | 21.0 ab | 17.1 b | 14.1 b | 3.8 d | 20.1 a | 22.3 b |
| WW7 | na | 9.6 c | na | na | 11.0 b | na | 13.9 cd |
| PD1 | na | na | 11.3 c | na | 18.3 a | na | 16.7 c |
| 2-79 | 14.5 ab | 23.0 ab | 14.3 bc | 14.3 b | 11.4 b | 8.3 b | 13.8 cd |
| Q8r1 | 10.0 c | 18.5 bc | 16.7 b | 23.1 a | 20.0 a | 7.5 b | 12.8 de |
Inhibition index = [y/(x + y)](100), for which x is the distance from the center of the fungal plug to the leading edge of the non-inhibited fungus, and y is the distance from the center of the bacterial colony to the inhibited edge of the fungus. Values are the averages of two experiments (n = 8). na, no activity. Letters indicate significant (P < 0.05) differences among means within each column, based on Fisher's LSD (Statistix, Tallahassee, FL).
Variances between three experiments were unequal based on Bartlett's test; a representative experiment is shown (n = 4).
No activity on full-strength PDA but partial activity on ¼X PDA.
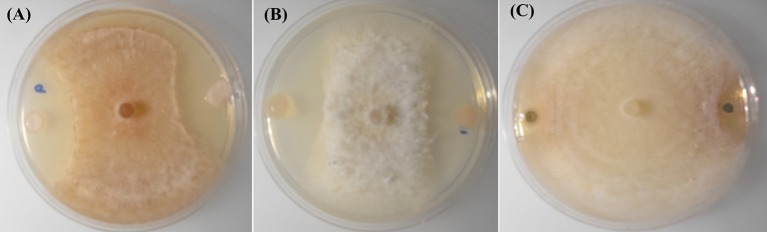
Figure 1.
In-vitro inhibition of R. solani AG-8 (Plate A), and Ggt (Plate B) by poplar endophytes; WP40 and WP42 respectively; with reference to inhibition of Ggt by the positive control strain, Pf 2-79 (Plate C).
Rapid assays to detect cyclic lipopeptides and volatile inhibitory compounds
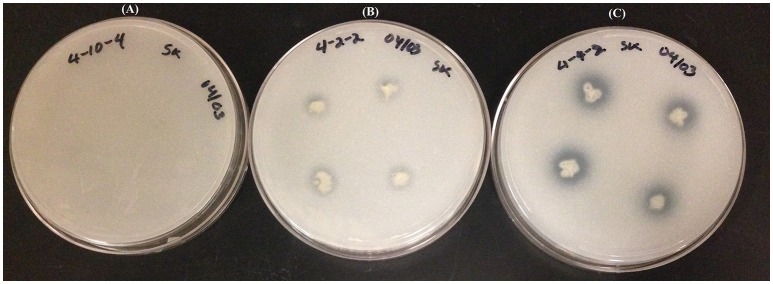
Cyclic lipopeptide surfactant production was detected in R. aquatilis 4-3-1 and P. fluorescens 2-79 (positive control) in drop collapse assays. Droplets of whole-cell cultures and non-diluted culture supernatants, but not 1:10 supernatant dilutions, exhibited pronounced spreading and flattening on Parafilm, evidence of surfactant activity.
Volatile inhibitor activity was tested for endophyte strains in which the leading edge of the R. solani AG-8 colony usually did not reach the Petri plate perimeter relative to the no-endophyte control in inhibition index studies. Small but significant (P < 0.05) decreases in pathogen colony radii were observed for endophyte strains 4-2-2, 4-4-6, and WP40 but not for 4-5-3 or WP41 (Table 4). Reference strains, P. protegens Pf-5 and Pseudomonas fluorescens 2-79 that harbored loci for hydrogen cyanide (HCN) production, and endophyte 4-10-4 inhibited R. solani AG-8 in only one of the two experiments, indicating the transient and variable nature of the volatile activity. As expected, strain WP41 was negative. It was not known if strain Q8r1-96 would produce inhibitory amounts of HCN in our assay; it also was negative. There was no correlation between inhibition in the I plate and dual plate assays. For instance, strains 4-4-6 and 4-10-4 were active in the I plate assays but not in dual plate assays, and vice versa for strains 4-3-2 and WP41. Our findings suggest that multiple mechanisms of inhibition are involved in suppressing R. solani AG-8 in vitro.
Table 4.
Evidence for production of volatile inhibitory compounds in bacterial endophytes.
| Strain1 | Medium2 | Colony radius (cm)3 | |
|---|---|---|---|
| Expt 1 | Expt 2 | ||
| PDA control | PDA | 34.4 a | 20.2 a |
| WP 4-2-2 | PDA | 33.7 bcd | Not tested |
| WP 4-4-6 | PDA | 33.2 de | 19.3 bc |
| WP 4-5-3 | PDA | 34.0 abc | 20.0 a |
| WP 4-10-4 | PDA | 34.2 abc | 19.0 c |
| WP40 | PDA | 33.0 e | 19.3 bc |
| WP41 | PDA | 34.2 ab | 19.8 ab |
| Pf Q8r1-96 | PDA | 34.0 abc | 19.7 ab |
| Pf -5 | PDA | 33.5 cde | 20.0 a |
| MG/L control | MG/L | 34.7 a | 31.3 a |
| WP 4-3-2 | MG/L | 34.2 a | 30.8 a |
| Pf 2-79 | MG/L | 34.3 a | 29.8 b |
Controls were done with medium only; WP41 was included as a negative endophyte control.
Preferred medium for rapid growth.
Radius of the R. solani AG8 colony 5 d and 4 d after growth on I-plates for experiments 1 and 2, respectively. n = 3. Letters indicate significant (P < 0.05) differences among means within each column, based on Fisher's LSD (Statistix, Tallahassee, FL).
Comparative analysis of genomic regions related to the synthesis of anti-Rhizoctonia metabolites in Burkholderia species (WP40 and WP42)
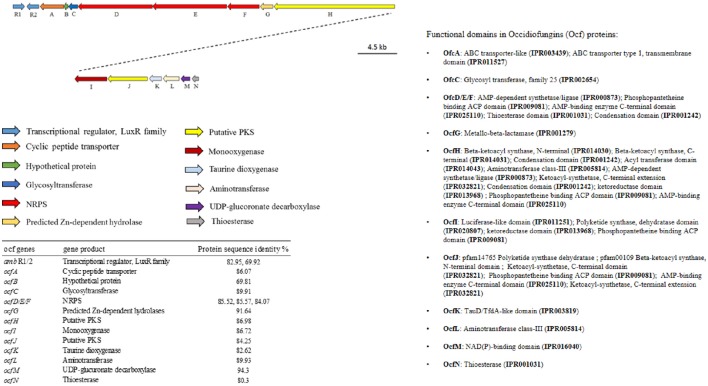
Among the endophytic strains used in this study, only the genomes of WP40 and WP42 had been sequenced previously. These were analyzed to assess the presence of known gene clusters involved in the synthesis of secondary metabolites with antifungal activities. A syntenic block for the 56-kb ocf gene cluster of B. contaminans MS14, a known antifungal strain was detected in the genomes of WP40 and WP42 (Figure 2). As reported in Gu et al. (2009a), the ocf gene cluster consists of 15 open reading frames (orf) involved in the biosynthesis and secretion of occidiofungin, a cyclic glycopeptide, which has been reported to inhibit the growth of R. solani, F. culmorum, P. ultimum and other plant and human pathogens (Lu et al., 2009). The ocf region was annotated in antiSMASH as hybrid NRPS-Type-1 PKS gene cluster that contains three orf each of which encodes for a non-ribosomal peptide synthetase (NRPS), and a coding sequence for a Type 1 PKS. However, gene clusters such as pyrrolnitrin (prnABCD), HQSs (hmqABCDEFG), AFC-BC11 and PKSs genomic island, involved in the synthesis of known antifungal compounds, are absent in WP40 and WP42. Moreover, a putative hcnABC gene cluster involved in the synthesis of HCN was also detected in the WP40 and WP42 genomes (Figure 3). The synthesis of HCN seems to be a common feature of plant growth promoting Burkholderia strains. Furthermore, genome annotation of PD1 revealed the presence of chitinase and secreted protein annotated as chitin deacetylase, which may play a role to inhibit the fungal growth.
Figure 2.
Annotation and graphical representation of the ocf gene cluster in WP42 [Locus tags: EX20DRAFT_04106 (ambR1)–04091 (ocfN)] and WP40 [Locus tags: Ga0008008_11691 (ambR1)–116106 (ocfN)]. The same colors were used for open reading frames (arrow) encoding for proteins with the same function (arrows). The right pane shows the protein domain.
Figure 3.
Protein alignment of putative hcn biosynthetic genes in WP40 (Locus tag: Ga0008008_101290–101292), and WP42 (Locus Tag: EX20DRAFT_00138–00140) with HcnA, B, and C from P. fluorescens Pf-5. The conserved amino acid residues of Fe-S binding sites, in HcnA, and ADP-binding motif, in HcnB and HcnC, important for hydrogen cyanide synthases function (Laville et al., 1998; Ryall et al., 2008), are indicated with black triangles.
IAA production
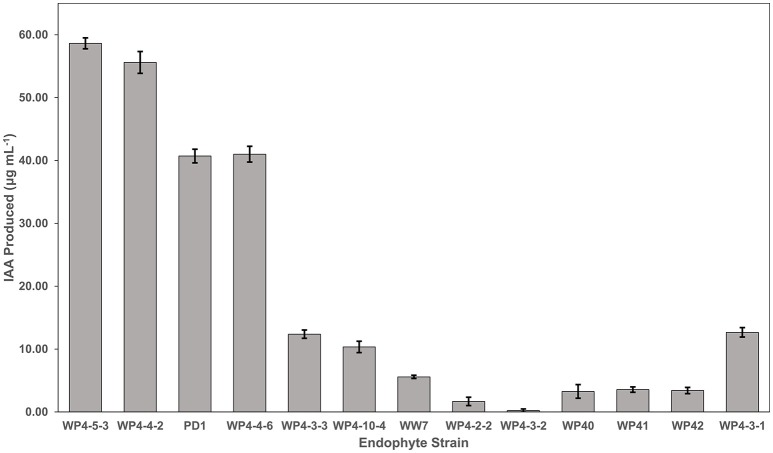
The amount of IAA produced by the different endophytes when supplemented with L-tryptophan varied between the different strains, generally along species lines (Figure 4). The two strains producing the highest auxin levels were both Rahnella aquatilis strains (WP4-5-3 and WP4-4-2). The next two highest producers were both Pseudomonas species (WP4-4-6 and PD1). The two Curtobacterium strains (WP4-4-2 and WP4-10-4) and the yeast strain (WP4-3-1) produced moderate levels of the phytohormone. All five of the Burkholderia strains produced the least amount of the auxin. A negligible amount (<1.30 μg ml-1 of endophyte culture) of IAA was observed for all the endophyte strains without addition of L-tryptophan (data not shown).
Figure 4.
IAA production by the Salicaceae endophytes. Strains were grown in triplicate for 4 days in TYC broth containing 0.1% L-tryptophan. The IAA amount produced by each endophyte strain was calculated using the standard curve.
Phosphate solubilization
Tricalcium phosphate/NBRIP plates were used for the phosphate solubilization assay. This assay was done once but strains were tested in quadruplicate positions on individual petri plates. The halo area surrounded by endophyte colonies was a zone of phosphate solubilization, which was assessed by calculating PSI as described earlier (Khan et al., 2015). Several strains solubilized tricalcium phosphate more effectively than others (Figure 5). The best performing strains were WP40, WP41, WP42, WP 4-4-2, WP 4-5-3, and scored more than 1.5 PSI. Some strains; WP 4-2-2, WP 4-3-2, WP 4-4-6, PD1, and WW7 showed a relatively inconspicuous halo area. No solubilization was observed by strains WP 4-3-1, WP 4-3-3, and WP 4-10-4.
Figure 5.
Tricalcium phosphate (NBRIP) plates showing the phosphate solubilization gradient: Plate (A); no solubilization, Plate (B); moderate solubilization, and Plate (C); high solubilization.
Siderophore production
CAS agar plates were used to observe the siderophore activity of Salicaceae endophytes. The orange halo area surrounded by endophyte colonies was measured to assess the siderophore production in-vitro. Poplar endophytes; WP40, WP41, WP42, WP 4-2-2, WP 4-3-2, WP 4-4-2 and WP 4-5-3, and willow endophyte WW7 showed siderophore activity creating the orange halo area contiguous with colony growth (Figure 6). No siderophore activity was observed by strains, WP 4-3-1, WP 4-4-6, WP 4-10-4, and PD1. The largest orange halo area; 7.539 cm2 was observed in willow endophyte, WW7.
Figure 6.
Area of orange halo (cm2) displayed by different Salicaceae endophytes in CAS agar plates. Histograms that do not share a letter are significantly different at <0.05 probability level.
Plant growth promoting features in poplar Burkholderia (WP40 and WP42) genomes
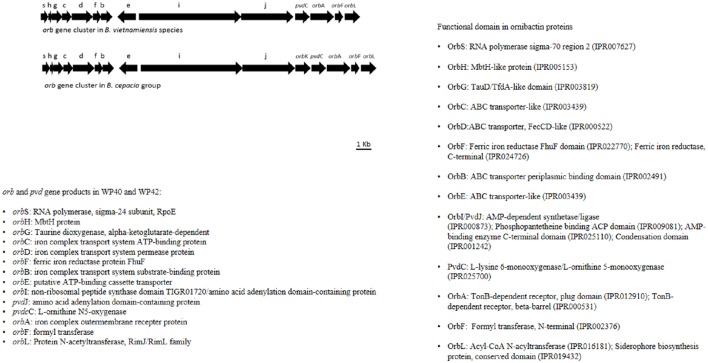
Plant associated (endophytic or rhizospheric) Burkholderia species have promising plant growth promoting properties (BNF, phosphate solubilization, siderophore production, degradation of aromatic compounds, and phytohormone production). From a genome analysis of poplar endophytic Burkholderia strains WP40 and WP42, it was revealed that they carry putative genes that are responsible for the above mentioned characteristics related to plant growth promotion. As observed for other Burkholderia strains, the nifHDK operon was detected in WP40 and WP42 along with 1-aminocyclopropane-1-carboxylate (ACC) deaminase coding sequence. In addition, these strains, WP40 and WP42, have the pyrroloquinoline quinone (pqqBCDE) operon that is essential to solubilize rock phosphates in soil. An interesting feature of WP40 and WP42 is the presence of a non-canonical ornibactin (orb) gene cluster, which encodes for biosynthesis of siderophore compounds. Compared to other orb gene clusters, this non-canonical cluster is present in all core genes but lacks N-acetyltransferase coding sequence orbK, which is not essential in the synthesis of ornibactins (Franke et al., 2014). Interestingly, this gene arrangement has never been observed and seems to be unique in the Burkholderia species (Figure 7).
Figure 7.
Annotation of orb and pvd genes in WP42 [Locus tags: EX20DRAFT_02566(orbS)–02579 (orbL)], and WP40 [Locus tags: Ga0008008_110136 (orbS)–110149 (orbL)], and B. cepacia groups.
Acetylene reduction assay
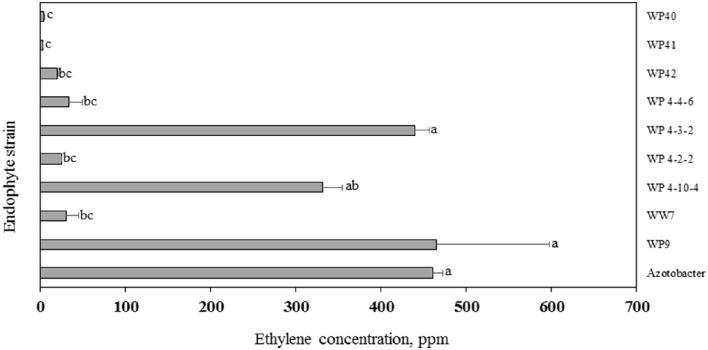
In the ARA, the activity of the nitrogenase enzyme leads to reduction of acetylene gas to ethylene, which is monitored through gas chromatography. The amount of ethylene provides an estimate of N fixed by diazotrophs. More than 300 ppm concentration of ethylene was produced by strains WP9, WP 4-3-2, WP 4-10-4, WP 4-4-6, and positive control Azotobacter sp. (Figure 8). A relatively smaller amount of ethylene was produced by endophyte strains WP40, WP41, WP42, WP 4-4-2, and WW7.
Figure 8.
Acetylene reduction assay. Ethylene produced by endophyte strains after 24 h of exposure to acetylene. Histograms that do not share a letter are significantly different at <0.05 probability level.
Analysis of the nitrogenase subunit gene, nifH
Endophyte strains WP40, WP41, WP42, WP 4-4-2, and WP 4-5-3 were found positive to universal nifH, and nifH primers. Strains WP 4-4-2 and WP 4-5-3 were positive to nifH-b1.
Discussion
In this study, we characterized the bio-control potential of Salicaceae endophytes including other microbial properties, which are associated with plant growth promotion. We used in-vitro microbiological techniques to observe the phenotypic functionality of these endophytes related to plant growth promotion and suppression of different plant pathogens, and genome analyses to understand the possible underlying mechanisms for these functions. The main objective of this study was to investigate whether any of the poplar or willow endophytes have antagonistic activities over the growth of soil borne plant pathogens including R. solani AG-8, F. culmorum, Ggt, and P. ultimum. Previous studies suggested that endophytes are vital for poplar and willow plants to colonize the flood prone areas of the Snoqualmie River, which are largely dominated by sand and rock gravels (Doty et al., 2005, 2009, 2016). With the large number of available strains from this environment, it is important to focus the research on those with the greatest potential of providing plant health benefits. No previous studies tested for antagonistic activities of these endophytes on plant pathogens. Among the 55 poplar and 4 willow endophyte strains used in the 2005 to 2016 studies, ten poplar endophytes and one willow endophyte used here showed antagonistic activities against R. solani AG-8, F. culmorum, Ggt, and P. ultimum. These strains were then further tested for additional growth promoting activities including IAA production, phosphate solubilization, siderophore production, and BNF.
Irrespective of the different types of media, all endophyte strains grew robustly. Pseudomonas species produced distinct yellow pigments in MG/L. The media containing glutamate or mannitol are known to enhance yellow pigment production in Pseudomonas (Osawa et al., 1963). The yellow pigment has the potential to chelate the iron from the environment, which is a required nutrient for bacterial growth (Meyer and Abdallah, 1978). The dual plate assays displayed variable antagonistic responses against F. culmorum, Ggt, P. ultimum, and R. solani AG-8. However, the endophyte Burkholderia spp. (WP40, WP41, and WP42) had antagonistic activities across all four tested pathogens. More importantly, the antagonistic performance of these strains was equally as impressive as that of the positive control Pseuodomonas strains. It was demonstrated that Burkholderia produced a glycolipopeptide named occidiofungin, which has an inhibitory effect on various fungal pathogens (Gu et al., 2009a,b). From a genome analysis of WP40 and WP42, a gene cluster responsible for biosynthesis of an ocf -like cyclic peptide was discovered. Among various genes embedded within that 56-kb ocf gene cluster, ambR1 was reported as a key regulatory gene controlling the biosynthesis of occidiofungin (Gu et al., 2009b), which is also conserved in the genomes of poplar Burkholderia.
Liquid chromatography-mass spectrometry and nuclear magnetic resonance spectroscopy of poplar endophytes WP40 and WP41 were conducted to analyze for the anti-fungal compounds, especially occidiofungin. However, the peaks that correspond to occidiofungin mass were not detected in the spectrum originated from WP40 and WP41 (data not shown). The anti-fungal compounds might not be synthesized to detectable levels when grown under medium away from the fungus. The strain WP 4-3-1 showed the evidence of CPL synthesis but did not show any antagonistic effect on dual plate assays. Other strains that showed antagonistic effects were negative for CLP secretion assay but showed fungal growth suppression in volatile inhibitory and dual plate assays. From the genome analysis, the hcnABC locus that encodes for VOCs like HCN was detected in the genomes of WP40 and WP42, which corroborates the results of dual plate assays and the volatile inhibitory assay. Despite an evident inhibitory effect in fungal growth, studies regarding the effectiveness of Burkholderia strains to produce discrete amounts of HCN are dissonant (Ryall et al., 2008; Gilchrist et al., 2013). Thus, it is a possibility that the strong anti-fungal activities observed in WP40 and WP42 resulted from the combination of synthesizing volatile compounds like HCN, and glycopeptides like occidofungin. The antagonistic activities of bacterial VOCs against phytopathogenic fungi were discussed elsewhere (Weisskopf, 2013). Further functional analysis of putative anti-fungal genes would provide the necessary information about the mechanisms used by endophytes to arrest the growth of various plant pathogens, which would be also useful to formulate biofungicides.
The antagonistic properties of fungal endophytes of poplar have been investigated before (Busby et al., 2016); however, to our knowledge, our study is the first to analyze these properties of bacterial endophytes recovered from poplar and willow plants. In the past, studies were concentrated on soil bacteria or rhizospheric bacteria as a means of bio-control agents (Beneduzi et al., 2012; Santoyo et al., 2012). In contrast, the endophytic lifestyle of microorganisms could offer systemic tolerance against many plant pathogens as they colonize the entire plant body and stably persist. In addition, they use internal plant tissues for nutrition and multiplication, which excludes them from competition with other microbes present in the phyllosphere or rhizosphere. In our study, the bio-control potential of Salicaceae endophytes was not restricted only to Burkholderia species but also observed in Pseudomonas, Rahnella, and Curtobacterium indicating that poplar endophyte consortia have the potential to inhibit the growth of several plant pathogens. From earlier studies, it is concluded that the poor performance of bio-control in field conditions is due to poor root colonization by bio-control agents (Compant et al., 2005). The endophyte strains used in our study were isolated from poplar and willow branches. These endophytes can colonize both roots and shoots of a variety of plant species and may provide promising results in the field conditions to manage different soil pathogens. Previous studies have shown that these endophytes were competent to colonize the endosphere of rice and conifer seedlings (Kandel et al., 2015; Khan et al., 2015).
Some concerns have been raised about the use of Burkholderia strains in agriculture since some strains are associated with the Burkholderia cepacia complex (Bcc), opportunistic pathogens in cystic fibrosis (CF) patients (Chiarini et al., 2006). However, members of this genera are wide-spread in the environment including within plants (Perin et al., 2006; Compant et al., 2008). Interestingly, pathogenicity tests performed on B. vietnameiensis strains isolated from environmental niches were negative for pathogenicity (Bernier et al., 2003; Agnoli et al., 2012). On the other hand, most strains isolated from CF patients show a strong or moderate pathogenic behavior, suggesting that specific genetic traits might be unique and positively selected in those strains isolated from CF patients (Saini et al., 1999; Chu et al., 2002; Cieri et al., 2002). This indicates that a phylogenetic classification does not fully represent the pathogenic potential of a strain (Eberl and Vandamme, 2016) and that more studies aimed to understand the molecular effectors exploited by opportunistic pathogens of the Bcc complex should determined. Angus et al. (2014) reported that plant symbiotic Burkholderia species lack virulence-associated loci required in mammalian pathogenesis. We used comparative based genomic analysis to distinguish the secretion systems of poplar Burkholderia strains from known pathogenic B. strains. Key pathogenesis-related gene clusters encoding for components of the Type III, Type IV (Sajjan et al., 2008), Type VI-5 (Schwarz et al., 2010) and Type VI-1 (Burtnick et al., 2011) secretion systems were absent from the genomes of WP40 and WP42 (data not shown) in contrast to pathogenic Burkholderia strains, suggesting that the secretion systems of poplar Burkholderia strains are most likely deficient to initiate infection, and thus might be separated from pathogenic Bcc strains.
All the endophyte strains that were tested for IAA biosynthesis were positive in the presence of L-tryptophan, with the exception of WP4-3-2. We were unable to detect any notable amount of IAA in the absence of L-tryptophan. It is possible that the colorimetric method used in this study to quantify IAA may not be sensitive enough to detect low levels of IAA biosynthesis. However, similar findings were reported for endophytes isolated from a variety of other plant species (Shi et al., 2009; Xin et al., 2009a; Videira et al., 2012). L-tryptophan is commonly available in plant exudates (Kamilova et al., 2006; Hardoim et al., 2008). The requirement for exogenous precursor may be a reflection of the mutually beneficial interactions of the symbiont and the host, with the microbe converting a host metabolite to a growth promoting substance for the host (Kravchenko et al., 2004; Kamilova et al., 2006; Xin et al., 2009b). The poplar endophyte strains varied in the amount of auxin produced, with the Rahnella and Pseudomonas strains producing the highest levels and the Burkholderia strains producing the least. Another poplar endophyte, Burkholderia strain WPB, not included in this study, produced similar levels of the auxin, and stimulated root growth by 72% (Xin et al., 2009b); therefore, even low levels produced under these in vitro conditions can have in vivo a strong impact on the plant. The Rhodotorula graminis yeast strain, WP4-3-1, produced a moderate level of auxin. Other endophytic yeast strains of poplar have been reported to produce this phytohormone (Xin et al., 2009a). IAA synthesis produced by endophytes can stimulate root growth as well as may influence other developmental processes such as apical dominance, tropic responses, flowering, and fruiting (Spaepen and Vanderleyden, 2011; Spaepen, 2015). Since the endophytes were capable of IAA production even though they colonized aboveground tissue, this may support the hypothesis that endophyte-produced auxins may have these impacts in addition to increasing rooting. However, the locations of IAA production in planta by these strains have yet to be determined.
The majority of the endophyte strains having antagonistic activities also solubilized phosphorus in NBRIP medium containing insoluble phosphorus. Many past studies reported that endophytic or growth promoting soil bacteria solubilize inaccessible soil phosphorus into bioavailable forms, providing potential phosphorus resources for plants to uptake for their growth and development (Gamalero and Glick, 2011; Oteino et al., 2015). Furthermore, evidence of siderophores production was observed in many endophyte strains. Siderophore are iron chelating organic compounds produced by microbes or plants to accrue iron from environments. Plant growth promotion and relief of iron deficiency symptoms have been demonstrated through microbial siderophores in different crop plants (Ahmed and Holmström, 2014; Saha et al., 2015).
Endophyte strains WP40, WP41, WP42, WP 4-4-2, and WP 4-5-3 reduced acetylene into ethylene in the ARA, indicating N-fixation, and were found positive for the nifH gene by PCR. However, strains WP 4-3-2, WP 4-10-4, and WP 4-4-6 were ARA positive but none of these had a nifH PCR product. Although the primers are considered as “universal,” it is often the case that this primer set does not amplify nifH of diazotrophic strains (Bürgmann et al., 2004). Previous studies also reported the discrepancy in relationship between the nifH profiles and ARA results (Deslippe et al., 2005; Patra et al., 2006). Future studies for these strains would be to have the genomes sequenced. Furthermore, strains 4-4-2 and 4-5-3 were nifH positive but ARA negative. The in vitro culture conditions for ARA may not be favorable for all strains to show N-fixing activity. Nitrogenase activities would differ with the culture medium, growth conditions, and growth stages of bacteria (Lin et al., 2012) or maybe it is only active in planta.
The results from genome analyses corroborate the phenotypic observations for BNF, phosphate solubilization, IAA production, and siderophore production. Since we observed the nifHDK gene cluster encoding for the nitrogenase enzyme in WP40 and WP42, we can postulate that they can fix atmospheric N, which is potentially available to the host plants to assimilate. The ARA results of WP40 and WP42 also supports the hypothesis of BNF. In addition, the pqq operon involved in phosphate solubilization, and tryptophan-2-monooxygenase and ACC deaminase for phytohormone production/modulation were observed in the WP40 and WP42 genomes. It has been shown that the expression of pqqABCDE genes stimulates gluconic acid production, which solubilizes the insoluble phosphates (Richardson et al., 2009; Oteino et al., 2015). In addition, it is claimed that ACC deaminase producing bacteria help plants to mitigate the biotic and abiotic stresses by lowering the ethylene level in plants that would otherwise interfere with the plant physiological processes and eventually damage the entire plant body (Glick, 2014).
The utilization of microbial symbionts in crop cultivation offers an ecologically sound and sustainable way of farming. The results from in vitro assays suggest that Salicaceae endophytes have the potential to increase plant growth, possibly through the mechanisms of N-fixation, IAA production, phosphate solubilization, and siderophore production but may also protect the host plants from pathogen-induced plant diseases. Recent advances in molecular biology and functional genomics will help to expand the existing understanding of plant-endophyte interactions. Integration of multiple microbial strains as a single consortium can offer different benefits to the crop plants. Further investigations about outcomes of plant-endophyte interactions in different ecological settings and under field conditions would be beneficial to utilize the full plant growth promoting potential of endophytes.
Author contributions
Conceived and designed the experiments: SLK, SD, and PO. Performed the experiments: SLK, PMJ, NL, and KM. Analyzed the data: SLK, PMJ, AF, NL, KM, PO, GM, AH, SHK, and SD. Contributed reagents/materials/analysis tools: SLK, SD, PO, and AF. Wrote the paper: SLK, AF, PO, PMJ, and SD.
Funding
This study was supported by a grant from the USDA NIFA grant 2012-68002-19824, and USDA Project Number 2090 22000 016 00D and Washington Grain Commission grant 3061-4548 (PO).
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors sincerely thank Mr. Andrew W. Sher for performing PCR and sequence analysis of 16S rRNA gene of some of the endophyte strains. We thank Dr. Timothy Paulitz for the culture of Rhizoctonia solani AG-8, and for helpful suggestions on the manuscript. We also thank Iqbal Singh for F. culmorum isolate 70110023 and Dr. Weidong Chen for P. ultimum isolate 217. We are grateful to Dr. Liladhar Paudel for metabolite analyses.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00386/full#supplementary-material
References
- Agnoli K., Schwager S., Uehlinger S., Vergunst A., Viteri D. F., Nguyen D. T., et al. (2012). Exposing the third chromosome of Burkholderia cepacia complex strains as a virulence plasmid. Mol. Microbiol. 83, 362–378. 10.1111/j.1365-2958.2011.07937 [DOI] [PubMed] [Google Scholar]
- Ahmed E., Holmström S. J. (2014). Siderophores in environmental research: roles and applications. Microb. Biotechnol. 7, 196–208. 10.1111/1751-7915.12117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angus A. A., Agapakis C. M., Fong S., Yerrapragada S., Estrada-de los Santos P., Yang P., et al. (2014). Plant-associated symbiotic Burkholderia species lack hallmark strategies required in mammalian pathogenesis. PLoS ONE 9:e83779. 10.1371/journal.pone.0083779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beneduzi A., Ambrosini A., Passaglia L. M. (2012). Plant growth-promoting rhizobacteria (PGPR): Their potential as antagonists and biocontrol agents. Genet. Mol. Biol. 35, 1044–1051. 10.1590/S1415-47572012000600020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernabeu P. R., Pistorio M., Torres-tejerizo G., Santos P. E. L., Galar M. L., Boiardi J. L., et al. (2015). Colonization and plant growth-promotion of tomato by Burkholderia tropica. Sci. Hortic. (Amsterdam). 191, 113–120. 10.1016/j.scienta.2015.05.014 [DOI] [Google Scholar]
- Bernier S. P., Silo-Suh L., Woods D. E., Ohman D. E., Sokol P. A. (2003). Comparative analysis of plant and animal models for characterization of Burkholderia cepacia virulence. Infect. Immun. 71, 5306–5313. 10.1128/IAI.71.9.5306-5313.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee R. B., Singh A., Mukhopadhyay S. N. (2008). Use of nitrogen-fixing bacteria as biofertiliser for non-legumes: prospects and challenges. Appl. Microbiol. Biotechnol. 80, 199–209. 10.1007/s00253-008-1567-2 [DOI] [PubMed] [Google Scholar]
- Bürgmann H. F., Widmer W., Von Sigler, Zeyer J. (2004). New molecular screening tools for analysis of free-living diazotrophs in soil. Appl. Environ. Microbiol. 70, 240–247. 10.1128/AEM.70.1.240-247.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtnick M. N., Brett P. J., Harding S. V., Ngugi S. A., Ribot W. J., Chantratita N., et al. (2011). The Cluster 1 Type VI secretion system is a major virulence determinant in Burkholderia pseudomallei. Infect. Immun. 79, 1512–1525. 10.1128/IAI.01218-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busby P. E., Ridout M., Newcombe G. (2016). Fungal endophytes: modifiers of plant disease. Plant Mol. Biol. 90, 645–655. 10.1007/s11103-015-0412-0 [DOI] [PubMed] [Google Scholar]
- Cangelosi G. A., Best E. A., Martinetti G., Nester E. W. (1991). Genetic analysis of Agrobacterium. Meth. Enzymology 204, 384–397. [DOI] [PubMed] [Google Scholar]
- Chakraborty S., Newton A. C. (2011). Climate change, plant diseases and food security: an overview. Plant Pathol. 60, 2–14. 10.1111/j.1365-3059.2010.02411.x [DOI] [Google Scholar]
- Chen C., Bauske E. M., Musson G., Rodriguezkabana R., Kloepper J. W. (1995). Biological control of fusarium wilt on cotton by use of endophytic bacteria. Biol. Control 5, 83–91. 10.1006/bcon.1995.1009 [DOI] [Google Scholar]
- Chiarini L., Bevivino A., Dalmastri C., Tabacchioni S., Visca P. (2006). Burkholderia cepacia complex species: health hazards and biotechnological potential. Trends Microbiol. 14, 277–286. 10.1016/j.tim.2006.04.006 [DOI] [PubMed] [Google Scholar]
- Chu K. K., Davidson D. J., Halsey T. K., Chung J. W., Speert D. P. (2002). Differential persistence among genomovars of the Burkholderia cepacia complex in a murine model of pulmonary infection. Infect Immun. 70, 2715–2720. 10.1128/IAI.70.5.2715-2720.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cieri M. V., Mayer-Hamblett N., Griffith A., Burns J. L. (2002). Correlation between an in vitro invasion assay and a murine model of Burkholderia cepacia lung infection. Infect. Immun. 70, 1081–1086. 10.1128/IAI.70.3.1081-1086.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compant S., Clément C., Sessitsch A. (2010). Plant growth-promoting bacteria in the rhizo- and endosphere of plants: their role, colonization, mechanisms involved and prospects for utilization. Soil Biol. Biochem. 42, 669–678. 10.1016/j.soilbio.2009.11.024 [DOI] [Google Scholar]
- Compant S., Duffy B., Nowak J., Clément C., Barka E. A. (2005). Use of plant growth-promoting bacteria for biocontrol of plant diseases: principles, mechanisms of action, and future prospects. Appl. Environ. Microbiol. 71, 4951–4959. 10.1128/AEM.71.9.4951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compant S., Nowak J., Coenye T., Clément C., Essaid B. A. (2008). Diversity and occurrence of Burkholderia spp. in the natural environment. FEMS Microbiol. Rev. 32, 607–626. 10.1111/j.1574-6976.2008.00113.x [DOI] [PubMed] [Google Scholar]
- Darling A. C., Mau B., Blattner F. R., Perna N. T. (2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403. 10.1101/gr.2289704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruijn I., Raaijmakers J. M. (2009). Regulation of cyclic lipopeptide biosynthesis in Pseudomonas fluorescens by the ClpP Protease. J. Bacteriol. 191, 1910–1923. 10.1128/JB.01558-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslippe J. R., Egger K. N., Henry G. H. R. (2005). Impacts of warming and fertilization on nitrogen-fixing microbial communities in the Canadian high arctic. FEMS Microbiol. Ecol. 53, 41–50. 10.1016/j.femsec.2004.12.002 [DOI] [PubMed] [Google Scholar]
- Doty S. L., Dosher M. R., Singleton G. L., Moore A. L., Van Aken B., Stettler R. F., et al. (2005). Identification of an endophytic Rhizobium in stems of Populus. Symbiosis 39, 27–35. [Google Scholar]
- Doty S. L., Oakley B., Xin G., Kang J. W., Singleton G., Khan Z., et al. (2009). Diazotrophic endophytes of native black cottonwood and willow. Symbiosis 47, 23–33. 10.1007/BF03179967 [DOI] [Google Scholar]
- Doty S. L., Sher A. W., Fleck N. D., Khorasani M., Bumgarner R. E., Khan Z., et al. (2016). Variable nitrogen fixation in wild populus. PLoS ONE 11:e0155979. 10.1371/journal.pone.0155979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberl L., Vandamme P. (2016). Members of the genus Burkholderia: good and bad guys. F1000Res. 5(F1000 Faculty Rev):1007. 10.12688/f1000research.8221.1.eCollection%202016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Shen D., Song W. (2006). Rice endophyte Pantoea agglomerans YS19 promotes host plant growth and affects allocations of host photosynthates. J. Appl. Microbiol. 100, 938–945. 10.1111/j.1365-2672.2006.02843.x [DOI] [PubMed] [Google Scholar]
- Franke J., Ishida K., Hertweck C. (2014). Evolution of siderophore pathways in human pathogenic bacteria. J. Am. Chem. Soc. 136, 5599–5602. 10.1017/CBO9781107415324.004 [DOI] [PubMed] [Google Scholar]
- Gagne-Bourgue F., Aliferis K. A., Seguin P., Rani M., Samson R., Jabaji S. (2013). Isolation and characterization of indigenous endophytic bacteria associated with leaves of switchgrass (Panicum virgatum L.) cultivars. J. Appl. Microbiol. 114, 836–853. 10.1111/jam.12088 [DOI] [PubMed] [Google Scholar]
- Gamalero E., Glick B. R. (2011). Mechanisms used by plant growth-promoting bacteria in Bacteria in Agrobiology: Plant Nutrient Management, ed Maheshwari D. K.(Berlin; Heidelberg: Springer; ), 17–47. [Google Scholar]
- Gilchrist F. J., Sims H., Alcock A., Jones A. M., Bright-Thomas R. J., Smith D., et al. (2013). Is hydrogen cyanide a marker of Burkholderia cepacia complex? J. Clin. Microbiol. 51, 3849–3851. 10.1128/JCM.02157-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick B. R. (2014). Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 169, 30–39. 10.1016/j.micres.2013.09.009 [DOI] [PubMed] [Google Scholar]
- Gordon S. A., Weber R. P. (1951). Colorimetric estimation of inodoleacetic acid. Plant Physiol. 26, 192–195. 10.1016/0003-2697(76)90514-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindarajan M., Balandreau J., Kwon S.-W., Weon H.-Y., Lakshminarasimhan C. (2008). Effects of the inoculation of Burkholderia vietnamensis and related endophytic diazotrophic bacteria on grain yield of rice. Microb. Ecol. 55, 21–37. 10.1007/s00248-007-9247-9 [DOI] [PubMed] [Google Scholar]
- Gu G., Smith L., Wang N., Wang H., Lu S. E. (2009a). Biosynthesis of an antifungal oligopeptide in Burkholderia contaminans strain MS14. Biochem. Biophys. Res. Commun. 380, 328–332. 10.1016/j.bbrc.2009.01.073 [DOI] [PubMed] [Google Scholar]
- Gu G., Wang N., Chaney N., Smith L., Lu S.-E. (2009b). AmbR1 is a key transcriptional regulator for production of antifungal activity of Burkholderia contaminans strain MS14. FEMS Microbiol. Lett. 297, 54–60. 10.1111/j.1574-6968.2009.01653.x [DOI] [PubMed] [Google Scholar]
- Hane J. K., Anderson J. P., Williams A. H., Sperschneider J., Singh K. B. (2014). Genome sequencing and comparative genomics of the broad host-range pathogen Rhizoctonia solani AG8. PLoS Genet. 10, 11–12. 10.1371/journal.pgen.1004281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardoim P. R., van Overbeek L. S., Berg G., Pirttilä A. M., Compant S., Campisano A., et al. (2015). The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 79, 293–320. 10.1128/MMBR.00050-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardoim P. R., van Overbeek L. S., van Elsas J. D. (2008). Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol. 16, 463–471. 10.1016/j.tim.2008.07.008 [DOI] [PubMed] [Google Scholar]
- Howell C. R., Stipanovic R. D. (1980). Suppression of Pythium ultimum induced damping-off of cotton seedlings by Pseudomonas fluorescens and its antibiotic, pyoluteorin. Phytopathology 70, 712–715. [Google Scholar]
- Kamilova F., Kravchenko L. V., Shaposhnikov A. I., Azarova T., Makarova N., Lugtenberg B. (2006). Organic acids, sugars, and L-tryptophane in exudates of vegetables growing on stonewool and their effects on activities of rhizosphere bacteria. Mol. Plant. Microbe. Interact. 19, 250–256. 10.1094/MPMI-19-0250 [DOI] [PubMed] [Google Scholar]
- Kandel S. L., Herschberger N., Kim S. H., Doty S. L. (2015). Diazotrophic endophytes of poplar and willow for growth promotion of rice plants in nitrogen-limited conditions. Crop Sci. 55, 1765–1772. 10.2135/cropsci2014.08.0570 [DOI] [Google Scholar]
- Khan Z., Guelich G., Phan H., Redman R., Doty S. (2012). Bacterial and yeast endophytes from poplar and willow promote growth in crop plants and grasses. ISRN Agron. 2012:890280 10.5402/2012/890280 [DOI] [Google Scholar]
- Khan Z., Kandel S. L., Ramos D. N., Ettl G. J., Kim S., Doty S. L. (2015). Increased biomass of nursery-grown douglas-fir seedlings upon inoculation with diazotrophic endophytic consortia. Forests 6, 3582–3593. 10.3390/f6103582 [DOI] [Google Scholar]
- Khan Z., Roman D., Kintz T., delas Alas M., Yap R., Doty S. (2014). Degradation, phytoprotection and phytoremediation of phenanthrene by endophyte Pseudomonas putida, PD1. Environ. Sci. Technol. 48, 12221–12228. 10.1021/es503880t [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoth J. L., Kim S.-H., Ettl G. J., Doty S. L. (2013). Effects of cross host species inoculation of nitrogen-fixing endophytes on growth and leaf physiology of maize. GCB Bioenergy 5, 408–418. 10.1111/gcbb.12006 [DOI] [Google Scholar]
- Knoth J. L., Kim S.-H., Ettl G. J., Doty S. L. (2014). Biological nitrogen fixation and biomass accumulation within poplar clones as a result of inoculations with diazotrophic endophyte consortia. New Phytol. 201, 599–609. 10.1111/nph.12536 [DOI] [PubMed] [Google Scholar]
- Kravchenko L. V., Azarova T. S., Makarova N. M., Tikhonovich I. A. (2004). The effect of tryptophan of plant root metabolites on the phyto stimulating activity of rhizobacteria. Mikrobiologiia 73, 195–198. 10.1023/B:MICI.0000023982.76684.9d [DOI] [PubMed] [Google Scholar]
- Kwak Y.-S., Bakker P. A., Glandorf D. C., Rice J. T., Paulitz T. C., Weller D. M. (2009). Diversity, virulence, and 2,4-diacetylphloroglucinol sensitivity of Gaeumannomyces graminis var. tritici isolates from Washington state. Phytopathology 99, 472–479. 10.1094/PHYTO-99-5-0472 [DOI] [PubMed] [Google Scholar]
- Laville J., Blumer C., Von Schroetter C., Gaia V., Défago G., Keel C., et al. (1998). Characterization of the hcnABC gene cluster encoding hydrogen cyanide synthase and anaerobic regulation by ANR in the strictly aerobic biocontrol agent Pseudomonas fluorescens CHA0. J. Bacteriol. 180, 3187–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L., Li Z., Hu C., Zhang X., Chang S., Yang L., et al. (2012). Plant growth-promoting nitrogen-fixing enterobacteria are in association with sugarcane plants growing in Guangxi, China. Microbes Environ. 27, 391–398. 10.1264/jsme2.ME11275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewen P. C. (1984). Isolation of catalase-deficient Escherichia coli mutants and genetic mapping of katE. a locus that affects catalase activity. J. Bacteriol. 157, 622–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S.-E., Novak J., Austin F. W., Gu G., Ellis D., Kirk M., et al. (2009). Occidiofungin, a Unique Antifungal Glycopeptide Produced by a Strain of Burkholderia contaminans. Biochemistry 48, 8312–8321. 10.1021/bi900814c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoel J., Marta T., Albuquerque L. S., De Montaldo Y. C., Ubaldo J., De Oliveira L., et al. (2015). Potential of the endophytic bacteria (Herbaspirillum spp. and Bacillus spp.) to promote sugarcane growth. Aust. J. Crop Sci. 9, 754–760. [Google Scholar]
- Markowitz V. M., Chen I.-M. A., Palaniappan K., Chu K., Szeto E., Grechkin Y., et al. (2012). IMG: the integrated microbial genomes database and comparative analysis system. Nucleic Acids Res. 40, D115–D122. 10.1093/nar/gkr1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattos K. A., Pádua V. L. M., Romeiro A., Hallack L. F., Neves B. C., Ulisses T. M. U., et al. (2008). Endophytic colonization of rice (Oryza sativa L.) by the diazotrophic bacterium Burkholderia kururiensis and its ability to enhance plant growth. An. Acad. Bras. Cienc. 80, 477–493. 10.1590/S0001-37652008000300009 [DOI] [PubMed] [Google Scholar]
- Mavrodi O. V., Walter N., Elateek S., Taylor C. G., Okubara P. A. (2012). Suppression of Rhizoctonia and Pythium root rot of wheat by new strains of Pseudomonas. Biol. Cont. 62, 93–102. 10.1016/j.biocontrol.2012.03.013 [DOI] [Google Scholar]
- McSpadden Gardener B. B., Weller D. M. (2001). Changes in populations of rhizosphere bacteria associated with take-all disease of wheat. Appl. Environ. Microbiol. 67, 4414–4425. 10.1128/AEM.67.10.4414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer J. A., Abdallah M. A. (1978). The fluorescent pigment of Pseudomonas fluorescens: biosynthesis, purification and physicochemical properties. Microbiology 107, 319–328. [Google Scholar]
- Mitter B., Petric A., Shin M. W., Chain P. S., Hauberg-Lotte L., Reinhold-Hurek B., et al. (2013). Comparative genome analysis of Burkholderia phytofirmans PsJN reveals a wide spectrum of endophytic lifestyles based on interaction strategies with host plants. Front. Plant Sci. 4:120. 10.3389/fpls.2013.00120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nautiyal C. S. (1999). An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol. Lett. 170, 265–270. 10.1016/S0378-1097(98)00555-2 [DOI] [PubMed] [Google Scholar]
- Osawa S., Yabuuchi E., Narano Y., Nakata M., Kosono Y., Takashina K., et al. (1963). Pigment production by Pseudomonas aeruginosa on glutamic acid medium and gel filtration of the culture fluid filtrate. Japan J. Microbiol. 7, 86–95. [PubMed] [Google Scholar]
- Oteino N., Lally R. D., Kiwanuka S., Lloyd A., Ryan D., Germaine K. J., et al. (2015). Plant growth promotion induced by phosphate solubilizing endophytic Pseudomonas isolates. Front. Microbiol. 6, 1–9. 10.3389/fmicb.2015.00745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pageni B. B., Lupwayi N. Z., Akter Z., Larney F. J., Kawchuk L. M., Gan Y. T. (2014). Plant growth-promoting and phytopathogen-antagonistic properties of bacterial endophytes from potato (Solanum tuberosum L.) cropping systems. Can. J. Plant Sci. 94, 835–844. 10.4141/CJPS2013-356 [DOI] [Google Scholar]
- Passari A. K., Mishra V. K., Gupta V. K., Yadav M. K., Saikia R., Singh B. P. (2015). In vitro and in vivo plant growth promoting activities and dna fingerprinting of antagonistic endophytic actinomycetes associates with medicinal plants. PLoS ONE 10:e0139468. 10.1371/journal.pone.0139468 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Patra A. K., Abbadie L., Clays-Josserand A., Degrange V., Grayston S. J., Guillaumaud N., et al. (2006). Effects of management regime and plant species on the enzyme activity and genetic structure of N-fixing, denitrifying and nitrifying bacterial communities in grassland soils. Environ. Microbiol. 8, 1005–1016. 10.1111/j.1462-2920.2006.00992.x [DOI] [PubMed] [Google Scholar]
- Patten C. L., Glick B. R. (2002). Role of Pseudomonas putida indoleacetic acid in development of the host plant root system. Appl. Environ. Microbiol. 68, 3795–3801. 10.1128/AEM.68.8.3795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulitz T. C. (2006). Low input no-till cereal production in the Pacific Northwest of the U.S.: the challenges of root diseases. Eur. J. Plant Pathol. 115, 271–281. 10.1007/s10658-006-9023-6 [DOI] [Google Scholar]
- Paungfoo-Lonhienne C., Lonhienne T. G. A., Yeoh Y. K., Webb R. I., Lakshmanan P., Chan C. X., et al. (2014). A new species of Burkholderia isolated from sugarcane roots promotes plant growth. Microb. Biotechnol. 7, 142–154. 10.1111/1751-7915.12105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perin L., Martínez-Aguilar L., Castro-González R., Estrada-De Los Santos P., Cabellos-Avelar T., Guedes H. V., et al. (2006). Diazotrophic Burkholderia species associated with field-grown maize and sugarcane. Appl. Environ. Microbiol. 72, 3103–3110. 10.1128/AEM.72.5.3103-3110.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirttilä A. M., Joensuu P., Pospiech H., Jalonen J., Hohtola A. (2004). Bud endophytes of Scots pine produce adenine derivatives and other compounds that affect morphology and mitigate browning of callus cultures. Physiol. Plant. 121, 305–312. 10.1111/j.0031-9317.2004.00330.x [DOI] [PubMed] [Google Scholar]
- Poly F., Monrozier L. J., Bally R.. (2001). Improvement in the RFLP procedure for studying the diversity of nifH genes in communities of nitrogen fixers in soil. Res. Microbiol. 152, 95–103. 10.1016/S0923-2508(00)01172-4 [DOI] [PubMed] [Google Scholar]
- Raaijmakers J. M., Bonsall R. F., Weller D. M. (1999). Effect of population density of Pseudomonas fluorescens on production of 2,4-diacetylphloroglucinol in the rhizosphere of wheat. Phytopathology 89, 470–475. 10.1094/PHYTO.1999.89.6.470 [DOI] [PubMed] [Google Scholar]
- Radzki W., Gutierrez Mañero F. J., Algar E., Lucas García J. A., García-Villaraco A., Ramos Solano B. (2013). Bacterial siderophores efficiently provide iron to iron-starved tomato plants in hydroponics culture. Antonie Van Leeuwenhoek 104, 321–330. 10.1007/s10482-013-9954-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson A. E., Barea J. M., McNeill A. M., Prigent-Combaret C. (2009). Acquisition of phosphorus and nitrogen in the rhizosphere and plant growth promotion by microorganisms. Plant Soil 321, 305–339. 10.1007/s11104-009-9895-2 [DOI] [Google Scholar]
- Rodriguez R. J., Henson J., Van Volkenburgh E., Hoy M., Wright L., Beckwith F., et al. (2008). Stress tolerance in plants via habitat-adapted symbiosis. ISME J. 2, 404–416. 10.1038/ismej.2007.106 [DOI] [PubMed] [Google Scholar]
- Ryall B., Lee X., Zlosnik J. E., Hoshino S., Williams H. D. (2008). Bacteria of the Burkholderia cepacia complex are cyanogenic under biofilm and colonial growth conditions. BMC Microbial. 8:108. 10.1186/1471-2180-8-108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan R. P., Germaine K., Franks A., Ryan D. J., Dowling D. N. (2008). Bacterial endophytes: recent developments and applications. FEMS Microbiol. Lett. 278, 1–9. 10.1111/j.1574-6968.2007.00918.x [DOI] [PubMed] [Google Scholar]
- Rybakova D., Cernava T., Köberl M., Liebminger S., Etemadi M., Berg G. (2016). Endophytes-assisted biocontrol: novel insights in ecology and the mode of action of Paenibacillus. Plant Soil 405, 125–140. 10.1007/s11104-015-2526-1 [DOI] [Google Scholar]
- Saha M., Sarkar S., Sarkar B., Sharma B. K., Bhattacharjee S., Tribedi P. (2015). Microbial siderophores and their potential applications: a review. Environ. Sci. Pollut. Res. 22, 1–16. 10.1007/s11356-015-4294-0 [DOI] [PubMed] [Google Scholar]
- Saini L. S., Galsworthy S. B., John M. A., Valvano M. A. (1999). Intracellular survival of Burkholderia cepacia complex isolates in the presence of macrophage cell activation. Microbiology 145(Pt 12), 3465–3475. [DOI] [PubMed] [Google Scholar]
- Sajjan S. U., Carmody L. A., Gonzalez C. F., LiPuma J. J. (2008). A type IV secretion system contributes to intracellular survival and replication of Burkholderia cenocepacia. Infect. Immun. 76, 5447–5455. 10.1128/IAI.00451-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santi C., Bogusz D., Franche C. (2013). Biological nitrogen fixation in non-legume plants. Ann. Bot. 111, 743–767. 10.1093/aob/mct048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoyo G., Moreno-Hagelsieb G., del Carmen Orozco-Mosqueda M., Glick B. R. (2016). Plant growth-promoting bacterial endophytes. Microbiol. Res. 183, 92–99. 10.1016/j.micres.2015.11.008 [DOI] [PubMed] [Google Scholar]
- Santoyo G., del Orozco-Mosqueda C. M., Govindappa M. (2012). Mechanisms of biocontrol and plant growth-promoting activity in soil bacterial species of Bacillus and Pseudomonas: a review. Biocontrol Sci. Technol. 22, 855–872. 10.1080/09583157.2012.694413 [DOI] [Google Scholar]
- Schwarz S., West T. E., Boyer F., Chiang W. C., Carl M. A., Hood R. D., et al. (2010). Burkholderia Type VI secretion systems have distinct roles in Eukaryotic and Bacterial cell interactions. PLoS Pathog 6:e1001068. 10.1371/journal.ppat.1001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwyn B., Neilands J. B. (1987). Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160, 47–56. 10.1016/0003-2697(87)90612-9 [DOI] [PubMed] [Google Scholar]
- Sgroy V., Cassán F., Masciarelli O., Del Papa M. F., Lagares A., Luna V. (2009). Isolation and characterization of endophytic plant growth-promoting (PGPB) or stress homeostasis-regulating (PSHB) bacteria associated to the halophyte Prosopis strombulifera. Appl. Microbiol. Biotechnol. 85, 371–381. 10.1007/s00253-009-2116-3 [DOI] [PubMed] [Google Scholar]
- Shi Y., Lou K., Li C. (2009). Promotion of plant growth by phytohormone-producing endophytic microbes of sugar beet. Biol. Fertil. Soils 45, 645–653. 10.1007/s00374-009-0376-9 [DOI] [Google Scholar]
- Spaepen S. (2015). Plant hormones produced by microbes, in Principles of Plant-Microbe Interactions, ed Lugtenberg B.(Basel: Springer International Publishing; ), 247–256. [Google Scholar]
- Spaepen S., Vanderleyden J. (2011). Auxin and plant-microbe interactions. Cold Spring Harb. Perspect. Biol. 3, 1–13. 10.1017/CBO9781107415324.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-Moreno Z. R., Caballero-Mellado J., Coutinho B. G., Mendonça-Previato L., James E. K., Venturi V. (2012). Common features of environmental and potentially beneficial plant-associated Burkholderia. Microb. Ecol. 63, 249–266. 10.1007/s00248-011-9929-1 [DOI] [PubMed] [Google Scholar]
- Thomashow L. S., Weller D. M., Bonsall R. F., Pierson L. S. (1990). Production of the antibiotic phenazine-1-carboxylic acid by fluorescent Pseudomonas species in the rhizosphere of wheat. Appl. Environ. Microbiol. 56, 908–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma V. C., Singh S. K., Prakash S. (2011). Bio-control and plant growth promotion potential of siderophore producing endophytic Streptomyces from Azadirachta indica A. Juss. J. Basic Microbiol. 51, 550–556. 10.1002/jobm.201000155 [DOI] [PubMed] [Google Scholar]
- Videira S. S., de Oliveira D. M., de Morais R. F., Borges W. L., Baldani V. L. D., Baldani J. I. (2012). Genetic diversity and plant growth promoting traits of diazotrophic bacteria isolated from two Pennisetum purpureum Schum. genotypes grown in the field. Plant Soil 356, 51–66. 10.1007/s11104-011-1082-6 [DOI] [Google Scholar]
- Weber T., Blin K., Duddela S., Krug D., Kim H. U., Bruccoleri R., et al. (2015). antiSMASH 3.0—a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 43, W237–W243. 10.1093/nar/gkv437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisskopf L. (2013). The potential of bacterial volatiles for crop protection against phytophathogenic fungi, in Microbial Pathogens and Strategies for Combating Them: Science, Technology and Education, ed Mendez-Vilas A.(Badajoz: Formatex Research Center; ), 1352–1363. [Google Scholar]
- Weller D. M., Cook R. J., MacNish G., Bassett E. N., Powelson R. L., Petersen R. R. (1986). Rhizoctonia root rot of small grains favored by reduced tillage in the Pacific Northwest. Plant Dis. 70, 70–73. [Google Scholar]
- Xin G., Glawe D., Doty S. L. (2009a). Characterization of three endophytic, indole-3-acetic acid-producing yeasts occurring in Populus trees. Mycol. Res. 113, 973–980. 10.1016/j.mycres.2009.06.001 [DOI] [PubMed] [Google Scholar]
- Xin G., Zhang G., Kang J. W., Staley J. T., Doty S. L. (2009b). A diazotrophic, indole-3-acetic acid-producing endophyte from wild cottonwood. Biol. Fertil. Soils 45, 669–674. 10.1007/s00374-009-0377-8 [DOI] [Google Scholar]
- Yun M., Park C. G., Kim J. Y., Rock C. O., Jackowski S., Park H. W. (2000). Structural basis for the feedback regulation of Escherichia coli pantothenate kinase by coenzyme A. J. Biol. Chem. 275, 28093–28099. 10.1074/jbc.M003190200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.