Abstract
The anaerobic protozoan parasite Entamoeba histolytica has mitosomes that are mitochondria lacking some canonical functions and organelle DNA. Mitosomes play an important role in the life cycle of the parasite. The distribution of proteins in mitosomes is not uniform, and how mitosomes are maintained and retained is unknown. To answer these questions, we developed a transplant method for mitosomes with hemagglutinin-tagged protein into recipient cells containing mitosomes with Myc-tagged protein. Immunofluorescence staining showed that the two protein tags colocalized in single mitosomes in some recipient cells. These results suggest that our transplant method can be used in anaerobic protozoa and that donor mitosomes may obtain recipient proteins through fusion with other mitosomes or through de novo synthesis of proteins in recipient cells.
The protozoan parasite Entamoeba histolytica is a significant cause of disease worldwide, including 50 million cases of colitis and liver abscess and up to 100,000 deaths annually1. E. histolytica has a simple life cycle, existing as either an infectious cyst or an amoeboid trophozoite that ingests bacteria and food particles and reproduces by binary fission in the host intestine. This parasite is adaptive to anaerobic conditions and has mitosomes that have largely lost canonical mitochondrial functions such as the respiratory chain and TCA cycle2,3.
Mitosomes are found in some anaerobic/microaerophilic protists and fungi4. The features of mitosomes shared with mitochondria include a double membrane structure5 and use of a mitochondrial chaperonin6, but mitosomes lack organelle DNA7. Mitosomes in E. histolytica play a role in the sulfate activation pathway3. Repression of proteins for sulfate activation8 and protein transport to mitosomes9 causes cell growth retardation, suggesting that the functions and biogenesis of mitosomes are important for proliferation of E. histolytica. Hence, the quality of mitosomes has to be maintained, but the quality control systems for DNA-lacking mitochondria in anaerobic microorganisms are unknown.
In mitochondria of aerobic eukaryotes, quality control includes digestion by autophagy or mitophagy10,11 and complementation of mitochondrial components, including self DNA, by mitochondrial fusion12,13,14,15. These control systems are important for prevention of mitochondrial dysfunction, incompatibilities between proteins encoded in nuclear and mitochondrial genomes, and mitochondrial diseases16. The complementation mechanism has been discovered by transplants of mitochondria using microinjection12 and cell fusion13.
To understand the behavior of mitosomes in E. histolytica, we performed transplant of mitosomes by microinjection and observed the exogenous mitosomes. In this study, we carried out transplant experiment using E. histolytica strains expressing mitosomal proteins with epitope tags that can discriminate between mitosomes derived from donor and recipient. The results of transplants suggest possible maintenance systems in which exogenous mitosomes associate with endogenous mitosomes or receive de novo synthesized proteins in the recipient. This is the first report describing transplant of DNA-lacking mitochondria into an anaerobic microorganism.
Results
Confirmation of AS-Myc expression in a recipient cell
To discriminate mitosomes derived from donor and recipient, we prepared an E. histolytica strain expressing ATP sulfurylase (AS; XP_653570, an authentic mitosomal protein)3 with a Myc-epitope tag at the C-terminus. Expression of AS-Myc was confirmed by western blotting and indirect immunofluorescence staining (Methods, Fig. S1). The results indicated that AS-Myc could be used to identify mitosomes in the recipient.
Evaluation of mitosomes isolated using a modified method
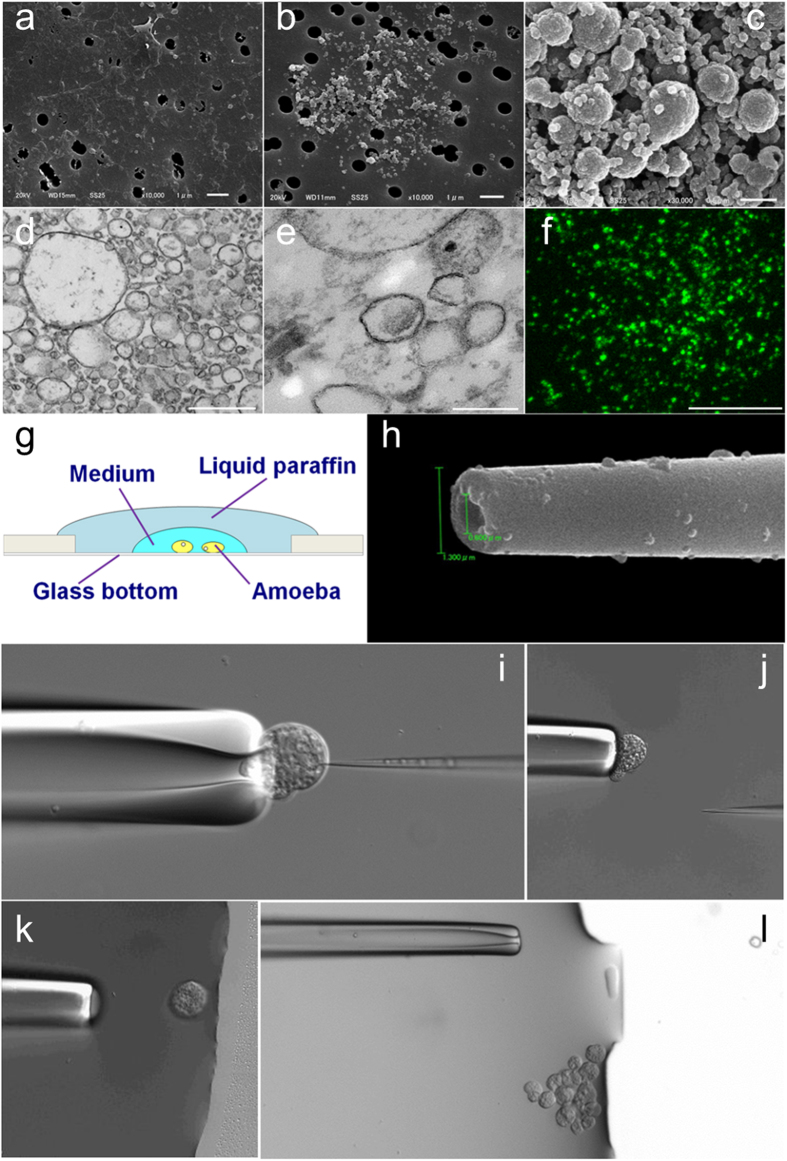
An E. histolytica strain expressing adenosine-5′-phosphosulfate kinase (APSK; XP_656278, a marker protein for Entamoeba mitosomes3) with a hemagglutinin (HA)-epitope tag (APSK-HA) was used to identify donor mitosomes for transplant. The mitosome fraction was obtained from cell lysate by the Percoll-gradient method3 with some modifications (see Methods). The main change was substitution of Percoll with Percoll-PLUS in gradient formation because Percoll showed lethal toxicity in cells after injection in preliminary experiments. In each fraction, the presence of Cpn60, a specific mitosome protein, was confirmed by Western blotting (Fig. S2). A scanning electron microscope (SEM) image of the contents of cell lysate before Percoll-PLUS gradient centrifugation is shown in Fig. 1a and images of the fraction after centrifugation are shown in Fig. 1b–f. Globular organelles of 0.2–0.7 μm diameter were concentrated in the fraction after centrifugation. The fraction was washed and concentrated by centrifugation (Fig. 1c). Transmission electron microscope (TEM) images (Fig. 1d,e) showed bilayer membrane structure, and direct immunofluorescence staining with anti-HA antibody also indicated spotted signals (Fig. 1f). The main diameter of these signals was <1 μm, similar to those in trophozoites (Fig. 2). These organelles were identified as mitosomes based on size, morphology, immunofluorescence staining and Cpn60 content in Western blotting. The concentration of the organelle was estimated to be 13.7 ± 3.8 per picoliter based on immunofluorescence images.
Figure 1. Microinjection of mitosomes into E. histolytica trophozoites.
Mitosomes were isolated by the Percoll-PLUS gradient method. (a) SEM image of the supernatant of cell lysate before gradient centrifugation. (b) SEM image of the mitosome fraction after gradient centrifugation. (c) SEM image of the fraction concentrated by centrifugation. (d–e) TEM images of concentrated fractions. (f) Fluorescence image of the concentrated fraction. Mitosomes with APSK-HA were stained using anti-HA-tag-Alexa Fluor 488. (g) Schematic illustration of the stage on the microinjection system. (h) SEM image of the tip of the injection capillary. (i–l), Time-lapse images during transplant of mitosomes (see Video S1). Bars indicate 1 μm in (a) and (b), 500 nm in (c) and (d), 200 nm in (e), and 10 μm in (f).
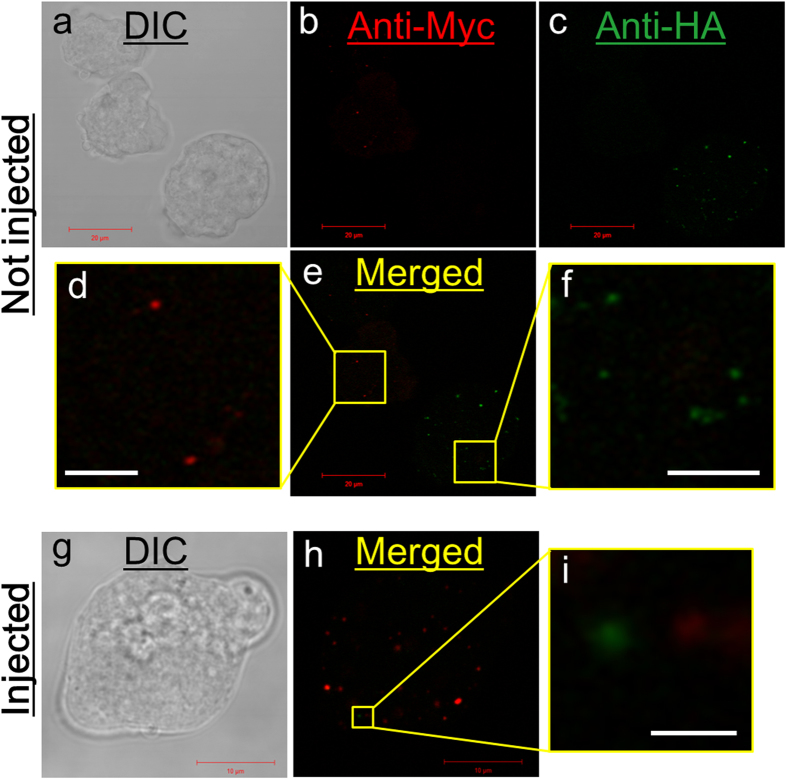
Figure 2. Detection of mitosomes by immunofluorescence staining in trophozoites.
(a–f) Trophozoites expressing AS-Myc and APSK-HA were co-cultured and stained by immunofluorescence with anti-Myc-Alexa594 and anti-HA-Alexa488 antibodies. (a) DIC image. (b,c) Fluorescence images of Alexa594 and Alexa488, respectively. (e) Merged image of (b) and (c). (d, f) Magnified images of the boxes in (e). (g–i) Trophozoite expressing AS-Myc after transplant with mitosomes containing APSH-HA. (g) DIC image. (h) Immunofluorescent image with anti-Myc-Alexa594 and anti-HA-Alexa488 antibodies. (i) Magnification of the box in (h). Bars indicate 20 µm in (a)-(c) and (e), 10 µm in (g) and (h), 5 µm in (d) and (f), and 1 µm in (i).
Transplant with mitosomes
Using an injection capillary with a 1.3–2.0 μm outer diameter and a 0.6–1.3 μm inner diameter on the tip (Fig. 1h), the transplant process (Fig. 1i–l) was performed by microinjection (see Methods and Video S1). The injected volume was estimated to be approximately 0.29–0.75 picoliter (see Supplemental information and Fig. S3). Therefore, the number of incorporated mitosomes was estimated to be 2.8–13 per recipient cell.
Just after injection, the recipient cell moved or a part of the cell became swollen on the tip of the holding pipette. Vomiting behavior of injected cells was not observed. Injected cells were carried to a restricted area on the dish (Fig. 1l) and some were transferred to another dish filled with fresh medium. After sealing, these cells were incubated at 37 °C and traced by tiling and time-lapse images taken at 30-min intervals (Video S2). The injected cells showed amebic motion and divided at about 20 h after injection. The daughter cells underwent the next division asynchronously at about 29 h and 34 h after injection. Entamoeba trophozoites sometimes require the assistance of a neighboring cell that mechanically severs the tubular bridge between two daughter cells17,18, but this midwife cell behavior was not observed in these divisions. The injected cells and their daughter cells showed regular binary fission. These results indicate that the injected cells are viable and retain the ability to proliferate.
Detection of donor mitosomes in recipient cells
Signals for APSK-HA were detected in recipient cells from three strains (Table 1) using direct immunofluorescence staining with anti-HA antibody. The diameter of the fluorescence signals ranged from 0.6 to 1.0 μm, the same size as that for signals in donor cells (Fig. S4). Injected cells with donor mitosomes were observed in approximately 23.5% of examined cells from strain HM-1:IMSS within 30 min after microinjection. The number of donor mitosomes in the recipient cell was scored as 0 to 15 per cell using this strain, and the density was estimated to be 0.88 donor mitosomes per examined cell. In Table 1, the transplant efficiency is expressed as the rate of cells with donor mitosomes per number of examined cells. These values were equal to or greater than those for transplant of mitochondria12 or exogenous cytoplasm19 in previous studies. The detection of donor mitosomes in recipient cells after microinjection indicates that our transplant method for mitosomes was successful.
Table 1. Efficiency of transplant with mitosomes containing APSK-HA into recipient strains.
| Recipient | Time* | N# | Cells |
Mitosomes |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of Injected | No. of Examined | No. of Detected | Total Rate | Mean | SD | SE | No. of Detected (merged) | Max. No. per cell | Density$ | |||
| HM-1: IMSS | 30 min | 4 | 120 | 34 | 8 | 0.24 | 0.27 | 0.23 | 0.11 | 30 | 15 | 0.88 |
| AS-Myc | 30 min | 10 | 371 | 76 | 17 | 0.22 | 0.25 | 0.18 | 0.06 | 22 (4) | 5 | 0.29 |
| 60 min | 8 | 313 | 58 | 10 | 0.17 | 0.09 | 0.12 | 0.04 | 32 (9) | 11 | 0.55 | |
| 120 min | 6 | 247 | 67 | 1 | 0.02 | 0.01 | 0.03 | 0.01 | 1 (0) | 1 | 0.02 | |
| G3 | 30 min | 3 | 121 | 24 | 13 | 0.54 | 0.58 | 0.24 | 0.14 | 42 | 6 | 1.75 |
Transplant efficiency is shown as the percentage of cells with donor mitosomes per number of examined cells. The number of incorporated organelles was estimated to be 2.8-13 per cell.
*Time indicates the processing time for microinjection and post-incubation until fixation.
#No. of Exp. (number of experiments) indicates the number of independent mitosome isolations.
$Number of detected mitosomes per number of examined cells.
Time-dependent decrease of donor mitosomes
We used a combination of APSK-HA and AS-Myc to identify donor and recipient mitosomes, respectively, in recipient cells. Immunofluorescence images of this mixture with strains expressing AS-Myc and APSK-HA are shown in Fig. 2a–f. Fluorescence signals were detected independently in each specimen by double staining. Each cell showed a single signal alone and these signals did not overlap. After injection with mitosomes from the strain expressing APSK-HA, the injected cell showed several signals for AS-Myc and a few signals for APSK-HA in a single layer on the Z-axis (Fig. 2g–i). When trophozoites expressing AS-Myc were used as recipient cells, the transplant efficiency (24.8 ± 18.1%, mean ± SD) was the same as that with strain HM-1:IMSS (27.1 ± 22.5%, p = 0.862). However, the transplant efficiency decreased with post-incubation time to 8.7 ± 12.2% and 1.3 ± 3.1% at 60 and 120 min after microinjection, respectively (the time includes the microinjection step). The efficiency differed significantly between 30 and 60 min (p = 0.04) and between 30 and 120 min (p = 0.003).
Staining patterns of mitosomes in recipient cells expressing AS-Myc
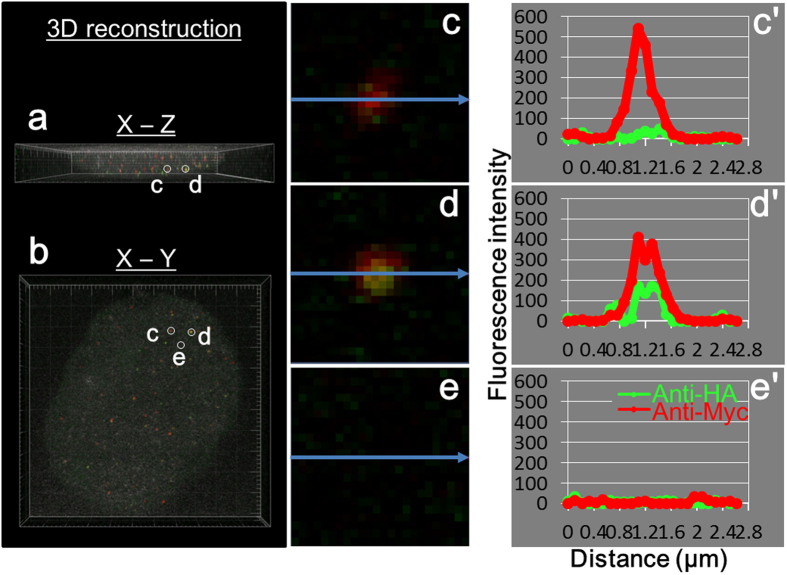
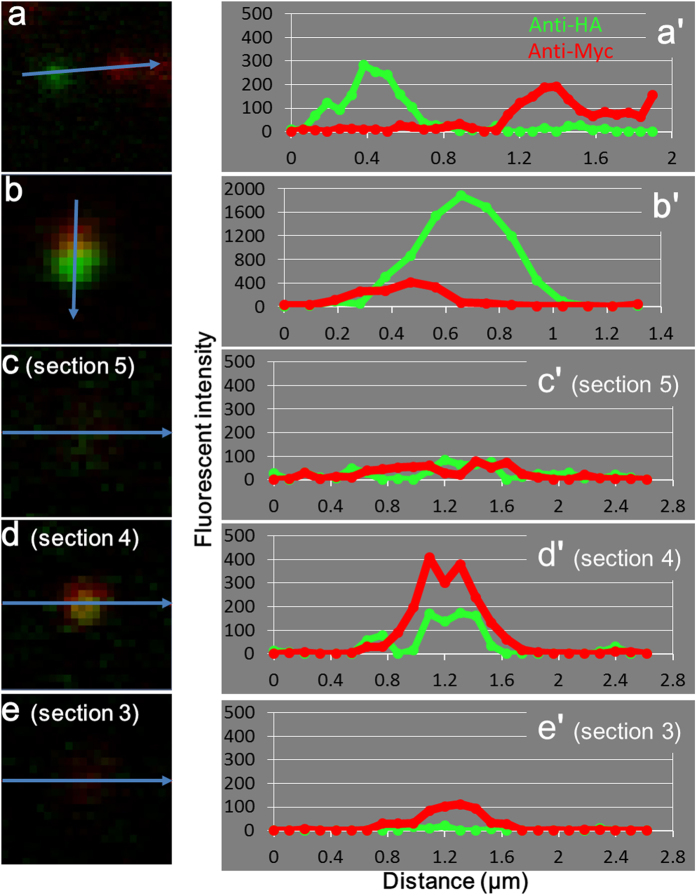
Recipient cells expressing AS-Myc after transplant of mitosomes containing APSK-HA were analyzed three-dimensionally (Fig. 3a and b, and Video S3). Donor and recipient signals were scattered in the cells. Figure 3 showed a recipient cell in the X-Z (Fig. 3a) and X-Y axis view (Fig. 3b). Figure 3c–e showed in an enlarged view, and the corresponding fluorescence intensities were Fig. 3c′–e′. In Fig. 3c, Myc-signal showed high fluorescence intensity and HA-signal was very low, similar to background noise (Fig. 3e). The image and intensities in Fig. 3c and c′ indicate that the mitosome contained AS-Myc, but not APSK-HA. In contrast, overlapping fluorescence signals for AS-Myc and APSK-HA were observed in a single mitosome (Fig. 3d) and both intensities were higher than the background level (Fig. 3d′ and e′).
Figure 3. Three-dimensional reconstruction of an injected cell and analysis of fluorescence intensity.
Z-stacking images were reconstructed by Imaris software. (a) X-Z and (b) X-Y images were shown and a rotated movie file (Video S3) was obtained from the same data. (c–e) Enlarged view of fluorescent images from (b). (c′–e′) Intensities correspond to (c–e) and are shown along arrows. Areas (c) and (d) contain mitosome signals. Background signals are collected in area (e).
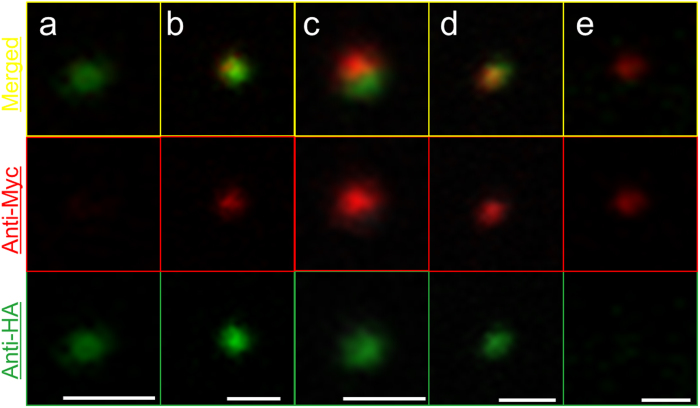
Various staining patterns of mitosomes are shown in Fig. 4. The mitosomes in Fig. 4a and e show a single fluorescent signal of AS-Myc and APSK-HA, whereas Fig. 4b–d show colocalization of two signals with different merged patterns. The two fluorescent signals in Fig. 4b and c are close, but do not overlap completely, and the green area is larger than the red area in both mitosomes. In contrast, the image in Fig. 4d shows an overlapped yellow region. Four and nine merged signals were detected from 76 cells within 30 min and from 58 cells within 60 min after injection (Table 1). Given the number of incorporated mitosomes in each cell (2.8–13), the rates of merged signals per total injected mitosomes was calculated as 0.4–1.9% and 1.1–5.5% within 30 min and 60 min after injection, respectively.
Figure 4. Fluorescence patterns of mitosomes.
Mitosomes from different injected cells are shown in each channel and as a merged view. Bars indicate 1 μm.
Analysis of fluorescence intensity showed three types of localization patterns of the two signals. In Fig. 5a, two mitosomes are shown with each signal independently localized in the recipient cell. This pattern of a single color was also shown in Fig. 2 and for mitosomes in Fig. 4a and e. In the pattern in Figs 4b,4c and 5b, fluorescence signals co-localize in single mitosomes, with the peaks of intensity from APSK-HA (donor) and AS-Myc (recipient) signals close together and partially overlapped. A single mitosome on three continuous Z-stack sections with an identical position on the X-Y axis is shown in Fig. 5c–e. This mitosome is identical to that in Fig. 3d and shows completely overlapped peaks on Z-stacking. These staining patterns were sometimes observed in a cell, such as for the mitosome in Fig. 4d. Colocalization of both fluorescence peaks demonstrates that proteins from donor and recipient were expressed in the same mitosome. These observations suggest that single mitosomes in transplanted cells contained APSK-HA and AS-Myc.
Figure 5. Analysis of fluorescent intensities in mitosomes.
Fluorescent intensities in mitosomes are shown along arrows. (a–e) Images of mitosomes. (a′–e′) Intensities corresponding to (a–e). Image (a) obtained from Fig. 2i. Images (c–e) were three continuous sections in 11 Z-stacking images of the single mitosome in Fig. 3d. Z-stacking images were taken by laser microscopy with 1.1-μm thickness sections and a 0.53-μm merge.
Comparison of transplant efficiency with HM-1:IMSS and G3
The transplant efficiency in strain G3 (57.6 ± 23.9%) was higher than that in HM-1:IMSS (27.1 ± 22.5%) within 30 min after microinjection, although the difference was not significant (p > 0.05). Strain G3 is a genetically modified avirulent strain from the virulent E. histolytica strain HM-1:IMSS20,21. Expression of mitosomal proteins occurs at similar levels in strains G3 and HM-1:IMSS based on transcript expression array profiles in AmoebaDB (http://amoebadb.org/amoeba, summarized in Table S1). Genes with different expression levels in these strains are listed in Tables S2 and S3. For example, AIG1 (avirulence inducing gene 1), which is similar to plant AIG genes involved in bacterial resistance22, is strongly expressed in strain G3 (Table S2). Amoebapore, which was silenced in establishment of strain G320,21, is markedly downregulated (Table S3).
Discussion
This study has four key findings: (1) microinjection can be used for transplant of mitosomes in the anaerobic protozoa, E. histolytica, (2) mitosomes derived from donor and recipient can be recognized in single cells using labeling with two epitope tags, (3) transplant efficiency decreases with time after microinjection, and (4) donor mitosomes in recipient cell show alternative characteristics with both proteins of donor and recipient.
Transplant of mitosomes by microinjection was applicable to cells in E. histolytica with high efficacy. Using the microinjection method, Flickinger et al.19 performed successful transplant of cytoplasm including mitochondria between free-living amoebas Amoeba discoides and Pelomyxa carolinensis. In our transplant experiments, we used an established method to isolate mitosomes3 that were of size 1–2 μm on fluorescence microscopy6 and were estimated to be <500 nm on high-resolution laser scanning sections7. The small organelles could pass through capillaries with similar diameters and are easily handled using a single recipient cell. In contrast to microinjection, cell fusion may transmit various contents of a donor cell to a transplant cell, and this method has not been established in Entamoeba. Therefore, microinjection is the best choice for transplant of organelles into Entamoeba species. As results shown in Table 1, all kinds of recipient cell using in this study showed more than 20% of transplant efficacy.
In addition, the combination with transfection techniques made it possible to identify the mitosomes by immunofluorescence staining to the expressed epitope-tags in marker proteins of E. histolytica. Although transplanted mitochondria are recognized by using polymorphism of mitochondrial DNA in many studies12,13,23,24,25, exogenous organelles could be detected without DNA probes in this method. Therefore, this is the first report of transplant with DNA-lacking mitochondria. This is also the first implementation of microinjection for the purpose of organelle transplant in anaerobic protozoa.
On the other hand, more than 40% of injected cells from all used strains showed no fluorescent signal from the donor within 30 min post-microinjection. Additionally, recipient cells derived from the strain expressing AS-Myc had reduced transplant efficiency with time, with a significant difference between 30 and 120 min after microinjection. In free-living amoebas Amoeba discoides and Pelomyxa carolinensis, which have mitochondria, recipient cells injected with exogenous cytoplasm expel the injected cytoplasm within 30 min post-injection, or exogenous organelles are segregated and digested by lysosomal enzymes in vacuole-like spaces between 6 h and 2 days post-injection19. Vomiting behaviors were not observed in any strains of E. histolytica in this study. There is a possibility that mitosomes from donors are digested by autophagosomes or lysosomes. Autophagy or mitophagy digests both damaged mitochondria10,11 and exogenous mitochondria derived from spermatozoa during fertilization in Caenorhabditis elegans26,27. Although the digestion mechanism of mitosomes is unknown in E. histolytica, we cannot exclude the possibility that mitosomes were digested by autophagy or mitophagy, similarly to mitochondria. Transplant with a larger number of donor mitosomes would allow observation of organelle degradation in E. histolytica.
Cells derived from strain G3 showed the highest efficacy among cells derived from all strains, although there were no significant differences among the cells (p > 0.05). A high fold change in strains HM-1:IMSS and G3 was found for AIG1 in transcript expression array profiles in AmoebaDB AIG family proteins are classified as stress response proteins that show high levels in comparative transcriptomic analysis between virulent and avirulent strains28. Thus, stress responses in E. histolytica might be involved in the success of transplant.
The most interesting observation in this study was the overlap of signals for donor and recipient proteins in single mitosomes in injected AS-Myc expressing cells (Figs 3, 4 and 5). This observation may give a clue to resolution of the question of how mitosomes are maintained and retained in E. histolytica because donor and recipient were derived from an identical strain. The present result shows that some donor mitosomes contain proteins from both the donor and recipient, which suggests that mitosomal proteins are compatible in donor and recipient cells. There are two possible explanations for this phenomenon. First, AS-Myc synthesized de novo in the recipient cell may be transported into donor mitosomes. We also cannot exclude the possibility that APSK-HA from broken or degraded injected mitosomes is imported into recipient mitosomes. However, this is unlikely because of the estimated small number of incorporated mitosomes of 2.8–13 per recipient cell. Second, complementation of proteins between donor and recipient mitosomes may occur by fusion within the recipient cell. Two types of mitochondrial fusion are known: complete fusion with sharing of mitochondrial DNA and detachable fusion, which is referred to as “kiss-and-run” fusion15,29. The overlap of signals in mitosomes (Figs 3d and 5c–e) and partially merged signals (Figs 4c and 5b) may represent the former and latter behavior, respectively. If mitosomes fuse to each other, the significance of fusion may be to exchange components between mitosomes because the distributions of several mitosome proteins, including AS and APSK, are not uniform based on immunostaining methods, which suggests heterogeneity of mitosomes in E. histolytica3,8. The quantity of these proteins in mitosomes may be a rate-limiting factor in the sulfate activation pathway, which is important for proliferation of E. histolytica8. Further analyses are required to examine this possibility, but fusion may contribute to maintenance of mitosome function through complementation.
Regardless of the mechanism, donor mitosomes must be acceptable to recipient proteins, which suggests that components are added to the donor mitosome. Moreover, the acceptance of mitosomal proteins from the recipient would lead to loss of identity of the donor mitosome. Therefore, the true number of transplanted mitosomes may be higher than the number determined in the study. This possibility does not negate the result that donor mitosomes decrease time-dependently in transplanted cells. Furthermore, exogenous mitosomes in the recipient cell are maintained by association with other mitosomes and/or by receiving mitosomal proteins synthesized de novo. Further development of the method used in this study to transplant DNA-lacking mitochondria will produce a primary tool to evaluate the behaviors of specialized organelles in anaerobic microorganisms.
Methods
Cell culture
Trophozoites of four strains
HM-1:IMSS30, strains expressing APSK-HA3 or AS-Myc, and G320,21 in Entamoeba histolytica were used in this study. These trophozoites were cultured axenically at 37 °C in BI-S-33 medium31 (donor) or YIMDHA-S medium32 (recipient) supplemented with 12% adult bovine serum. Medium for strains expressing APSK-HA and AS-Myc was supplemented with 20 μg/ml G418.
cDNA preparation
Total RNA was isolated from trophozoites of HM-1:IMSS clone 6 with Trizol® reagent (Life Technologies) and mRNA was purified using a GenEluteTM mRNA Miniprep Kit (Sigma, St. Louis, MO, USA). cDNA was synthesized from mRNA using SuperScriptTM III RNase H reverse transcriptase (Life Technologies) and oligo(dT)20 primer.
Plasmid construction
To generate an expression plasmid (pEhEx/AS-Myc) for E. histolytica AS tagged with Myc at the C-terminus, E. histolytica AS gene was PCR-amplified from cDNA using Phusion DNA polymerase (New England Biolabs) and specific primers (sense: 5′-GTC GGG ATC CAT GAG CAT TCA AGA AAA CTT AAA C-3′ and antisense: 5′-GTC GGG ATC CTT TCA TGG CAT CAC CAG TAG C-3′ (underlined bases indicate the BamH I restriction site). After BamH I digestion, the digested fragment was ligated with pEhEx/Myc33 digested by Bgl II using a Ligation-Convenience Kit (Nippongene, Tokyo, Japan).
Amoeba transformation
Transfection of trophozoites and selection and maintenance of transformants were performed based on the method of Nozaki et al34. Briefly, liposomes containing pEhEx/AS-Myc were made with 5 μg plasmid, Lipofectamine® and PlusTM reagent (both Life Technologies). The liposome was mixed into OPTI-MEM I medium (Life Technologies) with 5 mg/ml L-cysteine (Sigma) and 1 mg/ml ascorbic acid (Wako, Osaka, Japan). Approximately 5.0 × 105 trophozoites were incubated with the medium containing the liposome at 37 °C for 5 h. After incubation, trophozoites were transferred into culture test tubes with warmed YIMDHA-S medium supplemented with 15% adult bovine serum (Sigma). Drug selection by G418 was begun from 24 h later and medium was replaced by fresh medium with G418 every 24 h.
Mitosome isolation
Mitosomes were isolated by Percoll-gradient fractionation3 with modifications. Briefly, approximately 0.5–1.0 × 108 trophozoites were washed three times with PBS containing 2% glucose. After resuspending in SM buffer (10 mM MOPS-KOH/250 mM sucrose, pH 7.2) containing protease inhibitor cocktail (cOmplete Tablets, Mini EDTA-free, EASYpack, Roche, Mannheim, Germany) and 50 μM E-64 (Sigma), trophozoites were disrupted mechanically with a Dounce homogenizer. After centrifugation at 5,000 × g for 10 min at 4 °C, the supernatant was ultracentrifuged (Optima MAX-XP Ultracentrifuge, Beckman Coulter, Brea, CA, USA) on a gradient with Percoll-PLUS (GE Healthcare Bio-Sciences AB, Uppsala, Sweden). A total of 47–48 fractions (100 μl each) were collected from top to bottom in the tube. Two fractions with OD higher than 280 nm were mixed and washed three times with SM buffer without protease inhibitors by centrifugal filter units (Amicon Ultra-0.5 mL Ultacel-100K, Merck Millipore, Tullagreen, Carrigtwohill, Ireland). The fractions were diluted to an OD280 range of 1.5 to 2.5 with SM buffer without protease inhibitors, stored at 4 °C, and used within 8 h after preparation.
Microinjection devices
Glass thin-capillary tubes (GD-1 with filaments; Narishige Co., Tokyo, Japan) pulled on a P-97/IVF micropipette puller (Sutter Instrument Co., Novato, CA, USA) were used for transplant. After pulling, the capillary was bent at a 30° angle at 1.0–1.2 mm from the tip by a MF-1 micro-forge (Technical Products International, St. Louis, MO, USA). The injection capillary and a holding pipette were connected to an IM 300 Microinjector (Narishige) and hydraulic micromanipulator (NT-8, Narishige), respectively. The tip of the capillary and cells were visible on an inverted microscope (Observer Z1, Zeiss Co., Germany). The stage was kept at room temperature during microinjection. After injection, the stage was exchanged for a mechanical stage (ThermoPlate, MATS-555AXK and MATS-555RN, Tokai Hit Co., Shizuoka, Japan) to incubate cells at 37 °C. The whole injection device was placed on a vibration isolator system (ST-X, Showa Science Co., Tokyo, Japan).
Transplant by microinjection
The injection capillary was loaded with a mitosome fraction and stored at 4 °C before connection to the microinjection system. Recipient cells for transplant were cultured in YIMDHA-S medium18 at 37 °C and an aliquot was dropped onto a 50-mm glass bottom dish (MatTek Corp., Ashland, MA, USA) just before microinjection. To maintain anaerobic conditions and prevent drying, the medium on the dish was overlaid with liquid paraffin (Nacalai Tesque Inc., Kyoto, Japan) (Fig. 1g).
A single recipient cell was removed from the bottom by pushing with the holding pipette. Adhered recipient cells on the bottom of the dish became spherical by removing and holding. The removed cell was held on the tip of the pipette and injected with the mitosome fraction through the injection capillary for 2 s per injection with 9.0–11.0 psi pressure of nitrogen gas. The capillary for microinjection was pulled and shaped to a 1.3–2.0 μm outer diameter and a 0.6–1.3 μm inner diameter on the tip (Fig. 1h).
After injection, the cells were carried to a restricted area on the dish (Fig. 1l), and then released from the holding pipette. Microinjections were finished within 30 min in each trial, and 5 to 20 injected cells were collected from every trial. Recipient cells during microinjection were recorded by AxioVision 4.8 software (Zeiss), as shown in Fig. S1. Some injected cells were transferred to a 35-mm glass base dish (AGC Techno Glass Co., Tokyo, Japan) filled with medium. The dish was sealed with grease applied to the cover glass and incubated at 37 °C. The remaining cells were incubated in fresh YIMDHA-S medium at 37 °C and then fixed with 4% PFA within 2 h after microinjection.
Direct immunofluorescence staining
Cells and mitosomes after isolation were fixed with 4% PFA for 15 min and adhered to silane-coated glass slides (Muto Pure Chemicals Co., Tokyo, Japan) using a Shandon Cytospin 3 Centrifuge (Thermo Fisher Scientific, Waltham, MA, USA) at 800 rpm for 10 min. After washing with PBS for 15 min, specimens were immersed in blocking solution (1% BSA, 0.2% saponin in PBS) for 30 min at room, and then reacted with anti-HA-tag-Alexa Fluor 488 (M180-A48, MBL Co., Nagoya, Japan; 1/500 dilution in blocking solution) and/or anti-Myc-tag-Alexa Fluor 594 (M047-A59, MBL Co.; 1/200 dilution in blocking solution) at 4 °C overnight. After washing with 0.1% BSA in PBS for 15 min, glass slides were embedded with 10% glycerol in PBS containing 1.25 mg/ml DABCO and stored at 4 °C in the dark.
Electron microscopy
Mitosomes after isolation were subjected to electron microscopy. Mitosome fractions were pre-fixed with 2.5% glutaraldehyde in 0.05 M phosphate buffer (pH 7.6) for 1 h. For SEM, specimens were transferred to a sample folder (SEMpore. JEOL, Tokyo, Japan) and then post-fixed with 1% osmium tetroxide and dehydrated in a graded ethanol series (50, 70 and 90% ethanol, and anhydrous ethanol three times). The ethanol was substituted with t-butanol and the sample was freeze-dried using JFD-310 (JEOL). Specimens were observed at 15 kV accelerating voltage on a JSM-6510LV (JEOL) after osmium evaporation (Neoc-Pro, Meiwafosis Co., Tokyo, Japan). For TEM, specimens were adhered to a glass slide using a Shandon Cytospin 3 Centrifuge at 800 rpm for 10 min. After post-fixation and dehydration as for SEM specimens, samples were embedded in epoxy resin (Quttol-812, Nisshin EM Corp., Tokyo, Japan). The thin-sectioned specimens were observed at 80 kV accelerating voltage using a JEM1400 (JEOL) after staining with uranyl acetate and lead citrate.
Laser scanning microscopy
For visualization of mitosomes in cells after direct immunofluorescence staining, images were acquired in lambda (emission fingerprinting) and Z-stacking mode on a LSM 510 Meta confocal microscope equipped with a 488 nm Ar laser and a 543 nm He/He laser (Zeiss). Acquisitions were carried out at 499–745 nm using a C-Appochromat 63×/1.2 W corr. objective. Images were analyzed using LSM 510 Meta software (ZEN 2009 ver. 6.0.0.303, Zeiss). For three-dimensional analysis of cells, optical 1.1-μm sections with a 0.53-μm merge were reconstructed using Imaris software ver. 6.2.1. (Bitplane AG, Zurich, Switzerland).
Statistical analysis
The significance of differences was evaluated by Welch t-test, with p < 0.05 considered significant and p < 0.01 considered highly significant.
Additional Information
How to cite this article: Kazama, M. et al. Behavior of DNA-lacking mitochondria in Entamoeba histolytica revealed by organelle transplant. Sci. Rep. 7, 44273; doi: 10.1038/srep44273 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We thank Mr. Masayoshi Tokunaga, Dr. Nahoko Fukunishi and Dr. Chisa Okada in the Support Center for Medical Research and Education, Tokai University, for their help and technical advice in imaging analysis. We also thank Dr. Feng Meng for his help in amoeba transformation. This work was supported by grants from JSPS/MEXT KAKENHI (23117009, 24406013, 26460516, 16H05819 to H.T. and 15K19087 to T.M.).
Footnotes
The authors declare no competing financial interests.
Author Contributions M.K. performed most of the experiments. S.O. and K.Y. determined the transplant condition and assisted with microinjection. T.M. and K.N.T. established strains with transfection and assisted with mitosome isolation. T.N. and H.T. defined the research objectives. M.K., T.M. and H.T wrote the manuscript. All authors read and approved the manuscript.
References
- Stanley S. L. Jr. Amoebiasis. Lancet 361, 1025–1034 (2003). [DOI] [PubMed] [Google Scholar]
- van der Giezen M. Hydrogenosomes and mitosomes: conservation and evolution of functions. J Eukaryot Microbiol 56, 221–31 (2009). [DOI] [PubMed] [Google Scholar]
- Mi-ichi F., Abu Yousuf M., Nakada-Tsukui K. & Nozaki T. Mitosomes in Entamoeba histolytica contain a sulfate activation pathway. Proc. Natl. Acad. Sci. USA 106, 21731–6 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makiuchi T. & Nozaki T. Highly divergent mitochondrion-related organelles in anaerobic parasitic protozoa. Biochimie 100C, 3–17 (2014). [DOI] [PubMed] [Google Scholar]
- Ghosh S., Field J., Rogers R., Hickman M. & Samuelson J. The Entamoeba histolytica mitochondrion-derived organelle (crypton) contains double-stranded DNA and appears to be bound by a double membrane. Infect. Immun. 68, 4319–4322 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovar J., Fischer A. & Clark C. G. The mitosome, a novel organelle related to mitochondria in the amitochondrial parasite Entamoeba histolytica. Mol. Microbiol. 32, 1013–1021 (1999). [DOI] [PubMed] [Google Scholar]
- Leon-Avila G. & Tovar J. Mitosomes of Entamoeba histolytica are abundant mitochondrion-related remnant organelles that lack a detectable organellar genome. Microbiology-Sgm 150, 1245–1250 (2004). [DOI] [PubMed] [Google Scholar]
- Mi-ichi F., Makiuchi T., Furukawa A., Sato D. & Nozaki T. Sulfate activation in mitosomes plays an important role in the proliferation of Entamoeba histolytica. PLoS Negl. Trop. Dis. 5, e1263 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makiuchi T., Mi-ichi F., Nakada-Tsukui K. & Nozaki T. Novel TPR-containing subunit of TOM complex functions as cytosolic receptor for Entamoeba mitosomal transport. Sci. Rep. 3, 1129 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes L. C. & Scorrano L. Mitochondrial morphology in mitophagy and macroautophagy. Biochim. Biophys. Acta 1833, 205–212 (2013). [DOI] [PubMed] [Google Scholar]
- Kim I., Rodriguez-Enriquez S. & Lemasters J. J. Selective degradation of mitochondria by mitophagy. Arch. Biochem. Biophys. 462, 245–253 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- King M. P. & Attardi G. Injection of mitochondria into human cells leads to a rapid replacement of the endogenous mitochondrial DNA. Cell 52, 811–819 (1988). [DOI] [PubMed] [Google Scholar]
- Ono T., Isobe K., Nakada K. & Hayashi J. I. Human cells are protected from mitochondrial dysfunction by complementation of DNA products in fused mitochondria. Nature Genetics 28, 272–275 (2001). [DOI] [PubMed] [Google Scholar]
- Rolland S. G. et al. Impaired complex IV activity in response to loss of LRPPRC function can be compensated by mitochondrial hyperfusion. Proc. Natl. Acad. Sci. USA 110, E2967–76 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L. et al. Mitochondrial fusion provides an ‘initial metabolic complementation’ controlled by mtDNA. Cell Mol. Life Sci. 72, 2585–2598 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaoka M. et al. Complete repopulation of mouse mitochondrial DNA-less cells with rat mitochondrial DNA restores mitochondrial translation but not mitochondrial respiratory function. Genetics 155, 301–307 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biron D., Libros P., Sagi D., Mirelman D. & Moses E. Asexual reproduction - ‘Midwives’ assist dividing amoebae. Nature 410, 430–430 (2001). [DOI] [PubMed] [Google Scholar]
- Mukherjee C., Majumder S. & Lohia A. Inter-cellular variation in DNA content of Entamoeba histolytica originates from temporal and spatial uncoupling of cytokinesis from the nuclear cycle. PLoS Negl Trop. Dis. 3, e409 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flickinger C. J., Read G. A. & Kabana E. M. Structural responses of amoebae to the injection of heterologous cytoplasm. J. Cell. Sci. 45, 1–14 (1980). [DOI] [PubMed] [Google Scholar]
- Bracha R., Nuchamowitz Y. & Mirelman D. Transcriptional silencing of an amoebapore gene in Entamoeba histolytica: molecular analysis and effect on pathogenicity. Eukaryot. Cell. 2, 295–305 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujanover S., Katz U., Bracha R. & Mirelman D. A virulence attenuated amoebapore-less mutant of Entamoeba histolytica and its interaction with host cells. Int. J. Parasitol. 33, 1655–1663 (2003). [DOI] [PubMed] [Google Scholar]
- Abhyankar M. M. et al. Characterization of an Entamoeba histolytica high-mobility-group box protein induced during intestinal infection. Eukaryot. Cell. 7, 1565–1572 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada K. et al. Inter-mitochondrial complementation: Mitochondria-specific system preventing mice from expression of disease phenotypes by mutant mtDNA. Nature Medicine 7, 934–940 (2001). [DOI] [PubMed] [Google Scholar]
- Shitara H. et al. Selective and continuous elimination of mitochondria microinjected into mouse eggs from spermatids, but not from liver cells, occurs throughout embryogenesis. Genetics 156, 1277–1284 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K. et al. Influence of intergeneric/interspecies mitochondrial injection; parthenogenetic development of bovine oocytes after injection of mitochondria derived from somatic cells. J. Reprod. Dev. 58, 323–329 (2012). [DOI] [PubMed] [Google Scholar]
- Sato M. & Sato K. Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science 334, 1141–1144 (2011). [DOI] [PubMed] [Google Scholar]
- Sato M. & Sato K. Maternal inheritance of mitochondrial DNA by diverse mechanisms to eliminate paternal mitochondrial DNA. Biochim. Biophys. Acta 1833, 1979–1984 (2013). [DOI] [PubMed] [Google Scholar]
- Davis P. H., Schulze J. & Stanley S. L. Jr. Transcriptomic comparison of two Entamoeba histolytica strains with defined virulence phenotypes identifies new virulence factor candidates and key differences in the expression patterns of cysteine proteases, lectin light chains, and calmodulin. Mol. Biochem. Parasitol. 151, 118–128 (2007). [DOI] [PubMed] [Google Scholar]
- Liu X., Weaver D., Shirihai O. & Hajnoczky G. Mitochondrial ‘kiss-and-run’: interplay between mitochondrial motility and fusion-fission dynamics. EMBO J. 28, 3074–3089 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond L. S., Mattern C. F. & Bartgis I. L. Viruses of Entamoeba histolytica. I. Identification of transmissible virus-like agents. J. Virol. 9, 326–341 (1972). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond L. S., Harlow D. R. & Cunnick C. C. A new medium for the axenic cultivation of Entamoeba histolytica and other Entamoeba. Trans. R. Soc. Trop. Med. Hyg. 72, 431–432 (1978). [DOI] [PubMed] [Google Scholar]
- Kobayashi S. et al. Axenic cultivation of Entamoeba dispar in newly designed yeast extract iron gluconic acid dihydroxyacetone serum medium. J. Parasitol. 91, 1–4 (2005). [DOI] [PubMed] [Google Scholar]
- Nakada-Tsukui K., Tsuboi K., Furukawa A., Yamada Y. & Nozaki T. A novel class of cysteine protease receptors that mediate lysosomal transport. Cell. Microbiol. 14, 1299–1317 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozaki T. et al. Characterization of the gene encoding serine acetyltransferase, a regulated enzyme of cysteine biosynthesis from the protist parasites Entamoeba histolytica and Entamoeba dispar - Regulation and possible function of the cysteine biosynthetic pathway in entamoeba. J. Biol. Chem. 274, 32445–32452 (1999). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.