Abstract
Recent evidence indicates that bone marrow is a source of endothelial progenitor cells that are mobilized into the peripheral blood in response to cytokines or tissue injury. Previously, we showed that functional endothelial cells (ECs) can be clonally derived from phenotypically defined hematopoietic stem cells. To determine the EC potential of human bone marrow and peripheral blood stem cells, blood vessels in sex-mismatched transplant recipients were evaluated. EC outcomes were identified by using a combination of immunohistochemistry and XY interphase FISH. Donor-derived ECs were detected in the skin and gut of transplant recipients with a mean frequency of 2% and could readily be distinguished from CD45-expressing hematopoietic stem cells. None of the >4,000 ECs examined had more than two sex chromosomes, consistent with an absence of cell fusion. Y chromosome signals were not detected in sex-matched female recipients, excluding the vertical transmission of male cells. None of the recipients evaluated before hematopoietic engraftment demonstrated donor-derived ECs, indicating a close linkage between the recovery of hematopoiesis and EC outcomes. Transplantable bone marrow-derived endothelial progenitor cells may represent novel therapeutic targets for hematopoietic and vascular disease.
Keywords: endothelial cells, endothelial progenitor cells, FISH, hematopoietic stem cells
In addition to circulating blood cells, transplanted bone marrow has long been known to give rise to several hematopoietic-derived cell lineages including Kupffer cells in the liver (1), microglial cells in the central nervous system (2), and pulmonary alveolar macrophages (3). More recently, human allogeneic transplant studies have demonstrated the presence of donor markers in a variety of cell types including neurons (4, 5), Purkinje cells (6), gastrointestinal tract epithelium (7, 8), and hepatocytes (9, 10). Murine transplant studies also have reported donor-derived cells in diverse populations of nonhematopoietic cells (11–19). However, significant controversy exists as to whether these multilineage outcomes are the result of the physiologic differentiation of hematopoietic stem cells (HSCs) (20–22), fusion events between donor hematopoietic cells and host cells (23, 24), or technical artifacts associated with stem cell fate-mapping (25).
In contrast to the epithelial cell lineages, a close association between endothelial cells (ECs) and hematopoiesis was hypothesized >80 years ago (26, 27). Studies of the developing yolk sac and the dorsal aorta provided further evidence for a common precursor cell or hemangioblast (28, 29). Recent murine studies from our laboratory (30) and other laboratories (31) demonstrate that vascular endothelium and neovascularization can be clonally derived from phenotypically defined adult HSCs. We now extend these studies to sex-mismatched human female HSC recipients by using a blinded study design. Biopsy tissue from transplant recipients was evaluated by using a combination of immunohistochemistry (IHC) labeling for EC and hematopoietic markers and sequential interphase XY FISH. Donor-derived Y+ ECs were detected in the skin and gut of transplant recipients and could be readily distinguished from CD45+ hematopoietic cells. Recipients without evidence of hematopoietic engraftment did not have detectable donor-derived ECs, suggesting a close linkage between blood cell and EC outcomes.
Methods
Tissue Samples. Skin and endoscopic gut biopsies from female recipients of allogeneic bone marrow (BMT) or peripheral blood stem cells (PBSCs) were evaluated. Archived biopsy specimens originally obtained for diagnostic purposes were screened and analyzed in a blinded manner. Hematoxylin/eosin-stained sections were first reviewed (by C.L.C.) to ensure tissue integrity and the absence of significant graft-versus-host disease or other pathology. Coded biopsy specimens from 12 sex-mismatched female transplant recipients, 6 sex-matched female transplant recipients, 2 nontransplanted males, and 6 nontransplanted females were evaluated. Analysis of this tissue was approved by the institutional review board of Oregon Health and Science University.
Sequential IHC and XY FISH. To determine the sex-chromosome genotype of ECs and lymphocytes within tissue sections, we developed a sequential IHC/FISH approach. Sections were stained first with IHC for EC markers [CD31 or von Willebrand factor (VWF), with diaminobenzidine (DAB) chromagen] and a lymphocyte marker (CD45, with Vector Blue, Vector Laboratories), then processed for dual-color, interphase FISH for chromosomes X and Y. We found that the IHC chromagens were relatively resistant to the pepsin digestion, high-temperature treatments, and detergent washes required for FISH. Moreover, the DAB generated an epifluorescent signal that was readily observed through the red channel on a confocal microscope (1024 ES, Bio-Rad). Details on each step in the protocol are provided below.
IHC. Five-micrometer sections of paraffin-embedded skin and gut biopsy specimens were deparaffinized and rehydrated. Antigen retrieval was performed with 0.01 M citrate buffer (Citra HK086-9K, BioGenex Laboratories, San Ramon, CA) in a vegetable steamer unit for 30 min. Wash buffer was prepared with Tris-buffered saline containing 0.1% Tween 20 (BP337, Fisher) and 0.01% sodium azide (S 2002, Sigma). Slides were washed once in wash buffer, then incubated in blocking buffer (1% BSA in PBS; P 3688, Sigma) for 5 min at room temperature. The slides were then incubated with monoclonal mouse anti-human CD31 Ab (clone JC70A, DAKO) at a dilution of 1:150 in blocking buffer or polyclonal rabbit anti-human VWF (A0082, DakoCytomation, Carpinteria, CA) at a dilution of 1:4,000 in blocking buffer for 45 min at room temperature. Sections were rinsed and incubated for 30 min with an appropriate biotinylated secondary Ab in blocking buffer (anti-mouse IgG plus IgM at 1:400, BA-2000 plus BA-2020, or anti-rabbit at 1:400, BA-1000, Vector Laboratories). Slides were then treated with a 3% hydrogen peroxide solution to block endogenous peroxidase (5 min). After additional rinses, the slides were incubated with avidin–biotin complex (ABC Elite kit, PK-6100, Vector Laboratories), and staining was visualized by using DAB (K3466, DakoCytomation). The slides were then placed in PBS and incubated at 60°C for 30 min to remove the previous antibodies, preventing cross-reaction in the second step. To detect tissue leukocytes, a second IHC staining reaction was performed on the same tissue sections. The blocking-buffer step was repeated. Specimens were incubated with monoclonal mouse antileukocyte common antigen (CD45, LCA) (clone PD7/26 and 2B11, DAKO) at a dilution of 1:400 for 45 min, followed by incubation with biotinylated goat anti-mouse Ab IgG (BA-2020, Vector Laboratories) for 30 min. At this point, the slides were rinsed in Tris-buffered saline (pH 8.2) containing levamisole without sodium azide (one drop per 5 ml; SP-5000, Vector Laboratories) and incubated for 30 min with streptavidin alkaline phosphatase (AP) conjugate (SA-AP, Southern Biotechnology Associates) at a dilution of 1:800 in the same TBS buffer. Lastly, the Vector Blue Substrate kit (SK-5300, Vector Laboratories) was used to detect the alkaline phosphatase.
fish. Interphase FISH was carried out by using the CEP X,Y DNA probe kit (Vysis, Downers Grove, IL). Pretreatment was in 8% sodium thiocyanate solution for 20 min at 80°C, followed by rinsing and incubation for 15 min at 37°C in 0.2% pepsin (porcine stomach mucosa; 3,660 units per mg) in 0.2 M HCl. After washing, tissue sections were denatured in 70% formamide/2× SSC (1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7) at 73°C for 5 min. Slides were dehydrated with graded alcohols for 30 s each and air-dried. Seven microliters of probe mixture was added to one target area of tissue and immediately covered with glass coverslips. Slides were placed in a humidified chamber at 42°C for 16 h. After hybridization, slides were washed twice in 4× SSC/0.1% Nonidet P-40 and 2× SSC at 46°C for 8 min each. Sections were air-dried in the dark and counterstained with 10 μl of DAPI II (Vysis) before coverslipping. The X and Y chromosomes were visualized directly by using standard fluorescence filters for SpectrumOrange (Cy3) and SpectrumGreen (FITC), respectively. A DAPI II (Vysis) nuclear counterstain (blue color) mounting medium was used to determine that positive X and Y signals were colocalized to the same nucleus and, thus, the same cell.
Enumeration of Donor-Derived ECs. Tissue sections were systematically examined by two independent observers and photographed with a microscope (Axiophot, Zeiss) using a true-color camera (AxioCam, Zeiss) and standard epifluorescence filters for FITC, Cy3, and DAPI (Zeiss). Images were digitally combined by using axiovision 4.0 (Zeiss). Vascular endothelium was identified by morphology and the intensity/density of DAB staining. The total number of donor-derived ECs at ×1,250 magnification was obtained by counting all X and Y chromosomes on nuclei in cells that were both CD31+ and CD45- or VWF+ and CD45-. Colocalization of FISH signals in selected individual cells was confirmed by Z-stack analysis. ECs in contact with CD45+ hematopoietic cells were not enumerated.
Image Analysis. Fluorescent images of endothelium that contained male donor-derived ECs and X and Y chromosomes were obtained at ×1,250 by using a fluorescent microscope (Axioplan, Zeiss). Bright-field images of these ECs at the same magnification and field were immediately captured. Images were analyzed by using axiovision 4.0. To show true colocalization of FISH signals and EC markers in a single cell, Z-stack images were obtained by using a laser scanning microscope (1024 ES, Bio-Rad) and analyzed by using deltavision deconvolution software (Bio-Rad).
Results
Patient Characteristics. The Oregon Health and Science University BMT program database was used to identify sex-matched and sex-mismatched female BMT recipients. To ensure adequate tissue integrity, archived skin and gut biopsy specimens were reviewed (by C.L.C.) for significant graft-versus-host disease or other pathology. Suitable tissue samples were coded and analyzed in a blinded manner for the presence of Y+ ECs. Skin biopsies from 18 female recipients of BMT or PBSCs for the treatment of hematologic malignancies were studied. The characteristics of the 12 female recipients of sex-mismatched BMT (n = 7) or PBSC (n = 5) are shown in Table 1. The time from transplant to tissue biopsy ranged from 9 to 2,776 days (mean of 355 days). None of the specimens from the six recipients of female BMT or female PBSC had detectable Y chromosomes in any cell type. Nine of 12 sex-mismatched recipients demonstrated ECs containing a Y chromosome. Marrow chimerism studies were performed on seven of nine sex-mismatched transplant patients who demonstrated donor-derived endothelium. Six of these patients had 100% donor-derived hematopoiesis, whereas the remaining patient had relapsed acute leukemia with only 27.5% donor chimerism. The remaining three patients did not have a detectable Y chromosome in any cell type in the biopsy specimens. A review of the clinical records revealed that these three patients failed to show hematopoietic engraftment at the time of tissue biopsy.
Table 1. Patient characteristics and endothelial cell outcomes.
| UPN | Disease | Donor | Stem cell source | Days posttransplant | GVHD grade | Hematopoietic engraftment | Donor endothelium |
|---|---|---|---|---|---|---|---|
| 1 | CML | URD | BM | 73 | None | Yes | + |
| 2 | AML | URD | BM | 32 | None | Yes | + |
| 3 | AML | URD | BM | 252 | None | Yes | + |
| 4 | NHL | Sibling | PBSC | 50 | None | Yes | + |
| 5 | CML | Sibling | PBSC | 49 | I/II | Yes | + |
| 6 | AML | URD | PBSC | 9 | I/II | Yes | + |
| 7 | APL | URD | BM | 898 | None | Yes | + |
| 8 | AML | URD | PBSC | 81 | I/II | Yes | + |
| 9 | CML | Sibling | BM | 2,766 | None | Yes | + |
| 10 | AML | URD | BM | 14 | None | No | - |
| 11 | AML | URD | BM | 14 | None | No | - |
| 12 | AML | Sibling | PBSC | 18 | None | No | - |
Graft-versus-host disease (GVHD) prophylaxis: Patients 2, 3, and 11 received tacrolimus and methotrexate. All others received cyclosporine, methotrexate, and prednisone. Preparative regimens: Patients 2, 3, and 11 received fludarabine, cyclophosphamide, antithymocyte globulin, methylprednisolone, and total body irradiation. Patients 6 and 10 received busulfan/cyclophosphamide. All other patients received cyclophosphamide/total body irradiation. UPN, unique patient number; CML, chronic myeloid leukemia; AML, acute myeloid leukemia; NHL, non-Hodgkin's lymphoma; APL, acute promyelocytic leukemia; URD, unrelated donor; BM, bone marrow.
Detection of Donor-Derived ECs. To definitively identify the EC cell progeny of male donor-derived cells, we used sequential IHC and FISH. This combination of techniques allowed us to visualize X and Y chromosomes in phenotypically identified ECs and to then use Z-stack analysis to confirm the colocalization of these markers in an individual cell. With bright-field microscopy, perivascular and intravascular CD45+ leukocytes (Vector Blue) were easily distinguished from CD31- and VWF-expressing vascular endothelium labeled with DAB (Fig. 1A). Importantly, both Vector Blue and DAB staining patterns remained visible after the pepsin treatment and the hybridization procedures used to detect the X and Y chromosomes. In addition, the intense DAB signal associated with VWF staining of ECs produced an epifluorescent signal in the red channel that was readily detected by confocal microscopy (Fig. 1H). By using centromeric probes, Y chromosomes in DAPI-stained nuclei were clearly visualized as discrete green signals, whereas the X probes generated a characteristic red signal. FISH analysis revealed two sex chromosomes in 60% of ECs, which is the expected frequency given the 5-μm thickness of the sections used (8). This finding was confirmed by evaluating sequential images within a Z-stack (Fig. 2A).
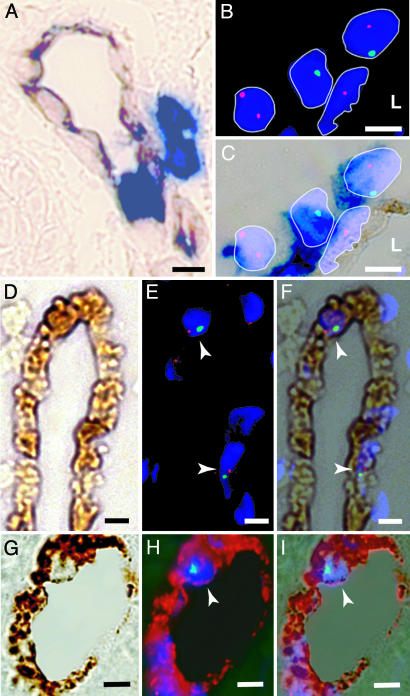
Fig. 1.
Detection of donor-derived ECs and hematopoietic cells in skin biopsies. (A) Skin sections from a sex-mismatched transplant recipient demonstrating the expression of CD31 (brown) and CD45 (blue). Both perivascular and intravascular CD45+ leukocytes are present. (B) Fluorescence image of interphase FISH showing X (red) and Y (green) chromosomes in four DAPI-stained nuclei (blue). Two donor-derived cells (XY, YO) and one host cell (XX) are shown behind an XO cell. (L, lumen). (C) Combined bright-field image of the FISH+ cells in B. DAPI-stained nuclei appear violet, and CD45 expression (dark blue) is present on all three perivascular cells. Because of the close proximity of these CD45+ perivascular cells, the CD45 status of the elongated nucleus in the vessel wall is considered indeterminate. (D–F) Detection of XY donor-derived CD31+ ECs. (D) Bright-field image of a vessel showing CD31 expression (brown) and an absence of CD45 expression (blue). (E) Fluorescence image of XY FISH demonstrating 2 XY cells (arrowheads, one red signal and one green signal) with DAPI-labeled nuclei (blue). (F) Merged image of D and E.(G–I) Detection of XY donor-derived VWF+ ECs in skin. (G) Bright-field image showing VWF expression (brown) and an absence of CD45 expression (blue). (H) Confocal image of the same vessel, showing Y (green) chromosome and DAPI-labeled nucleus (arrowhead, blue). Note that the intense VWF DAB signal produces a red epifluorescent signal. (I) Merged image of G and H. (Scale bar: 5 μm.)
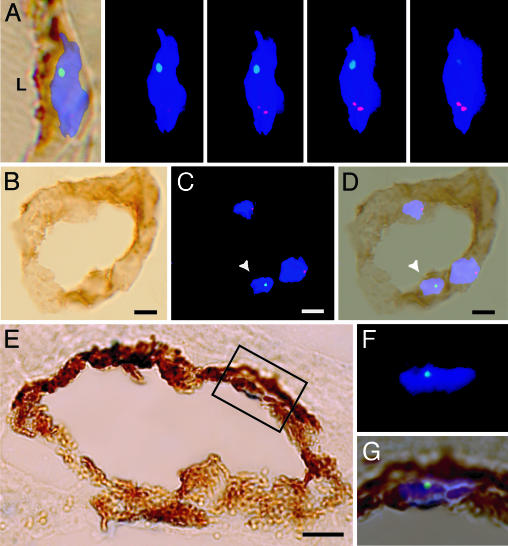
Fig. 2.
Donor-derived ECs in the gut. (A Left) A merged bright-field image of CD31 (brown) and an XY FISH image. A series of 4 Z-stack images of the same ECs taken at 0.5-μm intervals demonstrating an X chromosome (red) and a Y chromosome (green) in a DAPI+ nucleus (blue). (B) Bright-field image of a blood vessel in the gut (duodenum) demonstrating the expression of CD31 (brown) and an absence of CD45+ cells (blue). (C) Fluorescence image of the vessel in B with Y chromosome (green, arrowhead), X chromosome (red), and DAPI-stained nuclei (blue). (D) Merged image of B and C.(E) Bright-field image of a gastric blood vessel with VWF expression (brown) and an absence of CD45 expression (blue). (F) High magnification of the fluorescent image of the region indicated in E. (G) Merged image of E and F. (Scale bar: 5 μm.)
By combining the IHC for CD31 and CD45 with Y and X chromosome FISH, we found clear evidence of donor-derived Y+ ECs in all nine recipients that showed hematopoietic engraftment after sex-mismatched BMT or PBSC transplant (Table 2). Donor-derived ECs expressing CD31 or VWF also were readily detected in gut biopsies (Fig. 2 B–G). Cells of hematopoietic origin, including intraepithelial lymphocytes, were excluded by the presence of CD45 expression.
Table 2. Detection of Y+ donor endothelial cells.
| Endothelial cell number
|
|||||||
|---|---|---|---|---|---|---|---|
| Patient number | XX | XY | XO | OY | Total | %Y+ | %Y Corrected* |
| 1 | 134 | 3 | 91 | 2 | 230 | 2.2 | 2.8 |
| 2 | 177 | 5 | 116 | 4 | 302 | 3.0 | 3.8 |
| 3 | 138 | 8 | 106 | 2 | 254 | 3.9 | 5.0 |
| 4 | 175 | 2 | 111 | 2 | 290 | 1.4 | 1.8 |
| 5 | 196 | 3 | 122 | 0 | 321 | 0.9 | 1.1 |
| 6 | 196 | 7 | 106 | 2 | 311 | 2.9 | 3.7 |
| 7 | 152 | 5 | 100 | 2 | 259 | 2.7 | 3.4 |
| 8 | 114 | 4 | 112 | 0 | 230 | 1.7 | 2.2 |
| 9 | 173 | 5 | 130 | 2 | 310 | 2.3 | 2.9 |
| 10 | 135 | 0 | 114 | 0 | 249 | No Y | - |
| 11 | 142 | 0 | 68 | 0 | 210 | No Y | - |
| 12 | 103 | 0 | 98 | 0 | 201 | No Y | - |
“No Y” indicates no Y chromosomes detected in any cell type. (Patients 10-12 had a WBC count of <0.3.)
Correction factor of 1.27
Frequency of Y+ Donor-Derived ECs in Skin Biopsies. Each recipient tissue sample was assayed a minimum of two times by sequential IHC and FISH. The frequency of CD31+, VWF+, CD45-, or Y+ ECs in skin biopsies from 18 female transplant recipients was evaluated in a blinded manner by two independent observers. Only ECs with a DAPI-stained nucleus and at least one sex chromosome were enumerated. The common occurrence of a single sex chromosome is due to the plane of section examined, a finding consistent with previous reports (8). Z-stack images of X and Y chromosomes in a single EC nuclei obtained at 0.5-μm intervals illustrates this point (Fig. 2A). In male control skin biopsies, 79% of the total nuclei contained a single Y- chromosome. Consequently, the frequency of male donor-derived ECs represents a 21% underestimate. Y+ ECs were not detected in nine transplant recipients. On review of the clinical records, it was found that six of these patients had been transplanted with female donor cells. In the remaining three recipients who received sex-mismatched donor cells, Y chromosomes were not detected in any cell type, including intradermal lymphocytes (frequency of <1 in 10,000 cells, Table 2). None of these patients demonstrated hematopoietic engraftment, with WBC counts of <300 cells per μl. In the nine engrafted female patients who received sex-mismatched BMT or PBSC cells, 0.9–3.9% of the ECs showed a donor genotype (mean 2.3% ± 0.9%; Table 2). No regional predilection for the incorporation of endothelial progenitor cells was observed in vessels of either the skin or gut.
Donor-Derived ECs in the Gut. To determine whether donor-derived ECs were integrated into the endothelium of other tissues, biopsies of gastric and duodenal mucosa were examined (Fig. 2 B–G). Tissue samples determined to have minimal graft-versus-host disease or other pathology were evaluated by sequential IHC and FISH as described above. CD45+ lymphoid cells were readily detected and excluded from analysis. Y+, CD31+, VWF+, and CD45- ECs were found in the gut of four out of four patients examined, at a mean frequency of 1.7%, a level similar to that observed in skin biopsies.
Donor ECs Are Diploid. Recent studies of regenerating liver in a mouse model of hereditary tyrosinemia indicate that functional hyperdiploid hepatocytes can arise from the fusion of donor bone marrow-derived cells with recipient hepatocytes (23, 32). This important issue was addressed prospectively by enumerating the frequency of X and Y chromosome signals in all FISH-probed EC nuclei. Among the >4,000 EC nuclei examined in skin biopsies from 18 transplant receipts, none demonstrated more than two sex chromosomes. Although this finding does not rule out rare fusion events, it indicates that the vast majority of EC outcomes in the skin are not hyperdiploid.
Discussion
The results of this study demonstrate the presence of donor-derived endothelium in both the skin and gastrointestinal tract of allogeneic stem cell transplantation recipients. Donor ECs were detected at the time of early hematopoietic engraftment (day 9) and persisted for >7 years. A similar frequency of donor ECs was observed in recipients of bone marrow cells or PBSC. Of note, the frequency of EC outcomes may actually have been reduced in these patients, because they were all treated with a calcineurin inhibitor (cyclosporine or tacrolimus). These agents have been reported to impede the migration and incorporation of endothelial progenitor cells into vessels (33). Our findings are consistent with previous studies demonstrating endothelial progenitor cell activity in the blood in both experimental transplantation models and in vitro assays of circulating endothelial progenitor cells (34, 35). No donor ECs were identified in recipients who were evaluated before hematopoietic engraftment, consistent with the hypothesis that EC potential and hematopoiesis are functionally linked (27).
Using Y chromosome FISH to identify donor cells consistently leads to an underestimate of the frequency of Y+ cells because of the plane of section examined (Fig. 2A). Evaluation of three normal male control biopsy specimens indicated that 79% of EC nuclei were either XY or YO. By using a correction factor of 1.27, the frequency of donor-derived ECs in skin biopsies in our study ranges from 1.1% to 5.0%. The presence of Y chromosomes in females could theoretically result from blood-product transfusion (36) or by passive transfer through the fetal–maternal circulation (37). Typically, the frequency of Y+ cells posttransfusion is very low unless there is significant transfusion-related graft-versus-host disease (38). Similarly, a sensitive PCR assay often is required to detect the rare male cells acquired by fetal–maternal transfer. By contrast, our study has a detection level of ≈0.5% only. None of the six female recipients of female BMT or PBSC and none of the eight nontransplant female controls demonstrated Y chromosome signals in any cell type examined by interphase FISH. These results effectively eliminate concerns about the vertical transmission of the Y chromosome as an explanation of Y+ cells in our study. Furthermore, the biopsy specimens obtained before hematopoietic engraftment did not demonstrate any detectable Y signals despite transplantation of 1.9 × 1010-6.2 × 1010 mononuclear male donor cells. Taken together, these data indicate that the vertical transfer of Y chromosome-containing cells is very unlikely to be the mechanism responsible for the presence of male ECs.
Some early studies of the transdifferentiation of bone marrow-derived stem cells are now recognized as examples of fusion between populations of donor and host cells. Cell fusion has been reported as an important mechanism for generating hepatocytes (23, 32), and for repairing skeletal (39), epithelium (40), and cardiac muscle (24). However, our analysis of >4,000 EC nuclei, using quantitative centromeric X and Y probes, did not detect a single EC with more than two sex chromosomes. Although this result does not entirely rule out rare fusion events between donor hematopoietic cells and host ECs, or a fusion event followed by a reduction division (23), it clearly demonstrates that the great majority of endothelial outcomes in the skin are not hyperdiploid.
Previous work evaluating sex-mismatched transplant recipients reported a high frequency of donor-derived ECs in the bone marrow. Although Kvasnicka et al. (41) reported that 19–24% of bone marrow endothelium in transplant recipients was donor-derived, these investigators used morphology and the expression of CD34 as the only criteria for the identification of bone marrow ECs. The interpretation of these results is problematic, however, because significant numbers of total bone marrow mononuclear cells (1–2%) express CD34, and the majority of these cells are known to be hematopoietic progenitors (42). Blood vessel morphology in these bone marrow biopsies was relatively indistinct, raising the concern that perivascular CD34+ hematopoietic cells were inadvertently scored as ECs. (Of note, the patients examined in this study and our own patients all received myeloablative conditioning regimens. Consequently, the incorporation of donor cells into the endothelium probably did not occur under steady-state conditions.)
Another group of investigators identified ECs in the lung by using CD31 and CD45 expression in combination to exclude pulmonary alveolar macrophages and passenger leukocytes (43). Serial sections of lung biopsy specimens were obtained and examined by hematoxylin/eosin staining. Morphologically identified vascular structures were matched to an adjacent tissue section for the localization of CD31 and CD45 by immunofluorescence microscopy. FISH for the X and Y chromosomes was then performed on CD31 and CD45 double-stained sections. Unfortunately, the FISH procedure resulted in the complete loss of fluorescence of the CD31 and CD45 markers, so that the identification of putative Y+ ECs was made by morphology alone. This approach precluded the definitive colocalization of CD31 and the Y chromosome marker in a single cell by confocal microscopy. In our experience, combinations of lineage-specific cell markers and the absence of the expression of the panhematopoietic marker CD45 are required to distinguish nonhematopoietic cells from tissue-resident hematopoietic cells.
Our results also demonstrate that endothelial engraftment does not occur in the absence of initial hematopoietic engraftment, suggesting a functional linkage between bone marrow-derived hematopoiesis and EC potential. This finding is consistent with the report of human bone marrow-derived multipotent adult progenitor cells, a population that exhibits endothelial progenitor cell activity (44). Analysis of bone marrow-derived ECs in human BMT recipients also reveals significant donor-derived endothelial progenitor cell activity in the peripheral circulation (35). Furthermore, Pelosi et al. (45) showed that single human CD34+ KDR+ stem cells have the potential to differentiate into hematopoietic cells and ECs at the clonal level in vitro. Taken together, these results suggest that the close association between hematopoiesis and the vascular system originally identified during early development persists into adult life. Consequently, it will be important to study potential therapeutic roles of bone marrow-derived ECs in the setting of vascular and hematopoietic disease.
Acknowledgments
We thank Aurelie Snyder for assistance with confocal microscopy and Carolyn Gendron for assistance with histopathology. This work was performed in the Oregon Health and Science University Cancer Institute National Cancer Institute Pathology Core Facility and was supported by National Institutes of Health Grants HL 069133 and HD 39251 (to W.H.F.).
Author contributions: S.J., L.W., M.A., C.L.C., and W.H.F. designed research; S.J., L.W., M.A., D.A.A., and L.J.-M. performed research; S.J. contributed new reagents/analytic tools; S.J., L.W., M.A., D.A.A., L.J.-M., C.L.C., and W.H.F. analyzed data; and S.J., L.W., D.A.A., and W.H.F. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: BMT, allogeneic bone marrow; DAB, diaminobenzidine; EC, endothelial cell; HSC, hematopoietic stem cell; IHC, immunohistochemistry; PBSC, peripheral blood stem cell; VWF, von Willebrand factor.
References
- 1.Gale, R. P., Sparkes, R. S. & Golde, D. W. (1978) Science 201, 937-938. [DOI] [PubMed] [Google Scholar]
- 2.Unger, E. R., Sung, J. H., Manivel, J. C., Chenggis, M. L., Blazar, B. R. & Krivit, W. (1993) J. Neuropathol. Exp. Neurol. 52, 460-470. [DOI] [PubMed] [Google Scholar]
- 3.Venuat, A. M., Marinakis, T., Bourhis, J. H., Bayle, C. & Pico, J. L. (1994) Bone Marrow Transplant. 14, 177-179. [PubMed] [Google Scholar]
- 4.Mezey, E., Key, S., Vogelsang, G., Szalayova, I., Lange, G. D. & Crain, B. (2003) Proc. Natl. Acad. Sci. USA 100, 1364-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cogle, C. R., Yachnis, A. T., Laywell, E. D., Zander, D. S., Wingard, J. R., Steindler, D. A. & Scott, E. W. (2004) Lancet 363, 1432-1437. [DOI] [PubMed] [Google Scholar]
- 6.Weimann, J. M., Charlton, C. A., Brazelton, T. R., Hackman, R. C. & Blau, H. M. (2003) Proc. Natl. Acad. Sci. USA 100, 2088-2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okamoto, R., Yajima, T., Yamazaki, M., Kanai, T., Mukai, M., Okamoto, S., Ikeda, Y., Hibi, T., Inazawa, J. & Watanabe, M. (2002) Nat. Med. 8, 1011-1017. [DOI] [PubMed] [Google Scholar]
- 8.Spyridonidis, A., Schmitt-Graff, A., Tomann, T., Dwenger, A., Follo, M., Behringer, D. & Finke, J. (2004) Am. J. Pathol. 164, 1147-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Theise, N. D., Nimmakayalu, M., Gardner, R., Illei, P. B., Morgan, G., Teperman, L., Henegariu, O. & Krause, D. S. (2000) Hepatology 32, 11-16. [DOI] [PubMed] [Google Scholar]
- 10.Korbling, M., Katz, R. L., Khanna, A., Ruifrok, A. C., Rondon, G., Albitar, M., Champlin, R. E. & Estrov, Z. (2002) N. Engl. J. Med. 346, 738-746. [DOI] [PubMed] [Google Scholar]
- 11.Krause, D. S., Theise, N. D., Collector, M. I., Henegariu, O., Hwang, S., Gardner, R., Neutzel, S. & Sharkis, S. J. (2001) Cell 105, 369-377. [DOI] [PubMed] [Google Scholar]
- 12.Masuya, M., Drake, C. J., Fleming, P. A., Reilly, C. M., Zeng, H., Hill, W. D., Martin-Studdard, A., Hess, D. C. & Ogawa, M. (2003) Blood 101, 2215-2218. [DOI] [PubMed] [Google Scholar]
- 13.Orlic, D., Kajstura, J., Chimenti, S., Jakoniuk, I., Anderson, S. M., Li, B., Pickel, J., McKay, R., Nadal-Ginard, B., Bodine, D. M., et al. (2001) Nature 410, 701-705. [DOI] [PubMed] [Google Scholar]
- 14.Ferrari, G., Cusella-De Angelis, G., Coletta, M., Paolucci, E., Stornaiuolo, A., Cossu, G. & Mavilio, F. (1998) Science 279, 1528-1530. [DOI] [PubMed] [Google Scholar]
- 15.Bittner, R. E., Schofer, C., Weipoltshammer, K., Ivanova, S., Streubel, B., Hauser, E., Freilinger, M., Hoger, H., Elbe-Burger, A. & Wachtler, F. (1999) Anat. Embryol. 199, 391-396. [DOI] [PubMed] [Google Scholar]
- 16.Mezey, E., Chandross, K. J., Harta, G., Maki, R. A. & McKercher, S. R. (2000) Science 290, 1779-1782. [DOI] [PubMed] [Google Scholar]
- 17.Priller, J., Flugel, A., Wehner, T., Boentert, M., Haas, C. A., Prinz, M., Fernandez-Klett, F., Prass, K., Bechmann, I., de Boer, B. A., et al. (2001) Nat. Med. 7, 1356-1361. [DOI] [PubMed] [Google Scholar]
- 18.Lagasse, E., Connors, H., Al Dhalimy, M., Reitsma, M., Dohse, M., Osborne, L., Wang, X., Finegold, M., Weissman, I. L. & Grompe, M. (2000) Nat. Med. 6, 1229-1234. [DOI] [PubMed] [Google Scholar]
- 19.Theise, N. D., Badve, S., Saxena, R., Henegariu, O., Sell, S., Crawford, J. M. & Krause, D. S. (2000) Hepatology 31, 235-240. [DOI] [PubMed] [Google Scholar]
- 20.Anderson, D. J., Gage, F. H. & Weissman, I. L. (2001) Nat. Med. 7, 393-395. [DOI] [PubMed] [Google Scholar]
- 21.Lemischka, I. (2002) Exp. Hematol. 30, 848-852. [DOI] [PubMed] [Google Scholar]
- 22.Wagers, A. J., Sherwood, R. I., Christensen, J. L. & Weissman, I. L. (2002) Science 297, 2256-2259. [DOI] [PubMed] [Google Scholar]
- 23.Wang, X., Willenbring, H., Akkari, Y., Torimaru, Y., Foster, M., Al Dhalimy, M., Lagasse, E., Finegold, M., Olson, S. & Grompe, M. (2003) Nature 422, 897-901. [DOI] [PubMed] [Google Scholar]
- 24.Nygren, J. M., Jovinge, S., Breitbach, M., Sawen, P., Roll, W., Hescheler, J., Taneera, J., Fleischmann, B. K. & Jacobsen, S. E. (2004) Nat. Med. 10, 494-501. [DOI] [PubMed] [Google Scholar]
- 25.Jackson, K. A., Snyder, D. S. & Goodell, M. A. (2004) Stem Cells 22, 180-187. [DOI] [PubMed] [Google Scholar]
- 26.Sabin, F. R. (1920) Contrib. Embryol. 9, 213-262. [Google Scholar]
- 27.Bailey, A. S. & Fleming, W. H. (2003) Exp. Hematol. 31, 987-993. [DOI] [PubMed] [Google Scholar]
- 28.Haar, J. L. & Ackerman, G. A. (1971) Anat. Rec. 170, 199-223. [DOI] [PubMed] [Google Scholar]
- 29.Tavian, M., Coulombel, L., Luton, D., Clemente, H. S., Dieterlen-Lievre, F. & Peault, B. (1996) Blood 87, 67-72. [PubMed] [Google Scholar]
- 30.Bailey, A. S., Jiang, S., Afentoulis, M., Baumann, C. I., Schroeder, D. A., Olson, S. B., Wong, M. H. & Fleming, W. H. (2004) Blood 103, 13-19. [DOI] [PubMed] [Google Scholar]
- 31.Grant, M. B., May, W. S., Caballero, S., Brown, G. A., Guthrie, S. M., Mames, R. N., Byrne, B. J., Vaught, T., Spoerri, P. E., Peck, A. B., et al. (2002) Nat. Med. 8, 607-612. [DOI] [PubMed] [Google Scholar]
- 32.Willenbring, H., Bailey, A. S., Foster, M., Akkari, Y., Dorrell, C., Olson, S., Finegold, M., Fleming, W. H. & Grompe, M. (2004) Nat. Med. 10, 744-748. [DOI] [PubMed] [Google Scholar]
- 33.Hernandez, G. L., Volpert, O. V., Iniguez, M. A., Lorenzo, E., Martinez-Martinez, S., Grau, R., Fresno, M. & Redondo, J. M. (2001) J. Exp. Med. 193, 607-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takahashi, T., Kalka, C., Masuda, H., Chen, D., Silver, M., Kearney, M., Magner, M., Isner, J. M. & Asahara, T. (1999) Nat. Med. 5, 434-438. [DOI] [PubMed] [Google Scholar]
- 35.Lin, Y., Weisdorf, D. J., Solovey, A. & Hebbel, R. P. (2000) J. Clin. Invest. 105, 71-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee, T. H., Paglieroni, T., Ohto, H., Holland, P. V. & Busch, M. P. (1999) Blood 93, 3127-3139. [PubMed] [Google Scholar]
- 37.Bianchi, D. W., Zickwolf, G. K., Weil, G. J., Sylvester, S. & DeMaria, M. A. (1996) Proc. Natl. Acad. Sci. USA 93, 705-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Collins, R. H., Jr., Anastasi, J., Terstappen, L. W., Nikaein, A., Feng, J., Fay, J. W., Klintmalm, G. & Stone, M. J. (1993) N. Engl. J. Med. 328, 762-765. [DOI] [PubMed] [Google Scholar]
- 39.Camargo, F. D., Green, R., Capetenaki, Y., Jackson, K. A. & Goodell, M. A. (2003) Nat. Med. 9, 1520-1527. [DOI] [PubMed] [Google Scholar]
- 40.Spees, J. L., Olson, S. D., Ylostalo, J., Lynch, P. J., Smith, J., Perry, A., Peister, A., Wang, M. Y. & Prockop, D. J. (2003) Proc. Natl. Acad. Sci. USA 100, 2397-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kvasnicka, H. M., Wickenhauser, C., Thiele, J., Varus, E., Hamm, K., Beelen, D. W. & Schaefer, U. W. (2003) Leuk. Lymphoma 44, 321-328. [DOI] [PubMed] [Google Scholar]
- 42.Civin, C. I. & Small, D. (1995) Ann. N.Y. Acad. Sci. 770, 91-98. [DOI] [PubMed] [Google Scholar]
- 43.Suratt, B. T., Cool, C. D., Serls, A. E., Chen, L., Varella-Garcia, M., Shpall, E. J., Brown, K. K. & Worthen, G. S. (2003) Am. J. Respir. Crit. Care Med. 168, 318-322. [DOI] [PubMed] [Google Scholar]
- 44.Reyes, M., Dudek, A., Jahagirdar, B., Koodie, L., Marker, P. H. & Verfaillie, C. M. (2002) J. Clin. Invest. 109, 337-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pelosi, E., Valtieri, M., Coppola, S., Botta, R., Gabbianelli, M., Lulli, V., Marziali, G., Masella, B., Muller, R., Sgadari, C., et al. (2002) Blood 100, 3203-3208. [DOI] [PubMed] [Google Scholar]