Abstract
Heterogeneity among Helicobacter pylori strains in gastric epithelial adherence is postulated to contribute to pathogen fitness in the physiologically diverse human population. H. pylori adherence to ABO and Lewis b (Leb) blood group antigens in the human stomach is mediated by the blood group antigen-binding adhesin BabA. Approximately 70% of Swedish and U.S. H. pylori clinical isolates exhibit Leb binding, but here we show that the babA gene is present in each of 10 Leb-nonbinding strains. Fluorescence microscopy identified occasional bacterial cells with a Leb-binding phenotype in populations of Leb-nonbinding strains. Thus, nonbinding seemed to be a metastable phenotype. To model metastable transition into the virulence-associated Leb-binding mode, Leb-binding clones were isolated from nonadherent strains by panning with Leb-magnetic beads and characterized. Strain 17875 has two babA genes, babA1 (silent) and babA2 (expressed). We found that a babA2-cam derivative of strain 17875 regained Leb binding by recombination of the formerly silent babA1 gene into the expressed and partially homologous babB locus. The chimeric BabB/A adhesin binds Leb with an affinity similar to that of wild-type BabA adhesin, but its expression level was lower and was subject to phase variation through slipped-strand mispairing. Equivalent results were obtained with strain NCTC11638. We propose that adhesin metastability and heterogeneity contributes to bacterial fitness and results in some clones having potential for periodic activation and deactivation of virulence appropriate for intensity of the host response to infection.
Keywords: BabA, Hop, SabA, phase variation
Helicobacter pylori persistently infects the human stomach and causes chronic inflammation that can lead to peptic ulcer disease and gastric cancer (1). Both bacterial and host genetic determinants probably contribute to these various disease outcomes. Bacterial adherence is an important contributor to the vigor of infection and virulence. Functional receptors for H. pylori adherence include fucosylated ABO blood group antigens (2, 3) and sialyl-Lewis x/a antigens (4). These receptors are recognized by the blood group antigen-binding adhesin BabA, which binds Lewis b antigen (Leb) and related ABO antigens (3, 5), and the sialic acid-binding adhesin, SabA (4). To date, BabA–Leb is the best-characterized of H. pylori's several adhesin–receptor interactions (3, 5–7).
The local gastric environment changes during infection because of inflammatory responses and effector molecules produced by infecting H. pylori cells. H. pylori must have a remarkable capacity to adapt to such changes because infections tend to persist for life once established. Several reports have emphasized the high genetic diversity in H. pylori populations and the unusually high level of recombination both within and between strains (8–11). This genetic diversity ensures bacterial phenotype heterogeneity and suggests that subclones of any given bacterial lineage have adapted to alterations in host environment (12–15), including the changes in mucosal glycosylation patterns during acute and established chronic stages of persistent infection.
The BabA and the SabA adhesins belong to a large family of H. pylori outer-membrane proteins (Hop) (16). Some hop genes are duplicates, and others exhibit extensive homologies at their 5′ and 3′ ends that should facilitate frequent recombination between them (17, 18). For example, reference strain CCUG17875 (here denoted 17875) has two babA genes: babA2, from which the BabA adhesin is expressed, and babA1, which is silent because of an incomplete signal peptide (5). The babB gene is highly homologous to babA at its 5′ and 3′ ends but not at the central region that (in babA) determines the specificity of receptor binding (3, 5, 19). Other hop genes, babB included, contain repetitive sequence motifs that are prone to frameshift mutation and thereby metastable hop gene expression (17, 18, 20). Most disease-associated H. pylori strains express the BabA adhesin, whereas many of those implicated in more benign infections do not (5–7). Here we report that Leb-nonbinding strains also usually possess silent babA gene sequences, which can be activated by recombination into the babB locus. Once at this second locus, a BabB/A chimeric adhesin is expressed at lower levels and is subject to frameshift-based phase variation (ON/OFF switching). These changes in expression illustrate that bacterial adherence properties can be modulated by inclusion in expression loci of various strengths and plasticities. We propose that H. pylori populations use these mechanisms for quantitative modulation of adhesive properties during cycles of periodic activation and deactivation. Such flexibility, in turn, should contribute to the extraordinary persistence of H. pylori, compensate for changes in inflammation and other host responses, and impact significantly on the potential virulence of infection.
Methods
H. pylori Strains. CCUG17875, 17875ΔbabA1::cam, 17875ΔbabA2::cam (5), 17875ΔbabA1::kanΔbabA2::cam (4), 17875ΔbabB::cam (this study), NCTC11638 (21), and low-passage Swedish clinical isolates were obtained as sweeps of gastric biopsies (5). Reconstruction tests have shown that most of these sweeps are as highly uniform as if they were from single colonies. Bacteria were grown at 37°C in 5% O2/10% CO2 on Brucella agar. Escherichia coli strains were cultured in Luria broth/agar at 37°C. A babB knockout mutant was constructed in strain 17875. The babB gene was PCR-amplified by using the primers BF and A39R (Table 2, which is published as supporting information on the PNAS web site) and cloned in pBluescript SK+/- at the EcoRV site. The plasmid clone was then linearized with primers A14R and A36F (Table 2) and ligated with the camR gene (22). The location of the camR gene in babB was analyzed in the H. pylori transformants by using primers BF and A11R for babB (Table 2). The fragment could not be amplified in the 17875ΔbabB::cam mutant.
BabA Binding Activity, Affinity Analysis, and Biotinylation of Leb Glycoconjugate. Binding to Leb conjugate was analyzed essentially as described in ref. 5. Samples were assayed in duplicate. For Scatchard analysis, binding affinity and capacity were determined as described in ref. 3. Biotinylation of Leb-APD-HSA (Isosep, Tullinge, Sweden) was performed as described in ref. 4.
Fluorescence Microscopy. Bacterial cells were labeled with FITC (Sigma) as described in ref. 2. Cell density was adjusted to A600 = 0.2 with blocking buffer (PBS/0.05% Tween 20/1% BSA). A 1-ml sample of bacteria was incubated with 2 μg of biotinylated Leb for 2 h at 23°C, washed with blocking buffer, and finally incubated with Cy-3-streptavidin (1:10,000) (Sigma) for 1 h at 23°C. After washing, the 1-μl samples were analyzed in a Leica DM RBE microscope (magnification, ×400), which revealed 20–40 f luorescent Leb-binding cells. The Leb OFF→ON switch frequency (mean value of four samples) of strain 17875ΔbabA2::cam was manually quantified by counting the number of OFF→ON 17875ΔbabA2::cam clones present in samples and dividing them by the total number of bacterial cells determined by a count taken in a Petroff–Hausser chamber (CA Hausser and Son, Philadelphia).
Bio-Panning for Leb Binding. A 2-ml suspension of strain 17875ΔbabA2::cam (A600 = 0.5) was incubated with 5 μg of biotinylated Leb conjugate for 2 h at 37°C. The suspension was washed and mixed with 100 μg of streptavidin magnetic beads (Polysciences) for 2 h at 37°C. Bead-bound bacteria were recovered with a magnet and cultured on agar medium. Progeny bacterial cells were used in a second round of enrichment for Leb binding.
Colony Blotting to Identify Leb Binding. Bacteria were spread at a density of 500 colonies per plate, incubated, and transferred to nitrocellulose membranes (Bio-Rad), which were baked for 1 h at 70°C, washed in PBS/0.05% Tween 20, blocked in TBS buffer (50 mM Tris/150 mM NaCl/0.05% Tween 20, pH 7.4) plus 1.5% gelatin/1.0% BSA (buffer I) overnight at 4°C, incubated with 2 μg of biotinylated Leb in buffer I for 2 h at 23°C, and then incubated with streptavidin-peroxidase (Roche, Stockholm) (1:5,000) in buffer I for 1 h at 23°C, washed in TBS, and exposed to 4-chloro-1-naphtol as recommended by the manufacturer (Sigma). Leb-binding colonies were identified by blue color.
Immunoblot Analysis. One milliliter of bacterial culture (A600 = 1.0) was resuspended in 200 μl of sample buffer and denaturated at 100°C for 5 min. We separated 5–20 μl of suspension by electrophoresis on 7.5% Ready Gel and transferred the suspension to a 0.2-μm polyvinylidene difluoride membrane (PVDF, Bio-Rad). The membrane was blocked in 5% nonfat dry milk at 4°C overnight, followed by incubation for 1 h at 23°C with anti-BabA antibody Ak253 (1:6,000) (7), washed with TBS, incubated with anti-rabbit antibody horseradish peroxidase (1:2,000) (DAKO) for 1 h, and washed and developed with Super Signal (Pierce). To visualize the signals, we scanned the membranes with the Fluor-S MultiImager (Bio-Rad). Densities were measured by quantity one software (Bio-Rad).
ImmunoGold Analysis. An overnight bacterial culture from agar medium was resuspended in 10 mM Tris·HCl, pH 7.4/10 mM MgCl2 and applied to formvar-coated grids for 5 min at 23°C. The grids were incubated with anti-BabA antibody Ak253 (1:200) for 10 min at 23°C and for 10 min with a goat-anti-mouse Ab-gold (1:10) with 10-nm gold particles (GAR10, British Biocell International, Cardiff, U.K.). Grids were negatively stained with 1% sodium silicotungstate, pH 7.0, analyzed with a JEOL 1230 transmission electron microscope and documented by gatan msc (Link Analytical, Lidingö, Sweden).
DNA Isolation, PCR, Primers, and Probes. Genomic H. pylori DNA was isolated as described in ref. 23 from 12- to 20-h cultures and treated with RNase A. E. coli recombinant plasmids were isolated by using a Miniprep kit (Qiagen, Valencia, CA). PCR was performed by using Taq or Platinum Taq polymerase (Invitrogen) for 35 cycles: 30 sec at 94°C, 1 min at 50°C, 1 min (or 3 min if product was >3 kb) at 72°C, and, finally, with 15 min at 72°C. Primers A10F and A19R were used for babA PCR. Other primers used for PCR, sequencing, and probe construction are listed in Table 2.
Southern Blot Analysis (24). HindIII-digested DNA was separated by 2% agarose gel electrophoresis, blotted onto a Hybond-N (Amersham Pharmacia Biotech) and UV crosslinked. The membrane was soaked in 6× SSC (1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7) for 2 min and prehybridized in Church's buffer (0.5 M Na2HPO4,pH 7.2/1% BSA/1 mM EDTA/7% SDS) at 65°C and hybridized at 65°C overnight. Oligonucleotides A19R (babA) and A18R (babB) 32P-labeled with T4 polynucleotide kinase (Roche) and [γ-32P]-dATP [5,000 Ci/mmol (1 Ci = 37 GBq)] were used as probes. Signals were visualized by using storm 860 (Molecular Dynamics).
Northern Blot Analysis (25). H. pylori was grown for 12–20 h, and RNA was extracted, electrophoresed, and transferred to membranes. Oligonucleotides A19R (babA) and A18R (babB) were 32P-labeled with T4 polynucleotide kinase (Roche) and [γ-32P]-dATP (5,000 Ci/mmol) and used as probes. Signals were visualized by using storm 860 (Molecular Dynamics). Hybridization to 16S rRNA was carried out with a 32P-labeled fragment generated by PCR with primers 16SF and 16SR and [α-32P]-dATP (Amersham Pharmacia). Hybridization intensities were analyzed by image quant (Molecular Dynamics).
Primer Extensions (26). We 5′-end-labeled 15 pmol of primers A61R (babA2), A41R (babA), or A71R (babB) with [γ-32P]-dATP (5,000 Ci/mmol) and T4 polynucleotide kinase. Sequencing reactions were performed in parallel (Sequenase 2.0 kit, United States Biochemical) with recombinant plasmids carrying equivalent DNA fragments and the same primers as used for the primer extension reactions (A61R, A41R, and A71R). Signals were visualized by using storm 860 (Molecular Dynamics).
Sequencing of the babB/A Gene. This chimeric gene was amplified from genomic DNA of clones A1on1 and A1on2 using primers A64F and A39R. Nested PCR was performed with primers A10F–A25R, A21F–A30R, and A36F–A39R by using the A64F–A39R fragment as template. Fragments were cloned by using a TOPO TA Cloning kit (Invitrogen), and inserts were sequenced by using a DYEnamic ET terminator kit (Amersham Pharmacia). Sequences of upstream and 5′ regions were determined after amplification with primers A40F–A19R (babA1), F2F–A59R (babA2), and A64F–A18R (babB).
Results
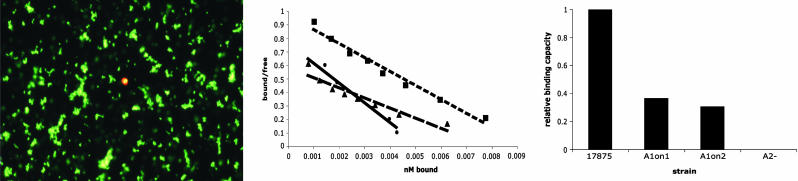
Metastability of Leb Binding. Approximately 70% of Swedish and U.S. H. pylori isolates can bind Leb (5, 7), but PCR tests of 10 randomly chosen Swedish strains that were nonbinding by radio immunoassay indicated that they carried babA sequences. Fluorescence microscopy indicated that each of the 10 strains contained rare cells that distinctly bound Leb conjugate (Fig. 1 Left). Bio-panning with a nonadherent engineered mutant of reference strain 17875 was used to examine this apparent metastability and to isolate Leb-binding derivative clones. Wild-type 17875 carries two babA genes, babA1 (silent because the 5′ end is nonfunctional) and babA2 (expressed) (5). A ΔbabA2::cam deletion/insertion mutation does not bind Leb. The frequency of 17875ΔbabA2::cam OFF→ON clones was calculated to ≈10-5 by manually counting the number of fluorescent Leb-binding cells by fluorescence microscopy and dividing by the total number of bacterial cells present in the sample (see Methods). To select for restoration of adherence, strain 17875ΔbabA2::cam was mixed with biotinylated Leb conjugate and streptavidin-coated magnetic beads, and Leb binders were captured with a magnet and identified by colony blotting. Dozens of Leb-binding clones were identified, and two, named A1on1 and A1on2, were further characterized. A Leb-nonbinding clone from the same biopanning experiment, A1off1, was used as a control.
Fig. 1.
Leb binding of OFF strain and ON variants. (Left) Fluorescence microscopy showing occasional Leb-binding bacterial cells detected with Cy-3-streptavidin and nonbinding cells with FITC. Hence, a bacterial cell with an OFF→ ON Leb-binding phenotype is detected by yellowish staining among green nonbinding cells of 17875ΔbabA2::cam. (Center) For affinity analyses according to Scatchard, the 125I-labeled Leb conjugate was diluted with unlabeled Leb conjugate and binding affinity was measured. Under these conditions, wild-type strain 17875 (•), A1on1 (▪), and A1on2 (▴) exhibited affinity constants of 1.5 × 10-11, 1.0 × 10-11, and 0.74 × 10-11 M-1, respectively. (Right) The full Leb-binding capacity was calculated for strains 17875, A1on1, and A1on2. The binding for A1on1 and A1on2 is shown relative to that of wild-type strain 17875.
Radio immuoassay of bacterial binding to 125I-Leb conjugate confirmed Leb binding by A1on1 and A1on2 (data not shown). Scatchard analysis of binding affinity, measuring the ratio of free versus bound 125I-Leb conjugate in a titration series, showed that A1on1 and A1on2 each bound 125I-Leb conjugate with affinities similar to those of ancestral strain 17875 (Fig. 1 Center). However, the calculated binding capacities were about 3-fold lower than those of strain 17875 (defined as 1.0 versus 0.36 for A1on1 and 0.30 for A1on2), which implies fewer adhesin molecules (Fig. 1 Right). Immunoblot analysis with BabA antibodies identified a protein the size of the BabA adhesin (75 kDa) in both A1on1 and A1on2 and absent in A1off1 (data not shown), but quantification of band intensities indicated only ≈30% as much protein in A1on1 and A1on2 as in 17875 (Fig. 2 Upper), and immunogold electron microscopy revealed fewer gold particles on A1on1 than on 17875 cells (Fig. 2 Lower). These three results (Scatchard, immunoblot, and immunogold electron microscopy) show that Leb-binding clones A1on1 and A1on2 express adhesins that are functionally equivalent to, but less abundant than, those of the wild-type 17875 ancestor.
Fig. 2.
Immunoblot and ImmunoGold electron microscopy analysis of OFF strain and ON variants. (Upper) Immunoblot analysis of whole-cell extracts by using Ak253 BabA antibodies. Lanes: 1, A1on1 (on1); 2, A1on2 (on2); 3, 17875ΔbabA2::cam (A2-); 4, wild-type 17875 (wt); 5, off phase shift of A1on1 (ON→ OFF). The relative amount of adhesin expressed in A1on1 and A1on2 was 0.3 as compared with the wild-type strain 17875 (1.0). (Lower) Electron micrographs of H. pylori. BabA and the BabB/A chimeric adhesin expressed on the bacterial surface is visualized by immunogold labeling with anti-BabA and 10-nm gold particles. Gold particles on five electron micrographs of each strain were counted, and the ratio of the wild-type 17875 to A1on1 was 0.25.
Recombination of babA1 into babB Restores Leb Binding. Southern blot hybridizations were used to assess whether the restoration of Leb binding could be explained by genetic rearrangements. HindIII-digested genomic DNA was blotted and hybridized with a probe corresponding to the highly specific central region of the babA gene (A19R). Two hybridizing fragments were detected from each of strains A1on1 and A1on2, one corresponding to the original babA1 locus and a second, novel fragment (see arrow in Fig. 3 Upper), which suggests babA sequence duplication. The high homologies between the 5′ and 3′ ends of babA and babB [HP0896 in strain 26695; JHP1164 (hopT) in strain J99; GenBank accession AF001389 in strain 17875] suggested recombination between these two loci as a possible explanation. An additional Southern blot showed that babB-specific sequences (probe A18R) had been lost from A1on1 and A1on2 (Fig. 3 Lower). PCR amplification with babA and babB primers (see Methods and Fig. 6, which is published as supporting information on the PNAS web site) showed that A1on1 and A1on2 each contained a recombinant babB/A DNA segment, that much of babB was lost, and that the silent wild-type babA1 gene was still present. Sequencing of regions upstream of babA1, babA2, and babB in A1on1 and A1on2 verified that adjacent sequences were still intact.
Fig. 3.
Southern blot analysis of the isolated A1on clones. Genomic DNA isolated from strains 17875 wild type (wt); A1on1 (on1); A1off (off1); A1on2 (on2), 17875ΔbabA1::cam (A1-); 17875ΔbabA2::cam (A2-); 17875ΔbabB::cam (B-); 17875ΔbabA2::camΔbabA1::kan (DM), digested with HindIII. (Upper) Hybridization to DNA of the 17875ΔbabA1::cam strain identified a babA2 specific fragment of 1,000 bp, whereas a 3,000-bp babA1-specific fragment was present in 17875ΔbabA2::cam. Both fragments are present in parent strain 17875 (wt). In the A1on clones, the babA1-specific fragment and an additional fragment of ≈900 bp were detected. (Lower) A 2,300-bp, babB-specific fragment was present in all strains except for the A1on clones and the babB knockout mutant, 17875ΔbabB::cam (B-).
To better understand these putative recombination events, we determined nucleotide sequences of the chimeric babB/A gene of A1on1 and A1on2. In each case, the first 47 bp were babB-specific, the following 66 bp were shared between both babA and babB, and the remaining sequence was babA-specific (GenBank accession nos. AY787038 and AY787039; see Fig. 7, which is published as supporting information on the PNAS web site). After these sequences were 1,150 bp of sequence identical to the babA1 locus and the babB locus, where the second crossover is likely to have occurred (900 bp from the 3′ ends of each gene, plus 250 bp of downstream untranslated sequence). These sequence data indicate that a copy of babA1 had recombined into the babB locus by using shared sequences at the 5′ and 3′ ends of these genes. An additional HindIII site (from the babB gene) present in the chimeric babB/A gene sequence predicts an 869-bp fragment in the chimeric gene that corresponds well to the size of the fragment obtained by Southern blot analysis (Fig. 3 Upper) and further supports our interpretation of babA1 recombination into babB. A few differences in the DNA sequences of the respective babB/A genes in A1on1 and A1on2 were identified that resulted in four and three amino acid divergences, respectively, relative to that of the wild-type sequence (Fig. 7). Recombination between babA1 and babB may have occurred by a nonreciprocal intragenomic event (gene conversion) or transformation with DNA from cells in the culture that had lysed spontaneously.
In further experiments, we isolated Leb-binding derivatives of the nonbinding strain NCTC11638 by bio-panning. This strain carries two silent babA1 genes but lacks introduced mutations or resistance markers. PCR and sequence analysis of Leb-binding clones similarly showed recombination of babA1 into the babB locus. (Fig. 8, which is published as supporting information on the PNAS web site).
babB Constitutes an Expression Locus. Both babA genes in strain 17875 could potentially code for the mature BabA adhesin, but babA1 lacks 10 bp in the first part of the ORF including the translational start codon (5). Northern blot transcript analysis revealed a 2,500-nt transcript in wild-type strain 17875 and its derivative 17875ΔbabA1::cam but no babA transcript in 17875ΔbabA2::cam or the 17875ΔbabA1::kanΔbabA2::cam double mutant (Fig. 4). This finding showed that only babA2 was transcribed. Its transcript of 2,500 nt corresponds to that of the babA gene. A 1,600-nt transcript was also present and may correspond to a specific cleavage product. Primer extension analysis identified a transcriptional start site 95 bp upstream of the translational start site of babA2 (Fig. 5). The babA2 transcriptional start site is preceded by a sequence (ATGACA-19-TATAAT) that is highly homologous to the consensus for E. coli σ70 promoters (TTGACA-15-19-TATAAT). The 160 bp immediately upstream of each babA gene are almost identical, except for four additional nucleotides (AAAA) between the -10 and -35 regions of the babA1 promoter. Thus, the defect in babA1 expression can be ascribed to an absence of translational and signal peptide-encoding sequences (5) and, probably, lack of transcription. Northern blot analysis showed that the relative amount of full-length babB/A mRNA was lower than that of babA2 mRNA (ratio 0.3:1) (Fig. 4 Top). This finding is in accord with a lower level of chimeric BabB/A adhesin and binding capacity of A1on1 and A1on2 derivatives relative to their wild-type ancestor. Further analysis with a babB-specific probe (Fig. 4 Middle) revealed a weak, babB 2,600-nt transcript in strain 17875 wild-type and babA mutant derivatives but not in A1on1 or A1on2 or in a babB knockout mutant control. Analysis of the primer extension product, although weak, identified the babB transcriptional start site 44 nt upstream of the translational initiation codon (Fig. 5). The babB transcriptional start site is preceded by the promoter sequence (TTATGC-19-GATAAG).
Fig. 4.
Northern blot analysis of total RNA isolated from strains 17875 wild type (wt), A1on1 (On1), A1on2 (On2), 17875ΔbabA1::cam (A1-), 17875ΔbabA2::cam (A2-), 17875ΔbabB::cam (B-), and 17875ΔbabA2::camΔbabA1::kan (DM) and hybridized with a babA-specific probe. (Top) A specific 2,500-nt transcript that corresponds to the size of the babA gene in wild-type 17875 and 17875ΔbabA1::cam mutant cells but not in the 17875ΔbabA2::cam mutant or the double mutant 17875ΔbabA2::camΔbabA1::kan. (Middle) The same filter hybridized with a babB probe identified a 2,600-nt babB transcript. A 1,600-nt transcript was present together with both the babA and the babB transcripts. (Bottom) The amount of RNA in each lane is visualized by hybridization with a 16S rRNA probe. The expression of the babA mRNA (Top) in A1on1 and A1on2 was quantified in relation to expression in strain 17875, for which the values from the 16S hybridization were used as references: 17875, 1.0; A1on1, 0.32; A1on2, 0.29.
Fig. 5.
Primer extension analysis. (Left) Analysis of the 5′ end of the babA2 transcripts. Total RNA was used for the primer extension with 32P-end-labeled oligonucleotide primers. babA2 extension products were analyzed from strain 17875 wild type (wt), 17875ΔbabA1::cam (A1-), 17875ΔbabA2::cam (A2-), and 17875ΔbabA2::camΔbabA1::kan (DM). (Center) Primer extension analysis of the babB operon. Total RNA was used for the primer extension with 32P-end-labeled oligonucleotide primer. babB extension products were analyzed from 17875 wild type and 17875ΔbabB::cam (B-). Sequence analysis of DNA corresponds to the respective upstream region and serves as a reference. The sequence analysis was performed with the same primer as used for the primer extension. (Right) The -10 and -35 promoter sequences are marked in the respective DNA sequences.
In summary, strain 17875ΔbabA2::cam gained the ability to express a chimeric, but functional, Leb-binding adhesin by recombination of silent babA1 sequences into the weaker transcribed babB gene, which allowed expression of a 3-fold reduced amount of Leb-binding chimeric BabB/A adhesin.
Leb Binding of the Chimeric BabB/A Adhesin Is Subject to Phase Variation. Whenever repetitive sequences are found, such as in many hop genes, phase variation through slipped-strand mispairing would provide a means of metastable ON/OFF switching of gene expression (17, 18, 20). The 5′ ends of both babB and chimeric babB/A contain a series of CT (cytosine-thymine) repeats, whereas no such repeats are found in babA. Accordingly, the stability of adherence of A1on1 was studied by colony blotting. Phase-shift variants that lacked the capacity to interact with Leb were obtained at a frequency of one per 200 colony-forming units, without prior bio-panning. Immunoblotting and immunogold electron microscopy showed that these OFF variants did not express chimeric BabB/A adhesin (Fig. 2 and data not shown), and sequence analysis revealed a change from eight CT repeats in the ON variants to seven or nine CT repeats in the OFF variants screened (Table 1). The gain or loss of one CT pair creates frame shifts that would cause loss of expression. By further colony screening of an A1on1→off phase-shift variant, we isolated revertants (A1on1→ off→onR) that had regained Leb-binding activity at a frequency of 5 × 10-3, the same as for ON→OFF switch variants. This frequency of reversible ON/OFF phase variation (5 × 10-3) is ≈500-fold higher than that of recombination of the silent babA1 into the babB locus (10-5). The ON revertants again contained eight CT repeats and, thus, a restored reading frame. Therefore, the Leb-binding adhesin expressed as a chimera after recombination into the babB locus is subject to reversible phase variation, thereby allowing significant phenotypic heterogeneity in previously uniform bacterial populations.
Table 1. CT repeats present in babB/A phase-shift variants.
| Origin | Phase variant | Leb binding | No. of CT repeats |
|---|---|---|---|
| 17875ΔbabA2::cam | A1on1 | + | 8 |
| A1on2 | + | 8 | |
| A1on1 | offA | - | 7 |
| A1on1 | offB | - | 7 |
| A1on2 | offA | - | 9 |
| A1on2 | offB | - | 9 |
| A1on2 | offC | - | 9 |
| offA | on1R | + | 8 |
| offA | on2R | + | 8 |
| offA | on3R | + | 8 |
Sequence analysis and the number of CT repeats present in ON and OFF variants. A change from eight CT repeats in ON variants to seven or nine CT repeats in OFF variants creates a frame-shift that brings the chimeric babB/A ORF out of frame.
Discussion
Abilities to cycle between nonadherence and adherence to epithelial surfaces and to dynamically change adherence strength is likely to help H. pylori establish and maintain chronic infections in the face of human diversity, immune responses, and disease progression (3, 4). Adherence promotes delivery of toxins and other effector molecules to target tissues and improves access to nutrients that leach from tissues when damaged (27), but it can be a mixed blessing in also exposing H. pylori to the brunt of bactericidal host responses. In principle, flexibility in adherence might have been achieved by classical regulatory mechanisms by using, for example, membrane sensors and transcription repressors or activators to monitor and cope with differences in the gastric milieu. However, our experiments illustrate that H. pylori uses another strategy to direct OFF→ON changes in BabA expression to become adherent and more virulent.
Each of the 10 nonadherent clinical isolates screened in our study contained babA sequences and were able to sport adherent cells at frequencies in the range of ≈10-5. To examine the basis of this heterogeneity, we selected Leb-binding bacterial clones from an engineered nonadherent derivative of reference strain 17875 (strain 17875ΔbabA2::cam) by enrichment with Leb magnetic beads (bio-panning) and colony blotting. The two clones analyzed in detail each had resulted from recombination of silent babA1 sequences into the babB locus. This recombination involved a patch of just 66 bp of matched sequence at the 5′ ends of these genes and a far longer homology (>1 kb) at their 3′ ends. This recombination event could have resulted from gene conversion or from transformation with DNAs released by spontaneous lysis of sibling cells in the culture. The frequency, although low (≈10-5), should be biologically significant, given the power of natural selection.
Leb-binding clones were also obtained from the naturally nonadherent strain NCTC11638 similarly by recombination of a silent babA sequence into an expressed babB locus to form a babB/A chimera. Chimeric babA/B genes were found in a population genetic survey, although they were not very common (28). Derivatives of human H. pylori strain J166 with formally equivalent replacements of babA by babB were found as the most common monkey-adapted type after experimental infection of each of several rhesus monkeys (19). A few of the other J166 derivatives had lost Leb binding by slippage in a CT repeat that happens to occur in this particular strain's babA gene. It is not known whether the adaptation of strain J166 to this primate model often involves selection against BabA, selection for elevated levels of BabB, or both. Assuming babB and babA each to be beneficial to H. pylori in particular hosts or stages of infection, the long-term survival of lineages in which particular bab genes have been lost by replacement, as witnessed here, and in strain J166 in monkeys (19) may depend on occurrence of at least transient mixed infection and reacquisition of lost bab genes by interstrain recombination.
Each of the naturally occurring Leb-nonbinding strain that we tested contained a quiescent but easily activated babA gene; therefore, we should expect babB/A chimeras to arise in a population. Scenarios in which some clones turn off and others reciprocally turn on adherence to particular receptors will create population heterogeneity sufficient to ensure availability of optimum binding phenotypes in response to different stages of infection. The plasticity of the BabA adhesin has also been illustrated by the finding that many H. pylori strains are “generalists” (able to bind ALeb and BLeb in addition to Leb) in contrast to “specialist” H. pylori strains that only bind Leb. Specialist H. pylori strains were transformed to the broader binding phenotype with genomic DNA from generalists strains and the formation of chimeric babA genes (3). These results also illustrate the power of recombination as an agent of dynamic change in adherence properties, which we suggest can contribute to the persistence of H. pylori in ever-changing gastric environments.
Expression of the BabA adhesin is believed to be regulated at the level of translation because of the lack of the translational start in babA1 (5). This hypothesis is further refined by the data presented here that indicate that the babA1 gene is transcriptionally silent, most likely because of four additional nucleotides between the -10 and -35 sites of pbabA1, which should diminish the strength of the promoter. The repetitive (A)n sequences between the -10 and the -35 sites should also be prone to slippage mutations and thereby allow changes in levels of transcription of downstream genes. Northern blot analysis also detected shorter distinct babA and babB transcripts in cells with functional bab genes. It is tempting to speculate that transcript cleavage may also be used to help fine-tune the level of expressed adhesin and Leb binding. The amount of expressed babA and babB/A transcript closely matched the level of expressed protein and the Leb-binding capacity. This feature probably reflects the strength of the promoters for which the babB promoter is weaker relative to that of babA2. Thus, BabA is more strongly expressed from the original pbabA2 compared with the babB promoter, which is in line with the idea of recombinations as mechanisms of generating subclones with altered levels of gene expression.
Synthesis of the adhesin encoded by the babB/A chimera was also prone to switch OFF and ON at frequencies at nearly 10-2, unlike that from the original babA gene. By flexible ON/OFF shifts in adherence, the bacterial population will contain cells that closely attach to the epithelium and others that better escape inflammation responses. Infection with H. pylori induces changes to mucosal glycosylation patterns, with up-regulation of inflammation association sialyl-Lewis antigens (4). Both sialylated and fucosylated antigens reciprocally change expression levels when fucosylation is down-regulated during early phases of infection but then again increase during persistent infection (A. Dubois, personal communication). Furthermore, higher infection loads of H. pylori were seen in patients with higher densities of Leb (29, 30). These results suggest that H. pylori infection promotes distinct shifts in mucosal glycosylation patterns and, in accord, selection of H. pylori clones with optimum adherence phenotype. We suggest that most H. pylori strains can evolve from benign to more virulent lifestyles whenever this would be advantageous to the bacterium. This flexibility merits consideration in relation to thinking of H. pylori as a beneficial commensals and the clinical implications that emerge from such an idea (31).
An improved understanding of the dynamics of these adhesins could be achieved by further investigating the time course of BabA and BabB expression during long-term experimental infection in nonhuman primates as well as by determining their prevalence in peptic ulcer disease and gastric cancer.
Supplementary Material
Acknowledgments
We thank R. Sjöström for excellent advice concerning the Scatchard analysis and I. Sjöström, U. Blank, and O. Kroupa for technical assistance. This work was supported by grants from the J. C. Kempe Memorial Foundation (to A.B.); the Kempestiftelserna, by Swedish Medical Research Council Grant 11218 and Swedish Cancer Society Grant 4101-B00-03XAB (to T.B.); National Institutes of Health Grants R01 DK53727, R0 DK63041, R01 A1316608, and P30 DK52474 (to D.E.B.); the B von Kantzows Foundation; the Tore Nilsson Medical Research Foundation; the Magnus Bergwall Research Foundation; and the Medical Faculty of Umeå University (A.A.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CT, cytosine-thymine; Leb, Lewis b antigen.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession numbers AY787038 and AY787039).
References
- 1.Cover, T., Berg, D. E., Blaser, M. J. & Mobley, H. (2001) in Principles of Bacterial Pathogenesis, ed. Groisman, E. A. (Academic, New York), pp. 509-558.
- 2.Borén, T., Falk, P., Roth, K. A., Larson, G. & Normark, S. (1993) Science 262, 1892-1895. [DOI] [PubMed] [Google Scholar]
- 3.Aspholm-Hurtig, M., Dailide, G., Lahmann, M., Kalia, A., Ilver, D., Roche, N., Vikström, S., Sjöstrom, R., Lindén, S., Bäckström, A., et al. (2004) Science 305, 519-522. [DOI] [PubMed] [Google Scholar]
- 4.Mahdavi, J., Sondén, B., Hurtig, M., Olfat, F. O., Forsberg, L., Roche, N., Ångström, J., Larsson, T., Teneberg, S., Karlsson, K.-A., et al. (2002) Science 297, 573-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ilver, D., Arnqvist, A., Ögren, J., Frick, I.-M., Kersulyte, D., Incecik, E. T., Berg, D. E., Covacci, A., Engstrand, L. & Borén, T. (1998) Science 279, 373-377. [DOI] [PubMed] [Google Scholar]
- 6.Gerhard, M., Lehn, N., Neumayer, N., Borén, T., Rad, R., Schepp, W., Miehlke, S., Classen, M. & Prinz, C. (1999) Proc. Natl. Acad. Sci. USA 96, 12778-12783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamaoka, Y., Souchek, J., Odenbreit, S., Haas, R., Arnqvist, A., Borén, T., Kodama, T., Osato, M. S., Gutierrez, O., Kim, J., et al. (2002) J. Clin. Microbiol. 40, 2244-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van der Ende, A., Rauws, E. A. J., Feller, M., Mulder, C. J. J., Tytgat, G. N. J. & Dankert, J. (1996) Gastroenterology 111, 638-647. [DOI] [PubMed] [Google Scholar]
- 9.Suerbaum, S., Smith, J. M., Bapumia, K., Morelli, G., Smith, N. H., Kunstmann, E., Dyrek, I. & Achtman, M. (1998) Proc. Natl. Acad. Sci. USA 95, 12619-12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kersulyte, D., Chalkauskas, H. & Berg, D. E. (1999) Mol. Microbiol. 31, 31-43. [DOI] [PubMed] [Google Scholar]
- 11.Israel, D. A., Salama, N., Krishna, U., Rieger, U. M., Atherton, J. C., Falkow, S. & Peek, R. M., Jr. (2001) Proc. Natl. Acad. Sci. USA 98, 14625-14630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuipers, E. J., Israel, D. A., Kusters, J. G., Gerrits, M. M., Weel, J., van der Ende, A., van der Hulsts, R. W. M., Wirth, H. P., Höök-Nikanne, J., Thompson, S. A., et al. (2000) J. Inf. Dis. 181, 273-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blaser, M. J. & Berg, D. E. (2001) J. Clin. Invest. 107, 767-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aras, R. A., Kang, J., Tschumi, A. I., Harasaki, Y. & Blaser, M. J. (2003) Proc. Natl. Acad. Sci. USA 100, 13579-13584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aras, R. A., Lee, Y., Kim, S.-K., Israel, D., Peek, R. M., Jr., & Blaser, M. J. (2003) J. Infect. Dis. 188, 486-496. [DOI] [PubMed] [Google Scholar]
- 16.Alm, R. A., Bina, J., Andrews, B. M., Doig, P., Hancock, R. E. W. & Trust, T. J. (2000) Infect. Immun. 68, 4155-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tomb, J.-F., White, O., Kerlavage, A. R., Clayton, R. A., Sutton, G. G., Fleischmann, R. D., Ketchum, K. A., Klenk, H. P., Gill, S., Dougherty, B. A., et al. (1997) Nature 388, 539-547. [DOI] [PubMed] [Google Scholar]
- 18.Alm, R. A., Ling, L.-S., Moir, D. T., King, B. L., Brown, E. D., Doig, P. C., Smith, D. R., Noonan, B., Guild, B. C., deJonge, B. L., et al. (1999) Nature 397, 176-180. [DOI] [PubMed] [Google Scholar]
- 19.Solnick, J. V., Hansen, L. M., Salama, N. R., Boonjukauakul, J. K. & Syvanen, M. (2004) Proc. Natl. Acad. Sci. USA 101, 2106-2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saunders, N. J., Peden, J. F., Hood, D. W. & Moxon, R. E. (1998) Mol. Microbiol. 27, 1091-1098. [DOI] [PubMed] [Google Scholar]
- 21.Akopyanz, N., Bukanov, N. O., Westblom, T. U., Kresovich, S. & Berg, D. E. (1992) Nucleic Acids Res. 20, 5137-5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang, Y. & Taylor, D. E. (1990) Gene 94, 23-28. [DOI] [PubMed] [Google Scholar]
- 23.Pitcher, D. G., Saunders, N. A. & Owen, R. J. (1989) Lett. Appl. Microbiol. 8, 151-156. [Google Scholar]
- 24.Sambrook, J., Fritsch, E. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY), 2nd Ed.
- 25.Arnqvist, A., Olsén, A., Pfeifer, J., Russell, D. G. & Normark, S. (1992) Mol. Microbiol. 6, 2443-2452. [DOI] [PubMed] [Google Scholar]
- 26.Arnqvist, A., Olsén, A. & Normark, S. (1994) Mol. Microbiol. 13, 1021-1032. [DOI] [PubMed] [Google Scholar]
- 27.Rhen, M., Eriksson, S., Clements, M., Bergström, S. & Normark, S. (2003) Trends. Microbiol. 11, 80-86. [DOI] [PubMed] [Google Scholar]
- 28.Pride, D. T. & Blaser, M. J. (2002) J. Mol. Biol. 316, 629-642. [DOI] [PubMed] [Google Scholar]
- 29.Heneghan, M. A., Moran, A. P., Feeley, K. M., Egan, E. L., Goulding, J., Connolly, C. E. & McCarthy, C. F. (1998) FEMS Immunol. Med. Microbiol. 20, 257-266. [DOI] [PubMed] [Google Scholar]
- 30.Sheu, B. S., Sheu, S. M., Yang, H. B., Huang, A. H. & Wu, J. J. (2003) Gut 52, 927-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blaser, M. J. & Atherton, J. C. (2004) J. Clin. Invest. 113, 321-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.