Abstract
During the development of human oral biofilm communities, the spatial arrangement of the bacteria is thought to be driven by metabolic interactions between them. Streptococcus gordonii and Veillonella atypica, two early colonizing members of the dental plaque biofilm, have been postulated to participate in metabolic communication; S. gordonii ferments carbohydrates to form lactic acid, which is a preferred fermentation substrate for V. atypica. We found that, during agar-plate coculture of these organisms, a signaling event occurs that results in increased expression of the S. gordonii α-amylase-encoding gene amyB. Confocal scanning laser microscopy of coculture flowcell-grown biofilms using human saliva as the sole nutrient showed that V. atypica caused S. gordonii to increase expression of a PamyB-′gfp transcriptional fusion in a spatially resolved fashion. In this open system, only those streptococci in mixed-species microcolonies containing V. atypica expressed GFP; nearby S. gordonii colonies that lacked V. atypica did not express GFP. In a closed system containing S. gordonii and V. atypica, flow cytometric analysis showed that S. gordonii containing the PamyB-′gfp reporter plasmid exhibited mean fluorescence levels 20-fold higher than did S. gordonii that had not been incubated with V. atypica. Thus, in a closed system where a diffusible signal can accumulate above a required threshold, interspecies signaling mediates a change in gene expression. We provide evidence that, in open systems like those that predominate in natural biofilms, diffusible signals between species are designed to function over short distances, on the order of 1 μm.
Keywords: bacterial communities, interspecies cell–cell signaling, oral bacteria
In nature, bacteria exist in multispecies communities, and signaling among the cells is thought to be part of community dynamics. Human dental plaque is a well recognized example of a natural multispecies bacterial community. A hallmark of the bacteria isolated from this community is the ability to coaggregate, which is defined as cell–cell interaction between genetically distinct organisms. It is hypothesized that coaggregation establishes spatial arrangement of bacteria to facilitate metabolic communication (1). Our usage of communication here is limited to interactions between species where at least one of the participating species elicits a measurable response. Evolutionary selection for the response is not implicit in this usage, although selection may occur in natural microbial ecosystems. For example, one such interaction between a streptococcus and an actinomyces results, in vitro, in a mutualistic relationship that allows growth of the two-species community under conditions in which neither species could grow alone (2). Furthermore, the occurrence of coaggregation-mediated interactions has been demonstrated in vivo (3), indicating that coaggregation may be critical for establishing mutualistic communities on the enamel surfaces of teeth.
Two early-colonizing species of the dental plaque biofilm, Streptococcus gordonii and Veillonella atypica, are an interesting pair of coaggregating organisms because their physiologies suggest that they participate in a metabolic interaction. S. gordonii generates energy by fermentation of sugars, yielding lactic acid as a major end product. V. atypica is unable to ferment sugars, but uses lactic acid as a preferred source of carbon and energy. Thus, a food chain could develop between these bacteria with the end-product of one organism serving as a source of energy for the other. This symbiosis has been demonstrated in vitro between Streptococcus mutans and Veillonella alcalescens (4). Furthermore, an ecological survey by our laboratory has shown that strains of veillonella exist in specific parts of the mouth that are environments for their streptococcal coaggregation partners (5).
Here we report a study of signaling within a dual-species human oral bacterial community. We show that V. atypica produces a signal that causes S. gordonii to increase expression of an α-amylase gene. In an open flow system, response to the signal occurred in only those S. gordonii cells located within a few micrometers of V. atypica. This observation has implications for understanding cell–cell signaling via diffusible signals in open, natural microbial systems.
Materials and Methods
Bacterial Strains and Culture Conditions. The sources and properties of bacterial strains and plasmids are presented in Table 1. For routine propagation, S. gordonii V288 and Actinomyces naeslundii strains T14V and PK606 were grown in brain–heart infusion (BHI) medium (Becton Dickinson) statically at 37°C. V. atypica strains PK1910 and PK1885 were grown in Todd–Hewitt broth (THB) (Difco) supplemented with lactic acid to 0.6% (THBL). Fusobacterium nucleatum ATCC10953 was grown in BHI medium supplemented with 0.25% ammonium glutamate. All above organisms were grown anaerobically by using the GasPak System (Becton Dickinson). Escherichia coli cultures were grown in LB medium at 37°C with shaking. Antibiotics (Sigma) were used at the following concentrations: for E. coli, 100 μg/ml ampicillin and 150 μg/ml erythromycin; for S. gordonii, 500 μg/ml kanamycin and 10 μg/ml erythromycin. When grown in the presence of erythromycin, E. coli was grown in BHI medium rather than LB.
Table 1. Bacterial strains and plasmids.
| Strain or plasmid | Relevant properties | Source |
|---|---|---|
| A. naeslundii strains | ||
| T14V | Coaggregation reference strain | 6 |
| PK606 | Coaggregation reference strain | 5 |
| F. nucleatum ATCC10953 | Coaggregation reference strain | ATCC |
| S. gordonii strains | ||
| V288 | Wild type | 7 |
| PK3248 | amyB mutant of V288 | This study |
| V. atypica strains | ||
| PK1910 | Wild type | 8 |
| PK1885 | Coaggregation mutant of PK1910 | 8 |
| Plasmids | ||
| pCM18 | Source of gfpmut3* | 9 |
| pPCR-Script Amp SK(+) | E. coli cloning vector, Ampr | Stratagene |
| pPCR-amyB | pPCR-Script Amp SK (+) containing amyB | This study |
| pDL276 | Source of kanamycin resistance gene aphAIII | 10 |
| pPamyB-′gfp | Contains gfp under control of amyB promoter | This study |
| pPE1007 | pTRKL2 derivative with XhoI site and MCS deleted | This study |
| pPE1010 | 'gfp reporter plasmid, Ermr | This study |
| pTRK-amyB | pTRKL2 containing amyB | This study |
| pTRKL2 | Low-copy streptococcal cloning vector | 11 |
Cloning and DNA Manipulation. Standard techniques were used for cloning and transformation (12, 13). Plasmid DNA was purified by using the QIA prep Spin Miniprep kit (Qiagen, Valencia, CA). DNA was extracted from agarose gels by using the QIA quick gel extraction kit. PCRs were performed by using Platinum Pfx DNA polymerase (Invitrogen).
The gfp reporter plasmid, pPE1010, was constructed by first PCR-amplifying the gfpmut3* gene and transcriptional terminators from pCM18 (9) by using primers 5′-gfp (5′-CGGAATTCGGATCCGGCTCGAGTTCATTAAAGAGGAGAAATTAAGCATG-3′) and 3′-term (5′-CGGAATTCAGCGGCGGATTTGTCCTACTCAG-3′) that incorporate EcoRI sites at each end of the PCR product and BamHI and XhoI sites 24 bases upstream of the gfp ATG start codon (restriction endonuclease sites are underlined). This PCR product contains a promoterless gfpmut3* followed by two transcriptional terminators. After digestion with EcoRI, the PCR product was cloned into the EcoRI site of pPE1007 to yield pPE1010. To construct the pPamy-′gfp fusion plasmid, primers BamHI-Pamy (5′-CGGGATCCGCTGCTAGCTCAGCTATCG-3′) and Pamy-XhoI (5′-CGCTCGAGTTAAATGCCATAGCCAACATCAT-3′) were used to amplify the region -325 to 174 relative to the ATG start codon of amyB. The PCR product was digested with BamHI and XhoI and cloned directionally into the BamHI and XhoI sites of pPE1010.
Sequence Analysis. The DNA sequence of the S. gordonii amyB gene was identified through a blast search of the preliminary sequence of the S. gordonii genome obtained from The Institute for Genomic Research web site (www.tigr.org) by using the sequence of the Streptococcus bovis amyB gene (14). Sequence comparisons were made by using the gap program of seqweb (version 2, Accelrys, San Diego).
Mutagenesis of amyB. An amyB mutant was constructed by using the PCR ligation mutagenesis technique (15). Segments of the amyB gene corresponding to bases -325 to 174 and 586–1088, relative to the ATG start codon, were PCR-amplified from S. gordonii V288 genomic DNA by using primers BamHI-Pamy and Pamy-XhoI, described above, and EcoRI-Cterm (5′-GGAATTCTATGACTATCTA ATGTATGCCG-3′) and Cterm-amy (5′-CATGGGAGGCCAGACTCTC-3′), respectively. The kanamycin resistance gene, aphAIII, was PCR-amplified from plasmid pDL276 (10) by using primers XhoI-aphAIII (5′-CGCTCGAGTGTGGTTTCAAAATCGGCTC-3′) and aphAIII-EcoRI (5′-GGAATTCCATCTAAATCTAGGTACTAAAAC-3′). The PCR products were digested with restriction enzymes (sites are underlined in primer sequences) and ligated together. The product of the ligation was used as template for a PCR with the 5′ primer of the first PCR, BamHI-Pamy, and the 3′ primer from the second reaction, Cterm-amy. The resultant PCR product was purified and transformed into S. gordonii V288. The transformation mixture was plated to brain–heart infusion medium containing 500 μg/ml kanamycin to select for isolates that had undergone double recombination. One of these isolates, PK3248, was chosen for further study.
Amylase Assays. Amylase activity was detected on agar plates containing THBL supplemented with 0.5% starch by applying cell suspensions to the agar surface and incubating anaerobically at 37°C for 36 h. Amylase activity was detected by the presence of an unstained zone around the bacterial colony after flooding the plates with iodine solution (0.2% I2/2% KI).
Amylase activity in culture supernatants was measured fluorimetrically by using the EnzChek amylase assay kit (Molecular Probes). Culture supernatants were passed through a 0.2-μm-pore-size filter and were then combined with the 4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-propionic acid (BODIPY FL)-starch substrate in accordance with the manufacturer's instructions. Fluorescence, an indicator of starch degradation, was measured every 30 s by using the Wallac Victor2 1420 multilabel counter (PerkinElmer). To normalize the amount of amylase activity to the number of S. gordonii cells in the culture, a sample of cells was taken from the suspensions before centrifugation and labeled with an anti-S. gordonii IgG-Alexa Fluor 488 conjugate (2). Fluorescence from the labeled cells was measured by using the multilabel counter, and the amount of fluorescence from each sample was used to derive conversion factors for normalizing enzyme activity with respect to the number of S. gordonii cells in each culture.
Growth and Analysis of in Vitro Biofilms. S. gordonii and V. atypica strains were grown to logarithmic phase in THB or THBL, respectively. Cells were harvested by centrifugation, washed with sterile 25% saliva (2), and resuspended in 25% saliva to an A600 of 0.05. For monospecies biofilms, cells were inoculated into saliva-conditioned flowcells as described (2), and flow was begun after a static adherence period of 15 min. For dual-species biofilms, a 15-min period of flow occurred between the sequential inoculations of the streptococci and the V. atypica cells (2). Sterile 25% saliva was supplied at a flow rate of 0.2 ml/min as the sole source of nutrient. Biofilms were stained for total biomass with the nucleic acid stain Syto-59 (Molecular Probes) and for V. atypica cells by primary immunofluorescence using Alexa Fluor 546-conjugated anti-V. atypica antibodies (16).
Flow Cytometric Analysis. Induction of expression from the amylase promoter in pPamy-′gfp was measured as GFP fluorescence by using a FACSCalibur flow cytometer (Becton Dickinson) with excitation at 488 nm. cellquest software (Becton Dickinson) was used for data analysis. S. gordonii containing the pPamy-′gfp reporter plasmid was grown in tryptone–yeast extract medium (TYE; 1% tryptone/0.5% yeast extract/0.3% K2HPO4) supplemented with 0.2% glucose. V. atypica was grown in THBL. One milliliter of each cell type was harvested and resuspended in a defined medium FMC (17), containing 1% glucose and adjusted to pH 7.3 by using 1 M K2HPO4. Samples for flow cytometry were diluted in PBS (pH 7.4) to ≈106 cells per ml before analysis.
Results
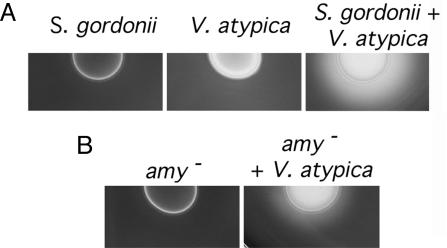
Induction of S. gordonii Amylase Activity During Growth with V. atypica. S. gordonii V288 and V. atypica PK1910 were grown separately and in coculture on agar plates containing starch. Growth of these organisms as a coculture resulted in starch hydrolysis, as detected by a zone of clearing around the colony upon iodine staining of the plates (Fig. 1A). No zone of starch hydrolysis was observed around colonies of either organism when grown separately. When A. naeslundii strains PK606 and T14V and F. nucleatum strain ATCC10953 were tested in coculture with S. gordonii, no starch hydrolysis was detected (data not shown), thereby suggesting that induction of starch hydrolyzing activity in S. gordonii is caused by a specific interaction with V. atypica.
Fig. 1.
Detection of amylase activity in starch plates. (A) Starch hydrolysis occurs in a coculture of S. gordonii and V. atypica (Right) but not when the species are grown separately. (B) Starch hydrolysis by the amyB mutant alone and in coculture with V. atypica.
Identification and Mutagenesis of the S. gordonii Amylase Gene. To identify potential amylase encoding genes in S. gordonii, a blast search of the preliminary genomic sequence of S. gordonii available from The Institute for Genomic Research was performed by using the sequences of the amylase gene from Streptococcus bovis 148 (14). A 1,452-bp ORF was identified on contig 4353. The deduced amino acid sequence of this gene was most similar (72% identical/79% similar) to AmyB, an intracellular α-amylase from S. bovis 148 (14). Based on this similarity, we have designated the S. gordonii gene amyB. The S. gordonii amylase sequence had a lower level of identity (59% identical/69% similar) to the intracellular amylase of Streptococcus mutans (18). Like the deduced amino acid sequence of AmyB from S. bovis and S. mutans, the S. gordonii AmyB lacked a signal sequence for protein export (14).
The region upstream of amyB contained sequences with high levels of identity to Gram-positive promoter elements. A sequence (5′-TTTGATAAAAT-3′) matching the extended -10 consensus (19, 20) (5′-TNTGNTATAAT-3′) at 10 of 11 bases was found at 65 bases upstream of the predicted start codon. This region also contained a 7-bp inverted repeat (5′-CGCAAACGTTTGCG-3′) matching the CRE consensus (21) (5′-WGNAASCGNWWNCA-3′) at 11 of 14 positions.
Construction and Characterization of an amyB Mutant. An amyB mutant, PK3248, was constructed by PCR ligation mutagenesis (15). The amyB mutant and the wild-type strain grew with the same doubling times on THB and tryptone–yeast extract medium. Starch plate assays showed that, compared to the wild-type strain, the amyB mutant was reduced in starch hydrolysis during coculture with V. atypica (Fig. 1B). The plasmid pTRK-amyB was transformed into strain PK3248 to complement the amyB mutation. PK3248 (pTRK-amyB) had a basal level of starch hydrolysis higher than wild-type. However, like the wild type, the zone of starch hydrolysis increased when the amyB mutant containing this plasmid was grown with V. atypica (data not shown). Although the amyB mutation was not in frame, it is unlikely to have a polar effect. The ORF adjacent and downstream of amyB is oriented in the opposite direction. In addition, the complemented mutant has restored amylase activity.
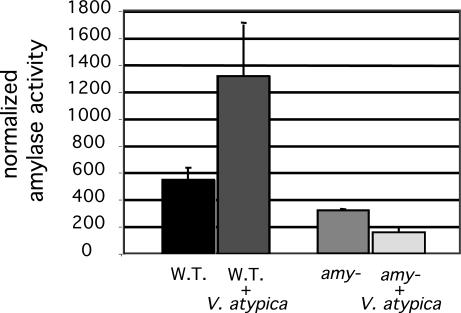
Although sequence data suggests that AmyB is not exported by use of a signal sequence, we initially identified the starch hydrolyzing activity of the enzyme extracellularly on starch plates. Therefore, examination of the effects of coculture with V. atypica, on amylase activity focused on activity found in culture supernatants. A quantitative, fluorescence-based amylase assay was used to measure the V. atypica-inducible amylase activity of the wild-type and amyB mutant strains of S. gordonii. Supernatants from the wild-type strain had two to three times more amylase activity when S. gordonii was grown in coculture with V. atypica than when it was grown alone (Fig. 2). The amyB mutant had basal levels of activity that were 60% of wild type. This remaining activity suggests the presence of a second starch-hydrolyzing enzyme. In contrast to the activity attributed to AmyB, the remaining starch hydrolyzing activity in the amyB mutant was reduced during coculture with V. atypica.
Fig. 2.
Amylase activity from culture supernatants. Activity is normalized for the fluorescence of labeled S. gordonii cells in each culture. Error bars represent standard deviations.
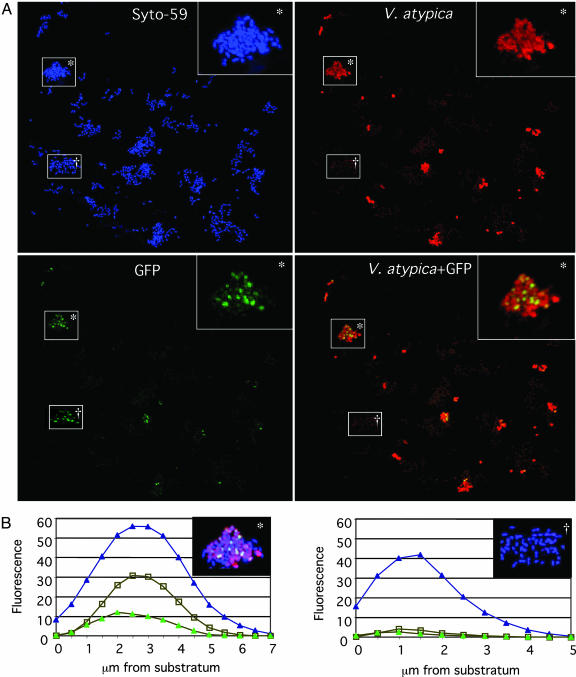
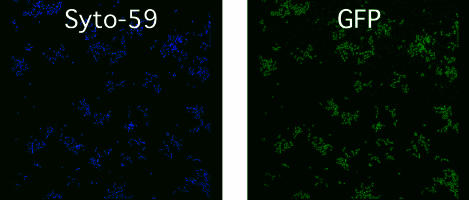
Juxtaposition with V. atypica in Dual-Species Open-System Biofilms Induces S. gordonii Amylase Expression. The possibility that growth of S. gordonii and V. atypica in a mixed species community results in increased transcription from the amylase promoter was investigated by using the gfp reporter plasmid, pPamy-′gfp, containing the amyB promoter region fused to a promoterless gfp. Coculture biofilms of S. gordonii (pPamy-′gfp) and V. atypica were grown with saliva as the sole source of nutrient. S. gordonii expressed GFP when in mixed-species microcolonies, but no GFP expression was seen when S. gordonii was not in association with V. atypica in either the dual-species biofilms (Fig. 3A) or in monospecies biofilms (Fig. 4). Examination of fields of view with each fluorescence channel separately showed that cells expressing GFP are adjacent to V. atypica cells. Quantitation of fluorescence in each optical slice comprising a maximum projection image showed that GFP expression occurred in the same optical slices where V. atypica cells were present, whereas no GFP was detectable in monospecies S. gordonii colonies (Fig. 3B). Only S. gordonii cells juxtaposed with V. atypica expressed GFP. S. gordonii cells at greater distances did not express GFP, suggesting that, in an open system such as a flowcell, signaling between these organisms occurred over only very short distances.
Fig. 3.
Confocal scanning laser microscopic analysis of dual-species biofilms. (A) Maximum projections (all confocal sections in a single field of view) of a single confocal stack showing fluorescence from Syto-59 (blue; all cells), Alexa Fluor 546-conjugated anti-V. atypica antibodies (red), and GFP (green; S. gordonii expressing amyB). The three fluorescence channels are shown separately and, in Lower Right, as overlay of GFP with V. atypica. (Inset) An enlargement of the boxed microcolony labeled with an asterisk. (B) Graphs of fluorescence intensity versus depth in a dual-species microcolony (Left) and a monospecies microcolony (Right) depicted in the upper right corner of each graph. The dual species microcolony is the same colony marked with an asterisk in A and is shown as an overlay of all three colors. The monospecies microcolony is labeled with a dagger in A. Microcolonies are shown as maximum projection images. Fluorescence of Syto-59 (blue triangles), Alexa Fluor 546-conjugated anti-V. atypica antibodies (open squares), and GFP (green triangles) are shown at each 0.5-μm-spaced optical slice of the confocal stack.
Fig. 4.
Confocal scanning laser microscopic analysis of S. gordonii (pPamy-′gfp) monospecies (uninduced control) biofilms. Maximum projection images of fluorescence from channels for the detection of Syto-59 (Left) and GFP (Right) are shown.
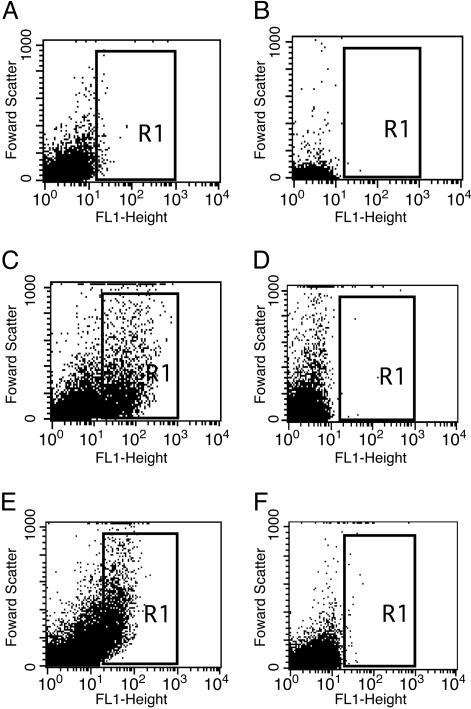
A Diffusible Signal Mediates Communication Between S. gordonii and V. atypica. It has been proposed that interspecies communication can be mediated by diffusible signaling molecules such as the autoinducer 2 (AI-2)-based quorum-sensing system (22, 23). To determine whether the basis of communication between S. gordonii and V. atypica could be a diffusible signal, we examined induction of the amylase gene in a closed system where signaling molecules could accumulate without being washed out of the flowcell, i.e., in batch cultures. S. gordonii (pPamy-′gfp) was incubated alone and in the presence of V. atypica PK1885 that is unable to coaggregate with S. gordonii (8). Fluorescence of GFP was measured by flow cytometry. Fluorescence from monocultures of V. atypica and S. gordonii (pPamy-′gfp) were similar, with a mean fluorescence of 3 units (Fig. 5 A and B). This level of fluorescence was identical to that seen in S. gordonii cells containing the promoterless gfp control plasmid, pPE1010 (data not shown). When S. gordonii (pPamy-′gfp) was incubated with V. atypica, an increase in fluorescence occurred in the coculture population (Fig. 5C), but not when the negative control, S. gordonii (pPE1010) was incubated with V. atypica (Fig. 5D). The cells with increased fluorescence had a mean fluorescence value of 66 units, indicating an increase in gfp expression from the amyB promoter during coculture with V. atypica.
Fig. 5.
Flow cytometric analysis of Pamy-directed GFP expression. S. gordonii (pPamy-′gfp) and V. atypica as monospecies cultures (A and B, respectively) and coculture (C). No increase in fluorescence was seen when S. gordonii containing the gfp reporter plasmid with no promoter, pPE1010, was incubated with V. atypica (D). (E and F) Fluorescence from S. gordonii (pPamy-′gfp) when separated by dialysis tubing from V. atypica (E) and sterile medium (F). Fluorescence (FL1-height) is graphed logarithmically on the x axis. Forward scatter (y axis) is indicative of particle size.
To verify that communication occurred via a diffusible signal rather than contact with V. atypica, fluorescence from S. gordonii (pPamy-′gfp) was measured in cells that were separated from V. atypica by dialysis tubing (14,000-kDa molecular mass cutoff). After 4 h of incubation, S. gordonii (pPamy-′gfp) became fluorescent when separated from V. atypica cultures (Fig. 5E), and no fluorescence was observed in cultures when sterile medium was used in place of the V. atypica culture (Fig. 5F). This result indicates that cell–cell communication between S. gordonii and V. atypica does not require contact of cells in the closed system of a batch culture and points to the existence of a diffusible factor mediating this interaction.
Discussion
Here we present evidence of interspecies communication between S. gordonii and V. atypica, two early colonizing species of the human dental plaque biofilm. Although we have not yet identified the mechanism of this interaction, effective signaling required that the organisms be in very close proximity during growth in the open system of the flowcell. However, when grown in a closed system, such as a batch culture, proximity was not required for signaling. This suggests the involvement of a diffusible signal that acts over very short distances in a flowing environment, but can reach a threshold level sufficient for induction in a closed system and result in cell-cell communication without contact between cells. With the exception of the archetypal model system of cell–cell signaling in myxobacteria (24, 25), distances over which signaling molecules act are not routinely examined; this is partially because most investigations have involved closed systems (batch cultures), where signal accumulates to the threshold concentration by default. However, in open systems, signal can be transposed from its point-of-origin by fluid flow. Not only could steep signal-concentration gradients result, but signal would also be constantly removed from the system. It has been proposed that, in natural environments, the function of quorum-sensing molecules may be to determine the extent of diffusion and mixing in the cell's immediate environment rather than to assess the size of the bacterial population (26). In the natural dental plaque environment, it is likely that flow of saliva would prevent signaling over distances exceeding a few micrometers. Under such circumstances, only cells in the immediate vicinity of the signal generation have the opportunity to experience a threshold signal concentration. In the present experiments, it is clear that streptococcal colonies located only a few micrometers away from veillonellae do not induce amylase expression, whereas those directly abutting veillonellae do. It is conceivable that, rather than producing a signal, veillonellae remove a signal produced by streptococci. If we assume that the salivary flow is sufficient to remove signal in the absence of veillonellae, then all streptococcal cells should be expressing GFP. However, GFP is produced only when veillonellae are in direct contact with S. gordonii. Consequently, the ability to coaggregate may be critical to establish the juxtaposition required for signaling when growing in dental plaque.
Others have shown that interspecies communication in biofilms includes the induction of gene expression in communities growing on aromatic compounds (27). In this case, signaling was shown between two organisms that were using the same carbon source for growth. Induction was attributed to the detection of a leaked shared pathway intermediate. In further studies, this signaling was shown to influence the positioning of cells within the community (28). Because S. gordonii and V. atypica use different substrates and, consequently, different pathways for energy generation and growth, induction in this system is likely to be based on a signal other than a shared metabolic intermediate. Communication among oral bacteria includes coaggregation and production of autoinducer-2 (AI-2) molecules (1). S. gordonii is known to produce AI-2 (29, 30); however, we have been unable to detect AI-2- or acyl-homoserine lactone-based signals in V. atypica cultures. This observation suggests that the signal supplied by V. atypica to S. gordonii is either a different type of molecule than those described in previously studied quorum sensing systems, or that the signal is similar, but not detectable by the available bioassays. Other compounds that could serve as signals could include propionate and acetate, the end-products of lactic acid metabolism by V. atypica. A response caused by change in pH mediated by the organic acids produced by V. atypica is unlikely because signaling occurred under highly buffered conditions. Lastly, we have attempted to induce reporter expression in S. gordonii by exposure to supernatants from V. atypica cultures; however, these experiments yielded equivocal results. Thus, the mechanism of this interaction may involve additional features other than signal production by one organism and signal sensing by the other.
This example of interspecies communication results in the expression of the S. gordonii amylase gene, amyB. Comparison of the S. gordonii amyB sequence to those of S. mutans (18) and S. bovis (14) suggest a specific intracellular function in breakdown of stored carbohydrate polymer. However, we have also detected extracellular amylase activity in both the starch plate assay and the quantitative fluorescence-based assay when cells are grown with V. atypica. We cannot exclude the possibility that this enzyme could function extracellularly during coculture of these organisms. The fermentation of either stored carbohydrate polymer or exogenous carbohydrate by S. gordonii would result in the formation of lactic acid, the preferred substrate for energy generation by V. atypica. Consequently, expression of the S. gordonii amyB would be beneficial to V. atypica when growing in close proximity, emphasizing the relevance of signaling over very short distances. We propose that juxtaposition is required for effective interspecies signaling in natural, open systems.
Author contributions: P.G.E., R.J.P., and P.E.K. designed research; P.G.E. and R.J.P. performed research; P.G.E., R.J.P., and P.E.K. analyzed data; and P.G.E., R.J.P., and P.E.K. wrote the paper.
Abbreviations: THB, Todd–Hewitt broth; THBL, THB supplemented with lactic acid.
References
- 1.Kolenbrander, P. E., Andersen, R. N., Blehert, D. S., Egland, P. G., Foster, J. S. & Palmer, R. J., Jr. (2002) Microbiol. Mol. Biol. Rev. 66, 486-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palmer, R. J., Kazmerzak, K. M., Hansen, M. C. & Kolenbrander, P. E. (2001) Infect. Immun. 69, 5794-5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palmer, R. J., Jr., Gordon, S. M., Cisar, J. O. & Kolenbrander, P. E. (2003) J. Bacteriol. 185, 3400-3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mikx, F. H. & Van der Hoeven, J. S. (1975) Arch. Oral Biol. 20, 407-410. [DOI] [PubMed] [Google Scholar]
- 5.Hughes, C. V., Kolenbrander, P. E., Andersen, R. N. & Moore, L. V. (1988) Appl. Environ. Microbiol. 54, 1957-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cisar, J. O., Kolenbrander, P. E. & McIntire, F. C. (1979) Infect. Immun. 24, 742-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Macrina, F. L., Wood, P. H. & Jones, K. R. (1980) Infect. Immun. 28, 692-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hughes, C. V., Roseberry, C. A. & Kolenbrander, P. E. (1990) Arch. Oral Biol. 35, 123S-125S. [DOI] [PubMed] [Google Scholar]
- 9.Hansen, M. C., Palmer, R. J., Jr., Udsen, C., White, D. C. & Molin, S. (2001) Microbiology 147, 1383-1391. [DOI] [PubMed] [Google Scholar]
- 10.Dunny, G. M., Lee, L. N. & LeBlanc, D. J. (1991) Appl. Environ. Microbiol. 57, 1194-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Sullivan, D. J. & Klaenhammer, T. R. (1993) Gene 137, 227-231. [DOI] [PubMed] [Google Scholar]
- 12.Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A. & Struhl, K. (1990) Current Protocols in Molecular Biology (Greene, New York).
- 13.Lunsford, R. D. (1995) Plasmid 33, 153-157. [DOI] [PubMed] [Google Scholar]
- 14.Satoh, E., Uchimura, T., Kudo, T. & Komagata, K. (1997) Appl. Environ. Microbiol. 63, 4941-4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lau, P. C., Sung, C. K., Lee, J. H., Morrison, D. A. & Cvitkovitch, D. G. (2002) J. Microbiol. Methods 49, 193-205. [DOI] [PubMed] [Google Scholar]
- 16.Hughes, C. V., Andersen, R. N. & Kolenbrander, P. E. (1992) Infect. Immun. 60, 1178-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Terleckyj, B., Willett, N. P. & Shockman, G. D. (1975) Infect. Immun. 11, 649-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simpson, C. L. & Russell, R. R. (1998) J. Bacteriol. 180, 4711-4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sabelnikov, A. G., Greenberg, B. & Lacks, S. A. (1995) J. Mol. Biol. 250, 144-155. [DOI] [PubMed] [Google Scholar]
- 20.Voskuil, M. I. & Chambliss, G. H. (1998) Nucleic Acids Res. 26, 3584-3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hueck, C. J., Hillen, W. & Saier, M. H., Jr. (1994) Res. Microbiol. 145, 503-518. [DOI] [PubMed] [Google Scholar]
- 22.Miller, M. B. & Bassler, B. L. (2001) Annu. Rev. Microbiol. 55, 165-199. [DOI] [PubMed] [Google Scholar]
- 23.Bassler, B. L., Greenberg, E. P. & Stevens, A. M. (1997) J. Bacteriol. 179, 4043-4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Igoshin, O. A., Welch, R., Kaiser, D. & Oster, G. (2004) Proc. Natl. Acad. Sci. USA 101, 4256-4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaiser, D. (2003) Nat. Rev. Microbiol. 1, 45-54. [DOI] [PubMed] [Google Scholar]
- 26.Redfield, R. J. (2002) Trends Microbiol. 10, 365-370. [DOI] [PubMed] [Google Scholar]
- 27.Møller, S., Sternberg, C., Andersen, J. B., Christensen, B. B., Ramos, J. L., Givskov, M. & Molin, S. (1998) Appl. Environ. Microbiol. 64, 721-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christensen, B. B., Haagensen, J. A., Heydorn, A. & Molin, S. (2002) Appl. Environ. Microbiol. 68, 2495-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McNab, R., Ford, S. K., El-Sabaeny, A., Barbieri, B., Cook, G. S. & Lamont, R. J. (2003) J. Bacteriol. 185, 274-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blehert, D. S., Palmer, R. J., Jr., Xavier, J. B., Almeida, J. S. & Kolenbrander, P. E. (2003) J. Bacteriol. 185, 4851-4860. [DOI] [PMC free article] [PubMed] [Google Scholar]