Abstract
Galanin is a neuropeptide with a wide variety of biological functions. Few nonpeptide ligands, capable of activating galanin receptors, are available today. Based on known pharmacophores of galanin and the tripeptidomimetic galnon, a combinatorial library was formulated, synthesized, and screened against the galanin receptor. An active compound, galmic, was identified and tested in vitro and in vivo for its affinity and efficacy at galanin receptors. The present work describes the total synthesis of galmic, the synthesis of its oxazole precursors, the coupling of the building blocks into a linear trimer, and the macrolactamization reaction.
Keywords: galnon, mimetic
Galanin is a 29-aa-long (30 aa in humans) neuroendocrine peptide, whose physiological functions are regulated by G protein-coupled receptors. Galanin is found throughout the central and peripheral nervous systems and the gastrointestinal tract (1–4). The effects of this peptide include inhibition of insulin release (5–7), inhibition of neurotransmiters involved in regulation and memory acquisition (8), modulation of food intake (9, 10) and sexual behavior (11), as well as effects on controlling pain threshold (12, 13) and in the pathogenesis of Alzheimer's disease (14).
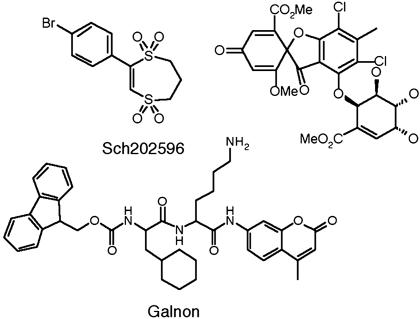
Studies involving l-Ala-substituted analogues of galanin have identified Trp-2, Asn-5, and Tyr-9 as well as the N-terminal amino group as being critical determinants for receptor binding. These amino acids are located in the N-terminal helical part of galanin that it is made up of the same 15 aa in all species from which it was isolated (15, 16). Fragments of galanin have shown low binding affinity but these peptides are vulnerable to the action of the peptidases and unable to cross the blood–brain barrier (17, 18). Two nonpeptide antagonists of the galanin receptor have been reported, spirocoumaranon (Sch202596) (19) and 2,3-dihydro-2-(4-methylphenyl)-1,4-dithiepin-1,1,4,4-tetroxide (20) (see Fig. 1), but there is only one nonpeptide galanin receptor ligand, galnon, that shows agonist activity (21).
Fig. 1.
Nonpeptide ligands of the galanin receptor.
In this study, we report an example of a nonpeptide subtype selective agonist for the galanin receptor, galmic, discovered by application of a combinatorial library approach (22).
Methods
Experimental. Reactions were performed under an argon atmosphere, NMR spectra were recorded on a Bruker DRX 600 spectrometer (unless otherwise mentioned), and the chemical shifts are given in ppm relative to tetramethylsilane. The spectra were referenced to deuterated solvents indicated in brackets in the analytical data. High-resolution MALDI-Fourier transform MS (FTMS) measurements were performed on an IonSPec (Lake Forest, CA) FTMS mass spectrometer by using 2,5-dihydroxybenzoic acid as the matrix. For TLC, silica plates IB2-F (J. T. Baker) were used. For column chromatography, silica gel 60, 230–400 mesh (Merck) was used. Preparative chromatographic plates and silica gel 60 F254 (20 × 20 cm, 0.5 mm) were used. Anhydrous solvents and chemicals were used as purchased from commercial suppliers.
General Procedures. (i) Peptide coupling conditions. To a stirred solution of acid compound (1 eq) and the trifluoroacetic acid (TFA) salt of the amino compound (1 eq) in N,N-dimethylformamide (DMF) (0.2 M) were added diphenylphosphine azide (1.2 eq) or 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDCI) (1.2 eq), and N-hydroxybenzotriazole (HOBt) (1.2 eq) at –20°C followed by the slow addition of N,N-diisopropylethylamine (DIEA) (3.6 eq). Stirring was continued for 20 h while the mixture was warmed up to room temperature (RT). Solvent was evaporated, and the residue was dissolved in EtOAc and extracted with 5% aqueous HCl, saturated NaHCO3 solutions, and brine. The organic layer was dried over MgSO4 and concentrated in vacuo. Purification was accomplished by chromatography on silica gel by using EtOAc/hexanes as eluent.
(ii) Hydrolysis of the methyl esters. Method a. The methyl ester was dissolved in a NaOH 5% solution in ethanol while cooling at 0°C. After 10 min, the reaction was completed and the solution was acidified with 5% HCl solution until pH = 1. Then EtOAc was added and the two phases separated; the organic layer was dried over MgSO4 and evaporated in vacuo to yield the corresponding acid.
Method b. The methyl ester was dissolved in acetone (0.1 M) and a solution of LiOH (1 eq) in H2O was added while cooling at 0°C. After 10 min, the reaction was completed and the solution was acidified with 5% HCl solution until pH = 1. Then EtOAc was added, and the organic layer was separated, dried over MgSO4, and evaporated in vacuo to yield the corresponding acid.
(iii) Synthesis of oxazole building blocks. To a stirred solution of dipeptide (1 eq) in dichloromethane (DCM) (0.1 M) was added Dess-Martin periodinane (1.5 eq) at 0°C. The ice bath was removed after 30 min and the mixture was stirred for 4 h longer at RT. To this mixture saturated Na2S2O3 and NaHCO3 solutions were added and stirring was continued for 45 min. Then the phases were separated and the organic layer was washed with brine, dried over MgSO4, and concentrated in vacuo. The amidoketone was purified by flash chromatography using a mixture of EtOAc/hexanes (1:2) as eluent. To a stirred solution of the ketone in DCM (0.01 M), triethylamine (4 eq) and 4-dimethylaminopyridine (DMAP) (0.1 eq) were added. The mixture was stirred for 1 h and then triphenylphosphine (1 eq) and iodine (1 eq) were added at 0°C. The cooling bath was removed after 20 min, and stirring was continued at RT for 2 h. The solvent was removed in vacuo, the residue was dried overnight and diluted with diethylether, and the triphenylphosphine oxide was filtrated. Pure oxazole was obtained after chromatography on silica gel by using EtOAc/hexanes (1:4) as eluent.
(iv) Deprotection of t-butyloxycarbonyl (Boc). Protected compound was dissolved in anhydrous DCM (0.1 M) and TFA (50–100 eq) was added slowly at RT. Stirring was continued until TLC showed the consumption of all starting material (usually 0.5–3 h) then the mixture was concentrated in vacuo (EtOAc or MeOH was coevaporated several times to remove traces of TFA) to yield the TFA salt of the pure deprotected amino compound.
(v) Deprotection of benzyl esters. A solution of the corresponding benzyl compound (1 eq) in EtOAc and Pd/C (10%) was stirred under a hydrogen atmosphere for 3–5 h, then filtered through Celite. The filtrate was evaporated, yielding the corresponding acid.
(vi) Synthesis of the pentafluorophenylester. To a solution of carboxylic acid (1 eq) in EtOAc (0.1 M), pentafluorophenol (1.1 eq) and N,N-dicyclohexylcarbodiimide (1.1 eq) were added while cooling at –20°C. The reaction mixture was allowed to warm to RT overnight, filtered from urea, and evaporated.
(vii) Macrolactamization reaction. The linear trimer (1 eq) was injected dropwise into a solution of DMAP (3 eq) in toluene at 95°C. After the addition, the macrocycle was the only product detected by TLC. Solvent was evaporated and residue was purified by chromatography on silica gel by using EtOAc/hexanes as eluent.
Synthesis of the Library Mixtures. To a solution of the triacid platform 1 (1 eq) and the amines cyclohexylmethylamine 2 (1 eq), 9-fluorenylmethylamine 3 (1 eq) and εt-Boc-l-lysine 4 (1 eq) or the amino acids Trp (1 eq), Asn (1 eq), and Tyr (1 eq) in DMF (0.2M) were added to EDCI (1.2 eq), and HOBt (1.2 eq) at –20°C followed by the slow addition of DIEA (3.6 eq). Stirring was continued for 20 h while allowing the mixture to warm up to RT. Solvent was removed in vacuo, and the obtained residue was washed with 10% HCl and saturated NaHCO3 solutions to remove any unreacted starting material. The mixtures were treated with 50% TFA in DCM and stirred for 1 h to deprotect the Boc side chains. After this time reaction mixtures were dried by evaporation and filtrated upon precipitation with diethyl ether.
The libraries were analyzed by HPLC-MS for characterization. Experiments were performed on an IonSPec FTMS mass spectrometer operated in the electrospray ionization mode. The column used was a Biobasic C4 with a flow rate of 1.5 ml/min. The gradient was of 20% acetonitrile (MeCN) (0.05% TFA) in H20 (0.05% TFA) for 2 min followed by a gradient of 50% MeCN (0.05% TFA) in H20 (0.05% TFA) for 18 min and then holding for another 20 min. The wavelength used for the detection was 213 nm. The mass range was m/z from 500 to 1,300.
The peaks corresponding to the expected products were detected and mixture was screened for 125I-galanin displacement activity in equilibrium binding assay. The mixture was then deconvoluted by HPLC under the same conditions described above. Fractions were collected every 2 min. In total, 20 fractions were further purified. Fractions were collected, analyzed by HPLC-MS, freeze-dried, and screened for 125I-galanin displacement activity.
Synthesis of Diester 13. To a solution of 60 g (0.24 mol) of carboxylic acid 12 in 1 liter of DCM at RT, 55.5 g (1.1 eq) of N,N-dicyclohexylcarbodiimide, 30 ml of isopropanol (1.5 eq), and 3 g (0.1 eq) of DMAP were added. The reaction mixture was stirred for 20 h at RT. The solution was filtered and concentrated. The residue was purified by flash chromatography on silica gel (AcOEt/hexanes, 1:3) to give 59 g (84%) of a white solid. 1H NMR-300 (CDCl3): 5.96 (s, NH); 5.02 (m, 1H); 3.75 (s, 3H); 1.71 (s, 3H); 1.42 (s, 9H); 1.22 (pseudo t, J = 4.5 Hz, 6H); 13C NMR-150 (CDCl3): 169.39; 168.11; 153.83; 80.03; 70.10; 62.83; 53.04; 28.10; 21.30; 21.26; 19.35. MALDI-FTMS [M+Na]+: expected, 312.1417; observed, 312.1415.
Synthesis of Ester 14. To a solution of 59 g (0.2 mol) of diester 13 in 560 ml of tetrahydrofuran (THF), 11 g (1.3 eq) of lithium hydroxide in 280 ml of water was added at 0°C. The reaction mixture was allowed to warm up to RT for 30 min. THF was evaporated under reduced pressure (T < 35°C), and the residue was acidified with 400 ml of 5% HCl. The carboxylic acid was extracted with 3 × 400 ml of Et2O, and the organic phase was separated, washed with 200 ml of brine, dried over MgSO4, and concentrated to give 56 g (98%) of carboxylic acid 14 as an viscous oil. 1H NMR-300 (CDCl3): 12.35 (s broad, 1H, OH); 6.00 (s, 1H, NH); 5.18 (m, 1H); 1.74 (s, 3H); 1.43 (s, 9H); 1.23 (d, J = 6.3 Hz, 6H). MALDI-FTMS [M+Na]+: expected, 298.1261; observed, 298.1261.
Synthesis of Peptide 15a. The coupling of the amino acid 14 (17 g) with cyclohexylmethylamine 2 following general procedure i (diphenylphosphine azide) afforded the corresponding amide (15.5 g, 68%) as a white powder. 1H NMR-300 (CDCl3): 6.37 (pseudo s, NH); 6.07 (s, NH); 5.07 (hept, J = 6.3 Hz, 1H); 3.19 (m, 1H); 3.05 (m, 1H); 1.76 (s, 3H); 1.68 (m, 7H); 1.46 (s, 9H); 1.27 (d, J = 7.8, 3H); 1.25 (d, J = 7.8, 3H); 1.18 (m, 2H); 0.92 (m, 2H). 13C NMR-75 (APT, CDCl3): 171.2 (+); 167.9 (+); 154.2 (+); 70.1 (–); 62.8 (+); 46.1 (+); 37.9 (–); 30.6 (+); 28.2 (–); 27.3 (–); 26.2 (+); 25.7 (+); 22.6 (–); 21.4 (–). MALDI-FTMS [M+Na]+: expected, 393.2360; observed, 393.2360.
Synthesis of Peptide 15b. To a solution of 17 g (61.75 mmol) of carboxylic acid 5 in 340 ml of tetrahydrofuran at –20°C was added 6.8 ml (1 eq) of N-methylmorpholine and 8 ml (1 eq) of isobutylchloroformate. The reaction mixture was allowed to warm up to 0°C in 30 min. A suspension of 13.6 g of 9-fluorenylamine hydrochloride (1 eq) in 70 ml DMF was added followed by 6.8 ml (1 eq) of N-methylmorpholine. The reaction mixture was stirred at RT for 5 h. The solvent was evaporated, and 850 ml of EtOAc was added. The organic phase was washed with 2 × 170 ml of water, 170 ml of HCl 5%, and 170 ml of saturated solution of NaCl, dried over MgSO4, evaporated, and purified by flash chromatography on silica gel (EtOAc/hexanes, 1:4; Rf = 0.24) to give 18 g (65%) of a white solid. 1H NMR-300 (CDCl3): 7.70 (dd, J1 = 3 Hz, J2 = 7.2 Hz, 2H); 7.50 (pseudo t, J = 8.1 Hz, 2H); 7.39 (pseudo q, J = 6.6 Hz, 2H); 7.33–7.23 (m, 2H); 6.53 (d, J = 8.7 Hz, 1H); 6.20 (d, J = 9 Hz, 1H); 6.09 (s, 1H); 5.04 (hept, J = 6.6 Hz, 1H); 1.84 (s, 3H); 1.47 (s, 9H); 1.26 (d; J = 6.3, 3H); 1.24 (d; J = 6.3, 3H). MALDI-FTMS [M+Na]+: expected, 461.2045; observed, 461.2045.
Synthesis of Amino Acid 16a. The isopropyl ester 15a (30 g) was hydrolyzed following general procedure ii (method a) to yield the acid (26 g, 98%) as a white powder. 1H NMR-300 (CDCl3): 7.54 (s, 1H); 7.03 (s, 1H); 5.73 (s, 1H); 3.24 (m, 1H); 3.05 (m, 1H); 1.72 (m, 4H); 1.68 (s, 3H); 1.54 (m, 1H); 1.41 (s, 9H); 1.21 (m, 3H); 0.95 (m, 2H). 13C NMR-75 [routine + distortionless enhancement by polarization transfer (DEPT), CDCl3]: 46.4 (CH2); 37.7 (CH); 30.7 (CH2); 28.0 (CH3); 26.2 (CH2); 25.7 (CH2); 25.2 (CH3). MALDI-FTMS [M+Na]+: expected, 351.1890; observed, 351.1895.
Synthesis of Amino Acid 16b. The isopropyl ester 15b (18 g) was hydrolyzed following general procedure ii (method a) to yield the acid (16 g, 98%) as a white powder. 1H NMR-300 (CDCl3): 7.69 (m, 2H); 7.60–7.20 (m, 6H); 7.04 (s, 1H); 6.19 (s, 1H); 6.16 (s, 1H); 1.74 (s; 3H); 1.40 (s, 9H). MALDI-FTMS [M+Na]+: expected, 419.1577; observed, 419.1572.
Synthesis of Dipeptide 17a. The dipeptide 17a was obtained following general procedure i (EDCI) in 80% yield (31 g) as a white solid. 1H NMR-300 (CDCl3): 7.54 (s, 1H); 7.03 (s, 2H); 5.73 (s, 1H); 3.24 (s, 1H); 3.05 (s, 1H); 1.72 (m, 5H), 1.68 (s, 3H) 1.54 (m, 1H); 1.41 (s, 9H); 1.21 (m, 3H); 0.95 (m, 2H). 13C NMR-75 (routine + DEPT, CDCl3): 46.40 (CH2); 37.69 (CH); 30.72 (CH2); 28.10 (CH3); 26.25 (CH2); 25.73 (CH2); 25.23 (CH3). MALDI-FTMS [M+Na]+: expected, 542.2837; observed, 542.2837.
Synthesis of Dipeptide 17b. The dipeptide 17b was obtained following general procedure i (EDCI) in 78% yield (18.8 g) as a white solid. 1H NMR-300 (CDCl3): 7.88 (s, 1H); 7.67 (d, J = 7.8 Hz, 2H); 7.58 (pseudo t, J = 5.6 Hz, 2H); 7.38 (pseudo t, J = 6.3 Hz, 2H); 7.31–7.23 (m, 7H); 6.17 (d, J = 8.7 Hz, 1H); 6.09 (s, 1H); 5.21 (d, J = 12 Hz, 1H); 5.08 (d, J = 12 Hz, 1H); 4.55 (dd, J1 = 8.4 Hz, J2 = 3 Hz, 1H); 4.39 (m, 1H); 3.20 (s, 1H); 1.87 (s, 3H); 1.48 (s, 9H); 1.24 (d, J = 6.6 Hz, 3H). 13C NMR-150 (CDCl3): 169.78; 143.71; 143.57; 140.37; 134.98; 128.43; 128.31; 128.14; 128.02; 127.51; 127.45; 119.70; 67.87; 67.19; 62.89; 60.35; 58.122; 55.03; 28.18; 21.03; 20.11; 14.20. MALDI-FTMS [M+Na]+: expected, 610.2524; observed, 610.2524.
Synthesis of Oxazole 8a. The oxazole 18a was prepared following general procedure iii (15.6 g, 81%) as a colorless oil. 1H NMR (acetone): 7.16–7.51 (m, H-ar, 5H); 6.66 (s, NH, 1H); 5.35 (m, CH2, 1H); 3.07–3.10 (m, CH2, 1H); 3.02–3.03 (m, CH2, 1H); 2.86 (s, CH, 1H); 2.57 (s, CH3, 3H); 1.59–1.66 (m, CH, CH2, 5H); 1.66 (s, CH3, 3H); 1.37 [s, (CH3)3, 9H]; 1.11–1.22 (m, CH2, 4H); 0.84–0.86 (m, CH2, 2H). 13C NMR (CD3OD): 174.77; 173.91; 156.80; 81.29; 62.76; 47.22; 39.22; 31.92; 28.76; 27.63; 27.11; 22.01. MALDI-FTMS [M+Na]+: expected, 422.20.19; observed, 422.2062. The Boc group was deprotected following general procedure iv to afford 8a in a 95% yield.
Synthesis of Oxazole 8b. The oxazole 18b was prepared following general procedure iii (15.8 g, 87%) as a colorless oil. 1H-NMR (CDCl3): 7.59 (d, J = 7.4 Hz, H-ar, 2H); 7.15–7.63 (m, H-ar, 11H); 6.84 (d, J = 7.6 Hz, NH, 1H); 6.47 (s, NH, 1H); 6.10 (d, J = 8.7 Hz, CH, 1H); 5.13 (s, CH2, 2H); 2.52 (s, CH3, 3H); 1.97 (s, CH3, 3H); 1.34 [s, C(CH3)3, 9H]. 13C-NMR (CDCl3): 169.59; 161.67; 161.37; 157.35; 154.05; 143.84; 143.68; 140.68; 140.66; 135.58; 128.85; 128.68; 128.45; 128.43; 127.82; 127.79; 127.63; 125.11; 124.91; 120.32; 120.14; 120.06; 80.55; 66.69; 59.38; 55.31; 28.31; 12.40. MALDI-FTMS [M+Na]+: expected, 590.1788; observed, 590.1731. The Boc group was deprotected following general procedure iv to afford 8b in a 95% yield (15.3 g).
Synthesis of the Dimer 19. The oxazoles 7 (1.36 g) and 8a (1.37 g) were coupled following general procedure i (EDCI) to afford the dimer 19 in 70% yield (1.67 g). 1H NMR (CDCl3): 8.41 (s, NH, 1H); 7.32–7.42 (m, H-ar, 5H); 6.84 (t, J = 5.9 Hz, NH, 1H); 6.08 (s, NH, 1H); 5.36 (s, CH2, 2H); 3.77 (s, CH3, 3H); 3.07–3.12 (m, CH2, 2H); 2.53 (s, CH3, 3H); 2.52 (s, CH3, 3H); 2.02 (s, CH3, 3H); 1.97 (s, CH3, 3H); 1.58–1.66 (m, CH, 6H); 1.44 [s, (CH3)3, 9H]; 1.15–1.24 (m, CH, 3H); 0.79–0.83 (m, CH, 2H). 13C-NMR (CDCl3): 191.86; 171.18; 170.06;. 168.27; 161.61; 160.96; 158.78; 157.27; 154.55; 154.10; 135.81; 129.2; 128.70; 128.59; 128.53; 128.51; 128.47; 128.43; 127.87; 80.65; 66.69; 60.46; 58.95; 58.34; 55.23; 53.77; 53.72; 46.44; 37.82; 31.67; 30.70; 30.63; 28.30; 26.41; 25.83; 24.19; 21.11; 14.29; 12.45; 11.88. MALDI-FTMS [M+Na]+: expected, 732.3215; observed, 732.3212.
Synthesis of the Trimer 20. The benzyl ester of the dimer 19 (1.67 g) was deprotected following general procedure v to give the corresponding acid that was coupled with 8b following general procedure i (EDCI). The trimer 20 was obtained in 60% yield (1.50 g) as a colorless foam. 1H NMR (CDCl3): 8.53 (s, NH, 1H); 8.46 (s, NH, 1H); 7.63 (t, J = 8.0 Hz, 2H); 7.46 (d, J = 7.5 Hz, 1H); 7.35 (t, J = 8.0 Hz, 1H); 7.31–7.14 (m, 4H); 6.91 (d, J = 8.0 Hz, 1H); 6.66 (broad s, 1H); 6.12 (d, J = 8.6 Hz, 2H); 5.20 (d, J = 3.8 Hz, 2H); 3.84 (s, 3H); 3.12 (m, 2H); 2.52 (s, 3H); 2.50 (s, 3H); 2.49 (s, 3H); 2.09 (s, 3H); 2.03 (s, 3H); 1.96 (s, 3H); 1.66–1.58 (m, 7H); 1.22–1.08 (m, 2H); 1.38 [s, (CH3)3, 9H]; 0.92–0.76 (m, 2H). 13C NMR (CDCl3): 170.02; 169.48; 168.36; 161.63; 161.00; 160.83; 160.73; 160.50; 157.37; 155.76; 143.47; 143.34; 140.70; 140.62; 135.55; 129.09; 129.04; 128.95; 128.90; 128.64; 128.60; 128.57; 128.49; 128.36; 127.93; 127.79; 127.74; 124.88; 124.67; 120.19; 120.15; 66.59; 60.41; 58.88; 58.54; 58.34; 55.45; 55.16; 53.71; 46.42; 37.75; 30.70; 30.63; 29.72; 28.25; 26.39; 25.82; 23.80; 23.75; 22.62; 21.05; 18.93; 14.23; 12.40; 11.87. MALDI-FTMS [M+Na]+: expected, 1091.4485; observed, 1091.4474.
Synthesis of the Pentafluorophenyl Ester 21. The benzyl ester of the trimer 20 (1.50 g) was deprotected following general procedure v to give the corresponding acid that was coupled with pentafluorophenol following general procedure vi. Deprotection of the Boc group following general procedure iv afforded the trimer 21 in a 82% overall yield (1.20 g). 1H NMR (CDCl3): 9.45 (t, J = 5.4 Hz, NH, 1H); 9.01 (s, NH, 1H); 8.76 (s, NH, 1H); 7.73 (d, J = 2.5 Hz, 1H); 7.70 (d, J = 2.5 Hz, 1H); 7.52 (d, J = 7.5 Hz, 1H); 7.46–7.25 (m, 6H); 6.62 (d, J = 8.4 Hz, 1H); 6.15 (d, J = 8.2 Hz, 1H); 3.86 (s, 3H); 3.34 (m, 2H); 2.73 (s, 3H); 2.50 (s, 3H); 2.49 (s, 3H); 2.23 (s, 3H); 2.14 (s, 3H); 2.10 (s, 3H); 1.67–1.42 (m, 7H); 1.24–0.76 (m, 4H). MALDI-FTMS [M+Na]+: expected, 1067.3340; observed, 1067.3341.
Synthesis of the Cyclic Product 6. Following general procedure vii the cyclic product 6 was obtained in 72% yield (710 mg) as a colorless foam. 1H NMR (CDCl3): 8.99 (s, NH, 1H); 8.92 (s, NH, 1H); 8.88 (s, NH, 1H); 7.66 (d, J = 7.5 Hz, 2H); 7.58 (d, J = 7.5 Hz, 1H); 7.36 (m, 3H); 7.29–7.19 (m, 2H); 6.70 (d, J = 9 Hz, 1H); 6.59 (t, J = 6 Hz, 1H); 6.24 (d, J = 9 Hz, 1H); 3.76 (s, 3H); 3.15 (m, 1H); 2.98 (m, 1H); 2.75 (s, 3H); 2.68 (s, 3H); 2.66 (s, 3H); 2.19 (s, 3H); 2.10 (s, 3H); 2.03 (s, 3H); 1.70–1.40 (m, 7H); 1.22–1.08 (m, 2H); 0.92–0.76 (m, 2H). 13C NMR (CDCl3): 168.36; 168.25; 167.53; 160.29; 160.23; 160.09; 160.03; 159.15; 156.26; 156.04; 155.76; 143.95; 143.79; 140.84; 140.79; 128.94 (CH); 128.89 (CH); 128.44; 128.20; 127.79 (CH); 127.77 (CH); 125.45 (CH); 124.91 (CH); 120.23 (CH); 120.16 (CH); 60.68; 60.55; 59.03; 55.53 (CH); 53.98 (CH3); 46.53 (CH2); 38.08 (CH); 30.93 (CH2); 26.74 (CH2); 26.13 (CH2); 26.12 (CH2); 22.90 (CH3); 21.88 (CH3); 21.76 (CH3); 12.25 (CH3); 12.21 (CH3); 12.15 (CH3). MALDI-FTMS [M+Na]+: expected, 883.3385; observed, 883.3385.
Synthesis of 5. The methyl ester 6 (190 mg) was hydrolyzed following general procedure ii (method b) to give the corresponding acid in 72% yield (135 mg). 1H NMR-300 (CDCl3): 8.96 (s, NH, 1H); 8.92 (s, NH, 1H); 8.89 (s, NH, 1H); 7.68 (s broad, 1H); 7.62 (d, J = 8.1 Hz, 2H); 7.55 (d, J = 6.9 Hz, 1H); 7.36–7.16 (m, 5H); 6.75 (d, J = 9 Hz, 1H); 6.60 (pseudo t, J = 6.3 Hz, 1H); 6.21 (d, J = 9 Hz, 1H); 3.14 (m, 1H); 2.92 (m, 1H); 2.73 (s, 3H); 2.67 (s, 3H); 2.62 (s, 3H); 2.17 (s, 3H); 2.07 (s, 3H); 2.03 (s, 3H); 1.65–1.35 (m, 7H); 1.20–1.00 (m, 2H); 0.90–0.72 (m, 2H). 13C NMR-75 (routine + DEPT, CDCl3): 176.65; 171.00; 168.14; 167.19; 160.53; 160.04; 159.97; 159.79; 158.58; 156.00; 155.82; 143.59; 143.39; 140.49; 128.646 (CH); 128.55 (CH); 128.11 (CH); 128.06 (CH); 127.72 (CH); 127.52 (CH); 127.46 (CH); 125.10 (CH); 124.59 (CH); 119.89 (CH); 119.84 (CH); 60.30; 60.26; 59.06; 55.24 (CH); 46.24 (CH2); 37.73 (CH); 30.58 (CH2); 26.42 (CH2); 25.79 (CH2); 22.32 (CH3); 21.58 (CH3); 21.46 (CH3); 11.93 (CH3); 11.90 (CH3); 11.88 (CH3). MALDI-FTMS [M+Na]+: expected, 869.3229; observed, 869.3221. The acid (130 mg) was coupled with εt-Boc-l-lysine following general procedure i to yield the corresponding amide in 85% yield (142 mg). 1H NMR-300 (CDCl3): 8.94 (s, NH, 1H); 8.90 (s, NH, 1H); 8.87 (s, NH, 1H); 7.64 (d, J = 7.5 Hz, 2H); 7.57 (d, J = 7.5 Hz, 1H); 7.38–7.16 (m, 5H); 7.05 (d, J = 7.5 Hz, 1H); 6.80 (m broad, 2H); 6.23 (d, J = 9 Hz, 1H); 4.80 (m broad, 1H); 4.51 (m, 1H); 3.58 (s, 3H); 3.16 (m, 1H); 3.06 (m broad, 2H); 2.98 (m, 1H); 2.75 (s, 3H); 2.67 (s, 6H); 2.18 (s, 3H); 2.11 (s, 3H); 2.10 (s, 3H); 1.88 (m, 2H); 1.72–1.10 (m, 15H); 1.43 (s, 9H); 0.92–0.76 (m, 2H). 13C NMR-75 (routine + DEPT, CDCl3): 171.67; 168.08; 167.29; 166.99; 160.19; 160.07; 159.96; 159.86; 159.81; 159.32; 155.80; 155.70; 143.60; 143.37; 140.42; 128.50; 128.44; 127.98; 127.83; 127.79; 127.45; 127.38; 125.00; 124.52; 119.82; 119.76; 60.60; 60.33; 60.27; 55.24 (CH); 52.62 (CH); 52.33 (CH3); 46.25 (CH2); 37.82 (CH); 31.71 (CH2); 30.70 (CH2); 30.65 (CH2); 29.37 (CH2); 28.52 (CH3); 26.47 (CH2); 25.87 (CH2); 22.36 (CH2); 21.77 (2 CH3); 21.39 (CH3); 12.03 (2 CH3); 11.96 (CH3). MALDI-FTMS [M+Na]+: expected, 1111.4859; observed, 1111.4843. The Boc group was deprotected following general procedure iv to yield 131 mg (100%) of 5 as a white powder. 1H NMR-300 (CDCl3): 8.96 (s, NH, 1H); 8.95 (s, NH, 1H); 8.89 (s, NH, 1H); 7.81 (s broad, 3H); 7.64 (d, J = 7.8 Hz, 2H); 7.56 (d, J = 7.5 Hz, 1H); 7.39–7.14 (m, 5H); 6.95 (t, J = 8.4 Hz, 2H); 6.22 (d, J = 9.3 Hz, 1H); 5.94 (s broad, 2H); 4.53 (m broad, 1H); 3.51 (s, 3H); 3.15 (s broad, 1H); 2.95 (m, 3H); 2.75 (s, 3H); 2.66 (s, 3H); 2.62 (s, 3H); 2.17 (s, 3H); 2.13 (s, 3H); 2.01 (s, 3H); 1.94 (m broad 1H); 1.80–1.30 (m, 14H); 1.16 (m, 2H); 0.86 (m, 2H). 13C NMR-75 (routine + DEPT, CDCl3): 171.43; 168.26; 168.03; 167.19; 161.21; 160.72; 160.46; 160.04; 159.86; 159.83; 159.12; 156.10; 156.01; 143.57; 143.42; 140.49; 140.43; 128.57 (CH); 128.53 (CH); 127.63 (CH); 127.48 (CH); 125.02 (CH); 124.59 (CH); 119.89 (CH); 119.84 (CH); 60.83; 60.27; 59.73; 55.24 (CH); 52.42 (CH3); 51.98 (CH); 46.28 (CH2); 39.65 (CH2); 37.69 (CH); 30.64 (CH2); 30.58 (CH2); 26.37 (CH2); 26.02 (CH2); 25.81 (CH2); 21.66 (CH3); 21.42 (CH3); 21.33 (CH3); 21.33 (CH2); 11.95 (CH3); 11.82 (CH3); 11.79 (CH3). MALDI-FTMS [M+H]+: expected, 989.4516; observed, 989.4514.
Results and Discussion
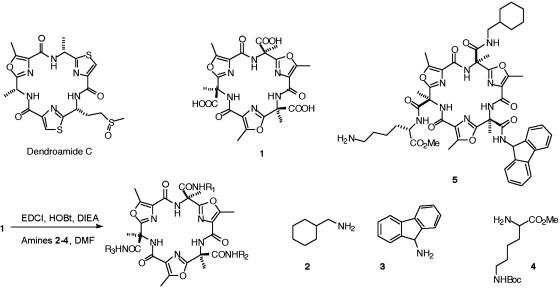
Synthesis of the Combinatorial Library. A solution phase-activated core approach was used for the synthesis of the combinatorial library. This approach can be divided into three components: the core, linker, and building blocks. As core molecule we used the triacid platform 1 developed in this research group (23–25), this triacid was inspired by a number of biologically active and peptide-derived marine natural products such as dendroamide C (26–30). These macrocycles feature oxazole rings linked by trans amide bonds. The most favored conformation features all lone pairs of oxazole nitrogens and the hydrogens of secondary amides directed to the center of the macrocycle, so the molecule is expected to be rigid and planar. The advantage of these molecules is that they can present structural diversity on a well defined rigid platform; the three side chains can act at the same time on the surface of the target.
Fig. 2 presents the synthesis of the libraries. The first step was the coupling of the triacid core molecule 1 with two sets of amines as building blocks: cyclohexylmethylamine 2, 9-fluorenylamine 3, and εt-Boc-l-lysine methyl ester 4 (based on the structure of the galnon) or the amino acids Trp, Asn, and Tyr (based on the pharmacophore groups of the galanin). We used EDCI in DMF as coupling reagent. After removing the solvent in vacuo, libraries were purified by washing the obtained residue with 10% HCl and saturated NaHCO3 solutions to remove any unreacted starting material. The mixtures were treated with 50% TFA in DCM and stirred for 1 h to deprotect the Boc side chains. After this time reaction mixtures were dried by evaporation and filtered upon precipitation with diethyl ether. All of the libraries were analyzed by HPLC-MS for initial characterization and screened for 125I-galanin displacement activity in equilibrium binding assay. The structure of the most active compound (structure 5, galmic or its cyclodiastereomer) was deduced reiteratively by deconvolution of the libraries. The different fractions were tested in the binding assay and the compounds present were identified by MS.
Fig. 2.
Synthesis of the libraries.
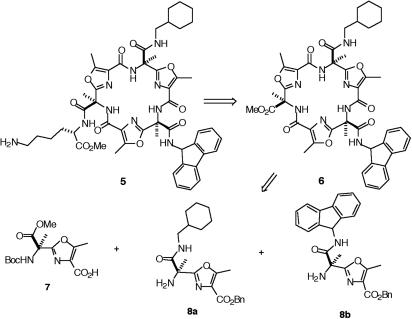
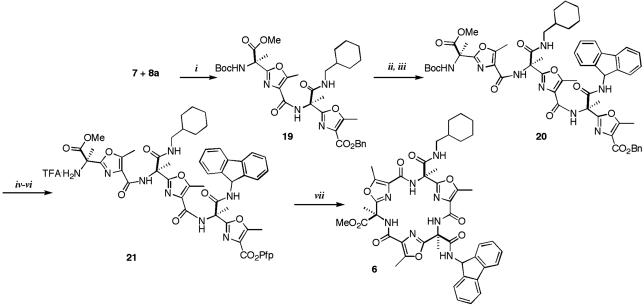
Synthesis of the Individual Compounds. The disconnection at the amide bonds of 5 leads to three oxazoles, each containing an amino and acid terminus and carrying the appropriate side chain. The exception is the amine 4 derivative that we decided to introduce as a protected lysine after the macrocyclization reaction, at the end of the synthesis (Fig. 3). This coupling is approximately the same reaction as was encountered during the synthesis of the active mixture. We chose the oxazole 7 as its precursor (23). The three different monomers 7, 8a and 8b, could be attached and successive steps of deprotection-coupling led to the cyclic product 6. Then the methyl ester could be hydrolyzed and coupled with εt-Boc-l-lysine 4 that, after Boc deprotection, afforded the enantiomerically pure galmic 5.
Fig. 3.
Retrosynthetic disconnection of galmic.
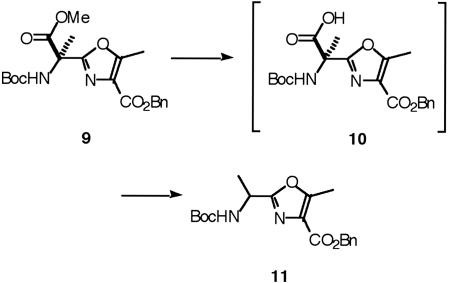
Following the procedure described (23), oxazole 9 was synthesized. The methyl ester of 9 was selectively hydrolyzed in the presence of the benzyl ester by using aqueous LiOH in acetone. Although acid 10 was detected, all our attempts to couple it with cyclohexylmethylamine 2 or 9-fluorenylamine 3 to give the corresponding oxazoles 8a and 8b were unsuccessful, and the decarboxylated product 11 was the only product isolated (Fig. 4).
Fig. 4.
Hydrolysis and decarboxylation of 9.
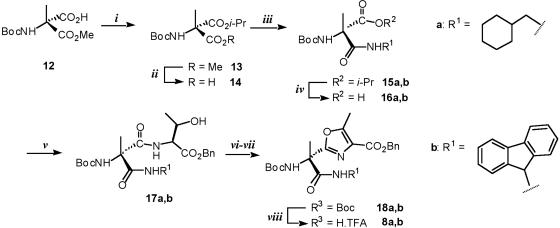
The oxazoles 8a and 8b can be traced back to a dipeptide, which consists of threonine coupled to an amino acid carrying the desired side chain. The synthesis of the oxazoles started with the transformation of optically active amino acid 12 (23) into the enantiopure amino acid 14, which was then coupled with the selected amines to give amides 15 a and b (Fig. 5). In both cases, the isopropyl ester was hydrolyzed under basic conditions and the resulting acid 16 was coupled with l-threonine by using EDCI as dehydrating reagent. Then after the oxidation of the hydroxy group by using Dess-Martin periodinane, the synthesis of oxazoles 18 a and b was accomplished in good yield by treating the corresponding amidoketone with PPh3 in the presence of I2 and Et3N (31). Following the conditions reported in the literature, the desired product was obtained in low yield (30–40%). We observed the formation of side products probably because of the competition between the two amide groups present in the molecule. The yields were significantly improved by reversing the addition of the reagents. We first formed the enolate by treatment of the ketone with triethylamine and DMAP in DCM. The mixture was allowed to react for 1 h at RT and then cooled to 0°C, then PPh3 was added followed by the slow addition of iodine. Monomers 8 a and b were obtained (after Boc deprotection with TFA) in excellent yield.
Fig. 5.
Synthesis of the oxazole monomers. Reagents and conditions: (i) i-PrOH, N,N-dicyclohexylcarbodiimide, DMAP, DCM, RT, overnight (84%); (ii) LiOH, tetrahydrofuran/H2O, RT, 30 min (98%); (iii) cyclohexylmethylamine 2, diphenylphosphine azide, DIEA, DMF, RT, overnight (68%) or 9-fluorenylamine 3, isobutyl chlorocarbonate, N-methylmorpholine, DMAP, DCM, –20°C to RT, 5 h (65%); (iv) NaOH 5%, EtOH, RT, 6 h (98%); (v) H-Thr-OBn, EDCI, HOBt, DIEA, DMF, RT, overnight (17a: 80%; 17b: 78%); (vi) Dess-Martin periodinane, DCM, RT, 1 h; (vii) triethylamine, DMAP, DCM, RT, 1 h then Ph3P, I2, 0°C to RT, 2 h (18a: 81%; 18b: 87%); (viii) TFA, DCM, RT, 3 h (95%).
Once prepared, the three monomers were taken through the successive steps of deprotection and coupling of the amines to acids to give linear trimer 21 (Fig. 6). Oxazoles 7 and 8a were coupled to form the protected dimer 19 by using EDCI. The hydrogenation of the benzyl ester and coupling with 8b afforded the linear trimer 20. Although there are several methods available for peptide cyclizations, in many examples the ring closure via the pentafluorophenyl ester method shows exceptional results (32, 33). Thus, benzyl ester 20 was transformed into a pentafluorophenyl ester, as depicted in Fig. 6. The Boc group was deprotected with TFA, and without purification the resulting residue was injected dropwise into a solution of DMAP in toluene at 95°C. The cyclic product was the only product detected by TLC, no polymerization was observed, and 6 could be obtained in a 72% yield after purification.
Fig. 6.
Synthesis of the linear trimer and macrocyclization step. Reagents and conditions: (i) EDCI, HOBt, DIEA, DMF (70%); (ii) H2/Pd-C (10%); (iii) 8b, EDCI, HOBt, DIEA, DMF (60%, two steps); (iv) H2/Pd-C (10%); (v) C6F5OH, N,N-dicyclohexylcarbodiimide, DCM; (vi) TFA, DCM, RT, 3 h (82%, three steps); (vii) DMAP, toluene, 95°C (72%).
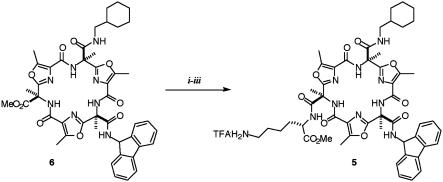
As we previously described, the lysine side chain was introduced in the last steps of the synthesis. After the hydrolysis of methyl ester 6 under basic conditions, the εt-Boc-l-lysine was coupled by using EDCI in DMF. Boc cleavage afforded the enantiomerically pure galmic (5) in good yield (Fig. 7).
Fig. 7.
Introduction of the lysine side chain. Reagents and conditions: (i) LiOH·H2O, acetone, RT, 30 min (72%); (ii) εt-Boc-l-lysine, EDCI, HOBt, DMF, DIEA (85%); (iii) TFA, DCM, RT, 3 h (quantitative).
Following a similar sequence, cyclodiastereomer 25 was obtained (see Supporting Text, which is published as supporting information on the PNAS web site, for more details).
In vitro assays showed that substituted macrocycle 5 (galmic) possessed a selective affinity for galanin receptor GalR1 at micromolar concentrations (Ki = 34.2 μM). In addition, in vivo experiments in rats showed that galmic penetrates the blood–brain barrier quickly and exhibits agonist-like properties that mimic the galanin neuropeptide (22). No such properties were found for cyclodiastereomer 25.
In summary, the synthesis of a small combinatorial library of molecules by using triacid platform 1 led to the discovery of galmic, a nonpeptide agonist of the galanin receptor GalR1. The total synthesis of galmic and its cyclodiastereomer allowed the definition of structure and provided sufficient material to study their biological properties in vivo.
Supplementary Material
Acknowledgments
We thank Dr. Wook Dong Cho for experimental assistance and The Skaggs Institute for support.
Author contributions: J.R. designed research; X.L. contributed new reagents/analytic tools; S.C.C., L.T., G.H., L.S., and T.B. performed research; and J.R. wrote the paper.
Abbreviations: Boc, t-butyloxycarbonyl; DEPT, distortionless enhancement by polarization transfer; DCM, dichloromethane; DIEA, N,N-diisopropylethylamine; DMAP, 4-dimethylaminopyridine; DMF, N,N-dimethylformamide; EDCI, 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide; FTMS, Fourier transform MS; HOBt, N-hydroxybenzotriazole; RT, room temperature; TFA, trifluoroacetic acid.
References
- 1.Tatemoto, K., Rokaeus, A., Jornvall, H., McDonald, T. J. & Mutt, V. (1983) FEBS Lett. 164, 124–128. [DOI] [PubMed] [Google Scholar]
- 2.Bersani, M., Johnsen, A. H., Højrup, P., Dunning, B. E., Andreasen, J. J. & Holst, J. J. (1991) FEBS Lett. 283, 189–194. [DOI] [PubMed] [Google Scholar]
- 3.Norberg, A., Sillard, R., Carlquist, M., Jornvall, H. & Mutt, V. (1991) FEBS Lett. 288, 151–153. [DOI] [PubMed] [Google Scholar]
- 4.Rokaeus, A. (1987) Trends Neurosci. 10, 158–164. [Google Scholar]
- 5.Tatemoto, T. J., Dupre, J., Tatemoto, K., Greenberg, G. R., Radziuk, J. & Mutt, V. (1985) Diabetes 34, 192–196. [DOI] [PubMed] [Google Scholar]
- 6.Amiranoff, B., Lorinet, A. M., Lagny-Pourmir, I. & Laburthe, M. (1988) Eur. J. Biochem. 177, 147–152. [DOI] [PubMed] [Google Scholar]
- 7.Sjoholm, A. A. & Efendic, S. (2001) Exp. Clin. Endocrinol. Diabetes 109, S109–S121. [DOI] [PubMed] [Google Scholar]
- 8.Wrenn, C. C. & Crawley, J. N. (2001) Prog. Neuropsychopharmacol. Biol. Psychiatry 25, 283–299. [DOI] [PubMed] [Google Scholar]
- 9.Crawley, J. N. (1999) Neuropeptides 33, 369–375. [DOI] [PubMed] [Google Scholar]
- 10.Kyrkouli, S. E., Stanley, B. G., Hutchinson, R., Seirafi, R. D. & Leibowitz, S. F. (1990) Brain Res. 521, 185–191. [DOI] [PubMed] [Google Scholar]
- 11.Bloch, G. J., Butler, P. C., Eckersell, C. B. & Mills, R. H. (1998) Ann. N.Y. Acad. Sci. 863, 188–205. [DOI] [PubMed] [Google Scholar]
- 12.Wiesenfeld-Hallin, Z., Xu, X.-J., Langel, U., Bedecs, K., Hökfelt, T. & Bartfai, T. (1992) Proc. Natl. Acad. Sci. USA 89, 3334–3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu, X. J., Hökfelt, T., Bartfai, T. & Wiesenfeld-Hallin, Z. (2000) Neuropeptides 34, 137–147. [DOI] [PubMed] [Google Scholar]
- 14.Hökfelt, T, Millhorn, D. & Seroogy, K. (1987) Experientia 43, 768–780. [DOI] [PubMed] [Google Scholar]
- 15.Land, T., Langel, U., Low, M., Berthold, M., Unden, A. & Bartfai, T. (1991) Int. J. Pept. Protein Res. 38, 267–272. [DOI] [PubMed] [Google Scholar]
- 16.Amiranoff, B., Lorinet, A. M., Yanaihara, N. & Laburthe, M. (1989) Eur. J. Pharmacol. 163, 205–207. [DOI] [PubMed] [Google Scholar]
- 17.Branchek, T. A., Smith, K. E., Gerald, C. & Walker, M. W. (2000) Trends Pharmacol. Sci. 21, 109–117. [DOI] [PubMed] [Google Scholar]
- 18.Floren, A., Land, T. & Langel, U. (2000) Neuropeptides 34, 331–337. [DOI] [PubMed] [Google Scholar]
- 19.Chu, M., Mierzwa, R., Truumees, I., King, A., Sapidou, E., Barrabee, E., Terracciano, J., Patel, M. G., Gullo, V. P. & Burrier, R. (1997) Tetrahedron Lett. 38, 6111–6114. [Google Scholar]
- 20.Scott, M. K., Ross, T. M., Lee, D. H., Wang, H. Y., Shank, R. P., Wild, K. D., Davis, C. B., Crooke, J. J., Potocki, A. C. & Reitz, A. B. (2000) Bioorg. Med. Chem. 8, 1383–1391. [DOI] [PubMed] [Google Scholar]
- 21.Saar, K., Mazarati, A. M., Mahlapuu, R., Hallnemo, G., Soomets, U., Kilk, K., Hellberg, S., Pooga, M., Tolf, B.-R., Shi, T. S., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 7136–7141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bartfai, T., Lu, X., Badie-Mahdavi, H., Barr, A. M., Mazarati, A., Hua, X., Yaksh, T., Haberhauer, G., Conde Ceide, S., Trembleau, L., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 10470–10475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haberhauer, G., Somogyi, L. & Rebek, J., Jr. (2000) Tetrahedron Lett. 41, 5013–5016. [Google Scholar]
- 24.Mink, D., Mecozzi, S. & Rebek, J., Jr. (1998) Tetrahedron Lett. 39, 5709–5712. [Google Scholar]
- 25.Somogyi, L., Haberhauer, G. & Rebek, J., Jr. (2001) Tetrahedron 57, 1699–1708. [Google Scholar]
- 26.Faulkner, D. (1999) J. Nat. Prod. Rep. 16, 155–198. [Google Scholar]
- 27.Pettit, G. R. (1994) Pure Appl. Chem. 66, 2271–2281. [Google Scholar]
- 28.Garson, M. G. (1993) Chem. Rev. 93, 1699–1733. [Google Scholar]
- 29.Davidson, B. S. (1993) Chem. Rev. 93, 1771–1791. [Google Scholar]
- 30.Fuesetani, N. & Matsunoga, S. (1993) Chem. Rev. 93, 1793–1806. [Google Scholar]
- 31.Wipf, P. & Miller, C. P. (1993) J. Org. Chem. 58, 3604–3606. [Google Scholar]
- 32.Schmidt, U., Utz, R., Lieberknecht, A., Griesser, H., Porzolli, B., Bahr, J., Wagner, K. & Fischer, P. (1987) Synthesis 3, 236–241. [Google Scholar]
- 33.Schmidt, U. & Utz, R. (1987) Angew. Chem. Int. Ed. Eng. 23, 725–726. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.