Abstract
Intercellular transfer of proteins across the immunological synapse is emerging as a common outcome of immune surveillance. We previously reported that target-cell MHC class I protein transfers onto natural killer (NK) cells expressing cognate killer Ig-like receptors (KIRs). We now show that, for both murine and human cells, target cells expressing inhibitory MHC class I ligands acquire cognate inhibitory NK receptors. Other cell-surface proteins, but not a cytoplasmic dye, also transferred from human NK cells to target cells across an inhibitory immunological synapse. The number of KIRs acquired from NK cells correlated with the level of expression of cognate MHC class I protein on target cells. Treatment with cytoskeletal inhibitors demonstrated that the target-cell cytoskeleton influences intercellular transfer of proteins in both directions. In contrast to constitutively expressed KIRs, a fraction of acquired KIRs could be removed by mild acid wash, demonstrating a difference between some of the acquired KIRs and constitutively expressed KIRs. An accumulation of phosphotyrosine at the location of the transferred KIRs implies a signaling capacity for NK cell proteins transferred to target cells. Thus, intercellular protein transfer between immune cells is bidirectional and could facilitate new aspects of immune cell communication.
Keywords: imaging, immunological synapse
Natural killer (NK) cell activity is regulated by a complex integration of negative and positive signals (1, 2). Cells failing to express, or expressing inadequate levels of, self-MHC molecules on their surface are lysed by NK cells, a concept known as the missing-self hypothesis (3). Receptor–ligand binding between MHC class I protein and an inhibitory NK receptor leads to an inhibitory signaling cascade by means of cytosolic immunoreceptor tyrosine-based inhibition motifs (4).
Recently, imaging intercellular communication between T, B, and NK cells and their respective target cells or antigen-presenting cells revealed that receptors commonly accumulate into supramolecular activation clusters at the immunological synapse (IS) (5, 6) and that effector cells are able to acquire proteins from target cells or antigen-presenting cells. For example, T cells acquire peptide/MHC class I proteins (7, 8) and costimulatory molecules (9) from surrounding cells, whereas B cells (10) and dendritic cells (DCs) (11) acquire antigen as well as peptide/MHC ligands from their neighbors. NK cells also acquire MHC class I proteins from a variety of cells in vitro and in vivo (12–14).
Several different mechanisms for specific intercellular protein transfer have been suggested, including proteolyic cleavage of proteins (15), exosome shedding (9, 16), or sharing of small pieces of membrane (8, 17). In addition, recent evidence suggests that proteins may also be able to transfer between cells across some distance, through membrane nanotubes (18). To date, protein acquisition by immune cells has been regarded as a unidirectional process from target cell or antigen-presenting cell to effector cell. Here, we report bidirectional transfer of proteins across the cell–cell contact in inhibitory murine and human NK–target-cell interactions.
Materials and Methods
Cells and Mice. The human Epstein–Barr virus-transformed cell line 721.221 (referred to as 221) and transfectants thereof have been described (19, 20). YTS, a subclone of the human NK tumor line YT (21), transfected to express KIR2DL1 (YTS/KIR2DL1), has been described (22). YTS transfected to express C-terminal GFP-tagged KIR2DL1 (YTS-TG) was a gift from D. Burshtyn (University of Alberta, Edmonton, AB, Canada) (23). A histogram of GFP expression in each transfectant had a single peak with a coefficient of variance of 50–63. Human cell lines were cultured at 37°C, in an atmosphere of 7.5% CO2 in RPMI medium 1640 supplemented with 10% FCS, 2 mM l-glutamine, 1× nonessential amino acids, 1 mM sodium pyruvate, 50 units/ml penicillin-streptomycin, 50 μM 2-mercaptoethanol (all from GIBCO/BRL, referred to as complete RPMI) containing 1.0 mg/ml G418 (GIBCO/BRL) or 0.7 μg/ml puromycin (Sigma) as appropriate. Human NK cells derived from peripheral blood were cultured and phenotyped as described (24).
EL-4, a murine lymphoma of B6 origin (25), was transfected to express H-2Dd protein tagged with GFP (EL4-Dd-GFP). EL4-Dd-GFP was negative for Ly49A (data not shown) and was cultured at 37°C, in an atmosphere of 7.5% CO2 in RPMI medium 1640, supplemented with 10% FCS, 2 mM l-glutamine, 50 units/ml penicillin-streptomycin, and 1.0 mg/ml G418. Untransfected EL4, but not EL4-Dd-GFP cells, were lysed by Ly49A+ NK cells (data not shown).
C57BL/6 (B6) mice expressing Ly49A under a modified CD2 promoter, B6VA49A, have been described (26). All mice were kept and bred at the Microbiology and Tumor Biology Center, Karolinska Institute, Stockholm, and animal experiments were approved by the Committee for Animal Ethics in Stockholm.
Murine NK Lymphokine-Activated Killer (LAK) Cultures. Spleens were homogenized in PBS, and the erythrocytes were lysed in 10 mM KHCO3/150 mM NH4Cl/0.1 mM EDTA, pH 8.0, on ice for 4 min. Cells were filtered, washed three times, and stained with anti-mouse CD3-FITC, anti-NK1.1-phycoerythrin, and anti-Ly49A-Alexa Fluor 633 for 40 min in PBS at 4°C. CD3-NK1.1+Ly49A+ cells were sorted by FACS and cultured for 4 days in MEM (α modification) supplemented with 10% FCS, 50 μM 2-mercaptoethanol, 10 mM Hepes buffer (GIBCO/BRL), 2 mM l-glutamine, and 1,000 units/ml IL-2 before use.
Antibodies. The following antibodies were all purchased from BD Pharmingen unless indicated: anti-mouse CD3-FITC (145–2C11), anti-NK1.1-phycoerythrin (PK136), anti-Ly49A (A1; YE1/48), anti-TNP (107.3, IgG1), anti-TNP (G155–178, IgG2a), anti-KIR2DL1 (EB6, Serotec), anti-phosphotyrosine (4G10, Upstate Biotechnology, Milton Keynes, U.K.), anti-CD56 (MY31), anti-GFP (JL8, Clontech), anti-human MHC class I (W6/32), anti-human MHC class II (TU39), anti-human CD54 (LB-2), anti-human CD53 (HI29), streptavidin Alexa Fluor 633 (Molecular Probes), Alexa Fluor 633 goat anti-mouse IgG (Molecular Probes), Cy5 goat anti-mouse IgG (Jackson ImmunoResearch), streptavidin-horseradish peroxidase (HRP) (Amersham Pharmacia), and HRP-goat anti-mouse IgG (Amersham Pharmacia).
Cell Labeling. For 1,1′dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine, 4-chlorobenzene-sulfonate salt (DiD) labeling, cells were incubated in 4 μg/ml DiD (Molecular Probes) in complete RPMI for 4 min at room temperature. Labeling of cells with PKH-26 (Sigma) was performed according to the manufacturer's instructions. Cells were biotinylated as described (13). For calcein labeling, cells were suspended at 106 cells per ml in complete RPMI with 20 ng/ml calcein AM ester (Molecular Probes) according to the manufacturer's instructions. All labeled cells were washed after labeling and rested in complete RPMI for 1 h at 37°C, in an atmosphere of 7.5% CO2 before use.
Kinetics of KIR2DL1-GFP Acquisition and Confocal Microscopy. A total of 5 × 105 YTS-TG cells were coincubated with 106 target cells in 30 μl of complete RPMI for various lengths of time, starting with the longest time point, at 37°C, in an atmosphere of 7.5% CO2. Cells were fixed with 4% paraformaldehyde for 15 min at 4°C, washed in PBS and analyzed by flow cytometry. Live target cells were quantified for their GFP expression by using the median fluorescence intensity (MFI) and the percent of inter-cellular transfer calculated as
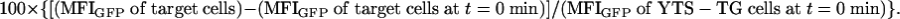 |
Cell staining and imaging by laser-scanning confocal microscopy (LSCM) was performed as described (24, 27). Protein clustering was assessed by plotting the fluorescence intensity around the cell membrane, where the intensity was at least twice the sum of the fluorescence in the unconjugated membranes the protein was scored as clustered at the IS as described (24). Transfer of proteins was assessed by imaging throughout the conjugate.
Intercellular Calcein Transfer. A total of 2.5 × 105 labeled or unlabeled 221/Cw6 target cells were incubated with 2.5 × 105 labeled or unlabeled YTS/KIR2DL1 cells at 37°C, in an atmosphere of 5% CO2 for 0 or 1 h, washed twice in PBS/2 mM EDTA/1% FCS, and analyzed by flow cytometry.
Western Blot Analysis. A total of 5 × 106 YTS-TG cells were coincubated with 107 DiD-labeled 221/Cw6 target cells at 37°C, in an atmosphere of 7.5% CO2 for 1 h. Cells were washed twice in PBS/1% FCS/2 mM EDTA to disrupt conjugates and populations separated by FACS (FACSVantage SE, Becton Dickinson). Cells were lysed in Laemmli sample buffer and stored at -20°C, and protein content was quantified by using a Coomassie R Plus protein assay reagent (Pierce). Fifty micrograms of protein, or 5 μg of control biotinylated YTS-TG protein, was separated by SDS/PAGE, transferred onto polyvinylidene di-fluoride membrane (Hybond P; Amersham Pharmacia Bio-sciences), and blocked overnight. The membrane was probed either with an anti-GFP mAb, followed by secondary HRP-goat anti-mouse IgG, or with streptavidin-HRP, and washed five times in PBS/0.1% Tween 20 (Sigma).
Intercellular Transfer Between Cells Treated with Cytoskeletal Inhibitors. To assess KIR2DL1-GFP transfer onto target cells, YTS-TG was coincubated with DiD-labeled 221/Cw6. To analyze transfer of HLA-C-GFP onto NK cells, DiD-labeled YTS/KIR2DL1 was mixed with 221/Cw6-GFP target cells. The following inhibitors were used: 5 μM latrunculin B (Sigma), 10 μM colchicine (Sigma), 10 μM cytochalasin B (Sigma), and 10 μM cytochalasin D (Sigma). Target or NK cells were treated for 1 h in complete RPMI supplemented with DMSO or a specific inhibitor dissolved in DMSO at 37°C, in an atmosphere of 7.5% CO2. Cells were washed four times with complete RPMI and mixed. For all inhibitor experiments, 5 × 104 NK cells were coincubated with 105 target cells for 0 or 1 h, and the amount of GFP on live DiD+ cells was analyzed by flow cytometry. The effect of drugs was expressed as a percentage change in the amount of constitutive intercellular transfer of protein according to
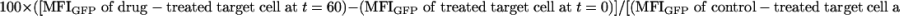 |
Correlating KIR2DL1-GFP Acquisition and Target-Cell MHC Expression. A total of 2.5 × 106 PKH-26-labeled 221/Cw6 target cells and 2.5 × 106 YTS-TG cells were coincubated in 100 μl of complete RPMI for 30 min at 37°C, in an atmosphere of 5% CO2. Three milliliters of RPMI was added, and conjugates were isolated by FACS, based on their high GFP and PKH-26 fluorescence. Next, conjugates were disrupted by incubation in PBS/0.5% BSA/5 mM EDTA for 40 min and stained with mAbs. Target cells were distinguished from NK cells by their high expression of PKH-26 and low expression of GFP. fcs assistant software (R. Hicks, FCS Press, Cambridge, U.K.) was used to export flow cytometric data for analysis.
Acid Wash. A total of 5 × 106 YTS-TG cells were coincubated with 107 PKH-26-labeled 221/Cw6 target cells in 200 μl of complete RPMI for 2 h at 37°C, in an atmosphere of 5% CO2. The sample was subjected to an acid wash as described (28). Cells were fixed in 4% paraformaldehyde for 15 min at 4°C and washed three times in PBS. For staining under nonpermeabilizing conditions, cells were blocked and stained in PBS/5% horse serum/3% BSA and washed with PBS. For permeabilizing conditions, cells were blocked and stained in 5% horse serum/3% BSA in Perm/Wash buffer (BD Pharmingen) and washed with PBS/0.1% Tween 20. Cells were blocked for 30 min at 4°C, incubated with 5 μg/ml anti-KIR2DL1 mAb for 30 min at 4°C, and washed three times with the appropriate buffer. The secondary Alexa Fluor 633 goat anti-mouse IgG was added for 20 min at 4°C followed by appropriate washes.
Results
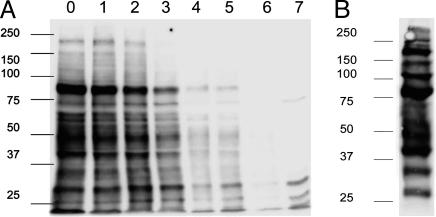
Multiple Proteins Transfer from YTS to Target Cells During Inhibitory Interactions. Previously, our laboratories and others demonstrated that MHC class I proteins transfer from target to NK cells during inhibitory interactions (12–14). To test whether proteins would also transfer from the NK to the target cell, the cell surface of a YTS transfectant expressing KIR2DL1 tagged with GFP at the C terminus (YTS-TG) was biotinylated and subsequently coincubated with 221/Cw6 target cells for 1 h. Biotinylation of effector cells did not affect NK cell cytotoxicity (data not shown). Target and effector cells were separated by FACS, and biotinylated proteins on target cells were detected by Western blotting. Multiple bands were revealed, demonstrating that several proteins transferred onto the target cell during inhibitory interactions with NK cells (Fig. 1A). Bands were still visible after 5 days of culture of target cells. By 6 days, bands were no longer seen, indicating that acquired proteins were gradually lost from the target cells. Analysis of a lysate from unconjugated biotinylated YTS-TG cells revealed a different set of protein bands (Fig. 1B), suggesting selection in the proteins that transfer from NK cells to target cells.
Fig. 1.
Multiple proteins transfer from NK to target cells during inhibitory interactions. (A) YTS-TG was biotinylated and subsequently coincubated with 221/Cw6 target cells for 1 h, after which target and effector cells were separated by FACS. At various times after separation from NK cells, target cell lysates were analyzed by Western blot with streptavidin-HRP. Lanes 0–6 correspond to days 0–6 after coculture and lane 7 is a control target-cell lysate. (B) Biotinylated YTS-TG cell lysates were analyzed by Western blot with streptavidin-HRP. Data shown are representative of four independent experiments. Numbers at left indicate molecular mass (kDa).
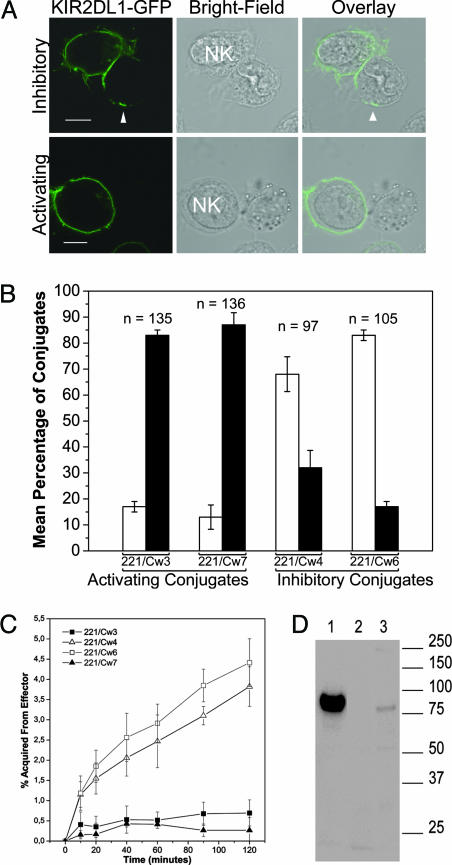
KIR2DL1 Transfers onto Target Cells Expressing a Cognate MHC Class I Ligand. To determine whether the inhibitory NK receptor KIR2DL1 was one of the proteins that transferred onto target cells, various target cells were coincubated with YTS-TG, and conjugates were imaged by LSCM. KIR2DL1-GFP was readily observed on target cells 221/Cw4 and 221/Cw6 that express inhibitory MHC ligands for KIR2DL1. In contrast, KIR2DL1-GFP did not cluster at the IS and transferred much less frequently to target cells lacking a ligand for KIR2DL1 (Fig. 2 A and B).
Fig. 2.
KIR2DL1-GFP transfers onto target cells expressing a cognate MHC class I ligand. (A) YTS-TG was coincubated with 221/Cw3, 221/Cw4, 221/Cw6, or 221/Cw7 for 1 h, fixed, and imaged. Representative fluorescence and bright-field images are shown for target cells expressing HLA-Cw6 (Upper)or expressing HLA-Cw3 (Lower). KIR2DL1-GFP clustering at the intercellular contact and patches of KIR2DL1-GFP fluorescence on the target cell (indicated by the arrowhead) could be observed in the conjugate with 221/Cw6 but not with 221/Cw3. (Scale bars, 8 μm.) (B) Conjugates were examined by LSCM and target cells were scored as either containing patches of KIR2DL1-GFP (white bars) or lacking KIR2DL1-GFP (black bars). The image is compiled from at least three independent experiments and the mean and SE are plotted. n, Number of conjugates. (C) Kinetics of KIR2DL1-GFP acquisition by 221/Cw3, 221/Cw4, 221/Cw6, or 221/Cw7 analyzed by flow cytometry. The mean and SE from six independent experiments are plotted. (D) Target cells, preincubated with YTS-TG, were probed by Western blotting with an anti-GFP mAb. Lane 1 is a total cell lysate of the YTS-TG transfectant to indicate the size of KIR2DL1-GFP, and lanes 2 and 3 contain lysates from 221/Cw3 and 221/Cw6, respectively, after coincubation and separation from NK cells. Numbers at right indicate molecular mass (kDa).
By flow cytometry, we assessed the kinetics of KIR2DL1-GFP transfer to target cells (Fig. 2C). After only 10 min of coculture, a significant amount of KIR2DL1-GFP had transferred onto target cells, and, after 2 h, target cells had acquired ≈4% of the fluorescence of NK cells. Target cells lacking a cognate MHC class I ligand, i.e., 221/Cw3 or 221/Cw7, did not acquire significant amounts of KIR2DL1-GFP.
To test whether the entire KIR2DL1-GFP protein transferred, target cells were separated from effector cells after 1 h of coincubation, lysed, and probed by Western blotting with an anti-GFP mAb. A band corresponding to the size of the entire KIR2DL1-GFP protein in 221/Cw6 (lane 3), but not 221/Cw3 (lane 2), indicated that the full KIR2DL1-GFP molecule transferred onto target cells expressing HLA-Cw6, a ligand for KIR2DL1 (Fig. 2D).
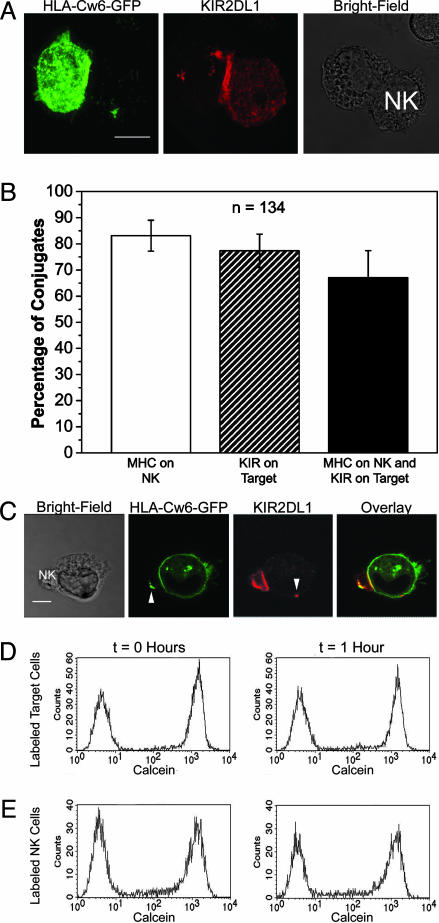
To exclude the possibility that the GFP tag on KIR2DL1 was responsible for the transfer of KIR2DL1, YTS-KIR2DL1 was coincubated with 221/Cw6-GFP for 1 h, fixed, and stained for KIR2DL1. In a 3D reconstruction of a single conjugate, anti-KIR2DL1 staining could be observed on the target cell (Fig. 3A), indicating that transfer was independent of the GFP tag. The simultaneous presence of HLA-Cw6-GFP on the NK cell and KIR2DL1 on the target cell was consistent with bidirectional protein transfer occurring within a single conjugate. This transfer occurred in ≈70% of conjugates where HLA-Cw6-GFP and KIR2DL1 clustered at the IS (Fig. 3B). In addition, target cells were readily seen to acquire KIR2DL1 from an NK cell clone derived from peripheral blood (Fig. 3C).
Fig. 3.
Transfer of KIR2DL1 receptors and MHC class I ligands, but not calcein, across an inhibitory NK cell IS. (A) A 3D reconstruction of fluorescence in a conjugate between YTS/KIR2DL1 and 221/Cw6-GFP shows HLA-Cw6-GFP fluorescence, and anti-KIR2DL1 staining as indicated. (Scale bar, 8 μm.) (B) Conjugates where both HLA-Cw6-GFP and anti-KIR2DL1 staining clustered at the IS were scored for the presence of HLA-Cw6-GFP on NK cells (white bar), KIR2DL1 on target cells (hatched bar), and both HLA-Cw6-GFP on NK cells and KIR2DL1 on target cells within the same conjugate (black bar). The mean and the SE for three independent experiments are shown. n, Number of conjugates imaged. (C) LSCM of a KIR2DL1+ KIR2DL2- NK cell clone derived from peripheral blood interacting with a 221/Cw6-GFP target cell immunostained for KIR. Image shown is representative of >30 micrographs. (Scale bar, 4 μm.) (D) Calcein-labeled 221/Cw6 target cells were incubated with equal numbers of unlabeled YTS/KIR2DL1 cells for 0 (Left) or 1 (Right) h and analyzed by flow cytometry. (E) Calcein-labeled YTS/KIR2DL1 NK cells were incubated with equal numbers of unlabeled 221/Cw6 for 0 (Left) or 1 (Right) h. Results are representative of three independent experiments.
A Cytoplasmic Dye Does Not Transfer in Either Direction During Inhibitory Interactions. To determine whether sharing of cytoplasm occurred between NK and target cells in inhibitory interactions, 221/Cw6 target cells and YTS/KIR2DL1 effectors were incubated at a 1:1 ratio, with either the target or effector cell previously labeled with calcein. After 1 h, no calcein fluorescence could be observed on either the unlabeled NK cells (Fig. 3D) or the unlabeled target cells (Fig. 3E), suggesting that there was no mixing of cytoplasm.
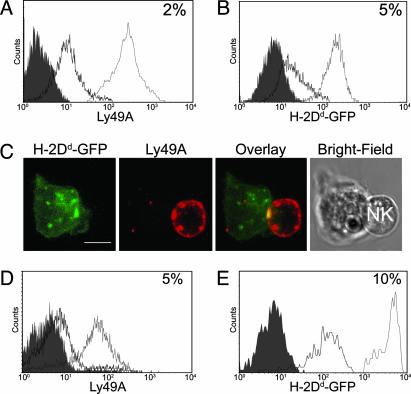
Murine Ly49A Transfers to Target Cells Expressing a Cognate MHC Class I Ligand. Because both human and murine NK cells are able to acquire MHC class I proteins from target cells, we set out to determine whether murine target cells expressing appropriate MHC ligands would be able to acquire the inhibitory NK cell receptor Ly49A. Sorted Ly49A+ LAK NK cells from B6 mice were cocultured with EL4 lymphoma cells expressing an inhibitory ligand for Ly49A, EL4-Dd-GFP. Coculture resulted in transfer of Ly49A receptors to target cells (Fig. 4A). Additionally, H-2Dd-GFP molecules were acquired by NK cells (Fig. 4B), as shown previously (13, 14). In a 3D reconstruction of a single conjugate, Ly49A receptors could be detected by mAb staining on the surface of target cells (Fig. 4C). Transfer of the inhibitory NK cell receptor Ly49A also occurred from naïve NK cells onto target cells (Fig. 4D). Over five independent experiments, the amount of transferred Ly49A ranged from 1% to 5% of the Ly49A expressed on the donor NK cell, the example shown in Fig. 4D being at the upper end of this range. Similarly, H-2Dd-GFP transferred from target cells to naïve Ly49A+ NK cells (Fig. 4E).
Fig. 4.
Bidirectional transfer of proteins across the inhibitory murine NK cell IS. Sorted Ly49A+ LAK NK cells were coincubated with EL4-Dd-GFP cells, stained with anti-Ly49A mAb, and analyzed by flow cytometry. (A) EL4-Dd-GFP target cells acquired Ly49A (black line) from LAK NK cells (gray line) compared with control noncoincubated target cells (filled histogram). (B) LAK NK cells acquired H-2Dd-GFP (black line) from EL4-Dd-GFP target cells (gray line) compared with LAK NK cells alone (filled histogram). (C) A 3D reconstruction of fluorescence in a conjugate between a Ly49A+ NK cell and EL4-Dd-GFP stained for Ly49A. (Scale bar, 4 μm.) (D) EL4-Dd-GFP cells acquired Ly49A (black line) from Ly49A+ splenocytes (gray line) in comparison to target cells which had not been cocultured (filled histogram). (E) Naive NK cells acquired H-2Dd-GFP (black line) from the target cells (gray line) compared with noncoincubated NK cells (filled histogram). Numbers in histograms represent the percentage acquired.
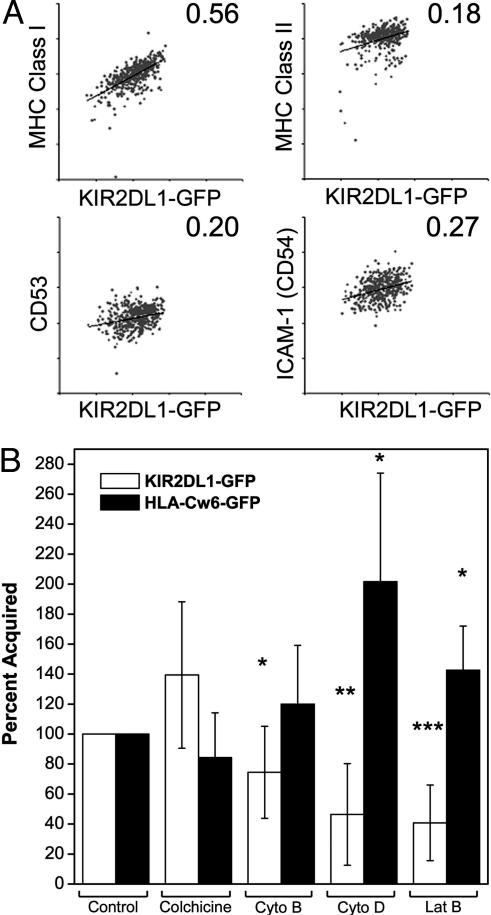
The Amount of KIR2DL1 Acquired Correlates with the Level of MHC Class I Expression on Target Cells. Acquisition of peptide/MHC protein by T cells is peptide dose-dependent (7, 29, 30). To determine whether the amount of KIR2DL1-GFP acquired by target cells correlated with the level of expression of some target-cell-surface proteins, conjugates of 221/Cw6 target cells with YTS-TG were isolated by FACS and stained with mAbs for target-cell proteins. On target cells that had been in contact with YTS-TG, the amount of acquired KIR2DL1-GFP was positively correlated with MHC class I expression but not with expression of MHC class II protein, CD53, or intercellular adhesion molecule 1 (Fig. 5A). This result is best seen by comparing the slope of a linear regression fitted to the plots of captured KIR2DL1-GFP against mAb staining (Fig. 5A).
Fig. 5.
Factors influencing the amount of KIR2DL1-GFP transfer to target cells. (A) Conjugates of YTS-TG- and PKH-26-labeled 221/Cw6 cells were isolated by FACS, disrupted with EDTA, and analyzed for expression of MHC class I, MHC class II, CD53, or CD54. Dot plots show the level of expression of MHC class I, MHC class II, CD53, and CD54 on PKH-26+ target cells plotted against the acquired KIR2DL1-GFP fluorescence. Results are representative of three independent experiments. Numbers in the top right corner of each dot plot represent the slope of the linear regression line fitted to the data points. (B) Treated target cells were coincubated with NK cells and analyzed by flow cytometry to assess changes in the amount of KIR2DL1-GFP transferred to target cells (white bars), or the effect on transfer of HLA-Cw6-GFP to NK cells (black bars). Cyto, cytochalasin; Lat B, latrunculin B. Results are shown as a percentage of acquisition in untreated samples. The mean and error bars for nine experiments are shown. Paired Student's t tests were performed. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
The Target-Cell Cytoskeleton Influences the Intercellular Transfer of Proteins in Both Directions. Transfer of proteins in both directions across the inhibitory IS was assessed after treatment of target cells with inhibitors affecting actin polymerization (cytochalasin B, cytochalasin D, and latrunculin B) or microtubule formation (colchicine). Treatment of 221/Cw6 with the cytoskeletal inhibitors cytochalasin D or latrunculin B, and perhaps, to some extent, cytochalasin B, reduced acquisition of KIR2DL1-GFP from the NK cell, suggesting that the target-cell cytoskeleton is important for KIR2DL1-GFP acquisition. Inhibiting the formation of microtubules did not affect the amount of KIR2DL1-GFP transferred (Fig. 5B). Also, treatment of the NK cell with cytochalasin B, cytochalasin D, and latrunculin B all reduced the amount of KIR2DL1-GFP acquisition by the target cell (data not shown). Thus, KIR2DL1-GFP transfer was reduced by inhibiting active actin polymerization in both the NK and the target cell, underlining the importance of their cytoskeleton in this phenomenon.
In the other direction, treatment of NK cells with the same inhibitors had no effect on the acquisition of HLA-Cw6-GFP by NK cells (data not shown). Interestingly, acquisition of target-cell HLA-Cw6-GFP by NK cells increased when target cells were treated with either latrunculin B or cytochalasin D, but not with colchicine or cytochalasin B (Fig. 5B), suggesting that the target-cell cytoskeleton influences the amount of HLA-Cw6-GFP transfer onto NK cells.
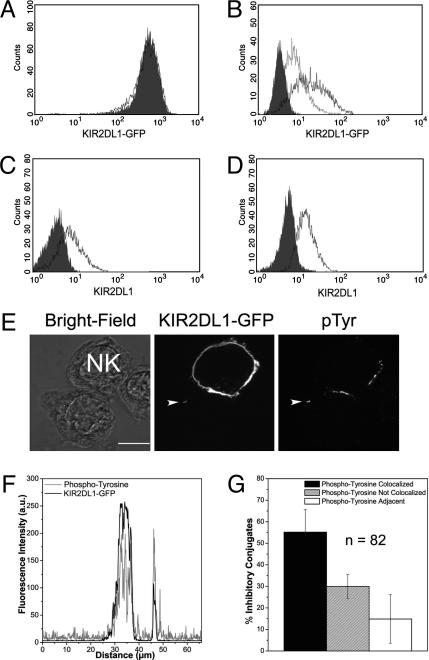
Orientation of Transferred KIR. To test whether transferred KIRs were embedded and how they were orientated in their new cell membrane, target cells were subjected to an acid wash. An acid wash did not reduce the fluorescence of KIR2DL1-GFP on NK cells (Fig. 6A), demonstrating that constitutively expressed KIR2DL1-GFP was resistant to acid treatment. However, an acid wash reduced the level of acquired KIR2DL1-GFP fluorescence on the target cell to 41 ± 16% of the initial level (Fig. 6B), showing that a fraction of acquired KIR2DL1-GFP was easily removed by an acid wash.
Fig. 6.
Orientation of transferred KIR and colocalization of transferred KIR with phosphotyrosine. (A) The fluorescence of KIR2DL1-GFP on the NK cell before (filled histogram) and after (black line) an acid wash. (B) Target cells that had acquired KIR2DL1-GFP (black line) were subjected to an acid wash (gray line). The filled histogram represents control acid-washed target cells. Acid-washed cells were stained with an isotype-matched control mAb (filled histogram) or anti-KIR2DL1 mAb (black line) under nonpermeabilizing (C) and permeabilizing (D) conditions. Results are representative of at least four independent experiments. (E) LSCM of YTS-TG conjugated to 221/Cw6 and stained for phosphotyrosine. (Scale bar, 8 μm.) (F) Fluorescence intensity around the target-cell membrane demonstrates colocalization of anti-phosphotyrosine staining (black line) with KIR2DL1-GFP (gray line) both at the synapse and at the patch of transferred KIR2DL1-GFP. (G) The location of anti-phosphotyrosine staining was scored in relation to transferred KIR2DL1-GFP in inhibitory conjugates. Shown is the frequency of colocalization in 82 conjugates. Mean and the SE from three different experiments are shown.
To determine the orientation of acid wash-resistant KIR2DL1-GFP proteins in the target-cell membrane, acid washed cells were fixed and stained for KIR2DL1 under both permeabilizing and nonpermeabilizing conditions. Under nonpermeabilizing conditions, a mAb targeting the extracellular portion of KIR2DL1 (EB6) was able to stain target cells, indicating that the extracellular epitope was accessible (Fig. 6C). Under permeabilizing conditions, anti-KIR2DL1 staining was consistently greater, with a fold increase of 1.5 ± 0.4, suggesting that a proportion of the acquired proteins were either located intracellularly or were inserted “upside down” in the new host membrane (Fig. 6D).
Colocalization of Transferred KIR with Phosphotyrosine. Patches of anti-phosphotyrosine clearly colocalized with patches of acquired KIR2DL1-GFP on the target cells, indicating the possibility for a functional consequence of transferred KIR (Fig. 6 E and F). Counting of conjugates revealed that in ≈50% of inhibitory conjugates, anti-phosphotyrosine staining was found to colocalize with at least some transferred KIR2DL1-GFP in the target cell (Fig. 6G).
Discussion
A common consequence of the formation of an IS by T, B, or NK cells is the acquisition of membrane fragments and/or proteins from their respective target cells or antigen-presenting cells (8–10, 12–15, 17, 29–31). We demonstrate here that target cells in an inhibitory interaction with an NK cell acquired several biotinylated NK cell-surface proteins, one of which was the NK cell receptor KIR2DL1, which is responsible for inhibition (Figs. 1, 2, 3). Transfer of the murine inhibitory receptor Ly49A was seen in similar experiments with mouse target cells (Fig. 4). KIR was only acquired by target cells expressing a cognate MHC class I ligand, and the amount of KIR acquisition by target cells depended on the level of target-cell MHC expression (Fig. 5). Acquired biotinylated proteins remained detectable in the target cell for up to 6 days after separation from NK cells, suggesting that target cells could possibly remain influenced by these proteins for some length of time. It is clear that a specific set of several proteins transferred from NK to target cells (Fig. 1), and thus, identifying them is an important next goal.
It was of interest that a fraction of acquired KIR2DL1-GFP was easily removed by a mild acid wash. Acid washes have been shown to remove mAb from the cell surface (32) or extracellularly bound proteins such as β2-microglobulin (33). However, loss of KIR2DL1-GFP fluorescence after acid treatment could also occur because of quenching of extracellular GFP by low pH (34). At this stage, our data do not allow us to conclude whether loss of KIR2DL1-GFP fluorescence is the result of acquired KIR having an altered conformation or different membrane anchoring, but it points to the fact that some of the acquired KIR is attached to the cell in a manner distinct from endogenously expressed KIR.
Most research in this area has focused on the molecular imprint that a target cell may leave on the effector cell perhaps because, in many instances, the target cell is lysed. However, in an inhibitory NK cell interaction, both the NK and the target cell will survive the encounter. The fact that phosphotyrosine staining colocalized with transferred KIR molecules suggests that a proportion of acquired proteins remain functional and that signaling molecules are able to associate with transferred KIR, or alternatively phosphorylated proteins transfer with KIR.
It is not obvious which cellular functions KIR would inhibit or which signaling molecules it would sequester in the target cells and what functional consequence this may have. One speculative function would be that proteins acquired by the target cells during inhibitory interactions would optimize immune surveillance by NK cells. For example, a target cell that has just been surveyed by an NK cell could be refractory to further NK cell surveillance for some time. A mechanism such as this could increase the efficiency of NK cell immune surveillance by eliminating repeated scanning of the same target cell.
Acknowledgments
We thank D. Burshtyn for the kind gift of YTS-TG and F. E. McCann, C. R. Almeida, D. Burshtyn, and B. Treanor for critical reading of the manuscript. This work was supported by a Eurogendis predoctoral award (to B.V.), grants from the Medical Research Council, the Biotechnology and Biological Sciences Research Council, the Department of Trade and Industry (to D.M.D.), the Human Frontier Science Program (to P.H. and D.M.D.), the Swedish Research Council, the Swedish Cancer Society, the Swedish Medical Doctor's Association, and the Clas Groshinsky Memory Foundation (to P.H.). Work in P.H.'s laboratory was performed within the Strategic Research Center for Studies of Integrative Recognition in the Immune System, sponsored by the Swedish Foundation for Strategic Research.
Author contributions: B.V., K.A., L.M.C., E.N.M.N.-t.H., G.S.W., P.H., and D.M.D. designed research; B.V., K.A., E.N.M.N.-t.H., and G.S.W. performed research; B.V., K.A., L.M.C., E.N.M.N.-t.H., G.S.W., P.H., and D.M.D. analyzed data; and B.V., P.H., and D.M.D. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: NK, natural killer; KIR, killer Ig-like receptor; IS, immune synapse; DiD, 1,1′dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine, 4-chlorobenzene-sulfonate salt; LSCM, laser-scanning confocal microscopy; MFI, median fluorescence intensity; EL4-Dd-GFP, EL4 transfected to express H-2Dd protein tagged with GFP; LAK, lymphokine-activated killer; HRP, horseradish peroxidase.
References
- 1.Lanier, L. L. (2003) Curr. Opin. Immunol. 15, 308-314. [DOI] [PubMed] [Google Scholar]
- 2.Yokoyama, W. M., Kim, S. & French, A. R. (2004) Annu. Rev. Immunol. 22, 405-429. [DOI] [PubMed] [Google Scholar]
- 3.Ljunggren, H. G. & Karre, K. (1990) Immunol. Today 11, 237-244. [DOI] [PubMed] [Google Scholar]
- 4.Long, E. O. (1999) Annu. Rev. Immunol. 17, 875-904. [DOI] [PubMed] [Google Scholar]
- 5.Kupfer, A. & Kupfer, H. (2003) Semin. Immunol. 15, 295-300. [DOI] [PubMed] [Google Scholar]
- 6.Davis, D. M. & Dustin, M. L. (2004) Trends Immunol. 25, 323-327. [DOI] [PubMed] [Google Scholar]
- 7.Huang, J. F., Yang, Y., Sepulveda, H., Shi, W., Hwang, I., Peterson, P. A., Jackson, M. R., Sprent, J. & Cai, Z. (1999) Science 286, 952-954. [DOI] [PubMed] [Google Scholar]
- 8.Stinchcombe, J. C., Bossi, G., Booth, S. & Griffiths, G. M. (2001) Immunity 15, 751-761. [DOI] [PubMed] [Google Scholar]
- 9.Hwang, I. & Sprent, J. (2001) J. Immunol. 166, 5099-5107. [DOI] [PubMed] [Google Scholar]
- 10.Batista, F. D., Iber, D. & Neuberger, M. S. (2001) Nature 411, 489-494. [DOI] [PubMed] [Google Scholar]
- 11.Russo, V., Zhou, D., Sartirana, C., Rovere, P., Villa, A., Rossini, S., Traversari, C. & Bordignon, C. (2000) Blood 95, 3473-3477. [PubMed] [Google Scholar]
- 12.Carlin, L. M., Eleme, K., McCann, F. E. & Davis, D. M. (2001) J. Exp. Med. 194, 1507-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sjostrom, A., Eriksson, M., Cerboni, C., Johansson, M. H., Sentman, C. L., Karre, K. & Hoglund, P. (2001) J. Exp. Med. 194, 1519-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zimmer, J., Ioannidis, V. & Held, W. (2001) J. Exp. Med. 194, 1531-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hudrisier, D. & Bongrand, P. (2002) FASEB J. 16, 477-486. [DOI] [PubMed] [Google Scholar]
- 16.Thery, C., Regnault, A., Garin, J., Wolfers, J., Zitvogel, L., Ricciardi-Castagnoli, P., Raposo, G. & Amigorena, S. (1999) J. Cell Biol. 147, 599-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tabiasco, J., Espinosa, E., Hudrisier, D., Joly, E., Fournie, J. J. & Vercellone, A. (2002) Eur. J. Immunol. 32, 1502-1508. [DOI] [PubMed] [Google Scholar]
- 18.Onfelt, B., Nedvetzki, S., Yanagi, K. & Davis, D. M. (2004) J. Immunol. 173, 1511-1513. [DOI] [PubMed] [Google Scholar]
- 19.Davis, D. M., Chiu, I., Fassett, M., Cohen, G. B., Mandelboim, O. & Strominger, J. L. (1999) Proc. Natl. Acad. Sci. USA 96, 15062-15067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mandelboim, O., Reyburn, H. T., Vales-Gomez, M., Pazmany, L., Colonna, M., Borsellino, G. & Strominger, J. L. (1996) J. Exp. Med. 184, 913-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoneda, N., Tatsumi, E., Kawano, S., Teshigawara, K., Oka, T., Fukuda, M. & Yamaguchi, N. (1992) Leukemia 6, 136-141. [PubMed] [Google Scholar]
- 22.Cohen, G. B., Gandhi, R. T., Davis, D. M., Mandelboim, O., Chen, B. K., Strominger, J. L. & Baltimore, D. (1999) Immunity 10, 661-671. [DOI] [PubMed] [Google Scholar]
- 23.Borszcz, P. D., Peterson, M., Standeven, L., Kirwan, S., Sandusky, M., Shaw, A., Long, E. O. & Burshtyn, D. N. (2003) Eur. J. Immunol. 33, 1084-1093. [DOI] [PubMed] [Google Scholar]
- 24.McCann, F. E., Vanherberghen, B., Eleme, K., Carlin, L. M., Newsam, R. J., Goulding, D. & Davis, D. M. (2003) J. Immunol. 170, 2862-2870. [DOI] [PubMed] [Google Scholar]
- 25.Gays, F., Unnikrishnan, M., Shrestha, S., Fraser, K. P., Brown, A. R., Tristram, C. M., Chrzanowska-Lightowlers, Z. M. & Brooks, C. G. (2000) J. Immunol. 164, 5094-5102. [DOI] [PubMed] [Google Scholar]
- 26.Fahlen, L., Oberg, L., Brannstrom, T., Khoo, N. K., Lendahl, U. & Sentman, C. L. (2000) Int. Immunol. 12, 215-222. [DOI] [PubMed] [Google Scholar]
- 27.Eleme, K., Taner, S. B., Onfelt, B., Collinson, L. M., McCann, F. E., Chalupny, N. J., Cosman, D., Hopkins, C., Magee, A. I. & Davis, D. M. (2004) J. Exp. Med. 199, 1005-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doucey, M. A., Scarpellino, L., Zimmer, J., Guillaume, P., Luescher, I. F., Bron, C. & Held, W. (2004) Nat. Immunol. 5, 328-336. [DOI] [PubMed] [Google Scholar]
- 29.Espinosa, E., Tabiasco, J., Hudrisier, D. & Fournie, J. J. (2002) J. Immunol. 168, 6336-6343. [DOI] [PubMed] [Google Scholar]
- 30.Hudrisier, D., Riond, J., Mazarguil, H., Gairin, J. E. & Joly, E. (2001) J. Immunol. 166, 3645-3649. [DOI] [PubMed] [Google Scholar]
- 31.Hwang, I., Huang, J. F., Kishimoto, H., Brunmark, A., Peterson, P. A., Jackson, M. R., Surh, C. D., Cai, Z. & Sprent, J. (2000) J. Exp. Med. 191, 1137-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Das, V., Nal, B., Dujeancourt, A., Thoulouze, M. I., Galli, T., Roux, P., Dautry-Varsat, A. & Alcover, A. (2004) Immunity 20, 577-588. [DOI] [PubMed] [Google Scholar]
- 33.Storkus, W. J., Zeh, H. J., III, Salter, R. D. & Lotze, M. T. (1993) J. Immunother. 14, 94-103. [PubMed] [Google Scholar]
- 34.Patterson, G. H., Knobel, S. M., Sharif, W. D., Kain, S. R. & Piston, D. W. (1997) Biophys. J. 73, 2782-2790. [DOI] [PMC free article] [PubMed] [Google Scholar]