Summary
Pseudomonas aeruginosa is an important life-threatening nosocomial pathogen that plays a prominent role in wound infections of burned patients. We designed this study to identify the isolates of P. aeruginosa recovered from burned patients at the genus and species level through primers targeting oprI and oprL genes, and analyzed their antimicrobial resistance pattern. Over a 2-month period, wound samples were taken from burned patients and plated on MacConkey agar. All suspected colonies were primarily screened for P. aeruginosa by a combination of phenotypic tests. Molecular identifications of colonies were done using specific primers for oprI and oprL genes. Bacterial isolates were recovered from burn wound infections. Based on phenotypical identification tests, 138 (34%) P. aeruginosa isolates were identified; whereas by molecular techniques, just 128 P. aeruginosa yielded amplicon of oprL gene using species-specific primers, verifying the identity of P. aeruginosa; the others yielded amplicon of oprI gene using genus-specific primers, confirming the identity of fluorescent pseudomonads. This study indicates that molecular detection of P. aeruginosa in burn patients employing the OprL gene target is a useful technique for the early and precise detection of P. aeruginosa. PCR detection should be carried out as well as phenotypic testing for the best aggressive antibiotic treatment of P. aeruginosa strains at an earlier stage. It also has significant benefits on clinical outcomes.
Keywords: Pseudomonas aeruginosa, burned patients, OprL, OprI
Abstract
Pseudomonas æruginosa (PA) est une bactérie nosocomiale potentiellement mortelle jouant un rôle majeur dans l’infection cutanée des brûlés. Nous avons étudié les PA (genre, espèce) isolés de patients brûlés, avec une PCR utilisant les gènes d’amorçage oprI et oprL, et avons analysé leurs profils de résistance. Des échantillons cutanés ont été prélevés chez des patients brûlés pendant une durée de 2 mois, et ensemencés sur gélose Mc Conkey. L’identification de PA a été faite selon les critères phénotypiques usuels. L’étude moléculaire a été réalisée par PCR utilisant les gènes d’amorçage oprI et oprL. Les identifications phénotypiques ont retrouvé 138 PA quand l’amplification d’oprI, spécifique de PA était positive 128 fois. Les 10 autres souches étaient des Pseudomonas fluorescents, caractérisés par l’amplification d’oprL. Cette étude montre que l’identification rapide de PA sur des échantillons de patients brûlés peut se faire par amplification du gène oprL et pourrait être utilisée concomitamment à la recherche des gènes codant les résistances bactériennes pour une adaptation plus rapide de l’antibiothérapie. On observe une amélioration du pronostic.
Introduction
Burn injury is one of the most common and devastating forms of trauma, and is a major public health problem worldwide. Infection of burns is common because the skin, which acts as a physical barrier against microbes, has been compromised. P. aeruginosa are the most common source of burn wound infections.1 These bacteria are known for their tendency to cause disease in immune compromised patients, such as those with AIDS, cystic fibrosis and burn patients, but rarely in healthy individuals.2 The accurate identification of P. aeruginosa and detection of their antimicrobial susceptibility are critical components of burn patient management.
Detection of P. aeruginosa colonisation is normally achieved by culture of wound swabbing on to artificial media. Typical isolation media for wound infections include blood agar and chocolate agar as well as selective agars such as Mac- Conkey agar and cetrimide-based media. Despite the large amounts of P. aeruginosa often present in the clinical samples of burn patients, isolation and identification may be challenging for microbiology laboratories. For example, other species of Pseudomonas, including Stenotrophomonas maltophilia, Achromobacter xylosoxidans and Ralstonia pickettii have been shown to grow on cetrimide-based media3 and may be indistinguishable from non-pigmented strains of P. aeruginosa. Difficulties in recognizing P. aeruginosa are compounded by difficulties in biochemical identification. Biochemical test kits such as API 20 NE are commonly used for identification;4 however, a high rate of misidentification of oxidase positive Gram negative rods including P. aeruginosa has been demonstrated using this system.5 In addition, the test requires the use of a pure bacterial subculture and a minimum incubation time of 48 h. Hence, identification using this method requires at least 3 days. Another limitation of conventional culture is that P. aeruginosa can be easily misidentified with closely related Gram-negative bacilli. The use of molecular techniques such as PCR could improve accurate and rapid identification of P. aeruginosa.6,7 L and I lipoproteins are two outer membrane proteins of P. aeruginosa which are responsible for inherent resistance of P. aeruginosa to antibiotics and antiseptics. As these proteins are found only in this organism, they could be a reliable factor for rapid identification of P. aeruginosa in clinical samples.8,9
In this study, we examined rapid and precise identification of P. aeruginosa strains isolated from burn patients hospitalized in a main burn center in Iran based on PCR amplification of I lipoprotein (OprI) for detection of genus and L lipoprotein (OprL) for detection of species of P. aeruginosa strains, and compared its performance to phenotypical and routine biochemical identification used in the laboratories.
Materials and methods
Qualitative conventional detection
The study was carried out over a 2-month period at a major center for burn patients in Tehran, Iran. Samples were obtained from burn wounds by swabbing. For routine phenotypical tests usually performed in clinical laboratories, we inoculated burn wound swabs primarily onto several selective media for the isolation of P. aeruginosa, including blood agar, MacConkey agar and Muller Hinton agar, and incubated them at 37ºC for 24-48 h. The isolates were presumptively identified by routine tests, including colony morphology and pigment formation on selective medium, positive oxidase test, glucose fermentation, hydrolysis of gelatin and growth at 42ºC.10,11
Molecular (PCR) detection
DNA extraction: in order to minimize contamination and hence the possibility of false positive results, all DNA isolation procedures were carried out in a room physically separated from that used to set up nucleic acid amplification reaction mixes and also from the post PCR room. Bacterial genomic DNA was extracted from all phenotypical and biochemical approved strains, as well as from the reference strain P. aeruginosa ATCC 27853, using the boiling method. For this purpose, depending on colony size, three to six colonies were picked from bacterial plates and mixed into 0.25 ml DNase/RNasefree water in sterile 1.5ml eppendorf tubes to obtain a turbid suspension of bacteria (~ 1–2 × 109 cells/ml). The cell suspensions were held in a boiling water bath for 10 minutes to lyse the cells, and then centrifuged at 10000g at 4°C for 10 minutes. Then, the supernatant was transmitted in sterile conditions into another tube and used as DNA template. Extracted DNA was stored at -20°C prior to PCR amplification.12
Primer selection
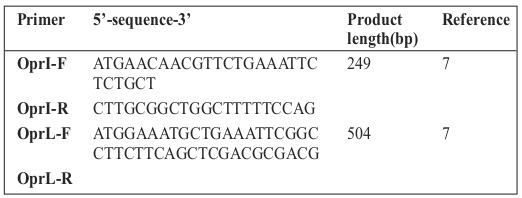
The primers used in this study are reported in Table I. PCR amplification of I lipoprotein (OprI) for detection of Pseudomonas genus and L lipoprotein (OprL) for detection of P. aeruginosa species was carried out for all phenotypically approved strains of P. aeruginosa.
Table I. Primers used in this study.
PCR amplification
All reaction mixes were set up in a PCR room separate from that used to extract DNA and from the amplification and post PCR room in order to minimize contamination. PCR was completed in adapted PCR micro centrifuge tubes according to the thermocycler used. Following optimization, reaction mixes (25 μl) were set up as follows: 11 μl DNase/RNase-free water, 8 μl 2x PCR Master Mix (1.5 mM mgcl2, Denmark), 0.5 μl of each set of primers (OprL or OprI) and 5 μl of DNA template. The reaction mixtures were subjected to the following empirically optimized thermal cycling parameters (Senso- Quest Labcycler thermocycler, Germany): in 94°C for 5 min followed by 30 cycles at 94°C for 1 min, 55°C for 1 min, 72°C for 1 min, finishing with a final extension at 72°C for 10 min. Positive (P. aeruginosaATCC 27853 DNA) and multiple negative (water) amplification controls were included in every set of PCR reactions.
Detection of amplicons
Following amplification, aliquots (10 μl) were removed from each reaction mixture and examined by electrophoresis (80 V, 45 min) in gels composed of 1% agarose in TBE buffer (40 mM Tris, 20 mM boric acid, 1 mM EDTA, pH 8.3). Gels were visualized under UV illumination using a gel image analysis system (UVitec, Cambridge, United Kingdom) and all images were archived. Where a band was visualized at the correct expected size for OprI, the specimen was considered positive for Pseudomonas genus, and if a band was visualized at the correct expected size for OprL loci, the specimen was considered positive for P. aeruginosa species.
Results
Over a 2-month period, bacterial samples were recovered from burn wound infections of individuals hospitalized in a major center for burn patients in Tehran, Iran. Our results showed that, based on the afore-mentioned routine phenotypic and biochemical tests, P. aeruginosa isolates were recovered from 138 (34%) burn patients; whereas using molecular techniques, there were just 128 (92.7%) positive samples for P. aeruginosa species and 10 (7.3%) samples verified as pseudomonas genus. PCR assays employing each primer pair produced DNA products of the predicted sizes (Figs. 1 and 2). The OprI and OprL amplicon genes were detected in all 128 P. aeruginosa isolates simultaneously. Table II shows the comparison of phenotypic and biochemical testing with the molecular detection of P. aeruginosa in samples from burn wound infections.
Fig. 1. PCR amplification using Pseudomonas genus-specific primers (Opr I gene), M = marker, line 1 = positive control, lines 2, 3, 4 = clinical isolates of Pseudomonas genus, line 5 = negative control.
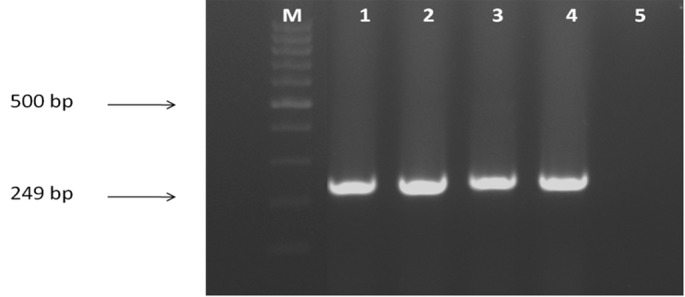
Fig. 2. PCR amplification using P. aeruginosa specific primers (Opr L gene), M = marker, line 1 = positive control, lines 2, 3, 4, 5, 6 = clinical isolates of P. aeruginosa, line 7 = negative control.
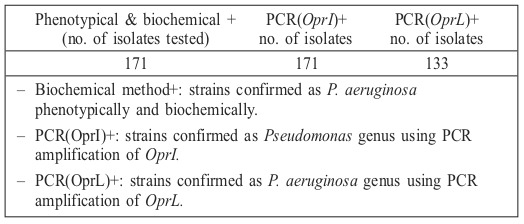
Table II. Comparison of phenotypic and biochemical tests with molecular detection of P. aeruginosa from burn wound infection samples.
Discussion
Bacterial infections in burn wounds are common and are difficult to control. In recent decades, following the introduction of antibiotic therapy, P. aeruginosa has emerged as one of the most problematic gram-negative bacteria in modern hospital settings. This organism is increasingly isolated as a nosocomial pathogen, resulting in high morbidity and mortality rates in burn patients, mechanically ventilated patients, and people with cystic fibrosis.13,14 The condition for patients infected by this bacterium is particularly problematic since the organism is intrinsically resistant to many drug classes and is able to acquire resistance to all effective antimicrobial drugs. Some studies carried out in Iran have also indicated that infections caused by multi drug resistant (MDR) P. aeruginosa are widespread among Iranian hospitals.15,16 Therefore accurate and rapid identification of P. aeruginosa and knowing the susceptibility pattern of this organism is important. This may avoid prolonged and sometimes unnecessary antibiotic treatments, which could be selected for other antibiotic resistant pathogens.12 Identification of P. aeruginosa has traditionally relied on phenotypic and biochemical methods. These tests take a long time to perform and require extensive hands-on work by the technologist, both for setup and for ongoing evaluation. Various methods have been developed to rapidly and accurately identify P. aeruginosa species as a medically important bacterium. According to our literature review, PCR has the potential to identify microbial species rapidly and precisely by amplification of gene sequences unique to a particular organism, 17 and several PCR based, DNA probe methods have been developed to detect various pathogens from clinical, water and food samples.18 Also in the case of P. aeruginosa, molecular methods have been reported to be superior to phenotypic methods for the identification of P. aeruginosa species.19
The outer membrane proteins of P. aeruginosa play an important role in the interaction of the bacterium with the environment.20 In this study, two PCR assays were performed individually for the molecular detection of two outer membrane lipoprotein genes, oprI and oprL, from wounds of burn patients.21 According to phenotypic and biochemical tests, 138 (34%) of the 385 recovered bacterial samples were verified as P. aeruginosa. However, our molecular tests, according to PCR results demonstrating the existence of oprI and oprL genes, showed that some of these phenotypically and biochemically approved isolates were pseudomonas genus and 128 (92.7%) isolates were P. aeruginosa species. Consequently there was almost complete agreement between molecular and conventional phenotypic and biochemical detection techniques regarding Pseudomonas genus, but not regarding P. aeruginosa species. This may account for potential phenotypic misidentification of P. aeruginosa, which has been recently described.22 Alternatively, discrepant results (PCR- /biochemical+) could reflect true P. aeruginosa colonization with a false negative culture result due to sample overgrowth by other bacteria, or to the presence of non-cultivable organisms or auxotrophic mutations in the organism.
Others have shown that OprI is conserved among members of fluorescent pseudomonads.23,24 De Vos et al., by designing a multiplex PCR assay based on oprI and oprL genes for molecular detection of P. aeruginosa, showed that specificity and sensitivity of the PCR assay were 74% and 100%, respectively.7Lavenir et al. also noted that all P. aeruginosa strains contained the oprI and oprL genes (sensitivity = 100%, specificity = 80%). Similarly, all of the 128 isolates in this study were remarkably positive for both OprI and OprL genes.25
Although our PCR and DNA sequence analyses revealed some isolates that had been misidentified by phenotypic testing, we must be clear in pointing out that our study was not designed to ascertain the frequency of misidentification of burned wound infection isolates nor to compare the relative accuracy of different phenotypic identification systems. Isolates used in this study most likely represent a biased set of atypical and difficult to identify isolates. Nevertheless, such isolates were well suited for a rigorous test of our PCR assays and represented strains for which molecular analysis would be expected to be most useful. Our study also restates that various non-aeruginosa pseudomonal species can occasionally be recovered from burn wound infection culture. Genotype- based identification methods circumvent the problem of variable phenotype to provide more accurate species identification.
Conclusion
It is important that primary diagnostic bacteriology methods have the ability to detect transient and early Pseudomonas colonization in burn patients, as early as possible, so that (i) aggressive antibiotic regimes may be reconsidered; (ii) the patient is managed optimally, in an attempt to avoid early biofilm formation and chronic colonization with P. aeruginosa and (iii) appropriate infection control precautions are considered.
Acknowledgments
Acknowledgements.The authors would like to acknowledge the Islamic Azad University for supporting this research.
References
- 1.Church D, Elsayed S, Reid O, Winston B, Lindsay R. Burn wound infections. Clin Microbiol Rev. 2006;19(2):403–434. doi: 10.1128/CMR.19.2.403-434.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lyczak JB, Cannon CL, Pier GB. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes and Infection. 2000;2:1051–1060. doi: 10.1016/s1286-4579(00)01259-4. [DOI] [PubMed] [Google Scholar]
- 3.Kodaka H, Iwata M, Yumoto S, Kashitani F. Evaluation of a new agar medium containing cetrimide, kanamycin and nalidixic acid for isolation and enhancement of pigment production of Pseudomonas aeruginosa in clinical samples. J Basic Microbiol. 2003;43:407–413. doi: 10.1002/jobm.200310264. [DOI] [PubMed] [Google Scholar]
- 4.Van Pelt C, Verduin CM, Goessens WHF, Vos MC. Identification of Burkholderia spp. in the clinical microbiology laboratory: comparison of conventional and molecular methods. J Clin Microbiol. 1999;37:2158–2164. doi: 10.1128/jcm.37.7.2158-2164.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wellinghausen N, Kothe JK, Wirths B, Sigge A, Poppert S. Superiority of molecular techniques for identification of Gram negative, oxidasepositive rods, including morphologically non typical Pseudomonas aeruginosa, from patients with cystic fibrosis. J Clin Microbiol. 2005;43:4070–4075. doi: 10.1128/JCM.43.8.4070-4075.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anuj SN, Whiley DM, Kidd TJ, Bell SC. Identification of Pseudomonas aeruginosa by a duplex real-time polymerase chain reaction assay targeting the ecfX and the gyrB genes. Diagn Microbiol Infect Dis. 2009;63(2):127–131. doi: 10.1016/j.diagmicrobio.2008.09.018. [DOI] [PubMed] [Google Scholar]
- 7.De Vos D, Lim A, Pirnay JP, Struelens M. Direct detection and identification of Pseudomonas aeruginosa in clinical samples such as skin biopsy specimens and expectorations by multiplex PCR based on two outer membrane genes, oprI and oprL. J Clin Microbiol. 1997;35:1295–1299. doi: 10.1128/jcm.35.6.1295-1299.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Douraghi M, Ghasemi F, Dallal MM, Rahbar M, Rahimiforoushani A. Molecular identification of Pseudomonas aeruginosa recovered from cystic fibrosis patients. J Prev Med Hyg. 2014;55(2):50–53. [PMC free article] [PubMed] [Google Scholar]
- 9.Osayande J. Easy Identification of Difficult-to-Type Pseudomonas Aeruginosa Clinical and Environmental Isolates. Internet J Microbiol. 2008;7(2):45–51. [Google Scholar]
- 10.Masuda N, Sakagawa E, Ohya S. Outer membrane proteins responsible for multiple drug resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:645–649. doi: 10.1128/AAC.39.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hall GS. Non fermenting Gram negative bacilli. Textbook of Diagnostic Microbiology. 2000:542–562. [Google Scholar]
- 12.Adabi M, Talebi-Taher M, Arbabi L, Afshar M. Spread of Efflux Pump Overexpressing-Mediated Fluoroquinolone Resistance and Multidrug Resistance in Pseudomonas aeruginosa by using an Efflux Pump Inhibitor. Infect Chemother. 2015;47(2):98–104. doi: 10.3947/ic.2015.47.2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forbes BA, Sahm DF, Weissfeld A. Bailey and Scott’s Diagnostic Microbiology. (St Louis) 2002 [Google Scholar]
- 14.Poh CL, Yeo CC. Recent advances in typing of Pseudomonas aeruginosa. J Hosp Infect. 1993;24:175–181. doi: 10.1016/0195-6701(93)90047-4. [DOI] [PubMed] [Google Scholar]
- 15.Adabi M, Talebi Taher M, Arbabi L, Afshar M. Determination of Antibiotic Resistance Pattern of Pseudomonas aeruginosa Strains Isolated from Patients with Burn Wounds. J Ardabil Uni Medl Scienc. 2015;15(1):66–74. [Google Scholar]
- 16.Shahcheraghi f, Feizabadi mm, Yamin v, Abiri r, Abedian Z. Serovar determination, drug resistance patterns and plasmid profiles of Pseudomonas aeruginosa isolated from burn patients at two hospitals of Tehran (IRAN) Burns. 2003;29(6):547–551. doi: 10.1016/s0305-4179(03)00142-6. [DOI] [PubMed] [Google Scholar]
- 17.Nikbin VS, Abdi-Ali A, Feizabadi MM, Gharavi S. Pulsed field gel electrophoresis & plasmid profile of Pseudomonas aeruginosa at two hospitals in Tehran, Iran. Indian J Med Res. 2007;126(2):146–151. [PubMed] [Google Scholar]
- 18.Saiki RK, Gelfand DH, Stoffel S, Scharf SJ. Primer directed enzymatic amplification of DNA with thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 19.Bej AK, Mahbubani MH, Dicesare JL, Atlas RM. Polymerase chain reaction gene probe detection of microorganisms by using filter-concentrated samples. Appl Environ Microbiol. 1999;1(57):3529–3534. doi: 10.1128/aem.57.12.3529-3534.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qin X, Emerson J, Stapp J, Stapp L. Use of real-time PCR with multiple targets to identify Pseudomonas aeruginosa and other non fermenting gram negative bacilli from patients with cystic fibrosis. J Clin Microbiol. 2003;4:4312–4317. doi: 10.1128/JCM.41.9.4312-4317.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hancock REW, Siehnel R, Martin N. Out membrane proteins of Pseudomonas. Mol Microbiol. 1990;4:1069–1075. doi: 10.1111/j.1365-2958.1990.tb00680.x. [DOI] [PubMed] [Google Scholar]
- 22.Cornelis P, Bouia A, Belarbi A, Guyonvarch A. Cloning and analysis of the gene for the major outer membrane lipoprotein from Pseudomonas aeruginosa. Mol Microbiol. 1989;3:421–428. doi: 10.1111/j.1365-2958.1989.tb00187.x. [DOI] [PubMed] [Google Scholar]
- 23.Kolmos HJ, Thuesen B, Nielsen SV, Lohmann M. Outbreak of infection in a burns unit due to Pseudomonas aeruginosa originating from contaminated tubing used for irrigation of patients. J Hos Infect. 1993;24(1):11–21. doi: 10.1016/0195-6701(93)90085-e. [DOI] [PubMed] [Google Scholar]
- 24.De Vos D, Lim A Jr, De Vos P, Sarniguet A. Detection of the outer membrane lipoprotein I and its gene in fluorescent and non fluorescent pseudomonads: implications for taxonomy and diagnosis. J Gen Microbiol. 1992;139:2215–2223. doi: 10.1099/00221287-139-9-2215. [DOI] [PubMed] [Google Scholar]
- 25.Lavenir R, Jocktane D, Laurent F, Nazaret S, Cournoyer B. Improved reliability of Pseudomons aeruginosa PCR detection by the use of the specific ecfx gene target. J Microbiol Methods. 2007;70:20–29. doi: 10.1016/j.mimet.2007.03.008. [DOI] [PubMed] [Google Scholar]


