Abstract
The complement system represents an evolutionary old and significant part of the innate immune system involved in protection against invading microorganisms. Here, we show that the anaphylatoxin C3a and its inactivated derivative C3a-desArg are antibacterial, demonstrating a previously unknown direct antimicrobial effect of complement activation. The C3a peptide, as well as functional epitopes in the sequence, efficiently killed the Gram-negative bacteria Escherichia coli, Pseudomonas aeruginosa, and the Gram-positive Enterococcus faecalis. In mice, a C3a-derived peptide suppressed infection by Gram-positive Streptococcus pyogenes bacteria. Fluorescence and electron microscopy demonstrated that C3a binds to and induces breaks in bacterial membranes. C3a was also found to induce membrane leakage of liposomes. These findings provide an interesting link between the complement system and antimicrobial peptides, which are two important branches of innate immunity.
Keywords: bacteria, inflammation, innate immunity
In contrast to adaptive immunity, innate immune effectors, such as phagocytes, complement, chemokines, and antimicrobial peptides (AMPs), provide a rapid and nonspecific host response (1). AMPs, which were originally described in silk worms (2), are important in the initial clearance of bacteria at biological boundaries that are susceptible to infection (3–5). However, many AMPs, such as defensins and LL-37, also possess chemotactic activity against leukocytes, T cells (6), and mast cells (7). Conversely, chemokines, which attract and activate specific leukocyte populations during inflammation and infection, were recently shown to exert direct antimicrobial functions (8). Thus, it is becoming increasingly clear that various protein and peptide mediators bridge the innate and adaptive immune systems.
In vertebrates, the complement system is activated by the classical, alternative, and lectin pathways, each converging at the step of C3 with release of multiple proteolytic fragments, including the anaphylatoxin C3a (9). C3a exerts multiple proinflammatory functions, involving histamine release from mast cells, smooth-muscle contraction, increased vascular permeability, and chemoattraction against mast cells (9). The biological effects of C3a are regulated by the plasma protease carboxypeptidase N, which cleaves off the C-terminal arginine to generate the inactive C3a-desArg peptide (9). Many AMPs, such as the human cathelicidin LL-37 (5, 10, 11), are able to interact with bacterial surface components like lipopolysaccharides, teichoic acid, peptidoglycans, and phospholipid groups (11). These interactions promote conformational changes, such as formation of an amphipatic helix, which in turn facilitate hydrophobic membrane interactions, oligomerization, and membrane destabilization and bacterial inactivation (11). The human C3a molecule (77 aa, 9,083 Da) is cationic (pI 11.3) and contains four α-helical regions (9). The structural features of C3a, which resemble those of AMPs, prompted us to investigate whether C3a and its inactivated form C3a-desArg exert antimicrobial effects. In this article, we show that C3a, C3a-desArg, and functional epitopes in the C3a-sequence function as “classical” AMPs, demonstrating a previously unknown direct antimicrobial effect of complement activation.
Materials and Methods
Biological Materials. Sterile wound fluids (WFs) were obtained from surgical drainages after mastectomy. Collection was performed for 24 h at 24–48 h after operation. WFs were centrifuged, aliquoted, and stored at -20°C. The use of human WF was approved by the Ethics Committee at Lund University (LU 708-01). Polymorphonuclear leukocytes (PMN) were obtained from blood samples drawn from healthy volunteers.
Peptides and Proteins. C3a and C3a-desArg were obtained from Calbiochem. The protein C3 was obtained from Reasearch Diagnostics (Flanders, NJ). The following peptides were synthesized by Innovagen (Lund, Sweden): LTE21, LRK26, LGE27, CNY21, CNY20, and LGL5 (see Fig. 3B); CNY21(S): CNYITELSSQHASASHLGLAR (see Fig. 5A); LL-37 and tetramethylrhodamine (TAMRA)-conjugated LRK26, LGE27, and CNY21. The purity (>95%) and molecular weight of these peptides was confirmed by MALDI-TOF MS analysis (Voyager, Applied Biosystems).
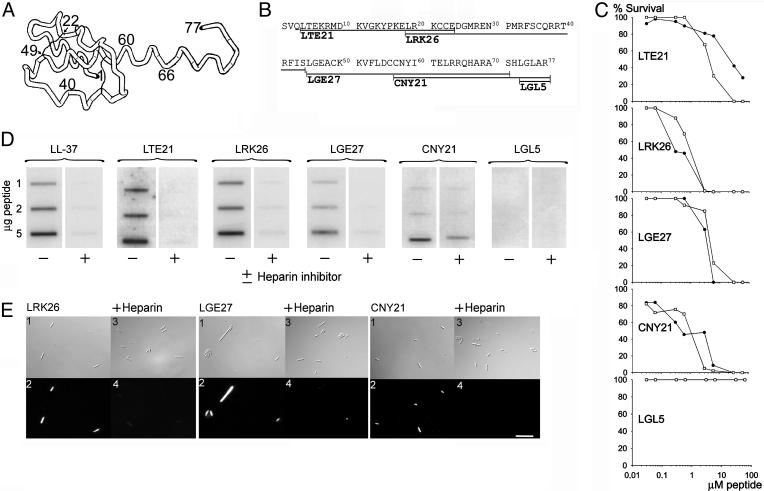
Fig. 3.
Activities of synthetic C3a-derived peptides. (A) Structure of the anaphylatoxin C3a peptide, modified from Hugli 1989 (9). (B) The synthetic peptides used in this study are indicated in the sequence. (C) Viable-count assays were performed by using both E. faecalis 2374 (•), and P. aeruginosa 27.1 (○). We incubated 2 × 106 cfu/ml bacteria in 50 μl with peptides at concentrations ranging 0.03–60 μM. For LGL5, the two graphs overlap. (D) Heparin-binding activity of the C3a-derived peptides. Peptides at indicated concentrations were applied to nitrocellulose membranes followed by incubation in PBS (containing 3% BSA) with iodinated (125I) heparin. LL-37 was used as positive control. Excess of unlabeled heparin (+) inhibited the binding of peptides to 125I-heparin. (E) Binding of TAMRA-labeled peptides to P. aeruginosa 27.1 and inhibition of binding by excess of heparin. Image 2 shows red fluorescence of bacteria (1 × 107 ml-1) stained with TAMRA-conjugated peptides (10 μg/ml-1), and image 4 shows bacteria incubated with heparin and TAMRA-conjugated peptides. Images 2 and 4 were recorded by using identical instrument settings. Images 1 and 3 are the corresponding Nomarski images. (Scale bar, 10 μm.)
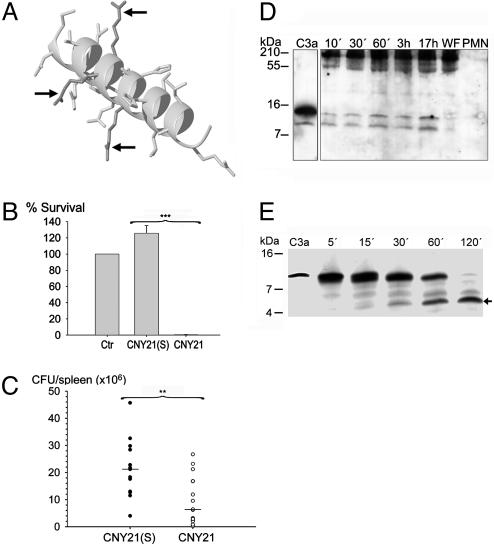
Fig. 5.
Antibacterial activities of CNY21 in vivo and generation of C3a-fragments ex vivo. (A) Space-filling model of CNY21 arranged as an α-helix. The arrows indicate positions of arginine residues that are replaced by serine residues in CNY21(S). (B) S. pyogenes bacteria were subjected to 10 μM CNY21(S) or CNY21 in 10 mM Tris (pH 7.4). At this dose, CNY21 was lethal to S. pyogenes whereas CNY21(S) had no effect; ***, P < 0.001 (n = 7). (C) C3a-derived peptides suppress bacterial dissemination to the spleen. Mice were i.p. injected with S. pyogenes bacteria, followed by i.p. injection with CNY21(S) (•) or CNY21 (○). The mice were killed 24 h after injection, and the total number of cfu in the spleen was determined for each mouse. Treatment with CNY21 yielded significantly lower bacterial numbers than treatment with CNY21(S). **, P = 0.006 (n = 14). Horizontal lines indicate the median value in each group. (D) Human WF (5 μl) was incubated at 37°C with lysed (0.3% Tween 20) neutrophils (corresponding to 9.4 × 104 cells) for the indicated time periods. Western blot analysis identified cleavage products recognized by polyclonal antibodies against the C3a-derived peptide LGE27. No C3a fragments were detected in WF or neutrophils (polymorphonuclear leukocytes, PMN). Molecular-mass markers are indicated on the left. (E) Neutrophil elastase degrades C3a into several fragments. C3a (2 μg) was treated with human neutrophil elastase (40 milliunits) for indicated time periods at 37°C and analyzed by SDS/PAGE (16.5% Tris·tricine gel). For control, 0.5 μg of C3a was used. Molecular mass markers are indicated on the left. The fragment indicated with an arrow was analyzed by N-terminal sequencing.
Microorganisms. Escherichia coli 37.4, Enterococcus faecalis 2374, and Pseudomonas aeruginosa 27.1 isolates, were obtained from patients with chronic venous ulcers. Streptococcus pyogenes AP1 (40/58) of the M1 serotype was provided by the World Health Organization Streptococcal Reference Laboratory in Prague.
Radial-Diffusion Assay (RDA). Essentially as described (12), bacteria were grown to midlogarithmic phase in 10 ml of full-strength (3% wt/vol) trypticase soy broth (TSB) (Becton Dickinson). The microorganisms were washed once with 10 mM Tris (pH 7.4). We added 4 × 106 bacterial colony-forming units (cfu) to 5 ml of the underlay agarose gel [0.03% (wt/vol) TSB/1% (wt/vol) low electroendosmosis type (EEO) agarose (Sigma)/0.02% (vol/vol) Tween 20 (Sigma)]. The underlay was poured into an 85-mm Petri dish. After agarose solidification, wells of 4 mm in diameter were punched, and 6 μl of test sample was added to each well. Plates were incubated at 37°C for 3 h to allow diffusion of the peptides. The underlay gel was then covered with 5 ml of molten overlay (6% TSB/1% low-EEO agarose in distilled H2O). Antimicrobial activity of a peptide is visualized as a zone of clearing around each well after 18–24 h of incubation at 37°C. The activity of the complement peptides C3a, C3a-desArg (50 and 10 μM), the C3 holoprotein (10 μM) (Fig. 1A), or the synthetic C3a-derived peptides LTE21, LRK26, LGE27, CNY21, and LGL5 (50 μM) were compared with the activity of the peptide LL-37. Minimum inhibitory concentration (MIC) values of C3a against P. aeruginosa and E. coli and of LL-37 against P. aeruginosa were determined by triplicate experiments by using five serial 2-fold dilutions (starting at 32 μM) of peptides. The logarithmic concentrations of peptides were plotted versus the respective diameter of inhibition zone. Linear regression using least squares was used to estimate MIC values. The antimicrobial activities of CNY21 and CNY20 were compared by measuring zones of clearance of 50 μM of respective peptide (n = 15). There was no statistical difference in activity between the two peptides. Inhibitory effects of heparin were examined by adding equimolar amounts (≈50 μM) of heparin to the peptides.
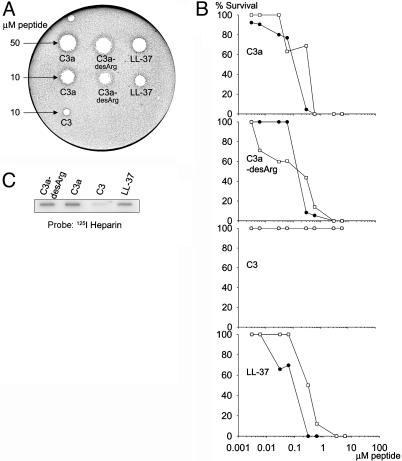
Fig. 1.
Antibacterial and heparin-binding effects of C3a and C3a-desArg. (A) Peptides were tested in RDA in low-salt conditions. E. coli isolate 37.4 (4 × 106 cfu) was inoculated in 0.1% TSB agarose gel. Each 4 mm-diameter well was loaded with 6 μl of peptide at the indicated concentration. The zones of clearance correspond to the inhibitory effect of each peptide after incubation at 37°C for 18–24 h. A negative control containing buffer (10 mM Tris, pH 7.4) was included in the well at the top left of the plate. This clear zone corresponds to the 4 mm well. (B) In viable-count assays antibacterial activities were seen against both E. faecalis isolate 2374 (•) and P. aeruginosa isolate 27.1 (○). We incubated 2 × 106 cfu/ml bacteria in 50 μl with peptides at concentrations ranging 0.003–6 μM. For C3, the two graphs overlap. (C) C3a and C3a-desArg were both able to bind heparin, whereas only weak binding was seen with C3. C3a and C3a-desArg (2 μg), C3, and LL-37 (2 and 5 μg) were applied to nitrocellulose membranes. These membranes were then incubated in PBS (containing 3% BSA) with iodinated (125I) heparin. LL-37 was used as positive control.
Viable-Count Analysis. E. faecalis, P. aeruginosa, and S. pyogenes bacteria were grown to midlogarithmic phase in Todd–Hewitt (TH) medium. Bacteria were washed and diluted in 10 mM Tris (pH 7.4) containing 5 mM glucose (Figs. 1B, 3C, and 4A) or in 10 mM Mes (pH 5.5) containing 5 mM glucose (Fig. 4A), with or without the addition of 0.15 M NaCl. Activity of 10 μM C3a and LL-37 was also tested against P. aeruginosa diluted in buffer with or without 20% human WF. Significance was determined by using the Holm–Sidak method; one-way repeated-measures ANOVA was performed, and we used the statistical software sigmastat (SPSS, Chicago) (Fig. 4B). The control peptide CNY21(S) was not active against S. pyogenes, whereas CNY21 killed the bacteria at 10 μM (P = 0.001, n = 7) (Fig. 5B). Bacteria (50 μl; 2 × 106 cfu/ml) were incubated, at 37°C for 2 h, with the complement peptides C3a and C3a-desArg; the holoprotein C3 or LL-37 at concentrations of 0.003–6 μM; or the synthetic peptides LTE21, LRK26, LGE27, CNY21, and LGL5 at concentrations of 0.03–60 μM. To quantify the bactericidal activity, serial dilutions of the incubation mixture were plated on TH agar, followed by incubation at 37°C overnight, and the number of cfu was determined.
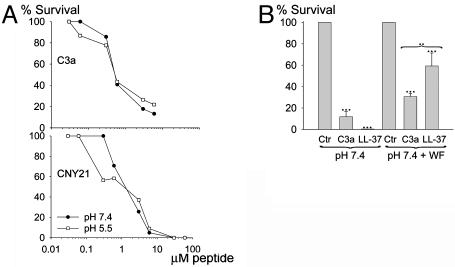
Fig. 4.
Antibacterial activities of peptides under physiological conditions. (A) Viable-count analysis of C3a and CNY21 in different buffers; 10 mM Tris (pH 7.4) (•) and 10 mM Mes (pH 5.5) (○), both containing 0.15 M NaCl. P. aeruginosa 27.1 (2 × 106 cfu/ml) were incubated in 50 μl with peptides at concentrations ranging 0.03–6 μM for C3a and 0.03–60 μM for CNY21. (B) P. aeruginosa bacteria were subjected to 10 μM C3a in 10 mM Tris (pH 7.4) containing 0.15 M NaCl in the presence or the absence of 20% WF. For comparison, LL-37 was used at the same concentration. ***, P < 0.001; **, P < 0.01.
Heparin-Binding Assay. The complement peptides C3a, C3a-desArg, and C3 (Fig. 1C) and the synthetic peptides LTE21, LRK26, LGE27, CNY21, and LGL5 (Fig. 3D) were tested for heparin-binding activities. LL-37 was used as positive control. C3a, C3a-desArg, C3 (2 μg), and the synthetic peptides (1, 2, and 5 μg) were applied onto nitrocellulose membranes (Hybond-C, Amersham Biosciences). Membranes were blocked (PBS, pH 7.4/3% BSA) for 1 h and incubated with radiolabeled heparin (≈10 μg/ml) (13). Unlabeled heparin (6 mg/ml) was added for competition of binding (Fig. 3D). The membranes were washed (3 × 10 min in 10 mM Tris, pH 7.4). A Bas 2000 radioimaging system (Fuji) was used for visualization of radioactivity.
Electron Microscopy. P. aeruginosa (16 × 106 cells per sample) were incubated for 2 h at 37°C with the complement factors C3a and C3a-desArg at ≈50% of their required bactericidal concentration (0.3 μM). The protein C3 (0.3 μM) was included as a control. Each sample was gently transferred onto poly-l-lysine-coated Nylaflo (GelmanSciences, Ann Arbor, MI) nylon membranes. The membranes were fixed in 2.5% (vol/vol) glutaraldehyde in 0.1 M sodium cacodylate (pH 7.2) for 2 h at 4°C and subsequently washed with 0.15 M cacodylate (pH 7.2). They were then postfixed with 1% osmium tetroxide (wt/vol) and 0.15 M sodium cacodylate (pH 7.2) for 1 h at 4°C, washed, and subsequently dehydrated in ethanol and further processed for Epon embedding. Sections were cut with a microtome and mounted on Formvar-coated copper grids. The sections were postfixed with uranyl acetate and lead citrate and examined in a Jeol 1200 EX transmission electron microscope operated at a 60-kV accelerating voltage.
Liposome Preparation and Leakage Assay. Dry lipid films were prepared by dissolving dioleoylphosphatidylcholine (Avanti Polar Lipids) (60 mol%) and cholesterol (Sigma) (40 mol%) in chloroform, and then removing the solvent by evaporation under vacuum overnight. Subsequently, buffer (10 mM Tris, pH 7.4) was added together with 0.1 M carboxyfluorescein (CF) (Sigma). After hydration, the lipid mixture was subjected to eight freeze–thaw cycles consisting of freezing in liquid nitrogen and heating to 60°C. Unilamellar liposomes, of ≈100 nm, were generated by multiple extrusions through polycarbonate filters (pore size, 100 nm) mounted in a LipoFast miniextruder (Avestin, Ottawa, Canada) at 22°C. Untrapped CF was then removed by two gel filtrations (Sephadex G-50) at 22°C with the Tris buffer as eluent.
In the liposome-leakage assay, self-quenching of CF was used. Thus, at 100 mM, CF is self-quenched, and the recorded fluorescence intensity from liposomes with entrapped CF is low. On leakage from the liposomes, released CF is dequenched, and hence, it fluoresces. The CF release was determined by monitoring the emitted fluorescence at 520 nm from a liposome dispersion (10 mM lipid in 10 mM Tris, pH 7.4). An absolute leakage scale is obtained by disrupting the liposomes at the end of the experiment by the addition of 0.8 mM Triton X-100 (Sigma), thereby causing 100% release and dequenching of CF. A Spex fluorolog 1650 0.22-m double spectrometer (Spex Industries, Edison, NJ) was used for the liposome-leakage assay.
Fluorescence Microscopy. P. aeruginosa bacteria were grown to midlogarithmic phase in TH medium. The bacteria were washed twice in 10 mM Tris, (pH 7.4). The pellet was dissolved to yield a suspension of 5 × 106 cfu/ml in the same buffer. We incubated 200 μl of the bacterial suspension with 1 μl of TAMRA-LGE27, TAMRA-LRK26, or TAMRA-CNY21 (2 mg/ml) on ice for 5 min and washed it twice in 10 mM Tris (pH 7.4). The bacteria were fixed by incubation on ice for 15 min and in room temperature for 45 min in 4% paraformaldehyde. The suspension was applied onto poly-l-lysine-coated cover glass, and bacteria were left to attach for 30 min. The liquid was poured away and the cover glass was mounted on a slide by mounting media (DAKO). Microscopy analysis was performed by using an Eclipse TE300 inverted fluorescence microscope (Nikon) equipped with a C4742–95 cooled charge-coupled device (CCD) camera (Hamamatsu, Bridgewater, NJ), a Plan Apochromat (×100 objective), and a high-numerical-aperture oil-condenser.
Bacterial Dissemination to the Spleen. S. pyogenes AP1 bacteria were grown to early logarithmic phase (OD620 ≈ 0.35), harvested, washed in 10 mM Tris (pH 7.4), diluted in the same buffer to 2 × 106 cfu/ml, and kept on ice until injection. We i.p. injected 500 μl of the bacterial suspension into female BALB/c mice. At 10 min after the bacterial injection, 0.5 mg of CNY21 or CNY21(S) in 10 mM Tris (pH 7.4) was i.p. injected into the mice. The injection was repeated after 6 h. After 24 h, the mice were killed, the spleens were removed and homogenized in 10 mM Tris (pH 7.4), and the number of cfu was determined. The P value was determined by using the Mann–Whitney U test.
Generation of C3a-Like Peptides in WF. Neutrophils were prepared by routine procedures (14) (Polymorphprep, Axis-Shield Po, Oslo) from blood obtained from healthy human donors. The cells were disrupted by freeze thawing and the addition of 0.3% Tween 20. The amount of neutrophils corresponding to 9.4 × 104 cells were incubated at 37° with 5 μl of human WF for different time periods (10 min, 30 min, 60 min, 3 h, and 17 h). Separate incubations (60 min) of neutrophils and WF were used as controls. The C3a (0.5 μg) peptide was added for size comparison. The materials were analyzed on 16.5% precast SDS/PAGE Tris·tricine gels (Bio-Rad) under reducing conditions. Proteins and peptides were transferred to nitrocellulose membranes (Hybond-C, Amersham Biosciences). Membranes were blocked by 3% (wt/vol) skimmed milk, washed, and incubated for 1 h with rabbit polyclonal LGE27 antibodies (1:1,500) (Innovagen), washed again, and subsequently incubated (1 h) with horseradish peroxidase-conjugated secondary antibodies (1:1,000). C3a proteins and/or fragments containing whole or parts of the LGE27 sequence were visualized by using the enhanced chemiluminescence (ECL) developing system (Amersham Biosciences).
Degradation of C3a with Human Neutrophil Elastase. C3a (2 μg, 0.2 mg/ml) was incubated with neutrophil elastase (40 milliunits) for increasing time periods (5, 15, 30, 60, and 120 min) at 37°C and were then analyzed by SDS/PAGE on a 16.5% Tris·tricine gel (Bio-Rad). For control, 0.5 μg of C3a was used.
Definition of C3a Cleavage Products. The final degradation products were reduced and alkylated by subsequent addition of 10–20 μl of 10 mM DTT (55°C, 30 min) and 10–20 μl of 110 mM iodoacetamide (20°C, 30 min). Peptides were purified by using C18 Ziptips (Millipore). Precrystallized 2,5-dihydroxybenzoic acid was provided on an Anchorchip target (Bruker Daltonik, Bremen, Germany). The purified peptides were eluted directly onto this matrix by using 50% acetonitrile/0.1% trifluoroacetic acid and allowed to cocrystallize. Spectrometry was carried out on a Reflex III MALDI-TOF mass spectrometer (Bruker Daltonik). The instrument was operated at an acceleration voltage of 26 kV in the positive-ion/delayed-extraction mode, and detection was performed in the reflector mode. Autolysis fragments of trypsin were used for calibration of the spectra that were summed of 75–100 single-shot measurements each. Peaks were assigned by using the findpept tool (www.expasy.org/tools/findpept.html). The detected signals corresponded to masses of 1,247.69, 2,177.97, and 3,068.47 Da and the peptides VQLTEKRMDK or QLTEKRMDKV, SLGEACKKVFLDCCNYIT, and SLGEACKKVFLDCCNYITELRRQHA, respectively. The cleavage products were also blotted from a 16.5% Tris·tricine gel onto PVDF membranes (Hybond-P, Amersham Biosciences). One major fragment (Fig. 5E, arrow) was cut out and sent for N-terminal sequencing at the Karolinska Institutet Protein Analysis Center (Stockholm). One fragment was identified by both methods, 44SLGEACKKVFLDCCNYITELRRQHA68.
Results and Discussion
To elucidate whether the anaphylatoxin C3a and its inactivated variant C3a-desArg possess antibacterial activity, we initially investigated effects on Escherichia coli (Fig. 1A). C3a, as well as C3a-desArg, were antibacterial in RDA. Noteworthy, C3a yielded a larger inhibition zone against E. coli than the classical AMP LL-37 in the low-salt conditions used. In viable-count assays, both C3a and C3a-desArg showed 80–100% killing at ≈0.6 μM, of the Gram-positive species E. faecalis and the Gram-negative P. aeruginosa (Fig. 1B). In these experiments, the holoprotein C3 did not exert any antibacterial effect. For comparison, the activity of the human AMP LL-37 is shown (Fig. 1A and B). In RDA, the MICs of C3a were determined to 0.70 μM and 0.75 μM against P. aeruginosa and E. coli, respectively. This result was comparable with the MICs obtained for LL-37 (1.03 μM for E. coli). The heparin-binding ability of the peptides was tested and the results showed that both C3a and C3a-desArg bind heparin (Fig. 1C). As recently shown, amphipaticity, cationicity, and helix structure are features that characterize heparin-binding peptides but also confer antimicrobial properties to this group of molecules (13). Therefore, the finding that C3a interacts with heparin at physiological conditions provides an additional link between C3a and many cationic AMPs.
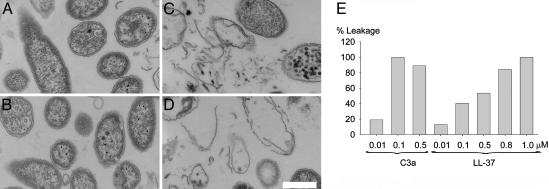
To examine whether C3a interacts with and generates breaks in bacterial plasma membranes, P. aeruginosa was incubated with C3a peptides at ≈50% of the required bactericidal concentrations, and analyzed by electron microscopy. Clear differences in the morphology of peptide-treated bacteria (Fig. 2C and D) in comparison with the control (Fig. 2A) and the holoprotein C3 (Fig. 2B) were demonstrated. C3a caused local perturbations and breaks along P. aeruginosa plasma membranes, and occasionally, intracellular material was found extracellularly. These findings were similar to those seen after treatment with the AMP LL-37 (13). To further analyze the effects of C3a on membranes, we used a liposome model to study membrane binding and permeabilisation (Fig. 2E). The activity of C3a was recorded by measuring fluorescence release of CF from dioleoylphosphatidylcholine liposomes. Addition of C3a caused increased fluorescence, and 100% liposome leakage was noted at ≈0.1 μM peptide. LL-37 yielded similar effects at ≈0.5 μM (Fig. 2E). Kinetic analysis showed that ≈80% of the maximum fluorescence was reached at ≈17 min for both peptides (at 0.5 μM) (data not shown).
Fig. 2.
C3a interacts with and generates breaks in bacterial plasma membranes. P. aeruginosa 27.1 was incubated with the holoprotein C3 and C3a peptides at 0.3 μM and analyzed with electron microscopy. Control (A), C3 (B), C3a (C), and C3a-desArg (D) are shown. (Scale bar, 0.5 μm.) (E) Effects of C3a and LL-37 on liposomes. The membrane permeabilizing effect was recorded by measuring fluorescence release of CF from liposomes. Values represents mean of double samples. A representative experiment of three is shown.
Studies have demonstrated that C3a contains four helical regions (8–15, 17–28, 36–43, and 47–66) (Fig. 3A) (9). To explore whether these helical epitopes of C3a were responsible for the antibacterial effects, we synthesized peptides spanning the four helices, including a peptide known to exert full anaphylatoxic activity, CNY21 (15), its inactive variant, CNY20, and the smallest peptide with any activity at all, LGL5 (16) (Fig. 3B). The experiments showed that these peptides indeed were antibacterial against E. faecalis and P. aeruginosa. Concentrations killing 80–100% of E. faecalis were 3–6 μM for LRK26, LGE27 (presented in ref. 13) and CNY21. LTE21 was less effective against this bacterium, and LGL5 showed no antimicrobial activity at all. Similar results were obtained for P. aeruginosa (Fig. 3C). Thus, in comparison with C3a, the C3a-derived peptides required an ≈10 times higher concentration for efficient killing of E. faecalis (Figs. 1B and 3C). Notably, CNY21, which has random conformation in aqueous solution (9, 15), adopts an α-helical and amphipatic conformation in trifluoroethanol/water (15), adding another structural and functional link between helical AMPs and this C3a-derived peptide. The antimicrobial activities of CNY21 and its desArg variant, CNY20 (devoid of anaphylatoxin activity), were compared in RDA by measuring zones of clearance by using 50 μM peptide (data not shown). There was no difference in activity between the two peptides, suggesting that the antimicrobial and anaphylactic functions are separate.
Like C3a and C3a-desArg, the C3a-derived antibacterial peptides bound radiolabeled heparin, and the binding was inhibited by an excess of unlabeled heparin (Fig. 3D). The binding of heparin to peptides LRK26 and LGE27 has been presented (13) but is shown here for completeness. Three peptides (LRK26, LGE27, and CNY21) were labeled with the fluorescent dye TAMRA and incubated with P. aeruginosa. As demonstrated by fluorescence microscopy analysis, the peptides were bound to the bacterial surface, and the binding was completely blocked by heparin (Fig. 3E). In antibacterial assays (RDA), heparin at equimolar amounts was able to abolish the antibacterial activities of the peptides (data not shown).
Activities of AMPs depend on salt concentration, pH, and the presence of plasma proteins. For example, the antimicrobial activities of defensins are inhibited by the presence of physiological salt (17), whereas the cathelicidin LL-37 is inhibited by plasma (18). Other AMPs, such as azurocidin and magainins, are potentiated by acidic conditions likely to occur in biological fluids following oxidative burst response of leukocytes (19). Hence, we examined the activities of C3a and related peptides in physiological salt. A minor decrease in the killing of P. aeruginosa was noted for C3a in 0.15 M NaCl at both pH 7.4 and 5.5 (Fig. 4A). No inhibition of bacterial killing in physiological salt was noted for CNY21 (Figs. 3C and 4A). LGE27 and LRK26 exerted only weak antibacterial activities in the same buffers (not shown). P. aeruginosa bacteria were also subjected to 10 μM C3a at physiological salt conditions in the presence of human WF (20%). For comparison, LL-37 was used at the same concentration (Fig. 4B). The peptides were active in both the absence and the presence of WF, but C3a was significantly more potent than LL-37 in the presence of WF. These experiments demonstrated that C3a has potent antibacterial effects under physiological conditions. However, the molecule exerts multiple proinflammatory functions, thus making it hard to dissect out a direct antibacterial role in vivo. Thus, the CNY21 peptide (Fig. 5A) and a variant, CNY21(S), in which three arginine residues were replaced with serines, rendering it totally devoid of antibacterial activity (Fig. 5B), were injected into mice infected by the Gram-positive bacterium S. pyogenes. Compared with the control peptide, treatment with CNY21 yielded significantly lower bacterial numbers in the spleen of the animals (P = 0.006) (Fig. 5C). This result, and the fact that both peptides contained the terminal anaphylatoxin determinant LGLAR (9), demonstrated a direct antibacterial role of a well defined helix-forming segment (15) of the C3a molecule.
Many AMPs, such as defensins and LL-37, are found at epithelial surfaces. Furthermore, Pasch et al. (20) have shown that keratinocytes constitutively produce C3 and that the production is up-regulated by inflammatory cytokines. In addition, neutrophil elastase and mast cell tryptase both release C3a-like peptides (21, 22). Acute human WF was therefore incubated with lysed neutrophils for various periods of time. Western blot analysis using an anti-LGE27 antibody showed that C3a-like peptides were generated over time (Fig. 5D). Also, we explored whether neutrophil elastase could further degrade C3a (Fig. 5E). C3a was treated with the enzyme for increasing periods of time, and the resulting peptide fragments were analyzed by MALDITOF MS and Edman degradation. The results showed that neutrophil elastase degraded C3a into several fragments. One fragment, 44SLGEACKKVFLDCCNYITELRRQHA68, containing a helical region of the C3a peptide, was identified by both methods. This peptide was synthesized and found to exert similar antibacterial effects as CNY21 and the other C3a-derived peptides (data not shown). This result indicates that, apart from C3a, additional antibacterial fragments of C3 may be generated during inflammation.
Significant amounts of C3a and C3a-desArg have been detected during sepsis (up to 0.5 μM in blood) (23), acute media otitis (≈0.7 μM in ear secretions) (24), and in epithelial lining fluids of patients at risk for adult respiratory distress syndrome (≈1.3 μM) (25). Most likely, even higher concentrations of C3a peptides occur at the site of complement activation. In blood the concentration of C3 is 5–11 μM, and therefore, local complement activation should induce generation of C3a and C3a-desArg at antibacterial concentrations. During wound healing and inflammation, local synthesis of C3 by monocytes and keratinocytes may constitute a significant additional source of C3a at epithelial surfaces (20, 26). Indeed, analogously to other AMPs such as defensins and LL-37 (27), C3a is present in psoriatic skin (28), which in part may explain the lower occurrence of bacterial infections in patients with psoriasis than in patients with atopic dermatitis (29). The fact that C3 deficiency is connected with increased susceptibility to bacterial infections in humans (30) as well as in animal models (31), is also compatible with the antibacterial effect of C3a revealed here. C3a homologues are found in lower organisms lacking an adaptive immune system (32), and it has been reported that a C3a-like peptide is proteolytically generated in serum from the tunicate Pyura stolonifera, after subjection to bacterial lipopolysaccharide (33). These findings together with the data presented here suggest that direct bacterial killing could be an original function of C3-related peptides. If so, this study shows that this function has been conserved during evolution, whereas the complement system of vertebrates has evolved to act in concert with adaptive immunity.
Acknowledgments
We thank Ms. Mina Davoudi, Ms. Maria Baumgarten, Ms. Lotta Wahlberg, and Ms. Lovisa Ringstad for expert technical assistance and Dr. Björn Walse (Active Biotech Research, Lund, Sweden) for the space-filling model of CNY21.This work was supported by grants from the Swedish Research Council (projects 7480, 13471, and 14379); the Royal Physiographic Society in Lund; the Welander-Finsen, Thelma-Zoegas, Groschinsky, Crafoord, Åhlen, Österlund, Lundgrens, Lions and Kock Foundations; and The Swedish Government Support for Clinical Research (ALF).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: AMP, antimicrobial peptide; WF, wound fluid; cfu, colony-forming units; CF, carboxyfluorescein; MIC, minimum inhibitory concentration; TSB, trypticase soy broth; RDA, radial-diffusion assay.
References
- 1.Fearon, D. T. & Locksley, R. M. (1996) Science 272, 50-53. [DOI] [PubMed] [Google Scholar]
- 2.Steiner, H., Hultmark, D., Engström, A., Bennich, H. & Boman, H. G. (1981) Nature 292, 246-248. [PubMed] [Google Scholar]
- 3.Schröder, J. M. & Harder, J. (1999) Int. J. Biochem. Cell Biol. 31, 645-651. [DOI] [PubMed] [Google Scholar]
- 4.Boman, H. G. (2000) Immunol. Rev. 173, 5-16. [DOI] [PubMed] [Google Scholar]
- 5.Zasloff, M. (2002) Nature 415, 389-395. [DOI] [PubMed] [Google Scholar]
- 6.De, Y., Chen, Q., Schmidt, A. P., Anderson, G. M., Wang, J. M., Wooters, J., Oppenheim, J. J. & Chertov, O. (2000) J. Exp. Med. 192, 1069-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niyonsaba, F., Iwabuchi, K., Someya, A., Hirata, M., Matsuda, H., Ogawa, H. & Nagaoka, I. (2002) Immunology 106, 20-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang, D., Chen, Q., Hoover, D. M., Staley, P., Tucker, K. D., Lubkowski, J. & Oppenheim, J. J. (2003) J. Leukocyte Biol. 74, 448-455. [DOI] [PubMed] [Google Scholar]
- 9.Hugli, T. E. (1989) Curr. Top. Microbiol. Immunol. 153, 181-208. [DOI] [PubMed] [Google Scholar]
- 10.Oren, Z., Lerman, J. C., Gudmundsson, G. H., Agerberth, B. & Shai, Y. (1999) Biochem. J. 341, 501-513. [PMC free article] [PubMed] [Google Scholar]
- 11.Tossi, A., Sandri, L. & Giangaspero, A. (2000) Biopolymers 55, 4-30. [DOI] [PubMed] [Google Scholar]
- 12.Lehrer, R. I., Rosenman, M., Harwig, S. S., Jackson, R. & Eisenhauer, P. (1991) J. Immunol. Methods 137, 167-173. [DOI] [PubMed] [Google Scholar]
- 13.Andersson, E., Rydengård, V., Sonesson, A., Mörgelin, M., Björck, L. & Schmidtchen, A. (2004) Eur. J. Biochem. 271, 1219-1226. [DOI] [PubMed] [Google Scholar]
- 14.Staali, L., Mörgelin, M., Björck, L. & Tapper, H. (2003) Cell. Microbiol. 5, 253-265. [DOI] [PubMed] [Google Scholar]
- 15.Lu, Z. X., Fok, K. F., Erickson, B. W. & Hugli, T. E. (1984) J. Biol. Chem. 259, 7367-7370. [PubMed] [Google Scholar]
- 16.Caporale, L. H., Tippett, P. S., Erickson, B. W. & Hugli, T. E. (1980) J. Biol. Chem. 255, 10758-10763. [PubMed] [Google Scholar]
- 17.Ganz, T. (2001) Semin. Respir. Infect. 16, 4-10. [DOI] [PubMed] [Google Scholar]
- 18.Wang, Y., Agerberth, B., Lothgren, A., Almstedt, A. & Johansson, J. (1998) J. Biol. Chem. 273, 33115-33118. [DOI] [PubMed] [Google Scholar]
- 19.Wright, J., Schwartz, J. H., Olson, R., Kosowsky, J. M. & Tauber, A. I. (1986) J. Clin. Invest. 77, 782-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pasch, M. C., Van Den Bosch, N. H., Daha, M. R., Bos, J. D. & Asghar, S. S. (2000) J. Invest. Dermatol. 114, 78-82. [DOI] [PubMed] [Google Scholar]
- 21.Taylor, J. C., Crawford, I. P. & Hugli, T. E. (1977) Biochemistry 16, 3390-3396. [DOI] [PubMed] [Google Scholar]
- 22.Schwartz, L. B., Kawahara, M. S., Hugli, T. E., Vik, D., Fearon, D. T. & Austen, K. F. (1983) J. Immunol. 130, 1891-1895. [PubMed] [Google Scholar]
- 23.Stove, S., Welte, T., Wagner, T. O., Kola, A., Klos, A., Bautsch, W. & Kohl, J. (1996) Clin. Diagn. Lab. Immunol. 3, 175-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Narkio-Makela, M., Teppo, A. M. & Meri, S. (2000) Laryngoscope 110, 1745-1749. [DOI] [PubMed] [Google Scholar]
- 25.Zilow, G., Joka, T., Obertacke, U., Rother, U. & Kirschfink, M. (1992) Crit. Care Med. 20, 468-473. [DOI] [PubMed] [Google Scholar]
- 26.Colten, H. R., Strunk, R. C., Perlmutter, D. H. & Cole, F. S. (1986) Ciba Found. Symp. 118, 141-154. [DOI] [PubMed] [Google Scholar]
- 27.Ong, P. Y., Ohtake, T., Brandt, C., Strickland, I., Boguniewicz, M., Ganz, T., Gallo, R. L. & Leung, D. Y. (2002) N. Engl. J. Med. 347, 1151-1160. [DOI] [PubMed] [Google Scholar]
- 28.Takematsu, H., Ohkohchi, K. & Tagami, H. (1986) Br. J. Dermatol. 114, 1-6. [DOI] [PubMed] [Google Scholar]
- 29.Christophers, E. & Henseler, T. (1987) Arch. Dermatol. Res. 279, Suppl., S48-S51. [DOI] [PubMed] [Google Scholar]
- 30.Alper, C. A. (1998) Exp. Clin. Immunogenet. 15, 203-212. [DOI] [PubMed] [Google Scholar]
- 31.Wessels, M. R., Butko, P., Ma, M., Warren, H. B., Lage, A. L. & Carroll, M. C. (1995) Proc. Natl. Acad. Sci. USA 92, 11490-11494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith, L. C., Clow, L. A. & Terwilliger, D. P. (2001) Immunol. Rev. 180, 16-34. [DOI] [PubMed] [Google Scholar]
- 33.Raftos, D. A., Robbins, J., Newton, R. A. & Nair, S. V. (2003) Comp. Biochem. Physiol. A Physiol. 134, 377-386. [DOI] [PubMed] [Google Scholar]