Abstract
CDK4/6 inhibitors have emerged as a powerful class of agents with clinical activity in a number of malignancies. Targeting the cell cycle represents a core attack on a defining feature of cancer. However, the mechanisms through which selective CDK4/6 targeted agents act has few parallels in the current pharmaceutical armamentarium against cancer. Notably, CDK4/6 inhibitors act downstream of most mitogenic signaling cascades, which have implications both related to clinical efficacy and resistance. Core knowledge of cell cycle processes has provided insights into mechanisms of intrinsic resistance to CDK4/6 inhibitors; however, the basis of acquired resistance versus durable response is only beginning to emerge. This review focuses on the mechanism of action and biomarkers to direct the precision use of CDK4/6 inhibitors and rationally-developed combination therapies.
Keywords: Cell Cycle, Cyclin D1, E2F, RB, Breast Cancer
TARGETING CYCLIN DEPENDENT KINASES FOR CANCER TREAMENT
Targeting the activity of cyclin dependent kinases (CDK) as a means to treat cancer is a concept that was first proposed when it became established that cell division is driven by these kinases [1, 2]. In spite of substantial knowledge related to cell cycle regulation and multiple efforts to develop drugs targeting CDK activity, only recently have there been significant successes with CDK inhibitors in the clinic. Selective CDK4/6 inhibitors (BOX one) have emerged as agents that could have broad clinical application, as evidenced by multiple recently reported clinical studies. The development of CDK4/6 inhibitors and the clinical outcomes to this point have been the subject of multiple excellent reviews [3–6]. Here, we focus on the biological response to CDK4/6 inhibitors, biomarkers, and the basis of resistance that will serve inform future clinical approaches.
BOX 1. Repertoire of CDK4/6 inhibitors.
There are a number of agents that can indirectly inhibit CDK4/6 activity. However, the current class of CDK4/6 directed agents consists of Palbociclib, Ribociclib and Abemaciclib[3, 88, 125]. These molecules were developed for specificity towards CDK4/6 and they exhibit specificity for CDK4/6 relative to other CDKs. Palbociclib and Ribociclib are very similar molecules structurally and exhibit similar kinase inhibitory activities with similar inhibitory activity against CDK4 and CDK6[3, 125]. They also exhibit similar toxicity profile, where the dose limiting toxicity for these agents is myelosuppression (neutropenia)[142–144]. Typically these agents are dosed with three weeks on treatment and one week of treatment discontinuation to recover neutrophil counts. In contrast, Abemaciclib has a different spectrum of inhibitory activity, with a preference toward CDK4[88]. The predominant toxicities of abemaciclib are diarrhea and fatigue[74]. Unlike the other two agents, the dosing of abemaciclib is continuous[74]. While the agents are considerably more selective for CDK4/6, they also can inhibit other kinases[88, 145, 146]. The extent to which these off-target events are of significance in the treatment of disease is not clear.
CDK4/6 IN CELL CYCLE CONTROL
Multiple mitogenic and growth inhibitory signals must be integrated to determine if it is appropriate for cells to divide. In metazoans, much of this regulation is directed at the kinases that mediate the earliest transitions in the G1 phase of the cell cycle (Fig. 1). While different cell types have different constellations of kinases, generally CDK4 and/or CDK6 activity is the point at which this integration occurs. These activity of these kinases is dependent on the synthesis, accumulation, and correct localization of their cognate cyclin partners [7–9]. Depending on the cell type, this could represent cyclin D1, D2, and/or D3. The tissue specific context for CDK4/6 and D-type cyclin function has been determined from extensive mouse studies[10]. For example, the deletion of cyclin D1 yields small body size and reduced mammary development in mice[11]. In contrast, deletion of cyclin D2 results in defective ovarian development and female infertility[12]. Similarly, CDK4 deletion is associated with small body size and multiple development defects, while CDK6 deletion results in largely hematopoietic phenotypes[10, 13]. These genes are under very complex regulatory controls. For example, cyclin D1 is a delayed early gene that is responsive to mitogenic signals[14, 15]; but protein accumulation is regulated by multiple mechanisms that impinge on stability and subcellular localization[16]. These mechanisms can have profound effects, as cyclin D1 half-life can fluctuate from ~30 minutes to 3.5 hours depending on a single phosphorylation event [17]. In parallel, with the regulation of the cyclin subunits, CDK4/6 kinases are regulated by phosphorylation events and the presence of physiological kinase inhibitory proteins[18]. These inhibitors are encoded by the CDKN2 gene family (e.g. CDKN2A and CDKN2B) and produce selective CDK-inhibitors for CDK4 and CDK6 (e.g. p16INK4a, p15INK4b)[18, 19].
Figure 1. Signaling integration through the CDK4/6-RB pathway.
Mitogenic signals and anti-proliferative signals are integrated to yield the expression of D-type cyclins and the activation state of CDK4/6. The active kinase complex can then initiate the phosphorylation of RB and the release from the E2F family of transcription factors. These actions can be counter-balanced by the activity of physiological CDK4/6 inhibitors exemplified by p16ink4a. The E2F family of transcription factors coordinates a gene expression program that is required for features of cell cycle progression, DNA replication, and mitosis. Thus, the pathway serves to integrate signal transduction in controlling cell division. Genetic events that impact directly on CDK4/6, the inhibitors of the active complex, or disease relevant substrates are shown. Data is summarized from TCGA data deposited in cbioportal (www.cbioportal.org) and impacts on multiple organi systems.
If mitogenic conditions are dominant, the regulatory pendulum will swing toward the accumulation of active CDK4/6-cyclin D complexes[7, 20]. The kinases then phosphorylate critical substrates to elicit progression through G1 and the commitment to progress through the cell cycle (Fig. 1). The first defined substrate for CDK4/6 was the RB tumor suppressor protein[15]. This protein contains 16 sites for potential phosphorylation by cyclin dependent kinases and can be phosphorylated on multiple sites by CDK4/6. It is believed that phosphorylation events elicit structural alterations in the RB protein that lessen its intrinsic growth suppressing activity and prime it for subsequent phosphorylation [21–26]. How this occurs remains a matter of debate; however, it is widely accepted that in the absence of CDK4/6 activity RB will not be phosphorylated. In this configuration RB and related proteins can elicit the inhibition of the E2F family of transcription factors[27, 28]. This event is particularly important as E2F controls a wide range of genes required for S-phase and mitotic progression [27, 29, 30]. This represents a simplified model of how CDK4/6 activity links mitogenic signaling to cell cycle progression (Fig. 1).
The importance of the appropriate regulation of CDK4/6 activity for normal tissue homeostasis is underscored by multiple genetic events in cancer that result in hyperactivation of CDK4/6 (Fig. 1). For example, cyclin D1 amplification, CDK4 amplification, and deletion of CDKN2A are common events in multiple human cancers [7, 9, 20]. These data served to credential CDK4/6 kinases and related cell cycle pathways as important targets for cancer therapy.
PHYSIOLOGICAL CDK4/6 INHIBITION
Under normal physiological conditions the inhibition of CDK4/6 activity arises as a consequence of limiting mitogen or intrinsic growth inhibitory signals. Inhibition of signaling pathways such as endocrine signaling, RAS/RAF/ERK, or EGFR/ERBB2 affects cell cycle in part by limiting the expression of D-type cyclins and CDK4/6 activity [31–36] (Fig. 2). Thus, limiting mitogenic signals physiologically or with therapeutic agents elicits inhibition of CDK4/6. However, mitogenic signaling has multiple collateral effects beyond the accumulation of CDK4/6 activity to promote cellular proliferation (e.g. increased metabolism, degradation of p27Kip1, etc.), and there are a myriad of bypass mechanisms that arise in cancer and ultimately overcome dependence on a single pathway. In the case of ER+ breast cancer, the estrogen dependence can be overcome through multiple mitogenic signaling pathways (e.g. deregulated HER2 or PI3K signaling)[37–39]. In KRAS or BRAF-driven tumors, MEK inhibition leads to compensatory PI3K/AKT activation that can circumvent the dependence on MEK for proliferation[40, 41]. Since CDK4/6 is downstream from these varied signaling pathways, it has been proposed to represent a therapeutic target for which there are fewer bypass mechanisms.
Figure 2. Oncogenic pathways impinging on CDK4/6.
Signaling pathways associated with tumorigenisis that represent common therapeutic targets are shown. These signaling pathways yield an induction of CDK4/6 activity through the stimulation of cyclin D expression. Representative therapeutic agents blunt this signaling, and yield cell cycle inhibition through multiple mechanisms including the attenuation of cyclin D1. In contrast the pharmaceutical inhibitors palbociclib, ribociclib and abemaciclib directly inhibit CDK4/6 activity to elicit the suppression of proliferation. Oncogenic stresses also impact on CDK4/6 activity. In this context, while mitogenic signaling is intact and D-type cyclins are expressed, CDK4/6 activity is inhibited through the action of endogenous inhibitors (e.g. p16ink4a).
There are few physiological of a selective CDK4/6 inhibitor, wherein upstream mitogenic/oncogenic signaling is maintained but cell cycle progression is inhibited. Perhaps the closest mechanism would be the response to “oncogenic stress” (Fig. 2). Deregulated oncogenic signals can drive cell cycle exit and a senescence-like phenotype[42–45], whereby cell division is blocked in spite of intact mitogenic signaling. This phenotype is thought to arise either from replication stress or reactive oxygen species that yield an induction of endogenous growth suppressive mechanisms[46–48]. Such stressors lead to physiological induction of endogenous CDK4/6 inhibitors; for example, CDKN2A encoding p16ink4a or CDKN2B encoding p15ink4b. This event is believed to represent a cornerstone tumor suppressive force in specific pre-neoplastic conditions[43, 44, 49]. Notably, oncogenic mutations in BRAF or NRAS that fuel melanoma are observed in melanocytic nevi; however, proliferation is restricted by the induction of endogenous CDK4/6 inhibitors[49–52]. Since oncogenic stress induces a DNA damage-like response, it likely elicits more complex effects on cell cycle than solely CDK4/6 inhibition. However, the potent function of p16ink4a in suppression of melanoma progression likely explains why germline mutations in CDKN2A are strongly associated with increased risk for melanoma. Furthermore, this model explains why loss of CDKN2A is prevalent in tumors types driven by BRAF or KRAS. The fact that endogenous CDK4/6 inhibitors can suppress proliferation of precursor lesions for years suggests that pharmaceutical mimicry of these tumor suppressors could represent a fruitful therapeutic approach.
PRECLINICAL STUDIES WITH CDK4/6 INHIBITORS
To date, preclinical studies have evaluated the single agent activity of CDK4/6 inhibitors in ~30 different cancer indications[3]. These tumors are driven by a plethora of high-potency oncogenic events as well as loss of various tumor suppressors that contribute to the malignant phenotype (Table 1). In general, the addition of the CDK4/6 inhibitor in these tumor types can limit cell cycle progression as long as there is an intact RB-pathway downstream. In solid tumor models the effect of CDK4/6 inhibition is cytostatic in cell culture and animal models with potential regression associated with the intrinsic tumor cell turnover. Inhibition of CDK4/6 is cytotoxic in select hetmatological malignancies, which is associated with a tissue selective impact of CDK4/6 on maintenance of cell viability[53]. From the analysis of the multiple cancer models and expanding clinical experience, rules guiding the use of CDK4/6 inhibitors in a more precise manner are beginning to emerge.
Table 1.
Summary of studies performed with CDK4/6 inhibitors in different tumor types
| Models interrogated for sensitivity to CDK4/6 inhibitors | Reference |
|---|---|
| Mantle Cell Lymphoma | [77, 113] |
| Acute Lymphoblastic Lymphoma | [53, 114] |
| Multiple Myeloma | [115] |
| Acute Myeloid Leukemia | [93] |
| Chronic Myelogenous Leukemia | [116] |
| Medulloblastoma | [117] |
| Neuroblastoma | [86] |
| Liposarcoma | [81, 82] |
| Rhabdomyosarcoma | [118] |
| Ewing Sarcoma | [119] |
| Synovial Sarcoma | [120] |
| Rhabdoid Tumors | [121] |
| MPNST | [122] |
| Gastric Cancer | [123] |
| Hepatocellular Carcinoma | [56] |
| Pancreatic Neuroendocrine | [124] |
| Colon Cancer | [125] |
| Esophageal Cancer | [126] |
| Pancreatic Ductal Adenocarcinoma | [80, 127] |
| Non small-cell lung cancer | [125, 128] |
| Prostate Cancer | [129] |
| Renal | [130] |
| Bladder | [131] |
| Melanoma | [132, 133] |
| Glioma | [92, 134, 135] |
| Squamous Cell Carcinoma of Head and Neck | [136] |
| Ovarian Cancer | [57] |
| Breast Cancer | [55, 58, 84, 137] |
MARKERS OF INTRINSIC RESISTANCE
Given all that is known about the cell cycle, an intact RB tumor suppressor downstream of CDK4/6 will be required for this class of agent to have effect. This dependence has been shown both with the endogenous CDK4/6 inhibitor p16ink4a and with pharmacological agents (Table 2) [54–60]. In the absence of RB there is no requirement for CDK4/6 to activate E2F-regulated genes (Fig. 1). This is particularly important as RB serves to couple CDK4/6 to many genes that are required for cell cycle progression (e.g. Cyclin A, Cyclin E, Cyclin B1) and that represent independent therapeutic targets (e.g. PLK1, DHFR, RNRII).
Table 2.
Cell cycle proteins that could be associated with response to CDK4/6 inhibition
| ACCEPTED RESISTANCE | RELEVANT TUMOR TYPES | REFERENCES |
|---|---|---|
| RB loss | Multiple | [55–58, 81, 92, 125] |
| p16INK4a high | RB-deficient tumors or HPV | [55–58, 130] |
| Viral oncoproteins of DNA tumor viruses | Cervical Cancer and SSCHN | [56, 90] |
| PUTATIVE RESISTANCE | RELEVANT TUMOR TYPES | |
| Cyclin E overexpression (Cyclin E1/E2 amplification) | Uterine (40%), Ovarian (20%), Bladder (15%) Metastatic prostate cancer (20%), Breast (15%) | [60, 71] |
| E2F overexpression (E2F3 amplification) | Bladder (30%), | [55, 56] |
| PUTATIVE SENSITIVITY | RELEVANT TUMOR TYPES | |
| Cyclin D1 amplification/translocation | MCL, Breast, Esophageal | [58, 113] |
| CDK4 amplification | Liposarcoma, GBM | [81, 82] |
| CDKN2A loss | Multiple | [135] |
However, there is another feature of the loss of RB which is often not considered in evaluating the sensitivity to CDK4/6 inhibitors. As shown a number of years ago, loss of RB function is associated with high-level expression of endogenous CDK4/6 inhibitors[19, 61, 62]. Specifically, loss of RB is speculated to induce its own replication stress that leads to the accumulation of p16ink4a [63]. This finding suggests that any tumor that expresses high levels of p16ink4a and is still progressing through the cell cycle will be resistant to CDK4/6 inhibition. This would include any HPV+ tumor where high levels of p16ink4a are routinely employed in the diagnosis (e.g. cervical cancer and squamous cell cancer of the head and neck) [19]. Since p16ink4a staining is used routinely in pathology labs, it represents an important tool for evaluating tumor sensitivity to CDK4/6 inhibitors (Table 2).
The combination of high p16ink4a expression with loss of RB expression has emerged as one of the more commonly used approaches to exclude or include patients on clinical trials with CDK4/6 inhibitors (e.g. NCT01536743, NCT02334527, NCT01976169). In several tumors types, where RB loss is rare, the use of these markers may not appear to be particularly important. This is the case with ER+/HER2- breast cancer, where RB genetic loss is rare (~3% of cases) and there has been essentially no use of RB-status for inclusion to CDK4/6 inhibitor-based therapy. However, it is important to acknowledge that there are fewer genetically characterized metastatic breast cases, and there is clear evidence that RB loss can occur more frequently with metastatic progression[64, 65]. These data reinforce the need to consider the status of the core RB pathway for targeting CDK4/6 in cancer therapy.
In spite of the seemingly obvious importance of RB and p16ink4a as biomarkers of the response to CDK4/6 inhibitors, there have been no clinical trials that evaluate the predictive status of these markers for two primary reasons. First, existing clinical trial strategies avoid tumors with a large number of RB-negative cases (ie. small cell lung cancer) or tumors driven by HPV (ie. cervical cancer). Additionally, many clinical trials actively exclude patients with RB-negative tumors. Second, since it can be viewed that treating patients with RB-deficient tumors with a CDK4/6 inhibitor is unethical, it is unlikely that conclusive clinical proof of these biomarkers will emerge.
PUTATIVE MARKERS OF RESISTANCE AND SENSITIVITY
Since cell cycle has been under study for many years a lot is known regarding how tumor cells can escape from CDK4/6 inhibitors (Table 2). Much of this work supersedes the current interest in pharmaceutical CDK4/6 inhibition and was based on mechanisms of cell cycle control by p16ink4a or RB. These data are based largely on preclinical data and the extent to which they apply to the clinic is unclear.
RESISTANCE MECHANISMS
A key feature of cell cycle inhibition at the level of CDK4/6 is the impact on downstream effectors through the RB pathway. Typically, the inhibition of CDK4/6 will elicit a suppression of CDK2-associated kinase activity. This event is believed to be mediated through the suppression of E2F-target genes including cyclin E1, cyclin E2, cyclin A2 [56, 66–68]. Thus, a CDK4/6 inhibitor has the net effect of suppressing multiple kinases that drive the G1/S transition, and mechanisms that uncouple CDK4/6 from these other cell cycle kinases ostensibly yields therapeutic failure.
Cyclin E amplification and protein overexpression is observed in multiple human tumors, including endometrial cancer and breast cancer (Table 2). In these tumor-types, cyclin E alterations are generally associated with poor prognosis [69]. Deregulation of cyclin E expression has been shown to compromise the response to CDK4/6 inhibition in multiple models [66, 70, 71]. Additionally, in certain settings of acquired resistance to CDK4/6 inhibition, deregulated cyclin E expression is functionally linked to resistance[72, 73]. What is less clear is whether cyclin E amplification, as observed in clinical specimens, is sufficient to bypass the activity of CDK4/6 inhibitors. This point is relevant when considering that certain luminal breast cancer cell lines that are sensitive to CDK4/6 inhibition harbor cyclin E amplification. In recently published studies, there was no evidence for cyclin E amplification as a determinant of the response to CDK4/6 inhibition [74], although this could simply reflect that the overall number of cases harboring this amplification was too small to detect the association. The regulation of cyclin E and its associated kinase is complex, and also reflects the activity of CDK inhibitors (e.g. p27Kip1) and proteolytic regulatory processes (e.g. FBXW7) that are also frequently dysregulated in cancer[75, 76]. Thus, it is perhaps too simple to only consider gene amplification without a more robust analysis of the entire pathway controlling cyclin E function.
The overexpression of E2F activating transcription factors represents a means to uncouple CDK4/6 inhibition from downstream gene-expression programs that drive DNA replication and mitosis [55, 56]. E2F3 amplification occurs in a number of tumor types, most notably bladder cancer, where E2F3 is amplified in up to 20% of cases. As is the case with cyclin E, the preclinical data has employed ectopic expression rather than a specific reliance on endogenous gene amplification events. Therefore, whether this event as occurs in cancer has bearing on response to CDK4/6 inhibition remains unknown.
Preclinical data has shown how how cyclin E or E2F3 amplification events could impact on the sensitivity to CDK4/6 inhibitors. However, how the TP53 tumor affects sensitivity to CDK4/6 inhibition is more obscure. From a fundamental feature of cell cycle control, TP53 loss is associated with a degree of CDK2 deregulation as a consequence of low levels of the TP53 target gene p21Cip1. Despite that fact, CDK4/6 inhibition has been shown to be effective in multiple tumor models that harbor TP53 mutation. However, in a recently reported clinical study, TP53 mutation was associated with poor clinical response to the CDK4/6 inhibitor Abemaciclib in breast cancer patients [74]. This finding could be due to the association of TP53 mutations with triple negative disease in the cohort analyzed. Therefore, additional analyses will be required to determine if TP53 mutation does represent an independent determinant of the response to CDK4/6 inhibition.
DETERMINANTS OF SENSITIVITY
While there has been a significant interest in determining means to direct CDK4/6 inhibitors in the clinic, the identification of specific markers for sensitivity remain unclear. Much of the prior work has focused on genetic events that would be associated with an addiction to CDK4/6 and represent common alterations in cancer (Table 2).
Tumors with deregulation of cyclin D1 by amplification or translocation have been extensively studied. In mantle cell lymphoma (MCL) translocations targeting cyclin D1 are considered driver events[77, 78]. Preclinical MCL models are sensitive to CDK4/6 inhibition, and clinically MCL is responsive to CDK4/6 inhibition[77]. Whether the response is directly reflective of the genetic alteration of the cyclin D1 gene is difficult to evaluate, as most patients harbor this hallmark genetic event. In other cancers, where there is diversity of cyclin D1 genetic deregulation, there is no positive data indicating the cyclin D1 is a predictive marker. In the Phase II Paloma 1 study there was a selection for patients with cyclin D1 amplification[79]. These patients harbored essentially the same outcome as an unselected patient population. Therefore, whether cyclin D1 amplification is a clear determinant of response remains unclear, although certain tumor types with a strong dependence on cyclin D1 alterations might be considered generally sensitive to CDK4/6 inhibition.
Loss of the CDK4/6 inhibitor CDKN2A is a frequent event in tumors, where there is substantial pressure to bypass oncogenic stress. Hence, there is a strong rationale for restoring the tumor suppressive features of CDKN2A from a pharmacological perspective. To date, CDKN2A loss does not predict sensitivity to CDK4/6 inhibitors in preclinical or clinical studies. In preclinical models with genetic loss of p16ink4a there is evidence for acquired resistance to CDK4/6 inhibitors[72, 80]. Similarly, in cancers that lose CDKN2A frequently there is little evidence for a direct correlation between response and loss of the tumor suppressor [74].
Amplification of CDK4 or CDK6 is observed in a number of cancers including sarcomas and glioblastoma multiforme. In liposarcoma the amplification of CDK4 is likely a driving event due to the frequency with which the amplification occurs. Consonantly, liposarcoma models are sensitive to CDK4/6 inhibition [81]. This feature was used to conduct a marker-driven Phase II trial[82]. The trial showed there was an increase in progression-free survival beyond the historical control, but since there was no control arm additional study will be required to determine if the amplification of CDK4 denotes selective sensitivity to CDK4/6 inhibitors. Selection for treatment based on CDK4 or CDK6 amplification is not without concern, as CDK6 amplification has been recently shown to drive resistance to CDK4/6 inhibitors in preclinical models[83].
In total review of predictive biomarkers for sensitivity to CDK4/6 inhibition, universal determinants have failed to emerge. Cyclin D1 amplification and loss of CDKN2A are mutually exclusive with loss of RB. However, due to the genetic complexity of cancer and potential mechanisms of acquired resistance the simple models of genetically encoded sensitivity may miss the mark. This is particularly relevant when considering the myriad of post-translational modifications that control cell cycle transitions. A notable example is in the comparison of melanoma, non small cell lung adenocarcinoma and colon cancer that frequently lose CDKN2A as the predominant genetic mechanism of CDK4/6 deregulation. Recently reported single agent data suggest that melanoma and lung adenocarcinoma have potential sensitivity to CDK4/6 inhibition, whilst colon cancer is largely resistant[74]. Thus, perhaps tissue-context, cell cycle plasticity, and co-occurring genetic events may trump simple genetic analysis of the pathway (outstanding question box). These considerations are significant as basket and umbrella trials have opened enrolling patients to treatment with CDK4/6 inhibitors based on CDKN2A loss, CDK4 or CDK6 amplification, and cyclin D1 amplification (BOX 2).
OUTSTANDING QUESTIONS BOX.
Biomarkers: Are genetic features that deregulate CDK4/6 activity in cancer effective biomarkers of response? Are there biomarkers of durable response to CDK4/6 inhibitors? How will biomarkers to direct CDK4/6 in combination therapy emerge?
Acquired Resistance: Does clinical acquired resistance represent the selection of existing clones? What drives the differing adaptive responses to CDK4/6 inhibition that are associated with acquired resistance in preclinical models? Can the ability of cell cycle to become CDK4/6-independet by predicted and selectively targeted therapeutically?
Combination Therapies: Are there general rules for selective combination therapies with CDK4/6 inhibition? Will there be selective combinations for each tumor type or is there a combination with CDK4/6 inhibitors that could work across multiple tumor types?
BOX 2. Umbrella and Basket trials with CDK4/6 inhibitors.
There are currently several basket and umbrella trials open using genetic features of tumors to assign treatment with CDK4/6 inhibitors. In the diagnosis agnostic basket-trial NCI-MATCH (NCT02465060) patients with tumors that harbor cyclin D1, D2 or D3 amplification can be treated with palbociclib. Correspondingly, the Novartis sponsored SIGNATURE trial (NCT02187783) enrolls patients with tumors containing either CDK4 amplification or mutation, CDK6 amplification or mutation, Cyclin D1 amplification, Cyclin D3 amplification, or p16ink4a loss to treatment with ribociclib. The Lung-MAP trial (NCT02154490) enrolls patients with recurrent metastatic squamous lung cancer who are “positive” for CDK 4/6, cyclin D1, cyclin D2, and cyclin D3 to treatment with palbociblib vs. a comparator arm of docetaxel. The MatchMel trial (NCT02645149) will match patients with metastatic melanoma to treatment with palbociclib if the tumors contain aberrations in cyclin D1, cyclin D3, CDK4 and p16ink4a genes. These study designs are based on the premise sthat the genetic aberrations present in the tumors will yield sensitivity to CDK4/6 inhibition.
CELL CYCLE EXIT AND SENESCENCE AS THERAPEUTIC ENDPOINTS
While predictive biomarkers for durable response to CDK4/6 inhibitors have been challenging to define, there are multiple potential pharmacodynamic markers. The suppression of the proliferative marker Ki67 has been shown to correlate with meaningful response to CDK4/6 inhibition in multiple preclinical models [84–88]. The suppression of Ki67 represents one of the key endpoints that are built into many ongoing neoadjuvant clinical trials (e.g. NCT02530424, NCT01723774, NCT02712723, NCT02441946). In addition, CDK4/6 inhibitors elicit the blockade of RB phosphorylation and down-regulation of a host of cell cycle regulatory genes (e.g. MCM7 and TopoIIα, as a consequence of RB activation and the inhibition of E2F[28, 77, 88]. Thus, there are multiple targets that could be measured to determine if the CDK4/6 inhibitors are acting on the target pathway; however, it remains generally undetermined how such acute responses correlate with long-term disease control.
It has been proposed that one of the key features of CDK4/6 inhibition is to elicit a cell cycle inhibitory response that mimics the intrinsic senescence biology. In multiple preclinical settings, it would appear that the durability of response to CDK4/6 inhibition is associated with the induction of a senescent-like phenotype[35, 55, 89, 90]. For example, in the context of melanoma models, CDK4/6 inhibition elicits a profound senescent-like arrest characterized [90]. In liposarcoma models, the continued presence of MDM2 and lack of ATRX in the context of CDK4/6 inhibition prevents the induction of senescence[89]. Importantly, the suppression of MDM2 was associated with improved response to CDK4/6 inhibition in a small collection of liposarcomas treated with palbociclib, suggesting that MDM2 downregulation could be an important pharmacodynamics marker related to senescence [89].
One of the principle caveats with senescent-like arrests is the extent to which they are biologically equivalent to true senescence. Senescence is defined by the irreversible cell cycle arrest and the acquisition of a specific senescence-associated secretory phenotype. While over-expression of p16 does not by itself induce senescence-associated secretory phenotype[91], recent studies in melanoma models suggested that treatment with CDK4/6 inhibitors induced this feature of senescence biology. In this setting, the arrest induced by CDK4/6 inhibition also appears to be largely irreversible. However, in other instances the senescent-like phenotype associated with treatment with CDK4/6 inhibitors is reversible and cells readily re-enter the cell cycle as soon as the drugs are withdrawn[92]. This also appears to be the case with single agent treatments in ER+/HER2- breast cancer (Ma et al., 2015 San Antonio Breast Cancer Symposium). While the biological state of senescence is very challenging to evaluate in clinical specimens[45], ongoing clinical studies are addressing the capacity of CDK4/6 inhibition to induce senescence in breast cancer specimens and define the relationship to overall clinical responses (e.g. NCT02008734). The findings from such studies will be important in deciphering biological markers of quiescence or senescence that could predict clinical sensitivity.
ADAPTATION AND ACQUIRED RESISTANCE
Whether the correct constellation of biomarkers will be able to predict the durable sensitivity to CDK4/6 inhibitors remains to be determined. This is particularly important to consider when evaluating the basis of acquired resistance that emerges. While CDK4/6 inhibition can have effect in the clinic, most patients will progress on treatment presumably through mechanisms of acquired resistance. Preclinical studies have suggested that resistance to CDK4/6 inhibitor can emerge through the selection for acquired mutation of RB1, amplification of cyclin E, amplification of CDK6, or suppression of CDK2 inhibitors (e.g. p27kip1 or p21cip1)[55, 73, 93]. These studies reinforce the concept that such events could represent predictive markers as described above.
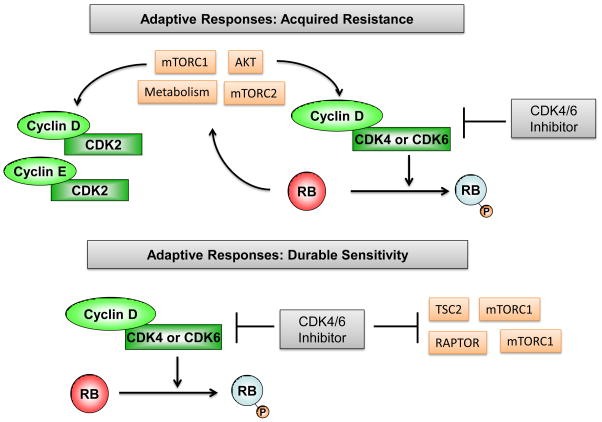
The adaptive response of mitogenic signaling pathways to CDK4/6 inhibition is complicated, but likely plays a critical role in the durability of response (Fig. 3). A growing body of recent work indicates that CDK4/6 inhibition can stimulate mitogenic signals that likely contribute to acquired resistance. The induction of AKT or MTOR activity as a response to CDK4/6 inhibition has been described in several settings. Specifically, CDK4/6 inhibition has been associated with an RB-dependent activation of AKT that is mediated by mTORC2[94]. In contrast, it has also been shown that CDK4/6 inhibition can induce metabolic reprogramming that acts to induce mTORC1[95]. These adaptive features of response to CDK4/6 inhibition are likely important for the increased expression of cyclin D1 that is observed following treatment with CDK4/6 inhibitors [55, 73, 96]. Work in breast cancer models has shown an upregulation of AKT signaling with pharmaceutical inhibition of CDK4/6 that is linked to the accumulation of cyclin D1 [73]. Similarly in pancreatic cancer models, the upregulation of both cyclin D1 and E can be abrogated by MTOR inhibitors[72]. These data suggest that net upstream mitogenic signaling can dictate the upregulation of cyclin expression with CDK4/6 inhibition. Importantly, the inhibition of these adaptive pathways improved durability of response [73, 97], and by selective knockdown approaches it was shown that cyclin D1 and E levels play a role in supporting acquired resistance in preclinical models [72]. How the levels of cyclin D1 could contribute to resistance in the presence of a CDK4/6 inhibitor is unclear, but it could facilitate overall leakiness of CDK4/6 kinase blockade, mediate complexes with CDK2, or titrate CDK2 inhibitors [72, 73, 96]. In contrast with the studies described above, other work indicates that CDK4/6 inhibition yields suppression of MTOR and/or AKT signaling to elicit durable response [87, 90]. Specifically, in models of HER2+ breast cancer CDK4/6 inhibition suppresses TSC2 phosphorylation resulting in diminished mTORC1 activity. In melanoma models, CDK4/6 inhibition elicits an inhibition of mTORC1 activity that is required for senescent-like arrest. In these models further suppression of MTOR/AKT signaling resulted in greater therapeutic effect. These combined findings suggest that the composite suppression of CDK4/6 activity in concert with inhibition of MTOR and/or AKT may represent a key biological feature of durable cell cycle exit and a particularly promising avenue for combination therapy.
Figure 3. Adaptive responses to CDK4/6 inhibition.
In multiple models the inhibition of CDK4/6 inhibition in the face of oncgenic signals results in increased cyclin D1 expression. The adaptive responses to CDK4/6 inhibition includes enhanced MTOR or PI3K activity are associated with acquired resistance which can be prevented through the use of combination treatment that incorporate CDK4/6 inhibitors with PI3K or MTOR inhibitors. In other settings the treatment with CDK4/6 inhibition elicits the inhibition of MTOR activity that is associated with durable response or senescence.
COMBINATION THERAPIES
In specific tumor types, such as MCL and liposarcoma, single agent CDK4/6 inhibitors have demonstrated activity that represents a meaningful advance in therapy refractory disease[77, 82]. Additionally, continuous dosing of abemaciclib was observed to have promising single agent activity in breast cancer, melanoma, and lung cancer [74]. These studies indicate that CDK4/6 inhibition by itself can suppress progression in advanced disease. This single agent experience to date seems to be best served with continuous dosing, presumably wherein the blockade of cell cycle represents the mechanism of action.
To date, the most significant successes of CDK4/6 inhibition in the clinic to date have largely occurred as a function of combination with endocrine therapy in context of metastatic ER+/HER2-disease[79, 98] (Table 3). In multiple reported studies CDK4/6 inhibitor combinations have demonstrated a highly significant improvement over single-agent letrpzole or fulvestrant treatment[99–101]. Endocrine therapy is known to suppress both CDK4/6 and CDK2 activity via mechanisms intrinsically distinct from pharmaceutical inhibition [102, 103], suggesting that in fact targeting various features of cell cycle and limiting the intrinsic mitogenic signaling may represent a critical therapeutic strategy. Correspondingly, high levels of E2F-regulated genes are associated with rapid resistance to endocrine therapy, commonly defining the luminal B subtype of breast cancer[104, 105]. Thus, one of the core features of CDK4/6 inhibitors is to suppress the proliferation of models that have developed resistance to endocrine therapy[35, 106]. Building on these studies, a number of therapeutic triplets that combine CDK4/6 inhibition with endocrine therapy and inhibition of PI3K or MTOR are currently ongoing (Table 3). The design of such studies is based on both preclinical work[73, 97] and the documented clinical efficacy of MTOR inhibitors with endocrine therapy[107].
Table 3.
Summary of clinical trials interrogating CDK4/6 drug combinations
| Drug Combinations in Clinical Trials | INDICATION | Preclinical Studies | Clinical Trial |
|---|---|---|---|
| Palbociclib+Endocrine Therapy | HR+/HER2- | [58, 138, 139] |
NCT02600923 NCT02668666 NCT02448771 NCT02679755 NCT02040857 NCT02917005 NCT02297438 NCT02690480 NCT01823835 |
| Ribociclib+Endocrine Therapy | HR+/HER2- |
NCT02586675 NCT02712723 NCT02941926 NCT02333370 NCT02632045 |
|
| Abemaciclib+Endocrine Therapy | HR+/HER2- |
NCT02747004 NCT02107703 NCT02675231 |
|
| Palbociclib+AZD2014+fulvestrant | HR+/HER2- | [73, 97] | NCT02599714 |
| Ribociclib+BYL719+Letrozole | HR+/HER2- | NCT01872260 | |
| Ribociclib+Fulvestrant+BYL719/BKM120 | HR+/HER2- | NCT02088684 | |
| Palbociclib+Everolimus+Exemestane | HR+/HER2- | ||
| Ribociclib+Everolimus+Exemestane | HR+/HER2- |
NCT02732119 NCT01857193 |
|
| Palbociclib+PI3K inhibitor+Fulvestrant | HR+/HER2- | NCT02389842 | |
| Palbociclib+Pembrolizumab+Letrozole, | HR+/HER2- | NA | NCT02778685 |
| Palbociclib+T-DM1 | HER2+ | [85] | NCT01976169 |
| Ribociclib+T-DM1 | HER2+ | NCT02657343 | |
| Pabocicblib+Paclitaxel | BCA | NA | NCT01320592 |
| Palbociclib+Paclitaxel | BCA | NCT01320592 | |
| Ribociclib+Paclitaxel | BCA | NCT02599363 | |
| Ribociclib+TACE | HCC | NA | NCT02524119 |
| Ribociclib following radiation therapy | Glioma | [92] | NCT02607124 |
| Ribociclib+ Docetaxel | PCA | NA | NCT02494921 |
| Palbociclib+Nab-Paclitaxel | mPDAC | NA | NCT02501902 |
| Palbociclib+Carboplain/Cisplatin | Solid Tumor | NA | NCT02897375 |
| Palbocicilb+5FU+Oxaliplatin | Solid Tumor | NA | NCT01522989 |
| Ribociclib+Cetuximab | SSCHN | NA | NCT02429089 |
| Palbociclib+ Cetuximab | SSCHN |
NCT02101034 NCT02499120 |
|
| Palbociclib+Trastuzumab | HER2+ | NCT02448420 | |
| Ribociclib+MEK162 | mMEL | [132] | NCT01781572 |
| Palbociclib +Androgen Deprivation | PCA | [129] | NCT02059213 |
| Ribociclib+Enzalutamide | PCA | NCT02555189 | |
| Palbocicilb+Ibrutinib | MCL | [140] | NCT02159755 |
| Ribociclib+ Ceritinib | Alk+NSCLC | NA | NCT02292550 |
| Palbociclib+MEK inhibitor | NSCLC-Solid Tumor | [111, 132, 141] | NCT02022982 |
| Ribociclib+Trametinib | Solid Tumor | NCT02703571 | |
| Ribociclib+ HDM201 | Liposarcoma | NA | NCT02343172 |
A number of additional combination approaches have been interrogated in preclinical models and are now the subject of ongoing clinical trials (Table 3). There are three fundamental features of CDK4/6 inhibition that are guiding most of the active combination clinical trials:
Combinations with chemotherapy: In has generally been reported that CDK4/6 inhibitors are antagonistic of chemotherapy regimens in preclinical models[108]. However, since chemotherapy is not delivered continuously, strategies positioning CDK4/6 inhibition between chemotherapy infusions are being employed in the clinic. The general concept behind this approach is the that CDK4/6 inhibition prevents the proliferation of residual clones of tumor cells that could escape the chemotherapy[109, 110]. This approach is attractive where the CDK4/6 inhibitors are also delivered discontinuously, such that the chemotherapy is delivered during the rest period from CDK4/6 inhibition.
Combination with active clinical regimens: The success of CDK4/6 inhibitors with endocrine therapy in ER+/HER2- breast has spawned a series of clinical trials, wherein the CDK4/6 inhibitor is positioned with standard of care targeted therapies in specific disease indications. For example, there are now trials of CDK4/6 inhibitors: with androgen antagonists in prostate cancer, with trastuzumab in HER2+ breast cancer, with EGFR inhibitors in squamous cell carcinoma of the head and neck, with MEK inhibitors in melanoma, and with ibrutinib in mantle cell lymphoma. In these indications it is expected that the CDK4/6 inhibitor will enhance durability of response to the standard-of-care agent. Typically, these clinical trials are supported by published preclinical data describing the mechanisms of cooperation (Table 3),
CDK4/6 selective combinations: In parallel with the approaches that are dependent on an existing standard of care strategy, multiple preclinical studies involving targeted or unbiased drug-screening approaches have defined combination therapies that are not commonly employed for a given indication. These studies have revealed profound cooperation of CDK4/6 inhibitors with PI3K inhibition, MTOR inhibition, and MEK inhibition[72, 73, 80, 97]. In many of these cases the cooperation leads to an enhanced cell cycle arrest phenotype. For example, combinations of CDK4/6 inhibitors with PI3K inhibitors in breast cancer models, or MEK inhibitors in colon cancer models yield potent cytostasis [73, 97, 111]. However, there are instances where it appears that the combinations result in a synthetic cytotoxic response. For example, the combination of MEK and CDK4/6 inhibitors in non-small cell lung cancer induces cell death in concert with cell cycle inhibition [112]. The exact mechanisms underlying each combinatorial sensitivity is likely conditioned by the underlying genetic features of the tumor, as the same drug combination can have differing effects based on the tumor model studied. Based on these preclinical studies, there are now several combination studies that are interrogating the activity of MEK and CDK4/6 inhibitors broadly. It is likely that more rationally developed combination trials with CDK4/6 inhibitors will emerge based on recent preclinical studies.
CONCLUDING REMARKS
CDK4/6 inhibition represents a fundamental approach to combat the deregulated proliferation that drives cancer phenotypes. The blockade of cell cycle with CDK4/6 inhibitors, while mitogenic signals remain engaged, represents a strange phenotypic state that conditions features of response versus acquired resistance. Rationally targeting the unique features of the response to CDK4/6 will provide important insights into combination treatments. Thus, there is significant promise that through a detailed understanding of the biology of CDK4/6 inhibitors more clinical successes will emerge (outstanding questions box).
To date, the only accepted marker for CDK4/6 inhibitors is loss of the RB tumor suppressor function, which is employed in clinical decision-making. This is a relatively rare event in many cancers, while genetic alterations of cyclin D1, CDK4, or CDKN2A that could be associated with sensitivity are more common. However, such events occur against a complex tumor genetic landscape that likely impacts on response to CDK4/6 inhibition. Currently, an integrated assessment of biomarkers of clinical sensitivity has not emerged and significant clinical research will be required to direct the use of CDK4/6 inhibitors based on genetic events in a given tumor. Holistic biomarker analysis that interrogates multiple features of cell cycle control in parallel with the genetics of the tumor will be required for accurate suites of predictive biomarkers to emerge.
At present, the basis for acquired resistance to CDK4/6 inhibitors in the clinic remains largely unknown. In preclinical models multiple alterations have been observed including loss of CDK2-inhibitors, cyclin E amplification, and RB loss. Interestingly, the extent to which CDK4/6 inhibition selects for pre-existing subclonal genetic alterations is not known. Additionally, there is progressive evidence that tumor cell cycles are not necessarily locked, and such plasticity allows for escape from CDK4/6 inhibitors. In this context, acquired resistance appears to be driven by AKT or MTOR signaling pathways that remain active in the presence of CDK4/6 inhibitors. However, there are distinct contexts where these pathways are shut-off that contributes to durable response. Thus, an important area of research is to identify how these disparate adaptive responses to CDK4/6 inhibition are determined and if can they be selectively targeted therapeutically.
While the future of most targeted therapy will be in combination, defining a general prescription is very challenging. At present endocrine therapy is the only proven doublet for the use of CDK4/6 inhibitors in the ER+/HER2- tumors. However, this leaves many tumors facing a potential plethora of possible combinations with CDK4/6 inhibitors and the possibility of selective triplet combinations. In spite of many outstanding preclinical studies, well-designed clinical trials will ultimately be required to delineate the ideal use of CDK4/6 inhibition for a given tumor. Furthermore, as the clinical focus with CDK4/6 inhibitors has shifted to combination therapy, it is highly likely that a key issue moving forward will not be to elucidate determinants for CDK4/6 inhibition in isolation, but rather to select for biomarkers of response to combination therapy. Together, such study will yield a precision approach for the use of CDK4/6 inhibitors in the clinic to improve cancer outcomes.
TRENDS BOX.
CDK4/6 inhibitors have a unique mechanism of action by eliciting cell cycle inhibition downstream of a myriad of oncogenic and tumor suppressive pathways.
Specific biomarkers of intrinsic resistance to CDK4/6 inhibition based on knowledge of core cell cycle signaling have emerged. However, the identification of biomarkers and biological features of durable response to CDK4/6 inhibitors clinical remain an ongoing challenge.
Acquired resistance to CDK4/6 inhibition emerges due to selection for alterations in core cell cycle signaling pathways. Additionally, adaptive features of mitogenic signaling can yield a degree of cell cycle plasticity to ultimately bypass the requirement of CDK4/6 activity for cell cycle progression.
Combination therapies target adaptive processes to elicit improved responses in preclinical models that have translated to improved clinical outcomes.
Acknowledgments
The authors thank Professor Andrew Koff at Memorial Sloan Kettering, and members of their research team for critical feedback on the manuscript. Sejing Chung, Renee Ramos, Robert Emmons, and Amanda Ruiz contributed to detailed editing of the manuscript. The authors acknowledge the vast contributions of all their colleagues in the field and regret any omissions. This article was submitted with a thought towards all of patients with cancer who have succumbed to their disease, including J.M.K. who died of metastatic breast cancer. The authors are supported by grants from the NIH/NCI and are involved in sponsored research and/or clinical trials supported by Pfizer, Novartis and Eli Lilly in reference to the use of CDK4/6 inhibitors.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sedlacek H, et al. Flavopiridol (L86 8275; NSC 649890), a new kinase inhibitor for tumor therapy. Int J Oncol. 1996;9:1143–1168. doi: 10.3892/ijo.9.6.1143. [DOI] [PubMed] [Google Scholar]
- 2.Meijer L. Chemical inhibitors of cyclin-dependent kinases. Prog Cell Cycle Res. 1995;1:351–363. doi: 10.1007/978-1-4615-1809-9_29. [DOI] [PubMed] [Google Scholar]
- 3.Asghar U, et al. The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat Rev Drug Discov. 2015;14:130–146. doi: 10.1038/nrd4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sherr CJ, et al. Targeting CDK4 and CDK6: From Discovery to Therapy. Cancer Discov. 2016;6:353–367. doi: 10.1158/2159-8290.CD-15-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Leary B, et al. Treating cancer with selective CDK4/6 inhibitors. Nat Rev Clin Oncol. 2016;13:417–430. doi: 10.1038/nrclinonc.2016.26. [DOI] [PubMed] [Google Scholar]
- 6.Dickson MA. Molecular pathways: CDK4 inhibitors for cancer therapy. Clin Cancer Res. 2014;20:3379–3383. doi: 10.1158/1078-0432.CCR-13-1551. [DOI] [PubMed] [Google Scholar]
- 7.Sherr CJ. D-type cyclins. Trends Biochem Sci. 1995;20:187–190. doi: 10.1016/s0968-0004(00)89005-2. [DOI] [PubMed] [Google Scholar]
- 8.Knudsen KE, et al. Cyclin D1: polymorphism, aberrant splicing and cancer risk. Oncogene. 2006;25:1620–1628. doi: 10.1038/sj.onc.1209371. [DOI] [PubMed] [Google Scholar]
- 9.Diehl JA. Cycling to cancer with cyclin D1. Cancer Biol Ther. 2002;1:226–231. doi: 10.4161/cbt.72. [DOI] [PubMed] [Google Scholar]
- 10.Sherr CJ, Roberts JM. Living with or without cyclins and cyclin-dependent kinases. Genes Dev. 2004;18:2699–2711. doi: 10.1101/gad.1256504. [DOI] [PubMed] [Google Scholar]
- 11.Sicinski P, et al. Cyclin D1 provides a link between development and oncogenesis in the retina and breast. Cell. 1995;82:621–630. doi: 10.1016/0092-8674(95)90034-9. [DOI] [PubMed] [Google Scholar]
- 12.Sicinski P, et al. Cyclin D2 is an FSH-responsive gene involved in gonadal cell proliferation and oncogenesis. Nature. 1996;384:470–474. doi: 10.1038/384470a0. [DOI] [PubMed] [Google Scholar]
- 13.Rane SG, et al. Loss of Cdk4 expression causes insulin-deficient diabetes and Cdk4 activation results in beta-islet cell hyperplasia. Nat Genet. 1999;22:44–52. doi: 10.1038/8751. [DOI] [PubMed] [Google Scholar]
- 14.Sherr CJ, et al. Regulation of CYL/cyclin D genes by colony-stimulating factor 1. Ciba Found Symp. 1992;170:209–219. doi: 10.1002/9780470514320.ch13. discussion 219–226. [DOI] [PubMed] [Google Scholar]
- 15.Matsushime H, et al. Identification and properties of an atypical catalytic subunit (p34PSK-J3/cdk4) for mammalian D type G1 cyclins. Cell. 1992;71:323–334. doi: 10.1016/0092-8674(92)90360-o. [DOI] [PubMed] [Google Scholar]
- 16.Diehl JA, et al. Glycogen synthase kinase-3beta regulates cyclin D1 proteolysis and subcellular localization. Genes Dev. 1998;12:3499–3511. doi: 10.1101/gad.12.22.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diehl JA, et al. Inhibition of cyclin D1 phosphorylation on threonine-286 prevents its rapid degradation via the ubiquitin-proteasome pathway. Genes Dev. 1997;11:957–972. doi: 10.1101/gad.11.8.957. [DOI] [PubMed] [Google Scholar]
- 18.Serrano M, et al. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 19.Witkiewicz AK, et al. The meaning of p16(ink4a) expression in tumors: functional significance, clinical associations and future developments. Cell Cycle. 2011;10:2497–2503. doi: 10.4161/cc.10.15.16776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sherr CJ. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 21.Hinds PW, et al. Regulation of retinoblastoma protein functions by ectopic expression of human cyclins. Cell. 1992;70:993–1006. doi: 10.1016/0092-8674(92)90249-c. [DOI] [PubMed] [Google Scholar]
- 22.Knudsen ES, Wang JY. Differential regulation of retinoblastoma protein function by specific Cdk phosphorylation sites. J Biol Chem. 1996;271:8313–8320. doi: 10.1074/jbc.271.14.8313. [DOI] [PubMed] [Google Scholar]
- 23.Knudsen ES, Wang JY. Dual mechanisms for the inhibition of E2F binding to RB by cyclin-dependent kinase-mediated RB phosphorylation. Mol Cell Biol. 1997;17:5771–5783. doi: 10.1128/mcb.17.10.5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rubin SM. Deciphering the retinoblastoma protein phosphorylation code. Trends Biochem Sci. 2013;38:12–19. doi: 10.1016/j.tibs.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Narasimha AM, et al. Cyclin D activates the Rb tumor suppressor by mono-phosphorylation. Elife. 2014:3. doi: 10.7554/eLife.02872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lundberg AS, Weinberg RA. Functional inactivation of the retinoblastoma protein requires sequential modification by at least two distinct cyclin-cdk complexes. Mol Cell Biol. 1998;18:753–761. doi: 10.1128/mcb.18.2.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Markey MP, et al. Unbiased analysis of RB-mediated transcriptional repression identifies novel targets and distinctions from E2F action. Cancer Res. 2002;62:6587–6597. [PubMed] [Google Scholar]
- 28.Knudsen ES, Witkiewicz AK. Defining the transcriptional and biological response to CDK4/6 inhibition in relation to ER+/HER2- breast cancer. Oncotarget. 2016 doi: 10.18632/oncotarget.11588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishida S, et al. Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol Cell Biol. 2001;21:4684–4699. doi: 10.1128/MCB.21.14.4684-4699.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cam H, Dynlacht BD. Emerging roles for E2F: Beyond the G1/S transition and DNA replication. Cancer Cell. 2003;3:311–316. doi: 10.1016/s1535-6108(03)00080-1. [DOI] [PubMed] [Google Scholar]
- 31.Musgrove EA, et al. Cyclins and breast cancer. Journal of mammary gland biology and neoplasia. 1996;1:153–162. doi: 10.1007/BF02013639. [DOI] [PubMed] [Google Scholar]
- 32.Musgrove EA, et al. Growth factor, steroid, and steroid antagonist regulation of cyclin gene expression associated with changes in T-47D human breast cancer cell cycle progression. Mol Cell Biol. 1993;13:3577–3587. doi: 10.1128/mcb.13.6.3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Filmus J, et al. Induction of cyclin D1 overexpression by activated ras. Oncogene. 1994;9:3627–3633. [PubMed] [Google Scholar]
- 34.Bhatt KV, et al. Mutant B-RAF signaling and cyclin D1 regulate Cks1/S-phase kinase-associated protein 2-mediated degradation of p27Kip1 in human melanoma cells. Oncogene. 2007;26:1056–1066. doi: 10.1038/sj.onc.1209861. [DOI] [PubMed] [Google Scholar]
- 35.Thangavel C, et al. Therapeutically activating RB: reestablishing cell cycle control in endocrine therapy-resistant breast cancer. Endocr Relat Cancer. 2011;18:333–345. doi: 10.1530/ERC-10-0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kalish LH, et al. Deregulated cyclin D1 expression is associated with decreased efficacy of the selective epidermal growth factor receptor tyrosine kinase inhibitor gefitinib in head and neck squamous cell carcinoma cell lines. Clin Cancer Res. 2004;10:7764–7774. doi: 10.1158/1078-0432.CCR-04-0012. [DOI] [PubMed] [Google Scholar]
- 37.Schiff R, et al. Breast cancer endocrine resistance: how growth factor signaling and estrogen receptor coregulators modulate response. Clin Cancer Res. 2003;9:447S–454S. [PubMed] [Google Scholar]
- 38.Dowsett M, et al. Growth factor signalling and response to endocrine therapy: the Royal Marsden Experience. Endocr Relat Cancer. 2005;12(Suppl 1):S113–117. doi: 10.1677/erc.1.01044. [DOI] [PubMed] [Google Scholar]
- 39.Osborne CK, Schiff R. Mechanisms of endocrine resistance in breast cancer. Annual review of medicine. 2011;62:233–247. doi: 10.1146/annurev-med-070909-182917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perna D, et al. BRAF inhibitor resistance mediated by the AKT pathway in an oncogenic BRAF mouse melanoma model. Proc Natl Acad Sci U S A. 2015;112:E536–545. doi: 10.1073/pnas.1418163112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Villanueva J, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18:683–695. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Serrano M, et al. Inhibition of ras-induced proliferation and cellular transformation by p16INK4. Science. 1995;267:249–252. doi: 10.1126/science.7809631. [DOI] [PubMed] [Google Scholar]
- 43.Collado M, et al. Tumour biology: senescence in premalignant tumours. Nature. 2005;436:642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- 44.Serrano M, et al. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 45.Sharpless NE, Sherr CJ. Forging a signature of in vivo senescence. Nat Rev Cancer. 2015;15:397–408. doi: 10.1038/nrc3960. [DOI] [PubMed] [Google Scholar]
- 46.Bartkova J, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 47.Bartkova J, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- 48.Serrano M, Blasco MA. Putting the stress on senescence. Current opinion in cell biology. 2001;13:748–753. doi: 10.1016/s0955-0674(00)00278-7. [DOI] [PubMed] [Google Scholar]
- 49.McNeal AS, et al. CDKN2B Loss Promotes Progression from Benign Melanocytic Nevus to Melanoma. Cancer Discov. 2015;5:1072–1085. doi: 10.1158/2159-8290.CD-15-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sviderskaya EV, et al. p16(Ink4a) in melanocyte senescence and differentiation. J Natl Cancer Inst. 2002;94:446–454. doi: 10.1093/jnci/94.6.446. [DOI] [PubMed] [Google Scholar]
- 51.Haferkamp S, et al. p16INK4a-induced senescence is disabled by melanoma-associated mutations. Aging Cell. 2008;7:733–745. doi: 10.1111/j.1474-9726.2008.00422.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Begg CB, et al. Lifetime risk of melanoma in CDKN2A mutation carriers in a population-based sample. J Natl Cancer Inst. 2005;97:1507–1515. doi: 10.1093/jnci/dji312. [DOI] [PubMed] [Google Scholar]
- 53.Sawai CM, et al. Therapeutic targeting of the cyclin D3:CDK4/6 complex in T cell leukemia. Cancer Cell. 2012;22:452–465. doi: 10.1016/j.ccr.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lukas J, et al. Retinoblastoma-protein-dependent cell-cycle inhibition by the tumour suppressor p16. Nature. 1995;375:503–506. doi: 10.1038/375503a0. [DOI] [PubMed] [Google Scholar]
- 55.Dean JL, et al. Therapeutic CDK4/6 inhibition in breast cancer: key mechanisms of response and failure. Oncogene. 2010;29:4018–4032. doi: 10.1038/onc.2010.154. [DOI] [PubMed] [Google Scholar]
- 56.Rivadeneira DB, et al. Proliferative suppression by CDK4/6 inhibition: complex function of the retinoblastoma pathway in liver tissue and hepatoma cells. Gastroenterology. 2010;138:1920–1930. doi: 10.1053/j.gastro.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Konecny GE, et al. Expression of p16 and retinoblastoma determines response to CDK4/6 inhibition in ovarian cancer. Clin Cancer Res. 2011;17:1591–1602. doi: 10.1158/1078-0432.CCR-10-2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Finn RS, et al. PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res. 2009;11:R77. doi: 10.1186/bcr2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lukas J, et al. Cyclin D1 is dispensable for G1 control in retinoblastoma gene-deficient cells independently of cdk4 activity. Mol Cell Biol. 1995;15:2600–2611. doi: 10.1128/mcb.15.5.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Knudsen KE, et al. The retinoblastoma tumor suppressor inhibits cellular proliferation through two distinct mechanisms: inhibition of cell cycle progression and induction of cell death. Oncogene. 1999;18:5239–5245. doi: 10.1038/sj.onc.1202910. [DOI] [PubMed] [Google Scholar]
- 61.Aagaard L, et al. Aberrations of p16Ink4 and retinoblastoma tumour-suppressor genes occur in distinct sub-sets of human cancer cell lines. Int J Cancer. 1995;61:115–120. doi: 10.1002/ijc.2910610120. [DOI] [PubMed] [Google Scholar]
- 62.Xiong Y, et al. Subunit rearrangement of the cyclin-dependent kinases is associated with cellular transformation. Genes Dev. 1993;7:1572–1583. doi: 10.1101/gad.7.8.1572. [DOI] [PubMed] [Google Scholar]
- 63.Tort F, et al. Retinoblastoma pathway defects show differential ability to activate the constitutive DNA damage response in human tumorigenesis. Cancer Res. 2006;66:10258–10263. doi: 10.1158/0008-5472.CAN-06-2178. [DOI] [PubMed] [Google Scholar]
- 64.Robinson D, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kumar A, et al. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nature medicine. 2016;22:369–378. doi: 10.1038/nm.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Knudsen ES, et al. Inhibition of DNA synthesis by RB: effects on G1/S transition and S-phase progression. Genes Dev. 1998;12:2278–2292. doi: 10.1101/gad.12.15.2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang HS, et al. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell. 2000;101:79–89. doi: 10.1016/S0092-8674(00)80625-X. [DOI] [PubMed] [Google Scholar]
- 68.Strobeck MW, et al. Restoration of retinoblastoma mediated signaling to Cdk2 results in cell cycle arrest. Oncogene. 2000;19:1857–1867. doi: 10.1038/sj.onc.1203510. [DOI] [PubMed] [Google Scholar]
- 69.Butt AJ, et al. Cell cycle machinery: links with genesis and treatment of breast cancer. Adv Exp Med Biol. 2008;630:189–205. doi: 10.1007/978-0-387-78818-0_12. [DOI] [PubMed] [Google Scholar]
- 70.Lukas J, et al. Cyclin E-induced S phase without activation of the pRb/E2F pathway. Genes Dev. 1997;11:1479–1492. doi: 10.1101/gad.11.11.1479. [DOI] [PubMed] [Google Scholar]
- 71.Caldon CE, et al. Cyclin E2 overexpression is associated with endocrine resistance but not insensitivity to CDK2 inhibition in human breast cancer cells. Mol Cancer Ther. 2012;11:1488–1499. doi: 10.1158/1535-7163.MCT-11-0963. [DOI] [PubMed] [Google Scholar]
- 72.Franco J, et al. CDK4/6 inhibitors have potent activity in combination with pathway selective therapeutic agents in models of pancreatic cancer. Oncotarget. 2014;5:6512–6525. doi: 10.18632/oncotarget.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Herrera-Abreu MT, et al. Early Adaptation and Acquired Resistance to CDK4/6 Inhibition in Estrogen Receptor-Positive Breast Cancer. Cancer Res. 2016 doi: 10.1158/0008-5472.CAN-15-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Patnaik A, et al. Efficacy and Safety of Abemaciclib, an Inhibitor of CDK4 and CDK6, for Patients with Breast Cancer, Non-Small Cell Lung Cancer, and Other Solid Tumors. Cancer Discov. 2016;6:740–753. doi: 10.1158/2159-8290.CD-16-0095. [DOI] [PubMed] [Google Scholar]
- 75.Miller JP, et al. p27kip1 protein levels reflect a nexus of oncogenic signaling during cell transformation. J Biol Chem. 2012;287:19775–19785. doi: 10.1074/jbc.M112.361972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Siu KT, et al. An integrated view of cyclin E function and regulation. Cell Cycle. 2012;11:57–64. doi: 10.4161/cc.11.1.18775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Leonard JP, et al. Selective CDK4/6 inhibition with tumor responses by PD0332991 in patients with mantle cell lymphoma. Blood. 2012;119:4597–4607. doi: 10.1182/blood-2011-10-388298. [DOI] [PubMed] [Google Scholar]
- 78.Bigoni R, et al. Characterization of t(11;14) translocation in mantle cell lymphoma by fluorescent in situ hybridization. Oncogene. 1996;13:797–802. [PubMed] [Google Scholar]
- 79.Finn Richard S, JPC, Lang Istvan, Boer Katalin, Bondarenko Igor M, Kulyk Sergey O, Ettl Johannes, Patel Ravindranath, Pinter Tamas, Schmidt Marcus, Shparyk Yaroslav V, Thummala Anu R, Voytko Nataliya L, Huang Xin, Kim Sindy T, Randolph Sophia S, Slamon Dennis J. Final results of a randomized Phase II study of PD 0332991, a cyclin-dependent kinase (CDK)-4/6 inhibitor, in combination with letrozole vs letrozole alone for first-line treatment of ER+/HER2- advanced breast cancer (PALOMA-1; TRIO-18) Cancer Res, AACR Supplement 2014 [Google Scholar]
- 80.Heilmann AM, et al. CDK4/6 and IGF1 Receptor Inhibitors Synergize to Suppress the Growth of p16INK4A-Deficient Pancreatic Cancers. Cancer Res. 2014;74:3947–3958. doi: 10.1158/0008-5472.CAN-13-2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang YX, et al. Antiproliferative Effects of CDK4/6 Inhibition in CDK4-amplified Human Liposarcoma in vitro and in vivo. Mol Cancer Ther. 2014 doi: 10.1158/1535-7163.MCT-14-0387. [DOI] [PubMed] [Google Scholar]
- 82.Dickson MA, et al. Phase II trial of the CDK4 inhibitor PD0332991 in patients with advanced CDK4-amplified well-differentiated or dedifferentiated liposarcoma. J Clin Oncol. 2013;31:2024–2028. doi: 10.1200/JCO.2012.46.5476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang C, et al. Acquired CDK6 amplification promotes breast cancer resistance to CDK4/6 inhibitors and loss of ER signaling and dependence. Oncogene. 2016 doi: 10.1038/onc.2016.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dean JL, et al. Therapeutic response to CDK4/6 inhibition in breast cancer defined by ex vivo analyses of human tumors. Cell Cycle. 2012;11:2756–2761. doi: 10.4161/cc.21195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Witkiewicz AK, et al. CDK4/6 inhibition provides a potent adjunct to Her2-targeted therapies in preclinical breast cancer models. Genes & cancer. 2014;5:261–272. doi: 10.18632/genesandcancer.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rader J, et al. Dual CDK4/CDK6 inhibition induces cell-cycle arrest and senescence in neuroblastoma. Clin Cancer Res. 2013;19:6173–6182. doi: 10.1158/1078-0432.CCR-13-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Goel S, et al. Overcoming Therapeutic Resistance in HER2-Positive Breast Cancers with CDK4/6 Inhibitors. Cancer Cell. 2016;29:255–269. doi: 10.1016/j.ccell.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gelbert LM, et al. Preclinical characterization of the CDK4/6 inhibitor LY2835219: in-vivo cell cycle-dependent/independent anti-tumor activities alone/in combination with gemcitabine. Investigational new drugs. 2014 doi: 10.1007/s10637-014-0120-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kovatcheva M, et al. MDM2 turnover and expression of ATRX determine the choice between quiescence and senescence in response to CDK4 inhibition. Oncotarget. 2015;6:8226–8243. doi: 10.18632/oncotarget.3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yoshida A, et al. Induction of Therapeutic Senescence in Vemurafenib-Resistant Melanoma by Extended Inhibition of CDK4/6. Cancer Res. 2016;76:2990–3002. doi: 10.1158/0008-5472.CAN-15-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Coppe JP, et al. Tumor suppressor and aging biomarker p16(INK4a) induces cellular senescence without the associated inflammatory secretory phenotype. J Biol Chem. 2011;286:36396–36403. doi: 10.1074/jbc.M111.257071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Michaud K, et al. Pharmacologic inhibition of cyclin-dependent kinases 4 and 6 arrests the growth of glioblastoma multiforme intracranial xenografts. Cancer Res. 2010;70:3228–3238. doi: 10.1158/0008-5472.CAN-09-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang L, et al. Pharmacologic inhibition of CDK4/6: mechanistic evidence for selective activity or acquired resistance in acute myeloid leukemia. Blood. 2007;110:2075–2083. doi: 10.1182/blood-2007-02-071266. [DOI] [PubMed] [Google Scholar]
- 94.Zhang J, et al. Inhibition of Rb Phosphorylation Leads to mTORC2-Mediated Activation of Akt. Mol Cell. 2016;62:929–942. doi: 10.1016/j.molcel.2016.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Franco J, et al. Metabolic Reprogramming of Pancreatic Cancer Mediated by CDK4/6 Inhibition Elicits Unique Vulnerabilities. Cell reports. 2016 doi: 10.1016/j.celrep.2015.12.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Paternot S, et al. The CDK4/CDK6 inhibitor PD0332991 paradoxically stabilizes activated cyclin D3-CDK4/6 complexes. Cell Cycle. 2014;13:2879–2888. doi: 10.4161/15384101.2014.946841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Vora SR, et al. CDK 4/6 Inhibitors Sensitize PIK3CA Mutant Breast Cancer to PI3K Inhibitors. Cancer Cell. 2014;26:136–149. doi: 10.1016/j.ccr.2014.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Turner NC, et al. Palbociclib in Hormone-Receptor-Positive Advanced Breast Cancer. The New England journal of medicine. 2015;373:209–219. doi: 10.1056/NEJMoa1505270. [DOI] [PubMed] [Google Scholar]
- 99.Hortobagyi GN, et al. Ribociclib as First-Line Therapy for HR-Positive, Advanced Breast Cancer. The New England journal of medicine. 2016;375:1738–1748. doi: 10.1056/NEJMoa1609709. [DOI] [PubMed] [Google Scholar]
- 100.Turner NC, et al. Palbociclib in Hormone-Receptor-Positive Advanced Breast Cancer. N Engl J Med. 2015 doi: 10.1056/NEJMc1510345. [DOI] [PubMed] [Google Scholar]
- 101.Finn RS, et al. The cyclin-dependent kinase 4/6 inhibitor palbociclib in combination with letrozole versus letrozole alone as first-line treatment of oestrogen receptor-positive, HER2-negative, advanced breast cancer (PALOMA-1/TRIO-18): a randomised phase 2 study. Lancet Oncol. 2015;16:25–35. doi: 10.1016/S1470-2045(14)71159-3. [DOI] [PubMed] [Google Scholar]
- 102.Prall OW, et al. c-Myc or cyclin D1 mimics estrogen effects on cyclin E-Cdk2 activation and cell cycle reentry. Mol Cell Biol. 1998;18:4499–4508. doi: 10.1128/mcb.18.8.4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Prall OW, et al. Estrogen-induced activation of Cdk4 and Cdk2 during G1-S phase progression is accompanied by increased cyclin D1 expression and decreased cyclin-dependent kinase inhibitor association with cyclin E-Cdk2. J Biol Chem. 1997;272:10882–10894. doi: 10.1074/jbc.272.16.10882. [DOI] [PubMed] [Google Scholar]
- 104.Desmedt C, Sotiriou C. Proliferation: the most prominent predictor of clinical outcome in breast cancer. Cell Cycle. 2006;5:2198–2202. doi: 10.4161/cc.5.19.3254. [DOI] [PubMed] [Google Scholar]
- 105.Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 106.Wardell SE, et al. Efficacy of SERD/SERM Hybrid-CDK4/6 Inhibitor Combinations in Models of Endocrine Therapy-Resistant Breast Cancer. Clin Cancer Res. 2015;21:5121–5130. doi: 10.1158/1078-0432.CCR-15-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Baselga J, et al. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. N Engl J Med. 2012;366:520–529. doi: 10.1056/NEJMoa1109653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Roberts PJ, et al. Multiple roles of cyclin-dependent kinase 4/6 inhibitors in cancer therapy. J Natl Cancer Inst. 2012;104:476–487. doi: 10.1093/jnci/djs002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Witkiewicz AK, et al. The retinoblastoma tumor suppressor pathway modulates the invasiveness of ErbB2-positive breast cancer. Oncogene. 2013 doi: 10.1038/onc.2013.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hashizume R, et al. Inhibition of DNA damage repair by the CDK4/6 inhibitor palbociclib delays irradiated intracranial atypical teratoid rhabdoid tumor and glioblastoma xenograft regrowth. Neuro Oncol. 2016;18:1519–1528. doi: 10.1093/neuonc/now106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ziemke EK, et al. Sensitivity of KRAS-Mutant Colorectal Cancers to Combination Therapy That Cotargets MEK and CDK4/6. Clin Cancer Res. 2016;22:405–414. doi: 10.1158/1078-0432.CCR-15-0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tao Z, et al. Coadministration of Trametinib and Palbociclib Radiosensitizes KRAS-Mutant Non-Small Cell Lung Cancers In Vitro and In Vivo. Clin Cancer Res. 2016;22:122–133. doi: 10.1158/1078-0432.CCR-15-0589. [DOI] [PubMed] [Google Scholar]
- 113.Marzec M, et al. Mantle cell lymphoma cells express predominantly cyclin D1a isoform and are highly sensitive to selective inhibition of CDK4 kinase activity. Blood. 2006;108:1744–1750. doi: 10.1182/blood-2006-04-016634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Choi YJ, et al. The requirement for cyclin D function in tumor maintenance. Cancer Cell. 2012;22:438–451. doi: 10.1016/j.ccr.2012.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Menu E, et al. A novel therapeutic combination using PD 0332991 and bortezomib: study in the 5T33MM myeloma model. Cancer Res. 2008;68:5519–5523. doi: 10.1158/0008-5472.CAN-07-6404. [DOI] [PubMed] [Google Scholar]
- 116.Nemoto A, et al. Specific Antileukemic Activity of PD0332991, a CDK4/6 Inhibitor, against Philadelphia Chromosome-Positive Lymphoid Leukemia. Mol Cancer Ther. 2016;15:94–105. doi: 10.1158/1535-7163.MCT-14-1065. [DOI] [PubMed] [Google Scholar]
- 117.Whiteway SL, et al. Inhibition of cyclin-dependent kinase 6 suppresses cell proliferation and enhances radiation sensitivity in medulloblastoma cells. J Neurooncol. 2013;111:113–121. doi: 10.1007/s11060-012-1000-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Saab R, et al. Pharmacologic inhibition of cyclin-dependent kinase 4/6 activity arrests proliferation in myoblasts and rhabdomyosarcoma-derived cells. Mol Cancer Ther. 2006;5:1299–1308. doi: 10.1158/1535-7163.MCT-05-0383. [DOI] [PubMed] [Google Scholar]
- 119.Kennedy AL, et al. Functional, chemical genomic, and super-enhancer screening identify sensitivity to cyclin D1/CDK4 pathway inhibition in Ewing sarcoma. Oncotarget. 2015;6:30178–30193. doi: 10.18632/oncotarget.4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Vlenterie M, et al. Targeting Cyclin-Dependent Kinases in Synovial Sarcoma: Palbociclib as a Potential Treatment for Synovial Sarcoma Patients. Ann Surg Oncol. 2016;23:2745–2752. doi: 10.1245/s10434-016-5341-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Katsumi Y, et al. Sensitivity of malignant rhabdoid tumor cell lines to PD 0332991 is inversely correlated with p16 expression. Biochem Biophys Res Commun. 2011;413:62–68. doi: 10.1016/j.bbrc.2011.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Perez M, et al. Efficacy of CDK4 inhibition against sarcomas depends on their levels of CDK4 and p16ink4 mRNA. Oncotarget. 2015;6:40557–40574. doi: 10.18632/oncotarget.5829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Huang S, et al. CDK4/6 inhibitor suppresses gastric cancer with CDKN2A mutation. Int J Clin Exp Med. 2015;8:11692–11700. [PMC free article] [PubMed] [Google Scholar]
- 124.Tang LH, et al. Attenuation of the retinoblastoma pathway in pancreatic neuroendocrine tumors due to increased cdk4/cdk6. Clin Cancer Res. 2012;18:4612–4620. doi: 10.1158/1078-0432.CCR-11-3264. [DOI] [PubMed] [Google Scholar]
- 125.Fry DW, et al. Specific inhibition of cyclin-dependent kinase 4/6 by PD 0332991 and associated antitumor activity in human tumor xenografts. Mol Cancer Ther. 2004;3:1427–1438. [PubMed] [Google Scholar]
- 126.Ismail A, et al. Early G(1) cyclin-dependent kinases as prognostic markers and potential therapeutic targets in esophageal adenocarcinoma. Clin Cancer Res. 2011;17:4513–4522. doi: 10.1158/1078-0432.CCR-11-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Liu F, Korc M. Cdk4/6 inhibition induces epithelial-mesenchymal transition and enhances invasiveness in pancreatic cancer cells. Mol Cancer Ther. 2012;11:2138–2148. doi: 10.1158/1535-7163.MCT-12-0562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Puyol M, et al. A synthetic lethal interaction between K-Ras oncogenes and Cdk4 unveils a therapeutic strategy for non-small cell lung carcinoma. Cancer Cell. 2010;18:63–73. doi: 10.1016/j.ccr.2010.05.025. [DOI] [PubMed] [Google Scholar]
- 129.Comstock CE, et al. Targeting cell cycle and hormone receptor pathways in cancer. Oncogene. 2013;32:5481–5491. doi: 10.1038/onc.2013.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Logan JE, et al. PD-0332991, a potent and selective inhibitor of cyclin-dependent kinase 4/6, demonstrates inhibition of proliferation in renal cell carcinoma at nanomolar concentrations and molecular markers predict for sensitivity. Anticancer Res. 2013;33:2997–3004. [PubMed] [Google Scholar]
- 131.Sathe A, et al. CDK4/6 Inhibition Controls Proliferation of Bladder Cancer and Transcription of RB1. J Urol. 2016;195:771–779. doi: 10.1016/j.juro.2015.08.082. [DOI] [PubMed] [Google Scholar]
- 132.Kwong LN, et al. Oncogenic NRAS signaling differentially regulates survival and proliferation in melanoma. Nature medicine. 2012;18:1503–1510. doi: 10.1038/nm.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Young RJ, et al. Loss of CDKN2A expression is a frequent event in primary invasive melanoma and correlates with sensitivity to the CDK4/6 inhibitor PD0332991 in melanoma cell lines. Pigment Cell Melanoma Res. 2014;27:590–600. doi: 10.1111/pcmr.12228. [DOI] [PubMed] [Google Scholar]
- 134.Barton KL, et al. PD-0332991, a CDK4/6 inhibitor, significantly prolongs survival in a genetically engineered mouse model of brainstem glioma. PLoS One. 2013;8:e77639. doi: 10.1371/journal.pone.0077639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wiedemeyer WR, et al. Pattern of retinoblastoma pathway inactivation dictates response to CDK4/6 inhibition in GBM. Proc Natl Acad Sci U S A. 2010;107:11501–11506. doi: 10.1073/pnas.1001613107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Beck TN, et al. EGFR and RB1 as Dual Biomarkers in HPV-Negative Head and Neck Cancer. Mol Cancer Ther. 2016;15:2486–2497. doi: 10.1158/1535-7163.MCT-16-0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Miller TW, et al. ERalpha-dependent E2F transcription can mediate resistance to estrogen deprivation in human breast cancer. Cancer Discov. 1:338–351. doi: 10.1158/2159-8290.CD-11-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Dean JL, et al. Therapeutic CDK4/6 inhibition in breast cancer: key mechanisms of response and failure. Oncogene. 29:4018–4032. doi: 10.1038/onc.2010.154. [DOI] [PubMed] [Google Scholar]
- 139.Thangavel C, et al. Therapeutically activating RB: reestablishing cell cycle control in endocrine therapy-resistant breast cancer. Endocr Relat Cancer. 18:333–345. doi: 10.1530/ERC-10-0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chiron D, et al. Cell-cycle reprogramming for PI3K inhibition overrides a relapse-specific C481S BTK mutation revealed by longitudinal functional genomics in mantle cell lymphoma. Cancer Discov. 2014;4:1022–1035. doi: 10.1158/2159-8290.CD-14-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lee MS, et al. Efficacy of the combination of MEK and CDK4/6 inhibitors in vitro and in vivo in KRAS mutant colorectal cancer models. Oncotarget. 2016;7:39595–39608. doi: 10.18632/oncotarget.9153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Flaherty KT, et al. Phase I, dose-escalation trial of the oral cyclin-dependent kinase 4/6 inhibitor PD 0332991, administered using a 21-day schedule in patients with advanced cancer. Clin Cancer Res. 2012;18:568–576. doi: 10.1158/1078-0432.CCR-11-0509. [DOI] [PubMed] [Google Scholar]
- 143.Schwartz GK, et al. Phase I study of PD 0332991, a cyclin-dependent kinase inhibitor, administered in 3-week cycles (Schedule 2/1) Br J Cancer. 2011;104:1862–1868. doi: 10.1038/bjc.2011.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Infante JR, et al. A Phase I Study of the Cyclin-Dependent Kinase 4/6 Inhibitor Ribociclib (LEE011) in Patients with Advanced Solid Tumors and Lymphomas. Clin Cancer Res. 2016 doi: 10.1158/1078-0432.CCR-16-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Chen P, et al. Spectrum and Degree of CDK Drug Interactions Predicts Clinical Performance. Mol Cancer Ther. 2016;15:2273–2281. doi: 10.1158/1535-7163.MCT-16-0300. [DOI] [PubMed] [Google Scholar]
- 146.Sumi NJ, et al. Chemoproteomics Reveals Novel Protein and Lipid Kinase Targets of Clinical CDK4/6 Inhibitors in Lung Cancer. ACS Chem Biol. 2015;10:2680–2686. doi: 10.1021/acschembio.5b00368. [DOI] [PMC free article] [PubMed] [Google Scholar]