Abstract
Vascular endothelial growth factor (VEGF) and its receptors have been implicated as key factors in tumor angiogenesis that are up-regulated by hypoxia. We evaluated the effects of DNA-binding small molecules on hypoxia-inducible transcription of VEGF. A synthetic pyrrole-imidazole polyamide designed to bind the hypoxia response element (HRE) was found to disrupt hypoxia-inducible factor (HIF) binding to HRE. In cultured HeLa cells, this resulted in a reduction of VEGF mRNA and secreted protein levels. The observed effects were polyamide-specific and dose-dependent. Analysis of genome-wide effects of the HRE-specific polyamide revealed that a number of hypoxia-inducible genes were down-regulated. Pathway-based regulation of hypoxia-inducible gene expression with DNA-binding small molecules may represent a new approach for targeting angiogenesis.
Keywords: gene regulation, hypoxia-inducible factor, polyamide
Angiogenesis, the induction of new blood vessels, is critical for growth and metastatic spread of solid tumors. It is tightly controlled by a number of specific mitogenic factors, among which vascular endothelial growth factor (VEGF) and its receptors play a central role. The levels of VEGF are up-regulated across a broad range of tumors and are involved in key aspects of cancer biology. A hallmark of many cancers, chronic hypoxia, in conjunction with activation of certain oncogenic signaling pathways, is responsible for the elevated levels of VEGF and is associated with invasion and altered energy metabolism (1).
In cells and tissues, hypoxia triggers a multifaceted adaptive response that is primarily driven by the heterodimeric hypoxia-inducible factor 1 (HIF-1) (2). Under normal dioxygen levels, the α-subunit of HIF-1 is successively hydroxylated at proline residue 564 (3), ubiquitinated, and then degraded by the ubiquitin–proteosome system. This process, mediated by the von Hippel–Lindau tumor suppressor protein (4), is responsible for controlling levels of HIF-1α and, as a result, the transcriptional response to hypoxia (5). Under hypoxic conditions, HIF-1α avoids hydroxylation and accumulates. Heterodimerization with its constitutively expressed binding partner, aryl hydrocarbon receptor nuclear translocator (ARNT) (6) and binding to a cognate hypoxia response element (HRE) (7) recruits the p300/CBP and SRC-1 family coactivators, which drive the expression of hypoxia-inducible genes. Among these are genes encoding angiogenic peptides such as VEGF and the platelet-derived growth factor B chain, as well as proteins involved in glucose metabolism, such as the glucose transporter GLUT1 (8, 9). Inhibition of VEGF, a downstream target of HIF, is sufficient to inhibit tumor growth in model systems (10).
We designed a sequence-specific DNA-binding molecule to inhibit binding of the HIF-1α/ARNT heterodimer to its cognate DNA sequence to down-regulate the expression of VEGF and other hypoxia-inducible genes. Because interaction of HIF with its cognate DNA sequence and subsequent transcriptional activation is a likely point of significant amplification of response, disruption of this interaction could represent a point of intervention in the hypoxia response pathway involving multiple genes.
To regulate the expression of endogenous genes, DNA-binding small molecules must permeate the cell, localize in the nucleus (11, 12), access chromatin (13–15), and bind DNA sequences with affinities and specificities sufficient to disrupt key regulatory proteins bound to genomic DNA (16–18). Synthetic oligomers containing N-methylpyrrole and N-methylimidazole amino acids conjugated to a fluorescein dye represent a modular molecular recognition toolkit with properties that satisfy these criteria. DNA sequence specificity is programmed by a simple code created by pairs of aromatic rings (19–22).
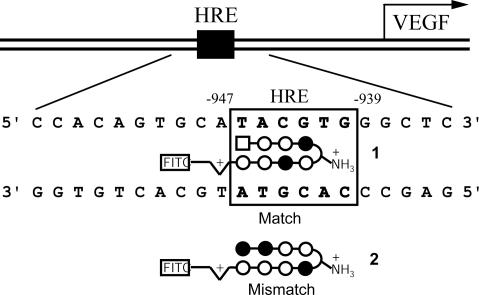
Although the VEGF gene encodes multiple splicing variants, analysis of its promoter revealed that a single HRE is located at nucleotide positions –947 to –939 (5′-TACGTG-3′) relative to the common transcription start site (Fig. 1) (23). We designed polyamide 1 to bind to the DNA sequence 5′-WTWCGW-3′ (where W = A or T) that encompasses the HRE site in the VEGF promoter according to the pairing rules (Figs. 1 and 2). A mismatch control polyamide 2, directed against an unrelated sequence 5′-WGGWCW-3′, was also synthesized.
Fig. 1.
Map of the VEGF promoter with the HRE site (Upper) and schematic representation of match polyamide 1 targeting the HRE and mismatch polyamide 2 designed for this study (Lower). Imidazole and pyrrole rings are represented as solid and open circles, respectively; 3-chlorothiophene is depicted as a square, and aliphatic linkers as curved lines. Half-diamonds with plus signs represent 3,3′-diamino-N-methyldipropylamine. FITC represents conjugated FITC (isomer I).
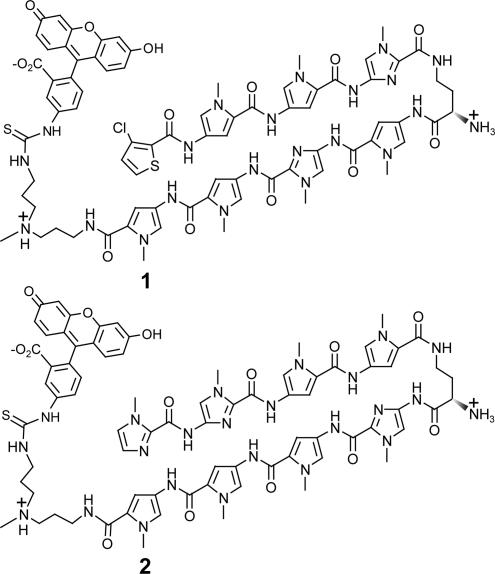
Fig. 2.
Structures of the polyamide–FITC conjugates 1 and 2.
Materials and Methods
Synthesis of Polyamides. Polyamides 1 and 2 were synthesized by solid-phase methods on Kaiser oxime resin (Nova Biochem) (24) and conjugated to FITC isomer I (11). The purity and identity of the polyamide-dye conjugates were verified by analytical HPLC, UV-visible spectroscopy, and MALDI-ToF MS.
Determination of DNA-Binding Affinities and Sequence Specificities. A 5′ 32P-labeled fragment was generated by PCR amplification of the site from the plasmid pGL2-VEGF-Luc by using primers 5′-CTC AGT TCC CTG GCA ACA TCT-3′ (VEGFP1) and 5′-TGG CAC CAA GTT TGT GGA GCT-3′ (VEGFP2) and isolated by nondenaturing gel electrophoresis (25). Quantitative DNase I footprint titration experiments were used to determine the binding affinities and specificities of polyamides 1 and 2 (25).
EMSA. The HIF1α/ARNT heterodimer was transcribed/translated in vitro by using Promega TNT kit according to the manufacturer's instructions. The double-strand oligonucleotide probe was prepared by annealing the two complementary strands 5′-GAC TCC ACA GTG CAT ACG TGG GCT CCA ACA GGT-3′ (HRE-EMSA1) and 5′-ACC TGT TGG AGC CCA CGT ATG CAC TGT GGA GTC-3′ (HRE-EMSA2). Before annealing, the HRE-EMSA1 oligonucleotide was 5′-end radiolabeled with γ-32-P-ATP (NEN) and T4 polynucleotide kinase, as described. The radiolabeled double-strand oligonucleotide probe was isolated by using a G25 Quickspin column (Boehringer Mannheim).
Polyamides were preincubated with the radiolabeled oligonucleotide in Z-buffer (100 mM KCl/25 mM Tris, pH 7.5/0.2 mM EDTA/20% glycerol/0.25 mg/ml BSA/0.05% Nonidet P-40/5 mM DTT/0.1 mg/ml PMSF/1.2 mM sodium vanadate) at 0°C for 30 min. Then the in vitro transcribed/translated protein mixture, diluted with the same buffer, was added, and the mixture was held on ice for an additional 30 min. Each time, the following controls were included: free oligonucleotide probe, probe with unprogrammed in vitro transcription/translation reaction mixture, and 100-fold excess of competing nonradiolabeled probe. The complexes were resolved on a 4% nondenaturing polyacrylamide gel and visualized with the Storm 820 Phosphorimager (Molecular Dynamics).
Cell Culture. The human cervical epithelial adenocarcinoma cell line HeLa (ATCC CCL-2) was maintained in DMEM as recommended by American Type Culture Collection. Cell growth and morphology were monitored by phase-contrast microscopy.
Confocal Microscopy. HeLa cells were trypsinized for 5–10 min at 37°C, centrifuged for 5 min at 2,000 rpm and 5°C in a Beckman Coulter Allegra 6R centrifuge, and resuspended in fresh medium to a concentration of 1.25 × 106 cells per milliliter. Incubations were performed by adding 150 μl of cells into culture dishes equipped with glass bottoms for direct imaging (MatTek, Ashland, MA). The cells were grown in the glass-bottom culture dishes for 24 h. The medium was then removed and replaced with 142.5 μl of fresh medium. Then 7.5 μl of the 100 μM polyamide solution was added, and the cells were incubated in a 5% CO2 atmosphere at 37°C for 10–14 h. Imaging was performed on a Zeiss LSM 5 Pascal inverted laser scanning microscope equipped with a ×40 oil-immersion objective lens. Analysis of images was performed as described (11).
Determination of the Relative mRNA and Protein Levels. RNA isolation. HeLa cells were plated in six-well dishes at a density of 6 × 105 in 1 ml of DMEM and allowed to attach for 16–20 h. Polyamides were added, and the cells were incubated for 48 h. The hypoxia conditions necessary for VEGF induction were created by incubation with 300 μM desferrioxamine mesylate (DFO) for 16–18 h (26, 27). Optionally, cells were tested for apoptosis by staining with annexin V. The medium was removed, and cells were washed with ice-cold PBS and immediately lysed with RLT buffer from the RNeasy kit (Qiagen, Chatsworth, CA) with 2-mercaptoethanol added. Further RNA isolation was carried out with the RNeasy kit as described in the manufacturer's manual. The isolated total RNA was quantified. The yields were 12–15 μg per well. Genomic DNA was digested by treatment with DNase I from a DNA Free kit (Ambion, Austin, TX), and DNase I was inactivated with bead-immobilized DNase I inactivation reagent (Ambion).
Reverse transcription. A 2.5-μg sample of total RNA was used to reverse-transcribe cDNA by using Powerscript II reverse transcriptase (BD Clontech) according to the manufacturer's protocol. Random hexamers and oligo-(dT)16 primers were used simultaneously in a 1:1 ratio. The total volume for each reverse transcription reaction was 20 μl.
Real-time quantitative RT-PCR analysis. Real-time quantitative RT-PCR analysis was performed by using the VEGF gene primers described below. The forward primer 5′-AGG CCA GCA CAT AGG AGA GA-3′ and reverse primer 5′-TTT CCC TTT CCT CGA ACT GA-3′ were used to amplify the 104-bp fragment from the 3′-translated region of VEGF. RNA was standardized by quantification of the β-glucuronidase gene as an endogenous control (28). The forward primer 5′-CTC ATT TGG AAT TTT GCC GAT T and reverse primer 5′-CCG AGT GAA GAT CCC CTT TTT A were used for this gene (29). Quantitative real-time RT-PCR was performed by using Applied Biosystems SYBR Green RT-PCR master mix according to the manufacturer's instructions. Temperature cycling and detection of the SYBR green emission were performed with an ABI 7300 real-time instrument by using Applied Biosystems Sequence Detection System, Version 1.2. Statistical analysis was performed on three independent experiments.
VEGF ELISA. Approximately 105 HeLa cells were split into 24-well plates. After 24 h, the cells were incubated with polyamides 1 or 2 (0.2 or 1 μM) for 32 h. Fresh polyamide was added followed by addition of DFO (300 μM) and further incubation for 16 h. The supernatant (100 μl) was used for the VEGF ELISA (R & D Systems), which was carried out according to the manufacturer's instructions.
Analysis of Gene Expression with Oligonucleotide Microarrays. Experiments were carried out at the Caltech Genome Expression Center. HeLa cells were split and plated in a manner similar to that in the RT-PCR experiments. Cultured cells were incubated for 48 h with 0.2 or 1 μM polyamide 1 or 2. Hypoxic conditions were induced by adding DFO to a final concentration of 300 μM. The cells were incubated with DFO for 12 h, and total RNA was collected as described for the RT-PCR experiments. After testing for quantity and quality, the total RNA was subjected to the Affymetrix protocols. Affymetrix Genechip Human Genome U133A microarrays were used in each experiment. The experiments were carried out in triplicate. Correlation between the replicates was >0.970. The data were analyzed with resolver, Ver. 3.0 (Rosetta Biosoftware, Seattle).
Results
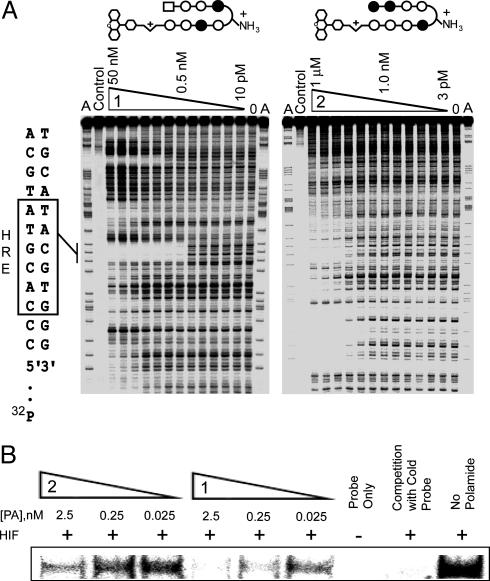
Binding Affinities and Specificities. Based on the pairing rules, match polyamide 1 targets sequences of the type 5′-WTWCGW -3′ (where W = A or T), whereas mismatch polyamide 2 targets sequences of the type 5′-WGGWCW-3′. The 3-chlorothiophene ring at the N terminus of polyamide 1 provides specificity for a T•A base pair (22). We mapped the detailed binding sites for both match and mismatch polyamides on the VEGF promoter fragment that encompasses the HRE. From DNase I footprint titrations, a Ka value of 6.3 × 109 M–1 was obtained for polyamide 1 at the HRE site (Fig. 3A). The mismatch polyamide 2 bound the HRE site with ≈100-fold lower affinity (Ka = 7.9 × 107 M–1). No match sites at 1.0 nM concentration could be found for polyamide 2 in the region of this DNA that can be resolved by gel electrophoresis.
Fig. 3.
Polyamide 1 binds HRE. (A) Storage phosphor autoradiograms from quantitative DNase I footprint titrations of polyamides 1 and 2. The boxed sequence (Left) represents the HRE site. For polyamide 1, lanes 1 and 16, A reaction: lane 2, intact DNA; lanes 3–14, DNase I digestion products in the presence of 50 nM, 20 nM, 10 nM, 5 nM, 2 nM, 1 nM, 500 pM, 200 pM, 100 pM, 50 pM, 20 pM, and 10 pM polyamide, respectively; lane 15, DNase I standard. For polyamide 2, lanes 1 and 16, A reaction: lane 2, intact DNA; lanes 3–14, DNase I digestion products in the presence of 1 μM, 300 nM, 100 nM, 30 nM, 10 nM, 3 nM, 1 nM, 300 pM, 100 pM, 30 pM, 10 pM, and 3 pM polyamide, respectively; lane 15, DNase I standard. (B) Storage phosphor autoradiogram from EMSA experiment with polyamides 1 and 2.
Disruption of the HIF–DNA Complex. We tested the ability of polyamides to inhibit the binding of HIF-1α/ARNT heterodimer to the HRE in an EMSA. The radiolabeled DNA fragment (24 bp) was first incubated with match or mismatch polyamide 1 or 2, respectively. After the subsequent addition of the in vitro translated HIF-1α/ARNT heterodimer, the resulting complexes were resolved on a nondenaturing polyacrylamide gel. Match polyamide 1 (0.25 nM) effectively inhibited binding of the heterodimer, whereas much less effect was observed for the mismatch polyamide 2 at concentrations as high as 2.5 μM (Fig. 3B). See also Fig. 7, which is published as supporting information on the PNAS web site.
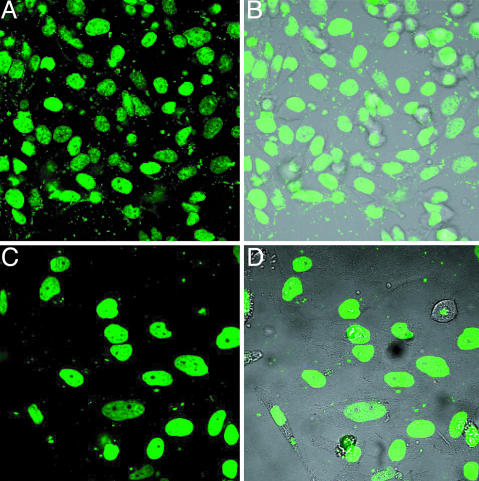
Uptake of Polyamides in Cultured HeLa Cells. The uptake of both polyamides by the HeLa cell line was examined by laser-scanning confocal microscopy. Previous studies indicated that the degree of cellular uptake and nuclear localization of polyamides containing an eight-ring sequence recognition core depends on the pyrrole/imidazole content of the core and varies for each cell line (11, 12). We find that both polyamides exhibit strong nuclear localization after incubation at 2 μM concentration for 12 h at 37°C in standard culture medium (Fig. 4).
Fig. 4.
Cellular localization of polyamides 1 (A and B) and 2 (C and D) in HeLa cells. (A and C) Fluorescence signals from polyamides. (B and D) Overlays of fluorescence signals with visible light images.
Effect of Polyamides on Cell Viability and Growth Rate. We examined whether prolonged incubation with polyamides affects cell viability. HeLa cells were incubated with polyamides at 1 μM concentration, trypsinized and counted at various time points (0–72 h) by using a hemocytometer. Measurements of cell growth rates indicate that polyamides at 1 μM in standard culture medium have no deleterious effects on cell growth and division (see Fig. 8, which is published as supporting information on the PNAS web site).
Analysis of Promoter Activity with Luciferase Assays. We used HeLa cells that had been stably transfected with a reporter plasmid VEGF-Luc containing the VEGF promoter upstream of luciferase CDNA. The experiments were carried out in a hypoxic chamber with 1% O2 to mimic closely the conditions of physiological hypoxia. Incubation with the match polyamide 1 resulted in a decrease of promoter activity in a dose-dependent manner, as indicated by decreased levels of luciferase activity. A negligibly small effect was observed for mismatch polyamide 2 (Fig. 9, which is published as supporting information on the PNAS web site).
As a specificity control, we constructed and used in parallel a nearly identical reporter VEGF-M1Luc where the HRE and surrounding sequences had been mutated to disfavor binding of HIF-1 (Fig. 10, which is published as supporting information on the PNAS web site). In these experiments and those that follow, the hypoxia mimetic compound DFO (26, 27) was used to stabilize HIF and activate HIF target genes. Cells were harvested after incubation with 300 μM DFO for 12–16 h.
Treatment of stably transfected HeLa cells with match polyamide 1 led to significant attenuation of hypoxia-inducible VEFG-Luc activity. By contrast, treatment with mismatch polyamide 2 resulted in only a modest decrease of VEGF-Luc activity. The VEGF-M1Luc promoter with a mutated HRE site showed no inducibility under hypoxic conditions. No effect of polyamides on the levels of luciferase activity in cells transfected with the mutant promoter was observed (Fig. 10). No obvious cytotoxicity was observed.
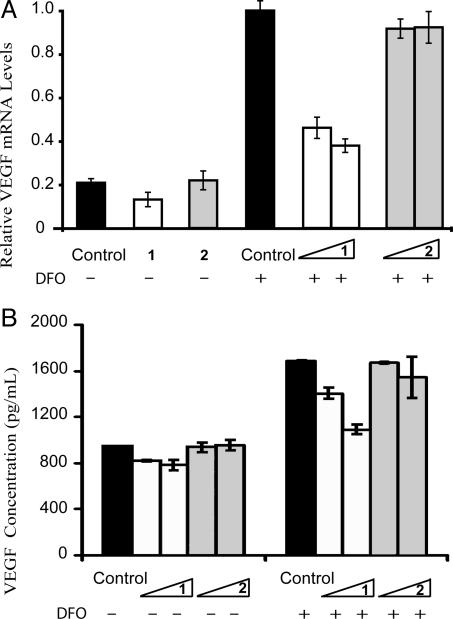
Suppression of Hypoxia-Inducible Transcription in Cultured Cells. We used real-time quantitative RT-PCR assays to evaluate the relative levels of VEGF mRNA in hypoxic HeLa cells treated with polyamides. In parallel, untreated cells were used as controls. Expression of β-glucuronidase was used as a control gene for determining the relative levels of transcription (29). After 48 h of incubation with polyamide 1, levels of VEGF expression were reduced in a dose-dependent manner (Fig. 5A). Polyamide 1 at 1 μM inhibits VEGF expression ≈60%, which is near the VEGF mRNA levels in the uninduced (normoxic) cells. Mismatch polyamide 2 shows minimal inhibition at either 0.2 μMor 1 μM concentrations.
Fig. 5.
Polyamide 1 blocks VEGF induction by hyopoxia. (A) Relative mRNA levels of expression of the VEGF gene as measured by real-time quantitative RT-PCR. (B) Levels of secreted VEGF protein as measured by ELISA. The final concentration of polyamides 1 and 2 was 0.2 or 1 μM. Noninduced polyamide concentrations in A were 1 μM for each.
ELISA was used to determine the levels of secreted VEGF. Total protein levels were monitored in parallel, to exclude the possibility of disruption of general transcriptional activity by the polyamides. Under normoxia, match polyamide 1 caused a modest decrease of the basal expression levels of VEGF, whereas mismatch polyamide 2 caused no decrease of VEGF levels. Under hypoxic conditions, polyamide 1 decreased levels of VEGF in a dose-dependent manner, whereas mismatch polyamide 2 had a minimal effect (Fig. 5B).
Genome-Wide Effects of Polyamides. The effects of polyamide treatment on nuclear transcription were monitored by global gene expression analysis by using Affymetrix high-density Uni-Gene 133A microarrays, which contain oligonucleotide sequences representing ≥20,000 annotated genes. HeLa cells were treated in triplicate with no polyamide, polyamide 1, or polyamide 2 at 1 μM and 0.2 μM concentrations, for 48 h. DFO was then added to a concentration of 300 μM for an additional 12–16 h and total RNA was collected. Purified RNA was treated and hybridized to the oligonucleotide microarrays according to established protocols.
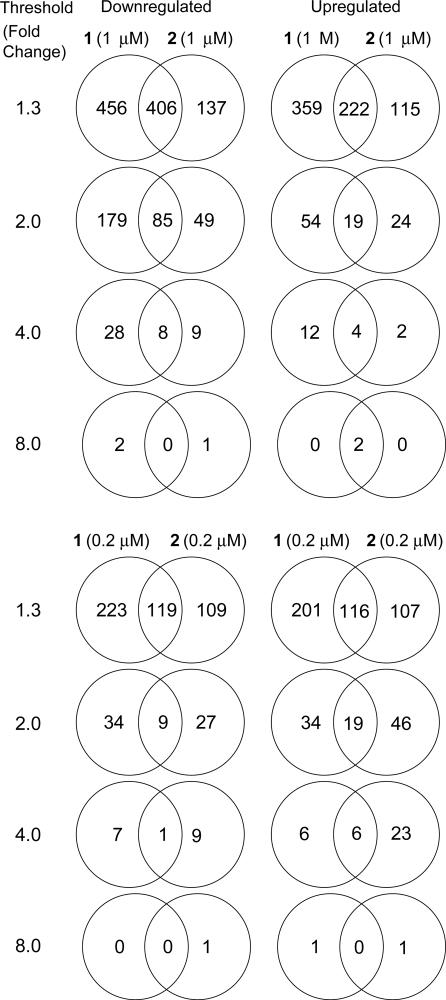
Fig. 6 lists the number of genes affected uniquely and similarly by polyamides 1 and 2 at 0.2 and 1 μM. At each threshold, there is a majority of genes uniquely affected by each polyamide, as well as a number of genes similarly affected by both polyamides. This is consistent with previous work suggesting that polyamides that target different DNA sequences can affect the expression of different sets of genes (13). At a threshold of 2.0-fold, 264 and 73 genes are down-regulated and up-regulated, respectively, in the presence of polyamide 1 at 1 μM. This represents only 1.5% of the interrogated genes. In the case of polyamide 2, <1.0% are affected at this threshold. These effects are surprising, given that a polyamide with a 6-bp binding site is expected to have ≥1.4 million match sites in a 3 billion-base pair genome. Genes affected at a threshold of 2.0-fold for each polyamide are listed in Tables 2 and 3, which are published as supporting information on the PNAS web site. Polyamides 1 and 2 at 0.2 μM affect the expression of fewer genes at each threshold level as compared with the 1 μM data sets. It should be noted that most genes down- and up-regulated by each polyamide at 0.2 μM are similarly affected in the 1 μM data set for each polyamide.
Fig. 6.
Venn diagrams representing the distribution of affected genes (P < 0.01) from the microarray experiments. The numbers outside the intersections represent genes uniquely affected by the individual polyamides.
Next, we analyzed differential expression levels of several hypoxia-inducible genes in the presence of polyamides 1 or 2 (Table 1). The expression of the main target gene, VEGF, is down-regulated by 1.34-fold with polyamide 1 and virtually unaltered with polyamide 2. These data parallel the RT-PCR experiments and luciferase experiments. Other hypoxia-inducible genes are also affected, albeit to a different extent. Remarkably, the microarray data indicated significantly down-regulated levels of the mRNAs corresponding to all three endothelin (ET) genes. In fact, the levels of ET-2 were >90% (13.6-fold) down-regulated with polyamide 1 as compared with the untreated controls. Interestingly, the controls treated with polyamide 2 show nearly 3-fold down-regulation of ET-2. This effect was validated by real-time quantitative RT-PCR of ET-2 mRNA levels, where 6.8-fold down-regulation was observed for polyamide 1 and 2.4-fold for polyamide 2. According to the microarray data, ET-1 was found to be down-regulated 2.4-fold by polyamide 1 and 1.26-fold by polyamide 2. Real-time quantitative RT-PCR measurements were generally consistent with a 1.5-fold down-regulation of ET-1 mRNA by polyamide 1 and no detectable down-regulation by polyamide 2. Recent studies indicate the emerging role of ETs in cancer (30). In addition, ET-2 has been recently implicated as an autocrine survival factor in hypoxic cells (31). We will defer a detailed discussion of the effects of polyamides 1 and 2 on the expression of the ET genes until a more thorough analysis of their regulation has been undertaken.
Table 1. Relative expression levels of selected HIF-inducible genes.
| Fold change
|
|||||
|---|---|---|---|---|---|
| Polyamide 1
|
Polyamide 2
|
||||
| GenBank accession no. | Annotated gene | 1 μM | 0.2 μM | 1 μM | 0.2 μM |
| Energy metabolism | |||||
| AI761561 | Hexokinase-2 | -1.3 | - | - | - |
| NM_005165.1 | Aldolase-C | - | - | - | - |
| Hormones/receptors | |||||
| J03241.1 | Transforming growth factor β3 | - | - | - | - |
| Vasoactive proteins | |||||
| AF022375.1 | VEGF | -1.35 | -1.4 | - | - |
| NM_002019.1 | VEGF receptor, Flt-1 | -1.5 | - | - | - |
| NM_001955.1 | ET-1 | -2.4 | -1.9 | -1.3 | - |
| NM_001956.1 | ET-2 | -13.2 | -2.2 | -2.8 | -2.0 |
| NM_000114.1 | ET-3 | -1.8 | - | - | - |
Fold change ≥ 1.2 and P ≤ 0.01.
Discussion
The expression of VEGF has received considerable attention because this potent mitogen can stimulate endothelial cell proliferation and migration in vitro (32, 33) as well as angiogenesis in vivo (34, 35). Elevated VEGF levels are associated with the progression of a variety of tumors and correlated to the outcome of cancer treatment (36, 37). To date, numerous attempts to block the activity of VEGF have been made, including the use of antibodies (38), soluble VEGF receptors (39), VEGF receptor antagonists (40), or degradation of the VEGF message through the use of antisense oligonucleotides (41) or by RNA interference (42, 43). The major focus of the previous studies was inhibition of a single target or a very limited number of targets. This work presents a pathway-specific approach where the expression of multiple genes is down-regulated by targeting a common transcription factor-binding site. Because there is some sequence variation within the consensus HRE site, we would anticipate that some, but not all, HIF-regulated genes would be affected by polyamide 1 programmed for 5′-WTWCGW-3′.
The details of oncogenic signaling pathways that give rise to the cancerous cellular phenotype continue to be elucidated. These signaling pathways involve a large number of proteins involved in signal transduction that ultimately converge upon a much smaller set of oncogenic transcription factors (44). Hence, targeting transcription factors with small molecules may be the most direct way of reversing the cancerous phenotype. Toward this goal, one can use small molecules to target critical protein–protein interactions between transcription factors and coactivators (45, 46). DNA-binding polyamides offer an alternate approach by interfering with protein–DNA interactions. However, selective gene regulation by programmable DNA-binding polyamides depends on a precise knowledge of cis-acting promoter elements and the trans-acting factors that bind them.
Our results indicate that polyamide–FITC conjugate 1, designed to target the HRE, can bind its cognate site with high affinity and specificity and is capable of disrupting binding of HIF-1 to HRE. The polyamide–FITC conjugate was localized in the nuclei of cultured HeLa cells with no deleterious effects on growth or replication rate. Analysis of the VEGF mRNA levels by real-time quantitative RT-PCR and secreted levels of VEGF by ELISA indicated reduction of the promoter activity in hypoxic cells resulting in the concomitant decrease of VEGF production to near its basal levels. Analysis of the genome-wide effects of the polyamide provided further insights into the transcriptional activity of multiple hypoxia-inducible genes. Because many biological responses are threshold-based, the overall decrease of the transcriptional activity to the basal levels could have pronounced downstream effects. Previous studies have shown that the magnitude of induction of VEGF mRNA in mice subjected to systemic hypoxia varies with tissue type but generally falls between 2- and 4-fold, consistent with the levels of induction measured in this study (47).
Supplementary Material
Acknowledgments
We thank the National Institutes of Health for support. Mass spectrometry analyses were performed in the Mass Spectrometry Laboratory of the Division of Chemistry and Chemical Engineering of California Institute of Technology, supported in part by the National Science Foundation Materials Research Science and Engineering program. Oligonucleotide microarray experiments were performed in the Millard and Muriel Jacobs Genetics and Genomics Laboratory at California Institute of Technology.
Author contributions: B.Z.O., G.-J.Z., W.G.K., and P.B.D. designed research; B.Z.O., G.-J.Z., J.M.K., and N.G.N. performed research; B.Z.O., N.G.N., W.G.K., G.-J.Z., and P.B.D. analyzed data; B.Z.O., G.-J.Z., N.G.N., W.G.K., and P.B.D. wrote the paper.
Abbreviations: HIF, hypoxia-inducible factor; ARNT, aryl hydrocarbon receptor nuclear translocator; HRE, hypoxia response element; DFO, desferrioxamine mesylate; ET, endothelin.
References
- 1.Kaelin, W. G. (2002) Genes Dev. 16, 1441–1445. [DOI] [PubMed] [Google Scholar]
- 2.O'Rourke, J. F., Pugh, C. W., Bartlett, S. M. & Ratcliffe, P. J. (1996) Eur. J. Biochem. 241, 403–410. [DOI] [PubMed] [Google Scholar]
- 3.Ivan, M., Kondo, K., Yang, H. F., Kim, W., Valiando, J., Ohh, M., Salic, A., Asara, J. M., Lane, W. S. & Kaelin, W. G. (2001) Science 292, 464–468. [DOI] [PubMed] [Google Scholar]
- 4.Kaelin, W. G. (2002) Nat. Rev. Cancer 2, 673–682. [DOI] [PubMed] [Google Scholar]
- 5.Maxwell, P. H., Wiesener, M. S., Chang, G. W., Clifford, S. C., Vaux, E. C., Cockman, M. E., Wykoff, C. C., Pugh, C. W., Maher, E. R. & Ratcliffe, P. J. (1999) Nature 399, 271–275. [DOI] [PubMed] [Google Scholar]
- 6.Wood, S. M., Gleadle, J. M., Pugh, C. W., Hankinson, O. & Ratcliffe, P. J. (1996) J. Biol. Chem. 271, 15117–15123. [DOI] [PubMed] [Google Scholar]
- 7.O'Rourke, J. F., Dachs, G. U., Gleadle, J. M., Maxwell, P. H., Pugh, C. W., Stratford, I. J., Wood, S. M. & Ratcliffe, P. J. (1997) Oncol. Res. 9, 327–332. [PubMed] [Google Scholar]
- 8.Forsythe, J. A., Jiang, B. H., Iyer, N. V., Agani, F., Leung, S. W., Koos, R. D. & Semenza, G. L. (1996) Mol. Cell. Biol. 16, 4604–4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okino, S. T., Chichester, C. H. & Whitlock, J. P. (1998) J. Biol. Chem. 273, 23837–23843. [DOI] [PubMed] [Google Scholar]
- 10.Underiner, T. L., Ruggeri, B. & Gingrich, D. E. (2004) Curr. Med. Chem. 11, 731–745. [DOI] [PubMed] [Google Scholar]
- 11.Best, T. P., Edelson, B. S., Nickols, N. G. & Dervan, P. B. (2003) Proc. Natl. Acad. Sci. USA 100, 12063–12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edelson, B. S., Best, T. P., Olenyuk, B., Nickols, N. G., Doss, R. M., Foister, S., Heckel, A. & Dervan, P. B. (2004) Nucleic Acids. Res. 32, 2802–2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dudouet, B., Burnett, R., Dickinson, L. A., Wood, M. R., Melander, C., Belitsky, J. M., Edelson, B., Wurtz, N., Briehn, C., Dervan, P. B. & Gottesfeld, J. M. (2003) Chem. Biol. 10, 859–867. [DOI] [PubMed] [Google Scholar]
- 14.Suto, R. K., Edayathumangalam, R. S., White, C. L., Melander, C., Gottesfeld, J. M., Dervan, P. B. & Luger, K. (2003) J. Mol. Biol. 326, 371–380. [DOI] [PubMed] [Google Scholar]
- 15.Edayathumangalam, R. S., Weyermann, P., Gottesfeld, J. M., Dervan, P. B. & Luger, K. (2004) Proc. Natl. Acad. Sci. USA 101, 6864–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gottesfeld, J. M., Neely, L., Trauger, J. W., Baird, E. E. & Dervan, P. B. (1997) Nature 387, 202–205. [DOI] [PubMed] [Google Scholar]
- 17.Dickinson, L. A., Gulizia, R. J., Trauger, J. W., Baird, E. E., Mosier, D. E., Gottesfeld, J. M. & Dervan, P. B. (1998) Proc. Natl. Acad. Sci. USA 95, 12890–12895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wurtz, N. R., Pomerantz, J. L., Baltimore, D. & Dervan, P. B. (2002) Biochemistry 41, 7604–7609. [DOI] [PubMed] [Google Scholar]
- 19.White, S., Szewczyk, J. W., Turner, J. M., Baird, E. E. & Dervan, P. B. (1998) Nature 391, 468–471. [DOI] [PubMed] [Google Scholar]
- 20.Kielkopf, C. L., White, S., Szewczyk, J. W., Turner, J. M., Baird, E. E., Dervan, P. B. & Rees, D. C. (1998) Science 282, 111–115. [DOI] [PubMed] [Google Scholar]
- 21.Dervan, P. B. & Edelson, B. S. (2003) Curr. Opin. Struct. Biol. 13, 284–299. [DOI] [PubMed] [Google Scholar]
- 22.Foister, S., Marques, M. A., Doss, R. M. & Dervan, P. B. (2003) Bioorg. Med. Chem. 11, 4333–4340. [DOI] [PubMed] [Google Scholar]
- 23.Tischer, E., Mitchell, R., Hartman, T., Silva, M., Gospodarowicz, D., Fiddes, J. C. & Abraham, J. A. (1991) J. Biol. Chem. 266, 11947–11954. [PubMed] [Google Scholar]
- 24.Belitsky, J. M., Nguyen, D. H., Wurtz, N. R. & Dervan, P. B. (2002) Bioorg. Med. Chem. 10, 2767–2774. [DOI] [PubMed] [Google Scholar]
- 25.Trauger, J. W. & Dervan, P. B. (2001) Methods Enzymol. 340, 450–466. [DOI] [PubMed] [Google Scholar]
- 26.Wood, S. M. & Ratcliffe, P. J. (1997) Int. J. Biochem. Cell Biol. 29, 1419–1432. [DOI] [PubMed] [Google Scholar]
- 27.Bianchi, L., Tacchini, L. & Cairo, G. (1999) Nucleic Acids. Res. 27, 4223–4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aerts, J. L., Gonzales, M. I. & Topalian, S. L. (2004) Biotechniques 36, 84–86, 88, 90–91. [DOI] [PubMed] [Google Scholar]
- 29.Hung, C. J., Ginzinger, D. G., Zarnegar, R., Kanauchi, H., Wong, M. G., Kebebew, E., Clark, O. H. & Duh, Q. Y. (2003) J. Clin. Endocrinol. Metab. 88, 3694–3699. [DOI] [PubMed] [Google Scholar]
- 30.Nelson, J., Bagnato, A., Battistini, B. & Nisen, P. (2003) Nat. Rev. Cancer 3, 110–116. [DOI] [PubMed] [Google Scholar]
- 31.Grimshaw, M. J., Naylor, S. & Balkwill, F. R. (2002) Mol. Cancer Ther. 1, 1273–1281. [PubMed] [Google Scholar]
- 32.Soker, S., Gollamudi-Payne, S., Fidder, H., Charmahelli, H. & Klagsbrun, M. (1997) J. Biol. Chem. 272, 31582–31588. [DOI] [PubMed] [Google Scholar]
- 33.Hanahan, D. & Folkman, J. (1996) Cell 86, 353–364. [DOI] [PubMed] [Google Scholar]
- 34.Plate, K. H., Breier, G., Weich, H. A. & Risau, W. (1992) Nature 359, 845–848. [DOI] [PubMed] [Google Scholar]
- 35.Leung, D. W., Cachianes, G., Kuang, W. J., Goeddel, D. V. & Ferrara, N. (1989) Science 246, 1306–1309. [DOI] [PubMed] [Google Scholar]
- 36.Gasparini, G., Toi, M., Gion, M., Verderio, P., Dittadi, R., Hanatani, M., Matsubara, I., Vinante, O., Bonoldi, E., Boracchi, P., et al. (1997) J. Natl. Cancer Inst. 89, 139–147. [DOI] [PubMed] [Google Scholar]
- 37.Kang, S. M., Maeda, K., Chung, Y. S., Onoda, N., Ogawa, Y., Takatsuka, S., Ogawa, M., Sawada, T., Nakata, B., Nishiguchi, Y., et al. (1997) Oncol. Rep. 4, 381–384. [PubMed] [Google Scholar]
- 38.Kim, K. J., Li, B., Winer, J., Armanini, M., Gillett, N., Phillips, H. S. & Ferrara, N. (1993) Nature 362, 841–844. [DOI] [PubMed] [Google Scholar]
- 39.Lin, P. N., Sankar, S., Shan, S. Q., Dewhirst, M. W., Polverini, P. J., Quinn, T. Q. & Peters, K. G. (1998) Cell Growth Differ. 9, 49–58. [PubMed] [Google Scholar]
- 40.Hennequin, L. F., Thomas, A. P., Johnstone, C., Stokes, E. S. E., Ple, P. A., Lohmann, J. J. M., Ogilvie, D. J., Dukes, M., Wedge, S. R., Curwen, J. O., et al. (1999) J. Med. Chem. 42, 5369–5389. [DOI] [PubMed] [Google Scholar]
- 41.Shi, W. & Siemann, D. W. (2002) Br. J. Cancer 87, 119–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang, L., Yang, N., Mohamed-Hadley, A., Rubin, S. C. & Coukos, G. (2003) Biochem. Biophys. Res. Commun. 303, 1169–1178. [DOI] [PubMed] [Google Scholar]
- 43.Reich, S., Fosnot, J., Kuroki, A., Tang, W. X., Yang, X. Y., Maguire, A., Bennett, J. & Tolentino, M. (2003) Mol. Vis. 9, 210–216. [PubMed] [Google Scholar]
- 44.Darnell, J. E. (2002) Nat. Rev. Cancer 2, 740–749. [DOI] [PubMed] [Google Scholar]
- 45.Kung, A. L., Wang, S., Klco, J. M., Kaelin, W. G. & Livingston, D. M. (2000) Nat. Med. 6, 1335–1340. [DOI] [PubMed] [Google Scholar]
- 46.Kung, A. L., Zabludoff, S. D., France, D. S., Freedman, S. J., Tanner, E. A., Vieira, A., Cornell-Kennon, S., Lee, J., Wang, B. Q., Wang, J. M., et al. (2004) Cancer Cell 6, 33–43. [DOI] [PubMed] [Google Scholar]
- 47.Marti, H. H. & Risau, W. (1998) Proc. Natl. Acad. Sci. USA 95, 15809–15814. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.