Abstract
Chronic periodontitis (CP) has a genetic component, particularly its severe forms. Evidence from genome-wide association studies (GWASs) has highlighted several potential novel loci. Here, the authors report the first GWAS of CP among a large community-based sample of Hispanics/Latinos. The authors interrogated a quantitative trait of CP (mean interproximal clinical attachment level determined by full-mouth periodontal examinations) among 10,935 adult participants (mean age: 45 y, range: 18 to 76 y) from the Hispanic Community Health Study / Study of Latinos. Genotyping was done with a custom Illumina Omni2.5M array, and imputation to approximately 20 million single-nucleotide polymorphisms was based on the 1000 Genomes Project phase 1 reference panel. Analyses were based on linear mixed models adjusting for sex, age, study design features, ancestry, and kinship and employed a conventional P < 5 × 10−8 statistical significance threshold. The authors identified a genome-wide significant association signal in the 1q42.2 locus (TSNAX-DISC1 noncoding RNA, lead single-nucleotide polymorphism: rs149133391, minor allele [C] frequency = 0.01, P = 7.9 × 10−9) and 4 more loci with suggestive evidence of association (P < 5 × 10−6): 1q22 (rs13373934), 5p15.33 (rs186066047), 6p22.3 (rs10456847), and 11p15.1 (rs75715012). We tested these loci for replication in independent samples of European-American (n = 4,402) and African-American (n = 908) participants of the Atherosclerosis Risk in Communities study. There was no replication among the European Americans; however, the TSNAX-DISC1 locus replicated in the African-American sample (rs149133391, minor allele frequency = 0.02, P = 9.1 × 10−3), while the 1q22 locus was directionally concordant and nominally significant (rs13373934, P = 4.0 × 10−2). This discovery GWAS of interproximal clinical attachment level—a measure of lifetime periodontal tissue destruction—was conducted in a large, community-based sample of Hispanic/Latinos. It identified a genome-wide significant locus that was independently replicated in an African-American population. Identifying this genetic marker offers direction for interrogation in subsequent genomic and experimental studies of CP.
Keywords: genetics, periodontal attachment loss, genomics, epidemiology, survey and questionnaires, observational study
Introduction
In periodontal health, the innate host defense system maintains homeostasis with the periodontal microbial community. This balance becomes disrupted when defects in the host immunoregulatory mechanisms shift the microbial community to a dysbiotic state (Hajishengallis 2015). Members of this highly pathogenic polymicrobial community operate synergistically to subvert the protective function of leukocytes in susceptible individuals. What ensues is an excessive release of proinflammatory cytokines and resistance to immune elimination. In this nonresolving inflammatory state, chronic periodontitis (CP) is characterized as the progressive and irreversible loss of the tooth’s attachment to the periodontal ligament and the destruction of connective tissue and alveolar bone. In the National Health and Nutrition Survey (2009 to 2012), the age-standardized prevalence of CP was 40% in non-Hispanic white, 60% in non-Hispanic black, and 68% in Hispanic adults (Eke et al. 2015). Lack of knowledge on the bacterial subversion of immunoinflammatory mechanisms in CP pathogenesis leading to oral dysbiosis has directed the search for genetic determinants that regulate immune and inflammatory responses. Genetic factors play a role in the etiology of CP, as evident by the age- and sex-adjusted heritability estimate of 59% in periodontal sites with ≥2 mm of attachment loss reported in a twin study (Michalowicz et al. 2000).
To date, 5 genome-wide association studies (GWASs) have been conducted for CP in European American (Divaris et al. 2013; Shaffer et al. 2014), German (Teumer et al. 2013), Japanese (Shimizu et al. 2015), and Korean populations (Hong et al. 2015). A variety of CP traits were examined in these GWASs, but no genome-wide significant loci have been identified for CP. However, several suggestive susceptibility loci have been reported. Encouragingly, suggestive evidence of an association in the chromosome 14q21 region was identified in 3 GWASs (Divaris et al. 2013; Shaffer et al. 2014; Shimizu et al. 2015), and the NPY locus has been reported by 2 (Divaris et al. 2013, (Freitag-Wolf et al. 2014). Finally, a recent Chinese study replicated prior findings of gene-centric associations of CP (Rhodin et al. 2014) for the FBXO38 and AP3B2 loci (Shang et al. 2015). This body of evidence has been generated among populations of primarily European and secondarily Asian ancestry, with Hispanic/Latinos being underrepresented. To address this gap, we conducted the first GWAS of CP among a large community-based sample of Hispanics/Latinos in the United States. We sought to identify CP susceptibility loci in this cohort and to examine their replication in independent cohorts of European Americans and African Americans.
Materials and Methods
Ethics Statement
The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) guidelines guided the reporting of this observational human research study, which was conducted according to the principles expressed in the Declaration of Helsinki. Participants provided informed consent, and the institutional review board at each field center approved the study protocols.
Discovery Population
The Hispanic Community Health Study / Study of Latinos (HCHS/SOL) is a multicenter, population-based prospective cohort study of individuals of Cuban, Dominican, Mexican, Puerto Rican, Central American, and South American ancestral origins. The purpose of the study, the details of the complex sampling design, and the implementation methods are published (Lavange et al. 2010; Sorlie et al. 2010). In brief, from 2008 through 2011, the HCHS/SOL investigators enrolled 16,415 Hispanic/Latino individuals aged 18 to 74 y from randomly selected households, using a stratified 2-stage area probability sample design in 4 U.S. communities: Bronx, New York; Chicago, Illinois; Miami, Florida; and San Diego, California.
Periodontal Assessment and CP Trait Definition (Interproximal Clinical Attachment Level)
Eighteen dental examiners conducted comprehensive periodontal examinations for participants not requiring prophylactic antibiotics. Probing pocket depth and recession were measured at 6 sites per tooth on all fully erupted teeth, excluding third molars. Clinical attachment level—quantified as the distance in millimeters from the cementoenamel junction (CEJ; a fixed, reproducible point) to the base of the sulcus—was determined from the sum of 2 measurements: the distance from the free gingival margin to the bottom of the pocket and the distance from the free gingival margin to the CEJ.
The methods used to derive the CP phenotype were identical in the discovery HCHS/SOL cohort and the dental Atherosclerosis Risk in Communities (ARIC) replication cohort. A comparison of the distributions of this phenotype in both populations (Appendix Table 1) shows the ARIC subjects to be older with few retained teeth. Analyses were restricted to periodontal measurements made at the 4 interproximal sites—mesiobuccal, distobuccal, mesiolingual, and distolingual. The rationale for selecting interproximal clinical attachment level (iCAL) alone, rather than measures that include probing pocket depth as the CP trait, is that pocket depth at a given site is a changeable measure and is likely to underestimate past destructive periodontal disease (Carlos et al. 1987). By contrast, iCAL provides a more reliable assessment of periodontal tissue destruction accumulated over a person’s lifetime (Albandar and Rams 2002), and so this CP trait is less prone to misclassification bias. Because 2 earlier GWASs had defined the CP trait according to the case classifications of the Centers for Disease Control and Prevention and American Academy of Periodontology (Divaris et al. 2013; Teumer et al. 2013), we also conducted a GWAS of CP using 4 case definitions from the same institutions (Eke et al. 2012). However, like these previous studies, our top hits did not reach genome-wide statistical significance (Appendix Figs. 1, 2).
Genotyping, Quality Control, and Imputation
In the HCHS/SOL, DNA was extracted from blood samples according to standard protocols. Participants were genotyped on the HCHS Custom 15041502 array (Illumina Omni2.5M + custom content). Quality control was conducted as described (Laurie et al. 2010; Conomos et al. 2016). In brief, samples were checked for annotated versus genetic sex discrepancies, gross chromosomal anomalies, missing call rates, contaminations, and batch effects. Single-nucleotide polymorphism (SNP) quality metrics included the Illumina/LA Biomed assay failure indicator, missing call rates, deviation from Hardy-Weinberg equilibrium, Mendelian errors, and duplicate sample discordance.
Genotypes were prephased with SHAPEIT2 (Delaneau et al. 2012) and imputed through IMPUTE2 (Howie et al. 2012; Howie et al. 2009) into the 1000 Genomes phase 1 reference panel (1000 Genomes Project Consortium 2012). For imputed SNPs, we calculated the imputation quality scores “info” and “oevar” (ratio of observed to expected variance of imputed dosage) for each SNP and excluded SNPs with oevar <0.3. Finally, SNPs with ≤30 counts of the expected or effective number of copies of the minor allele were excluded from analysis. A variant’s effective number of copies is approximately its minor allele count and was estimated as 2 × MAF × (1 – MAF) × N × oevar, where MAF is the minor allele frequency, N is the number of participants, and oevar is set to 1 for genotyped variants.
Replication Population
The ARIC cohort (1987 to present) is a prospective investigation of atherosclerosis and cardiovascular disease (Salem et al. 1978). At baseline (1987 to 1989), ~4,000 individuals were enrolled in ARIC from each of 4 U.S. communities: Forsyth County, North Carolina; Jackson, Mississippi; suburban Minneapolis, Minnesota; and Washington County, Maryland. Participants completed baseline interviews, laboratory measurements, and clinic examinations. Overall, 11,478 ARIC participants were European American and 4,314 were African American.
At ARIC visit 4 in 1996 to 1998, a dental study was conducted for dentate subjects not needing prophylactic antibiotics for periodontal probing. In the dental ARIC, the calibrated dental examiners collected clinical measures of probing pocket depth and gingival recession from 6 sites for all teeth present. In both the HCHS/SOL and the ARIC, the clinical attachment level was recorded as the measurement of the position of the soft tissue in relation to a fixed reference point: the CEJ. The clinical attachment level is determined by summing the probing pocket depth and the distance from the gingival margin to the CEJ. A complication can arise because of the varying position of the gingival margin. When the CEJ is coronal to the gingival margin or when the CEJ and gingival margin are at the same level, recession is simple to calculate. However, when the gingival margin extends over the CEJ toward the gingivae, there is no loss of clinical attachment, and recession is recorded as a negative number.
A total of 5,552 adults aged 52 to 74 y participated in the dental ARIC. A GWAS of moderate CP and severe CP was subsequently performed on 4,504 European Americans in the dental ARIC (Divaris et al. 2013).
GWAS Approach and Annotation
For the HCHS/SOL, we estimated the association of each SNP with iCAL using a linear mixed model regression (Conomos et al. 2016). Genetic models were adjusted for the fixed effects of sex, age, field center, cigarette use, sampling weights, genetic subgroup and population stratification via 5 genetic principal components representing ancestry, and the random effects of census block group, household, and kinship. A cubic root transformation was employed to account for the positively skewed iCAL trait so that the residuals approximated a normal distribution. A conventional P < 5 × 10−8 criterion was used for genome-wide significance, and a P < 5 × 10−6 threshold was used to denote suggestive association and to carry forward markers for functional annotation and replication to the ARIC study.
We used the GTEx (http://www.gtexportal.org) database to investigate possible regulatory expression quantitative trait loci in 43 tissues (GTEx Consortium 2015), including fibroblasts (no oral tissue–specific expression data), and the SCAN database (http://www/scandb.org) to examine associations with gene expression in lymphoblastoid cell lines of apparently healthy individuals of European (CEU) and African (YRI) ancestry (Zhang et al. 2015). Additionally, we used the National Cancer Institute’s LDlink (version 1.1, http://analysistools.nci.nih.gov/LDlink/; Machiela and Chanock 2015) to identify SNPs correlated with the ones highlighted in this study that are known or predicted regulatory elements in intergenic regions according to RegulomeDB criteria (Boyle et al. 2012; Appendix Table 3).
To test the replication of the prioritized loci (P < 5 × 10−5) in independent samples of European American and African- American participants, we examined SNP estimates of association generated by a similarly conducted GWAS analysis of iCAL (linear regression adjusting for study design features, age, sex, smoking, and 10 ancestry principal components) of the cube root–transformed mean iCAL. Our rationale for determining the number of tests for which to adjust in determining the replication P value was that the 2 ARIC replication populations (European American and African American) were independent. Hence, analysis should correct for 5 tests within each study population, resulting in a P < 0.01 replication statistical significance threshold.
We considered replication using 3 tiers of evidence: 1) significant association in the each ARIC replication sample after multiple-testing correction (Bonferroni correction accounting for the number of SNPs tested separately in ARIC European-American and African-American cohorts), 2) nominal association (P < 0.05) in the replication samples, and 3) directional concordance beyond what would be expected by chance alone (using a binomial test and a conventional P < 0.05 criterion). Furthermore, we explored whether the formerly reported region on chromosome 14 is associated with iCAL in the HCHS/SOL. Thus, we examined and report the association of 2 SNPs cited by previous GWASs: rs12883458 (Divaris et al. 2013) and rs3783412 (Shaffer et al. 2014).
Results
Within the genotyped HCHS/SOL population, 1,806 study participants with missing or incomplete periodontal information were excluded from analyses, along with 62 who had missing information on cigarette use or genetic subgroup covariate, leaving 10,935 participants in the sample (mean age: 45 y; range: 18 to 76 y). There were 253 European-American and 4 African-American ARIC participants excluded due to missing covariates, resulting in a replication analytic sample including 4,402 subjects (mean age: 63; range: 53 to 74 y) and 908 subjects (mean age: 61; range: 52 to 74 y), respectively.
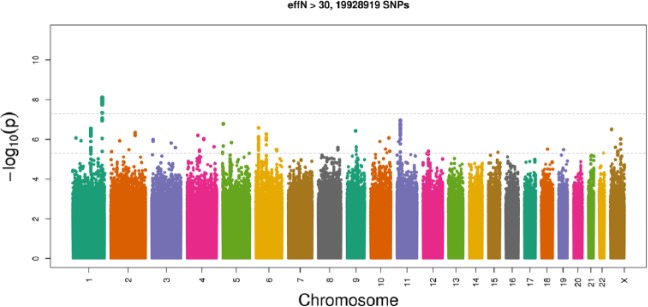
Approximately 20 million genotyped and imputed SNPs passed quality control filters and were included in the discovery GWAS. The genomic inflation factor regarding genotyped and imputed SNPs was low (λ = 0.990), and the quantile-quantile plot (Appendix Fig. 3) suggested virtually no residual population stratification. Only 1 locus in the 1q42.2 region (Fig. 1; lead SNP: rs149133391, MAF = 0.01; P = 7.9 × 10−9) demonstrated evidence of genome-wide statistically significant association. We found 4 loci showing suggestive evidence (P < 5 × 10−6) of association (Table 1): 11p15.1 (lead SNP: rs75715012, MAF = 0.09, P = 1.1 × 10−7), 5p15.33 (lead SNP: rs186066047, MAF = 0.003, P = 1.7 × 10−7), 6p22.3 (lead SNP: rs10456847, MAF = 0.33, P = 2.6 × 10−7), and 1q22 (lead SNP: rs79308117, intronic to ASH1L, MAF = 0.005, P = 2.9 × 10−7). All these SNPs were imputed with high quality (oevar ≥0.93). In Appendix Table 2, we provide additional functional annotation information for SNPs, highlighting these loci and proxy SNPs, along with supplemental annotation (http://genomewide.net/public/hchs_sol/periodontitis/meanALi/SOL_meanALi_topSNP_annotation.xlsx).
Figure 1.
Manhattan plot presenting the genome-wide association study results of chronic periodontitis (mean interproximal attachment level) in the HCHS/SOL study (n = 10,935) with SNP P values ordered by chromosomal position. The higher dashed line illustrates the genome-wide significance threshold (P = 5 x 10−8) and the lower dashed line denotes a line of suggestive association.
Table 1.
Genomic and Functional Context of Top SNPs in Loci with P < 5 × 10−6 for Association with Chronic Periodontitis in the HCHS/SOL Study (N = 10,935).
| Allele |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Region | SNP | Chr Positiona | Gene (Role) or Nearest Gene | Coded | Minor | MAF | b (SE)b | Oevarc | P Value |
| 1q42.2 | rs149133391 | 1: 231580785 | TSNAX-DISC1 (intron) | T | C | 0.011 | −0.139 (0.024) | 0.94 | 7.9 × 10−9 |
| 11p15.1 | rs75715012 | 11: 21627604 | NELL1 | G | A | 0.089 | 0.045 (0.008) | >0.99 | 1.1 × 10−7 |
| 5p15.33 | rs186066047 | 5: 3875660 | IRX1; LINC01017; LINC01019 | G | A | 0.003 | 0.225 (0.043) | 0.93 | 1.7 × 10−7 |
| 6p22.3 | rs10456847d | 6: 18954940 | LOC645157; RNF144B | C | G | 0.330 | −0.026 (0.005) | >0.99 | 2.6 × 10−7 |
| 1q22 | rs79308117e | 1: 155509388 | ASH1L (intron) | A | C | 0.005 | 0.178 (0.035) | >0.99 | 2.8 × 10−7 |
Chronic periodontitis: mean interproximal attachment loss continuous trait.
Chr, chromosomal; HCHS/SOL, Hispanic Community Health Study / Study of Latinos; MAF, minor allele frequency; SNP, single-nucleotide polymorphism.
Chromosome positions are based on GRCh38.p2 annotation release 107.
Per minor allele effect size based on the linear mixed additive genetic model; standard error in parentheses.
Imputation quality metric based on the ratio of observed to expected variance of imputed dosage.
Predicted as an expression quantitative trait locus in the SCAN database: RAB37 (CEU) P = 7 × 10−5; FSCN1 (YRI) P = 10−4. SNP in linkage disequilibrium with rs1334772 (P = 1.1 × 10−6): D′ = 0.94, r2 = 0.837 and rs1891657 (P = 3.6 × 10−6): D′ = 0.89, r2 = 0.746.
SNP in linkage disequilibrium with rs13373934 (P = 4.5 × 10−7): D′ = 1.0, r2 = 1.000 is a missense intron variant of ASH1L. Ser→Pro change, predicted to be “benign” by PolyPhen-2 with HumDiv score 0.107 (sensitivity: 0.93, specificity: 0.86).
We examined the “top SNP” in each of the 5 loci (Fig. 2) for replication among the ARIC European-American and African- American samples using a P < 0.01 association threshold for replication in each sample. We found no evidence of replication among European Americans, wherein 2 of the 5 SNPs (rs13373934 and rs186066047) were actually monomorphic. The genome-wide significant TSNAX-DISC1 locus in 1q42.2, which has a high imputation score in the HCHS/SOL, was replicated in the African-American sample (rs14913 3391: P = 9.1 × 10−3; see regional association plot, Appendix Fig. 4), although this SNP had low imputation quality score (0.44). A directionally consistent and nominally significant association was found for the 1q22 locus (rs13 373934: P = 4.0 × 10−2). Both SNPs reported by Divaris et al. (2013) and Shaffer et al. (2014) showed no evidence of association with iCAL (rs12883458: P = 0.12, rs3783412: P = 0.76). Given the low MAF of the lead SNP, we found limited evidence of genetic association for CP in this GWAS in the HCHS/SOL.
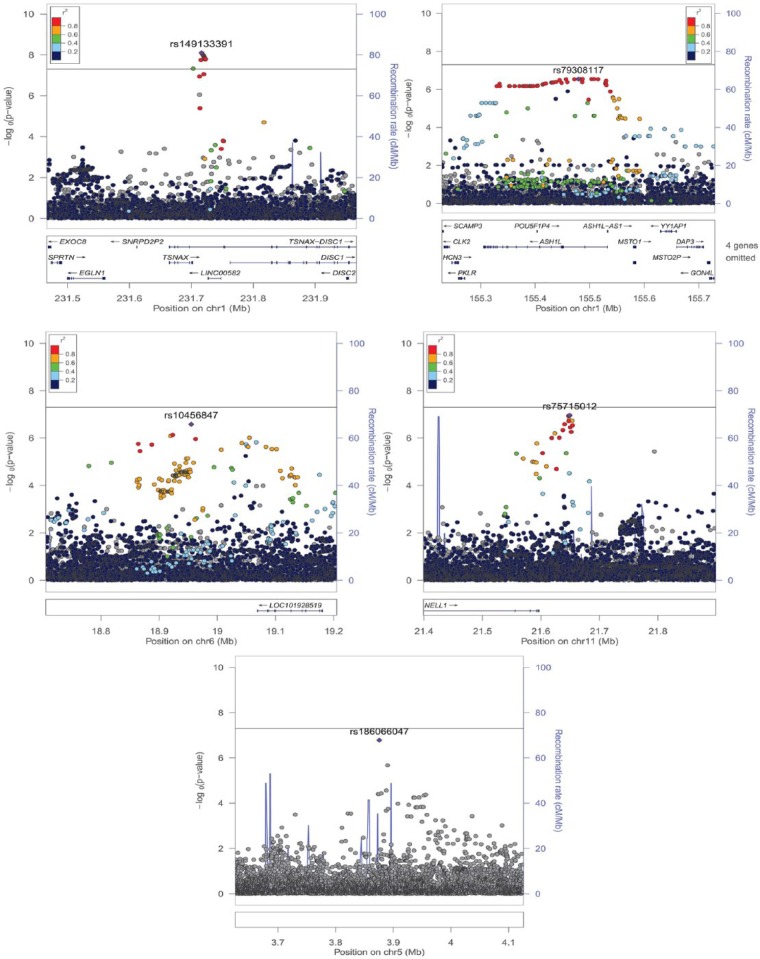
Figure 2.
Regional association plots generated with LocusZoom illustrating the five loci with the strongest (P < 5 x 10−6) association signals in the GWAS of chronic periodontitis in the HCHS/SOL cohort (n = 10,935). The left vertical axis corresponds to –log10 (P values) and the right vertical axis corresponds to recombination rates obtained by the AMR (admixed American) super-population of 1000 genomes via LocusZoom. Each circle depicts a SNP tested for association with chronic periodontitis in the HCHS/SOL. The position on the X-axis corresponds to genomic position, and the position on the Y-axis corresponds to each SNP’s –log10(P values). The top, or “lead”, SNP is colored in purple, while other variants are color-coded by their r2, a measure of LD, with the lead SNP. Gray circles are presented when LD information was unavailable for some SNPs. Regions flanking 250Kb each top SNP are presented. The horizontal gray line marks the genome-wide significance level 5 x 10−8.
The 5 loci from the HCHS/SOL discovery cohort are reported in Table 2, and estimates of association with iCAL and SNP information among the ARIC European-American and African- American samples for these top 5 loci are reported in Table 3.
Table 2.
Estimates of Association with Chronic Periodontitis and SNP Information (P < 5 × 10−6) from the HCHS/SOL Discovery Cohort.
| HCHS/SOL (N = 10,935)a |
|||||
|---|---|---|---|---|---|
| Region | SNP | g/ib | b | P Value | MAF |
| 1q22 | rs13373934 | g | 0.173 | 4.5 × 10−7 | G, 0.005 |
| 1q42.2 | rs149133391 | i | −0.139 | 7.9 × 10−9 | C, 0.011 |
| 5p15.33 | rs186066047 | i | 0.225 | 1.7 × 10−7 | A, 0.003 |
| 6p22.3 | rs10456847 | i | −0.026 | 2.6 × 10−7 | G, 0.330 |
| 11p15.1 | rs75715012 | i | 0.045 | 1.1 × 10−7 | A, 0.089 |
Chronic periodontitis: mean interproximal attachment loss continuous trait.
HCHS/SOL, Hispanic Community Health Study / Study of Latinos; MAF, minor allele frequency; SNP, single-nucleotide polymorphism.
Results based on linear mixed modeling of mean interproximal attachment loss (cubic root transformation) adjusting for 5 ancestry principal components, genetic subgroup, log sampling weight, relatedness including residence/household, age, sex, examination center, and smoking (never/former/current).
Genotyped or imputed SNP. Imputation quality metric based on the observed to expected dosage variance ratio after imputation.
Table 3.
Estimates of Association with Chronic Periodontitis and SNP Information among the ARIC European-American and African-American Samples for the 5 Loci Prioritized (P < 5 × 10−6) from the HCHS/SOL Discovery.
| European American (n = 4,402)a |
African American (n = 908)a |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Region | SNP | g/ib | b | P Value | MAF | Oevarc | g/ib | b | P Value | MAF | Oevarc |
| 1q22 | rs13373934 | i | Monod | i | 0.049 | 4.0 × 10−2 | G, 0.029 | 0.969 | |||
| 1q42.2 | rs149133391 | i | 0.040 | 3.2 × 10−1 | C, 0.004 | 0.595 | i | −0.124 | 9.1 × 10−3 | C, 0.019 | 0.444 |
| 5p15.33 | rs186066047 | i | Monod | i | 0.023 | 4.7 × 10−1 | A, 0.019 | 0.823 | |||
| 6p22.3 | rs10456847 | i | −0.007 | 8.5 × 10−2 | G, 0.460 | 1.000 | i | −0.007 | 4.4 × 10−1 | G, 0.222 | 1.000 |
| 11p15.1 | rs75715012 | i | −0.001 | 9.0 × 10−1 | A, 0.099 | 0.988 | i | −0.001 | 9.6 × 10−1 | A, 0.148 | 0.943 |
Chronic periodontitis: mean interproximal attachment loss continuous trait.
ARIC, Atherosclerosis Risk in Communities; HCHS/SOL, Hispanic Community Health Study / Study of Latinos; MAF, minor allele frequency; SNP, single-nucleotide polymorphism.
Results based on linear regression modeling of mean interproximal attachment loss (cubic root transformation) adjusting for 10 ancestry principal components, age, sex, examination center, and smoking (never/former/current); the analytic sample numbers reflect the exclusion of 253 European-American and 4 African-American ARIC participants due to missing covariates.
Genotyped or imputed SNP.
Imputation quality metric based on the observed to expected dosage variance ratio after imputation.
Monomorphic.
Discussion
In this first GWAS of CP in a Hispanic/Latino population, we found 1 locus (in the region of TSNAX-DISC1) with a genome-wide significant association signal and 4 others with suggestive evidence of association. The genome-wide significant association with iCAL as the CP trait was replicated among a community-based sample of African-American adults. However, the lead SNP had a low MAF (1% in the discovery sample and 2% in the replication sample) and a low imputation score in the African Americans. We found no evidence of replication to European Americans, which had an even lower MAF (0.004) than that in the HCHS/SOL. These findings offer novel insights into potential genetic influences on CP among a traditionally understudied segment of the population.
The region of TSNAX-DISC1 marked the only genome-wide significant locus in this study—one that was significantly associated with iCAL in the independent African-American sample. Of note, according to currently available information on the National Center for Biotechnology Information Entrez-Gene database, TSNAX-DISC1 encodes a nonsense-mediated mRNA decay candidate and is unlikely to make a functional protein. ASH1L—or (absent, small, or homeotic)-like (Drosophila)—encodes a member of the trithorax group of transcriptional activators. It is possible that polymorphisms in these loci may have some unknown functional or regulatory role. In fact, both TSNAX-DISC1 and ASH1L loci had SNPs in linkage disequilibrium (LD) with known or predicted functional roles. Nevertheless, the SNPs marking these loci had low minor allele frequencies; the former had low imputation quality in African Americans; and currently, no mechanistic evidence exists to support a functional role of these candidates.
The 5 markers identified in this study were mostly of low MAF; 2 are rare variants (MAF <0.01), and 1 is a low-frequency variant (MAF: 0.01 to 0.05). This is not surprising given that rare variants constitute the majority of polymorphic sites associated with complex traits in human populations (International HapMap 3 Consortium 2010; Marth et al. 2011; 1000 Genomes Project Consortium 2012). This means that these markers account for only a very small proportion of the risk for iCAL. Increasingly, studies of complex traits are finding that multiple, rare, and low-frequency variants independently yet collectively contribute to risk for the trait (for review, see Panoutsopoulou et al. 2013). Future genetic studies of CP will employ fine-mapping and functional annotation techniques to investigate the reported markers and other variants in strong LD with them.
In the search for candidate genes for CP in European and non-European ancestry groups, polymorphisms of the interleukin 1 cytokine family of gene variants, as well as interleukin 6 and tumor necrosis factor α genes, have been most extensively studied (Nikolopoulos et al. 2008; Karimbux et al. 2012; Wu et al. 2015). None of these candidate genes has been replicated by subsequent agnostic GWASs, including this present study.
For the significant as well as suggestive genomic regions, we acknowledge that these are almost assuredly markers of causal variation and not causal variants themselves. They may or may not represent genetic variation in a specific gene in that region. Although there may be some promising candidate genes in those regions, we do not know whether the significant “hits” are associated with that gene, regulate the expression of other distant genes, or represent neither of these possibilities.
This report is based on sizable study populations of well-characterized, community-dwelling participants, which is a rare instance in genetic epidemiologic studies of oral health. Nevertheless, even larger studies and consortia pooling available samples will be required to efficiently interrogate low-frequency variants and examine the generalization of the novel candidate loci across ancestral groups. Some promising results have already emerged from collaborative work in the Gene Lifestyle Interactions and Dental Endpoint Consortium, which recently reported on a Mendelian randomization study of the association between adiposity and CP (Shungin et al. 2015). Another promising approach into the genomic underpinning of CP is likely to be realized via a combined “deepening” of periodontal phenotypes with enrichment of biological intermediates paired with high-quality genotype and periodontal phenotype information (Offenbacher et al. 2016).
The evidence of replication in this report should be viewed with caution. First, the winner’s curse phenomenon is likely omnipresent in GWASs exploring relatively new traits (Lohmueller et al. 2003).
Second, the generalizability of loci across racial groups should be done with generally modest expectations. For example, only 8% of adult height loci transferred from European ancestry discovery to an independent African-American sample (Shriner et al. 2009), but this proportion increased to >70% when SNPs in LD with the genome-wide significant SNPs were considered. Yet, populations of African ancestry are characterized by reduced LD as compared with Europeans (Shriner et al. 2009), and this offers a potential advantage in efforts to fine-map replicated GWAS signals. It is noteworthy that 4 of 5 loci were directionally consistent in the ARIC African-American sample. Nevertheless, these findings were based on a sample size of <1,000 (n = 908). The top SNPs in 2 loci on chromosome 1 (1q22 and 1q42.2) that showed some evidence of replication in African Americans were low frequency (≤1% in HCHS and ≤3% in ARIC), and the lead marker of the formally replicated locus (rs149133391) had low imputation quality in ARIC. The HCHS/SOL discovery cohort and the ARIC replication cohort differ in their age distribution. Consequently, since iCAL increases over time, the trait distribution also differs between the studies. However, this difference should not lead to confounding bias, since age is not associated with genetic variants; rather, it may lead to lower power in the discovery stage.
In summary, this GWAS of a CP trait (iCAL) was conducted in a community-based study population of Hispanic/Latinos, and it identified a genome-wide significant locus that was associated with iCAL among an independent community-based African-American cohort. Although this SNP has not been mechanistically validated as a causal variant, these findings provide initial insights into several promising candidate loci for interrogation by additional genomic and experimental studies.
Author Contributions
A. E. Sanders, contributed to conception, design, data analysis, and interpretation, drafted and critically revised the manuscript; T. Sofer, contributed to design, data analysis, and interpretation, drafted and critically revised the manuscript; Q. Wong, contributed to design, data analysis, and interpretation, critically revised the manuscript; K.F. Kerr, contributed to design, critically revised the manuscript; C. Agler, contributed to design, data acquisition, and interpretation, drafted and critically revised the manuscript; J.R. Shaffer, J.D. Beck, S. Offenbacher, C.R. Salazar, and M.L. Marazita, contributed to design, critically revised the manuscript; K.E. North, C.C. Laurie, contributed to conception and design, critically revised the manuscript; R.H. Singer, contributed to design and data acquisition, critically revised the manuscript; J. Cai, contributed to design and data interpretation, critically revised the manuscript; T.L. Finlayson, contributed to design and data acquisition, critically revised the manuscript; K. Divaris, contributed to conception, design, data acquisition, and interpretation, drafted and critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Supplementary Material
Footnotes
A supplemental appendix to this article is published electronically only at http://jdr.sagepub.com/supplemental.
The Hispanic Community Health Study / Study of Latinos (HCHS/SOL) was carried out as a collaborative study supported by contracts from the National Heart, Lung, and Blood Institute (NHLBI) to the University of North Carolina (N01-HC65233), University of Miami (N01-HC65234), Albert Einstein College of Medicine (N01-HC65235), Northwestern University (N01-HC65236), and San Diego State University (N01-HC65237). The following institutes, centers, and offices contribute to the HCHS/SOL through a transfer of funds to the NHLBI: National Center on Minority Health and Health Disparities, the National Institute of Deafness and Other Communications Disorders, the National Institute of Dental and Craniofacial Research, the National Institute of Diabetes and Digestive and Kidney Diseases, the National Institute of Neurological Disorders and Stroke, and the Office of Dietary Supplements. The Genetic Analysis Center at the University of Washington was supported by the NHLBI and the National Institute of Dental and Craniofacial Research (contracts HHSN268201300005C AM03 and MOD03). Genotyping efforts were supported by the NHLBI (HSN 26220/20054C), the National Center for Advancing Translational Sciences (Clinical and Translational Science Institute; grant UL1TR000124), and the National Institute of Diabetes and Digestive and Kidney Diseases (Diabetes Research Center; grant DK063491). The Atherosclerosis Risk in Communities Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201 100007C, HHSN268201100008C, HHSN268201100009C, HHS N268201100010C, HHSN268201100011C, and HHSN2682011 00012C). The authors thank the staff and participants of the ARIC study for their important contributions. R01HL087641, R01HL 59367, and R01HL086694, the National Human Genome Research Institute (contract U01HG004402), the National Institutes of Health (contract HHSN268200625226C), the National Institute of Environmental Health Sciences (P30ES010126), and the National Institute of Dental and Craniofacial Research (R01DE11551, R01DE021418, and R01DE023836). Infrastructure was partly supported by the National Institutes of Health (UL1RR025005) and the NIH Roadmap for Medical Research (NCAT; UL1-RR025747). This manuscript has been reviewed by the HCHS/SOL Publications Committee for scientific content and consistency of data interpretation with previous HCHS/SOL publications, as well as the ARIC Study Publications Committee.
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- 1000 Genomes Project Consortium. 2012. An integrated map of genetic variation from 1,092 human genomes. Nature. 491(7422):56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albandar JM, Rams TE. 2002. Global epidemiology of periodontal diseases: an overview. Periodontol 2000. 29:7–10. [DOI] [PubMed] [Google Scholar]
- Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, Karczewski KJ, Park J, Hitz BC, Weng S, et al. 2012. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 22(9):1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlos JP, Brunelle JA, Wolfe MD. 1987. Attachment loss vs. pocket depth as indicators of periodontal disease: a methodologic note. J Periodontal Res. 22(6):524–525. [DOI] [PubMed] [Google Scholar]
- Conomos MP, Laurie CA, Stilp AM, Gogarten SM, McHugh CP, Nelson SC, Sofer T, Fernández-Rhodes L, Justice AE, Graff M, et al. 2016. Genetic diversity and association studies in US Hispanic/Latino populations: applications in the Hispanic Community Health Study/Study of Latinos. Am J Hum Genet. 98(1):165–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaneau O, Marchini J, Zagury JF. 2012. A linear complexity phasing method for thousands of genomes. Nat Methods. 9(2):179–181. [DOI] [PubMed] [Google Scholar]
- Divaris K, Monda KL, North KE, Olshan AF, Reynolds LM, Hsueh WC, Lange EM, Moss K, Barros SP, Weyant RJ, et al. 2013. Exploring the genetic basis of chronic periodontitis: a genome-wide association study. Hum Mol Genet. 22(11):2312–2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eke PI, Dye BA, Wei L, Slade GD, Thornton-Evans GO, Borgnakke WS, Taylor GW, Page RC, Beck JD, Genco RJ. 2015. Update on prevalence of periodontitis in adults in the United States: NHANES 2009 to 2012. J Periodontol. 86(5):611–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eke PI, Page RC, Wei L, Thornton-Evans G, Genco RJ. 2012. Update of the case definitions for population-based surveillance of periodontitis. J Periodontol. 83(12):1449–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitag-Wolf S, Dommisch H, Graetz C, Jockel-Schneider Y, Harks I, Staufenbiel I, Meyle J, Eickholz P, Noack B, Bruckmann C, et al. 2014. Genome-wide exploration identifies sex-specific genetic effects of alleles upstream NPY to increase the risk of severe periodontitis in men. J Clin Periodontol. 41(12):1115–1121. [DOI] [PubMed] [Google Scholar]
- GTEx Consortium. 2015. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 348(6235):648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G. 2015. Periodontitis: from microbial immune subversion to systemic inflammation. Nat Rev Immunol. 15(1):30–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong KW, Shin MS, Ahn YB, Lee HJ, Kim HD. 2015. Genomewide association study on chronic periodontitis in Korean population: results from the Yangpyeong health cohort. J Clin Periodontol [epub ahead of print 25 Jul 2015] in press. [DOI] [PubMed] [Google Scholar]
- Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. 2012. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 44(8):955–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie BN, Donnelly P, Marchini J. 2009. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 5(6):e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap 3 Consortium. 2010. Integrating common and rare genetic variation in diverse human populations. Nature. 467(7311):52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimbux NY, Saraiya VM, Elangovan S, Allareddy V, Kinnunen T, Kornman KS, Duff GW. 2012. Interleukin-1 gene polymorphisms and chronic periodontitis in adult whites: a systematic review and meta-analysis. J Periodontol. 83(11):1407–1419. [DOI] [PubMed] [Google Scholar]
- Laurie CC, Doheny KF, Mirel DB, Pugh EW, Bierut LJ, Bhangale T, Boehm F, Caporaso NE, Cornelis MC, Edenberg HJ, et al. 2010. Quality control and quality assurance in genotypic data for genome-wide association studies. Genet Epidemiol. 34(6):591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavange LM, Kalsbeek WD, Sorlie PD, Avilés-Santa LM, Kaplan RC, Barnhart J, Liu K, Giachello A, Lee DJ, Ryan J, et al. 2010. Sample design and cohort selection in the Hispanic Community Health Study/Study of Latinos. Ann Epidemiol. 20(8):642–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmueller KE, Pearce CL, Pike M, Lander ES, Hirschhorn JN. 2003. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat Genet. 33(2):177–182. [DOI] [PubMed] [Google Scholar]
- Machiela MJ, Chanock SJ. 2015. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 31(21):3555–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marth GT, Yu F, Indap AR, Garimella K, Gravel S, Leong WF, Tyler-Smith C, Bainbridge M, Blackwell T, Zheng-Bradley X, et al. 2011. The functional spectrum of low-frequency coding variation. Genome Biol. 12(9):R84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalowicz BS, Diehl SR, Gunsolley JC, Sparks BS, Brooks CN, Koertge TE, Califano JV, Burmeister JA, Schenkein HA. 2000. Evidence of a substantial genetic basis for risk of adult periodontitis. J Periodontol. 71(11):1699–1707. [DOI] [PubMed] [Google Scholar]
- Nikolopoulos GK, Dimou NL, Hamodrakas SJ, Bagos PG. 2008. Cytokine gene polymorphisms in periodontal disease: a meta-analysis of 53 studies including 4178 cases and 4590 controls. J Clin Periodontol. 35(9):754–767. [DOI] [PubMed] [Google Scholar]
- Offenbacher S, Divaris K, Barros SP, Moss KL, Marchesan JT, Morelli T, Zhang S, Kim S, Sun L, Beck JD, et al. 2016. Genome-wide association study of biologically-informed periodontal complex traits offers novel insights into the genetic basis of periodontal disease. Hum Mol Genet. 25(10):2113–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panoutsopoulou K, Tachmazidou I, Zeggini E. 2013. In search of low-frequency and rare variants affecting complex traits. Hum Mol Genet. 22(R1):R16–R21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodin K, Divaris K, North KE, Barros SP, Moss K, Beck JD, Offenbacher S. 2014. Chronic periodontitis genome-wide association studies: gene-centric and gene set enrichment analyses. J Dent Res. 93(9):882–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem MR, Toyama T, Wong AY, Jacobs HK, Bennett EJ. 1978. Haemodynamic responses to induced arterial hypotension in children. Br J Anaesth. 50(5):489–494. [DOI] [PubMed] [Google Scholar]
- Shaffer JR, Polk DE, Wang X, Feingold E, Weeks DE, Lee MK, Cuenco KT, Weyant RJ, Crout RJ, McNeil DW, et al. 2014. Genome-wide association study of periodontal health measured by probing depth in adults ages 18–49 years. G3 (Bethesda). 4(2):307–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang D, Dong L, Zeng L, Yang R, Xu J, Wu Y, Xu R, Tao H, Zhang N, et al. 2015. Two-stage comprehensive evaluation of genetic susceptibility of common variants in FBXO38, AP3B2 and WHAMM to severe chronic periodontitis. Sci Rep. 5:17882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu S, Momozawa Y, Takahashi A, Nagasawa T, Ashikawa K, Terada Y, Izumi Y, Kobayashi H, Tsuji M, Kubo M, et al. 2015. A genome-wide association study of periodontitis in a Japanese population. J Dent Res. 94(4):555–561. [DOI] [PubMed] [Google Scholar]
- Shriner D, Adeyemo A, Gerry NP, Herbert A, Chen G, Doumatey A, Huang H, Zhou J, Christman MF, Rotimi CN. 2009. Transferability and fine-mapping of genome-wide associated loci for adult height across human populations. PLoS One. 4(12):e8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shungin D, Cornelis MC, Divaris K, Holtfreter B, Shaffer JR, Yu YH, Barros SP, Beck JD, Biffar R, Boerwinkle EA, et al. 2015. Using genetics to test the causal relationship of total adiposity and periodontitis: Mendelian randomization analyses in the Gene-Lifestyle Interactions and Dental Endpoints (GLIDE) Consortium. Int J Epidemiol. 44(2):638–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlie PD, Avilés-Santa LM, Wassertheil-Smoller S, Kaplan RC, Daviglus ML, Giachello AL, Schneiderman N, Raij L, Talavera G, Allison M, et al. 2010. Design and implementation of the Hispanic Community Health Study/Study of Latinos. Ann Epidemiol. 20(8):629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teumer A, Holtfreter B, Völker U, Petersmann A, Nauck M, Biffar R, Völzke H, Kroemer HK, Meisel P, Homuth G, et al. 2013. Genome-wide association study of chronic periodontitis in a general German population. J Clin Periodontol. 40(11):977–985. [DOI] [PubMed] [Google Scholar]
- Wu X, Offenbacher S, Lόpez NJ, Chen D, Wang HY, Rogus J, Zhou J, Beck J, Jiang S, Bao X, et al. 2015. Association of interleukin-1 gene variations with moderate to severe chronic periodontitis in multiple ethnicities. J Periodontal Res. 50(1):52–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Gamazon ER, Zhang X, Konkashbaev A, Liu C, Szilágyi KL, Dolan ME, Cox NJ. 2015. SCAN database: facilitating integrative analyses of cytosine modification and expression QTL. Database (Oxford). 2015. pii:bav025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.