Abstract
MYO5A is a major actin-based vesicle transport motor that binds to one of its cargos, the melanosome, by means of a RAB27A/MLPH receptor. When one of the members of this receptor–motor complex is mutated, the melanosomes clump in the perinuclear region of the melanocyte and are transferred unevenly to the developing hair, leading to a dilution of coat color. Mutation of a fourth gene, dilute suppressor (dsu), suppresses this coat color dilution. MYO5A is required for the peripheral accumulation of melanosomes in melanocytes, but its role in melanosome transfer to neighboring keratinocytes and the hair is unknown. Here, we show that MYO5A is nonessential for melanosome transfer, although pigment incorporation into the hair in MYO5A-deficient mice is uneven, probably due to the clumping of melanosomes that occurs in the perinuclear region of mutant melanocytes. We also show that dsu is caused by a loss-of-function mutation in a unique vertebrate-specific protein that appears to function in an MYO5A-independent pathway to alter pigment incorporation into the hair. Therefore, dsu identifies a unique protein involved in pigmentation of the mammalian hair.
Keywords: melanosome transfer, myosin 5A, vesicle transport
In 1983, H. Sweet (1) reported the identification of one of the first suppressor mutations in mammals, dilute suppressor (dsu); dsu was identified by its ability to reverse the diluted coat color of mice carrying a hypomorphic Myo5a allele encoded at the dilute locus (dv/dv) to nearly WT (1). dsu is located on chromosome 1 and is unlinked to dilute, which is on chromosome 9. Subsequent studies showed that dsu is inherited in a semidominant manner; dv/dv, dsu/+ mice are lighter than dv/dv, dsu/dsu mice but darker than dv/dv, +/+ mice (2). dsu also suppresses the diluted coat color of mice carrying a null allele of dilute (dl20J). Thus, however dsu acts, it must somehow compensate for the loss of the dilute-encoded MYO5A motor (2). The MYO5A motor, when bound to melanosomes, allows them to be captured and locally transported on actin filaments in the melanocyte dendritic tips, a region of the melanocyte that is particularly dense in actin filaments, while an as yet undefined microtubule-based motor transports melanosomes to the dendritic tips where they can be captured on actin filaments by MYO5A. When MYO5A is mutated, the melanosomes are not captured, and they concentrate in the perinuclear region, the region of the cell with the highest concentration of microtubule tracks.
Two other mouse coat color mutations, ashen (ash) and leaden (ln), have phenotypes that are similar to dilute. This observation is not surprising because ash and ln encode RAB27A (3) and MLPH (4), respectively, two proteins that together constitute the receptor for MYO5A on the melanosome membrane (5–7). When one of these two proteins are mutated, MYO5A cannot bind to the melanosome, the melanosomes are not captured in the dendritic tips, and the melanosomes clump in the perinuclear region of the cell as they do in dilute melanocytes. dsu also suppresses the diluted coat color of ash and ln mice (8), although suppression is not as complete as with dilute.
dsu does not suppress the diluted coat color of 14 other mouse mutations that dilute pigment in other ways (9). Unexpectedly, however, dsu does suppress the ruby eye color of ruby-eye (ru) and ruby-eye-2 (ru2) mice to nearly black (9). Histological examination of the eyes of these mice indicates that dsu suppresses the eye color by increasing the overall level of pigmentation in the choroid but not the retinal pigment epithelium (9). Choroidal melanocytes, like those in the skin, are derived from the neural crest, whereas melanocytes in the retinal pigment epithelium are derived from the neuroectoderm. dsu therefore might act specifically on neural crest-derived melanocytes.
To determine whether dsu is a cell autonomous or nonautonomous suppressor, chimeric mice were generated by aggregating dv/dv embryos with dv/dv, dsu/dsu, c2J/c2J triple-mutant embryos (10). These chimera experiments suggested that dsu is a cell autonomous suppressor that works in the context of the melanocyte in the skin and is not a diffusible product.
In yeast, a high copy suppressor of MYO2, a homolog of mouse Myo5a, has been identified that encodes a kinesin-related gene, SMY1 (11). Myo2 protein (Myo2p) normally localizes to the sites of active growth, such as the bud tip and the mother-daughter neck, during cytokinesis (12) and has been implicated in the delivery of vacuoles to the emerging bud (13, 14). Smy1p does not require microtubules for suppression (15) but instead exerts its effects on Myo2p by means of its physical interaction with Myo2p (16).
Based on the yeast data, we hypothesized that dsu might result from a regulatory mutation that results in the up-regulation of another motor that somehow can compensate for the loss of the MYO5A motor. However, because dsu suppresses a dilute null allele, it must act differently from Smy1p. Here, we positionally clone dsu and show that it does not encode another motor. Instead, the WT product of dsu appears to function in a MYO5A-independent pathway that affects pigment incorporation in the developing hair rather than in actin capture and localized transport in the dendritic tips.
Materials and Methods
Mice. The mice used in these studies were maintained and propagated at the National Cancer Institute.
Genetic and Physical Mapping. Genetic and yeast artificial chromosome (YAC)-based maps of the dsu-critical region were generated as described in ref. 17. Bacterial artificial chromosomes (BACs) were identified by screening a 129/Sv BAC library (ResGen) with YAC end clones and YAC 414E2-derived markers (see below) from the dsu-critical region. A BAC contig was assembled as described in ref. 3.
BAC Complementation. BAC DNAs were prepared and injected into dsu/dsu, dv/dv zygotes as described in ref. 18. Transgenic (Tg) founders were identified on the basis of Southern blot analysis of genomic DNA isolated from tail biopsies (19), and complementation was assessed by visual inspection for reversion to the dv diluted coat color.
dsu cDNA Cloning. WT mouse melanocyte melan-a (20) randomand oligo(dT)-primed cDNA libraries (Stratagene) were screened with the mouse expressed sequence tag (EST) vl03b12.r1 (GenBank accession no. AA670621). Sequence from the positive clones corresponded to bases 448–2,291 of the WT dsu transcript, and the largest clone identified (RGA10a) spanned that entire region. RACE was performed to acquire more upstream sequence.
dsu Expression. Northern blots were purchased from OriGene Technologies (Rockville, MD) (adult tissues) or Clontech (embryonic tissues). dsu expression in mutant tissues was determined as described in ref. 4 by using tissues from 6-wk-old C57BL/6 and dsu/dsu mice as well as the melan-a cell line (20). Hybridization was performed as described in ref. 19 for Southern blotting by using the RGA10a clone as probe.
Western Blotting. Whole-cell extracts of primary melanocytes cultured from the skin of WT and dsu/dsu, dv/dv mice (>98% pure; ref. 21) were resolved on a 4–20% gradient SDS/PAGE, transferred to nitrocellulose, probed with a 1:3,000 dilution of anti-DSU antiserum, and processed for enhanced chemiluminescence-based Western blotting as described in ref. 22.
Harderian Gland Histology. Harderian glands from mouse pups that were euthanized on postnatal day 10–12 were dissected and fixed by overnight immersion in 4% paraformaldehyde and 2% glutaraldehyde in sodium cacodylate buffer (0.1 M, pH. 7.4). The tissue was rinsed thoroughly in cacodylate buffer, postfixed in 1% osmium tetroxide for 1 h at room temperature, and en bloc stained in 0.5% uranyl acetate in acetate buffer (0.1 M, pH 4.5). The tissue was dehydrated in graded ethanols and propylene oxide, infiltrated overnight in a 1:1 mixture of propylene oxide and epoxy resin, and embedded in pure resin that was then cured in a 55°C oven for 48 h. The cured block was trimmed, and semithin sections (0.5 μm) were cut and mounted on glass slides. Images of melanocytes were obtained by using a Zeiss Axiophot microscope attached to a Zeiss Axiocam digital camera running openlab 3.0 software.
Hair Preparation. Hair samples were plucked from the back of 4-wk-old mice and processed as described in ref. 23. They then were mounted by using Permount mounting medium (no. 17986-01, Electron Microscopy Sciences, Fort Washington, PA) and covered with a coverslip. After the mounting medium had completely polymerized, hair shafts were imaged under bright field optics with a Zeiss Axioplan microscope equipped with a ×40 phase objective.
Results and Discussion
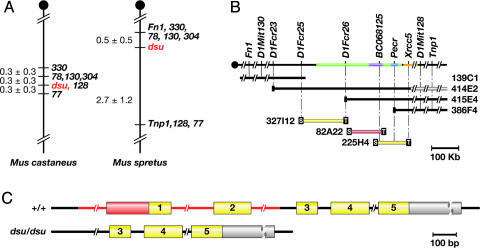
Genetic Map of the dsu-Critical Region. To initiate the positional cloning of dsu, we generated ≈1,600 (dsu/dsu, dv/dv × Mus castaneus) F1 × dsu/dsu, dv/dv progeny, which were phenotyped at weaning for dv by Southern blot analysis and for dsu by visual inspection (24). Seventy-five percent of the progeny from this cross carry an agouti allele inherited from the M. castaneus parent or were heterozygous for dv and had to be excluded because they could not be typed for dsu. Tail DNA from the remaining 417 animals that were homozygous dv and dsu then was typed for a series of simple sequence-length polymorphism markers that were predicted to map near dsu. These mapping studies defined a 0.6-cM dsu critical region flanked by D1Mit78, D1Mit130, and D1Mit304 on the proximal side and D1Mit77 on the distal side (Fig. 1A). We also generated 187 dsu informative animals from a (dsu/dsu, dv/dv × M. spretus) F1 × dsu/dsu, dv/dv cross and defined a 4-cM dsu-critical region (10) that was further narrowed to 3.2 cM (Fig. 1 A). Comparison of the two genetic maps showed that D1Mit78, D1Mit130, and D1Mit304 on one side and D1Mit77 on the other were located 0.6 cM apart in the M. castaneus cross but 3.2 cM apart in the M. spretus cross, implying a cold spot for recombination in the M. spretus cross or a hot spot in the M. castaneus cross. Two markers typed on the M. spretus cross, Fn1 and Tnp1, have been localized to the same 740-kb human YAC (25), although current mouse and human databases indicate that they are 1.5 Mb apart. Because 3.2 cM, the distance between Fn1 and Tnp1 in the M. spretus cross, corresponds to >5 Mb in the mouse, this infers a hot spot for recombination in the M. spretus cross.
Fig. 1.
Genetic and physical maps of dsu and the genomic structure dsu.(A) Genetic maps of dsu generated from an intersubspecific backcrosses to M. castaneus and an interspecific backcross to M. spretus, producing a dsu-critical interval of 0.6 and 3.2 cM, respectively, on mouse chromosome 1. (B) Physical map of the dsu critical region. YAC 414E2 is chimeric. Three BACs spanned the critical region, shown in green, which is ≈300 kb in length. The location of the markers listed, determined by using the May 2004 release of the mouse genome sequence (29), are as follows: D1Fcr23 (72, 242, 762–72, 245, 326), D1Fcr25 (falls within a 1.6-kb sequence gap located at 72,343,425–72,345,003), D1Fcr26 (72, 503, 480–72, 503, 649), intron of dsu represented by BC068125 (72, 633, 106–72, 633, 127), YAC 386F4 end clone contained within an intron of Pecr (72, 693, 607–72, 693, 628), and BAC 225H4 end clone contained within an intron of Xrcc5 (72, 769, 068–72, 769, 226). The extent of the genomic loci for BC068125 (dsu), Pecr, and Xrcc5 are shown in lavender, blue, and orange, respectively. BAC 82A22 (red) contains dsu.(C) Exon-intron structure of dsu in WT and mutant animals. The WT gene contains five exons. In dsu, the first two exons and surrounding noncoding region are deleted. The deleted region is shown in red. Coding regions are shown in yellow, and 5′ and 3′ UTR sequences are in orange and gray, respectively.
Physical Map of the dsu-Critical Region. Simple sequence-length polymorphism markers closely linked to dsu were used to screen a mouse YAC library (ResGen) (Fig. 1B). Several new unique-sequence probes from the YAC clones that spanned dsu were obtained and mapped back on the dsu genetic map. YAC 414E2 end clone (D1Fcr23) was the closest proximal marker and Xrcc5 the closest distal marker. Each segregated away from dsu by one recombinant in the M. spretus cross. These markers, in conjunction with other YAC end clones and unique-sequence probes, were used to screen a mouse BAC library. Additional unique-sequence probes from the BACs then were obtained and mapped back on the dsu genetic map. These studies defined a dsu-critical region that was spanned by three BACs and estimated to represent <500 kb of genomic DNA.
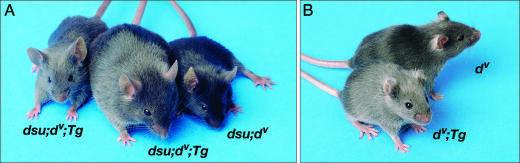
BAC Complementation. To further narrow the dsu-critical region, we performed BAC complementation. The three BACs that spanned the dsu-critical region were microinjected into dsu/dsu, dv/dv zygotes and the Tg founders examined for their coat color. In the case of BAC 82A22 (Fig. 1B), two BAC Tg founders were obtained that were gray in color (Fig. 2A and data not shown) unlike their nontransgenic littermates that were black in color due to the suppression of dv by dsu. BAC 82A22 was obtained from a WT BAC library and is therefore WT at dsu. Because this BAC reverses the suppression of dv by dsu, dsu most likely results from a loss-of-function mutation. Both BAC Tg founders were subsequently backcrossed to the dsu/dsu, dv/dv parent. Progeny that inherited the BAC were gray and not suppressed, whereas those that failed to inherit the BAC were black and suppressed (Fig. 2 A and data not shown). dsu is thus located on BAC 82A22.
Fig. 2.
BAC complementation and enhancement. (A) Suppression of dv by dsu is reversed in Tg mice carrying BAC 82A22 containing the WT dsu gene. A BAC Tg founder animal shown in the middle is homozygous for dv and dsu, carries BAC 82A22 and is dilute in color. Expression of the WT dsu gene thus reverses the suppression of dv by dsu. The BAC founder was crossed to a homozygous dv, dsu animal. The progeny animal on the left inherited the BAC and is dilute in color, whereas the progeny animal on the right did not inherit the BAC and is black because of suppression of dv by dsu.(B) BAC 82A22 acts as an enhancer of dv on a WT dsu background. The animal at the bottom is homozygous dv, WT dsu, and carries approximately four to eight copies of BAC 82A22. This animal is lighter gray in color than the animal at the top, which is homozygous dv, WT dsu, and lacks the BAC. Overexpression of the WT dsu gene thus enhances the dv phenotype.
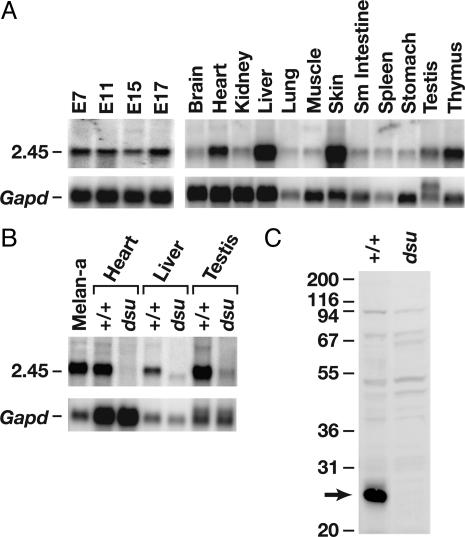
A dsu Candidate Gene. Sample sequencing of BAC 82A22 showed that it contained an unique EST (which is now represented by multiple ESTs in current databases) that is expressed in human fetal liver, spleen, mammary gland, and mouse blastocysts. No other intact genes encoding appreciable ORFs were identified on BAC 82A22, indicating that dsu is located in a gene-poor region of the genome. Northern blot analysis of embryonic and adult mouse tissues showed that this dsu candidate gene is expressed from embryonic days 7 to 17 and in all adult tissues examined with the highest level of expression in skin, heart, liver, testis, and thymus (Fig. 3A). This gene also is highly expressed in WT mouse melan-a melanocyte cells (Fig. 3B), consistent with chimera experiments suggesting that dsu encodes an autonomous suppressor that functions within the melanocyte (10).
Fig. 3.
Expression of dsu mRNA and protein in WT and mutant animals. (A) dsu mRNA expression in WT embryonic and adult mouse tissues. dsu is expressed throughout embryogenesis and in all adult tissues tested. (B) dsu is highly expressed in WT melan-a melanocytes in addition to heart, liver, and testis but is undetectable or very poorly expressed in dsu mutant tissues. (C) A polyclonal DSU Ab recognizes an ≈26-kDa protein in cultured WT melanocytes, which is absent in cultured dsu mutant melanocytes.
Sequence of the dsu Candidate Gene. Several dsu candidate gene cDNA clones were obtained from a melan-a cDNA library and sequenced, and the sequences were compared with the BAC sequence. These studies showed that the dsu candidate gene is encoded by five exons that are distributed over 53 kb of genomic DNA with an undefined 5′ UTR and a 1,648-nt 3′ UTR (Fig. 1C). Because the 5′ end of this gene is extremely GC-rich, we were unable to obtain the complete 5′ sequence by using cDNA library screening, 5′-RACE, or primer extension. From the BAC sequence, we were able to deduce a putative 5′ end with a good Kozak consensus sequence (26) for the initiator methionine, but we were unable to confirm it. blast searches did, however, identify a related human clone from a 5′-capped library derived from a 10-wk-old human embryo (GenBank accession no. AK000978). Translation of this sequence showed that it encoded a protein with 80% identity and 86% similarity to the mouse protein. The human gene contains six exons compared with five in the mouse and is located on chromosome 2 in a region syntenic with the mouse gene. The human initiation methionine aligns perfectly with the mouse initiation methionine predicted from the BAC sequence.
The dsu Candidate Gene Contains a Large Deletion in dsu Mice. Northern blot analysis of selected tissues from WT (C57BL/6) and dsu/dsu mutant mice showed that this candidate gene is not expressed, or is very poorly expressed, in mutant mice (Fig. 3B). This finding was subsequently confirmed by more sensitive RT-PCR experiments; no WT transcript could be detected in the mutant animals (data not shown). Western blot analysis of homozygous mutant dsu melanocytes by using a rabbit Ab raised against the entire DSU protein fused to GST (pGEX 2TEX) (27) showed that it also is not expressed at the protein level (Fig. 3C). Southern blot analysis by using a portion of exon 1 and all of exons 2 and 3 as probe indicated that dsu mutant animals carried some form of genomic rearrangement (data not shown). Genomic PCR and DNA sequencing studies showed that dsu animals carry a deletion, which begins in intron 2 and terminates 11.1-kb upstream of the initiator methionine (Fig. 1C). This deletion accounts for the lack of expression in dsu mice. These results are also consistent with the BAC complementation experiments, which suggest that dsu results from a loss-of-function mutation. This result was surprising because it indicates that loss of dsu function can compensate for the loss of an actin-based receptor–motor complex.
DSU Protein. Unfortunately, the sequence of dsu provides no clues about its function. WT dsu is predicted to encode a small 214-aa protein that is vertebrate specific. It is not present in yeast, worms, or flies, but it is present in all vertebrates examined. This finding is consistent with recent studies indicating that many pigment genes, which regulate vesicle trafficking, are vertebrate specific (28). DSU protein does not resemble any known motor proteins or any known transcription factors that might regulate motor activity. It also does not have any known functional domains. Although it is still possible that dsu loss might affect the expression of another motor that can substitute for the loss of MYO5A, our results raise the interesting possibility that dsu functions in a pathway separate from MYO5A.
dsu Is Dosage-Sensitive. dsu is semidominantly inherited (2), yet it results from a loss-of-function mutation. This characteristic suggests that dsu is dosage-sensitive: Loss of one copy of dsu partially suppresses the diluted coat color of dv/dv mice, whereas loss of two copies suppresses the coat to nearly WT. To determine whether overexpression of WT dsu might enhance the diluted coat color of dv mice, we bred animals from both of the dsu/dsu, dv/dv, BAC Tg/+ lines produced in the BAC complementation experiments to mice that were WT at dsu but homozygous for dv and then intercrossed the progeny and selected animals that were dv/dv, BAC Tg/BAC Tg and WT at dsu. The animals produced from both of the lines are lighter than homozygous dv mice that lack the BAC (Fig. 2B, data not shown). These mice carry four to eight extra copies of the WT dsu gene inherited on the BAC (data not shown). Overexpression of WT dsu therefore acts as an enhancer of dv.
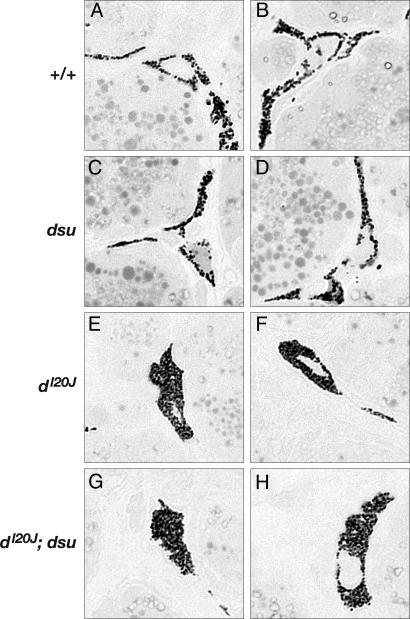
dsu Does Not Affect the Distribution of Melanosomes in dl20J Melanocytes. Histological examination of Harderian gland melanocytes has been the method of choice for examining melanocytes in mutant mice. Previous studies suggested that dsu loss can partially restore the peripheral distribution of melanosomes in Harderian gland melanocytes of d, ash, and ln mice, indicating that dsu loss somehow compensates for the loss of the MYO5A motor (8). Given our observations suggesting that WT dsu might function in a MYO5A-independent pathway, we decided to repeat these experiments, this time using a null allele of dilute (dl20J). Previous experiments used a hypomorphic allele (dv), which expresses a small amount of MYO5A protein. Thin sections of Harderian glands from WT mice contain many melanocytes with long thin dendritic processes that are filled with pigmented melanosomes (Fig. 4 A and B). The same is true of the Harderian glands of dsu/dsu mutant mice (Fig. 4 C and D), consistent with previous studies indicating that dsu has no obvious coat-color phenotype on its own. As expected, virtually all of the melanosomes in dl20J/dl20J melanocytes are clumped in the perinuclear region of the cell (Fig. 4 E and F). Surprisingly, dsu had no obvious effect on the peripheral distribution of melanosomes in dl20J/dl20J melanocytes (Fig. 4 G and H). The same is true for dl20J/dl20J, dsu/dsu skin melanocytes grown in culture (X.S.W. and J.A.H., unpublished results). To determine whether the differences seen in these experiments are due to the hypomorphic nature of the dv allele used in the published experiments, we repeated the experiments using the dv allele. Again, we failed to observe any obvious effect of dsu on the peripheral distribution of melanosomes in dv melanocytes (data not shown). Although it is unclear why previous experiments showed such an effect, the results presented here provide further support for the hypothesis that dsu operates in a MYO5A-independent pathway and dsu loss does not help restore MYO5A-mediated capture and local transport of melanosomes within the dendritic tips of mutant melanocytes.
Fig. 4.
dsu does not induce the peripheral distribution of melanosomes in Harderian gland melanocytes isolated from homozygous dl20J null mice. (A and B) Melanosomes in WT melanocytes (two examples shown) extend throughout the cytoplasm, reaching into the dendritic processes of the cell. (C and D) Compared with WT, the distribution of melanosomes is not altered in homozygous dsu mutant melanocytes. (E and F) Melanosomes are clumped around the perinuclear region of the cell in melanocytes that are homozygous for the dilute null allele, dl20J. (G and H) The abnormal distribution of melanosomes is not corrected by loss of dsu in dsu/dsu, dl20J/dl20J melanocytes. (Original magnification: ×100.)
The MYO5A Motor Is Not Essential for Melanosome Transfer to the Hair. The MYO5A motor is essential for the capture and local transport of melanosomes in dendritic tips (7), but its role in melanosome transfer to neighboring keratinocytes and the hair is unknown. If the MYO5A motor were essential for melanosome transfer to neighboring keratinocytes and the hair, then we would expect the hairs of dilute mice to be white, which they are not. To the contrary, studies completed >50 years ago suggest that the hairs of dv mice contain more pigment than the hairs of WT mice (30).¶ However, some of the pigment in dv hair is deposited in very large clumps (31), which have little effect on light absorption (30). This feature is thought to help explain why dv mice appear diluted. Given the experiments described here, we decided to reexamine the role of the MYO5A motor in melanosome transfer to the hair.
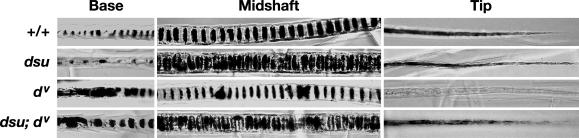
Hair samples were plucked from the backs of 4-wk-old +/+ and dv/dv mice and cleared by means of immersion in several different alcohol solutions before being mounted onto glass slides (23). Twenty hairs from each genotype then were scored blindly from the base to the tip. The base was scored for the presence or absence of pigment clumping. In the midshaft, the spacing of pigment bands (normal or small) was scored in addition to the presence or absence of pigment clumps. For the tip, pigment amounts were quantified visually and graded from – to +++ corresponding to their darkness. In WT hair, pigment is incorporated into the hair in a regularly repeated pattern, and the pigment extends all the way to the tip (+++ score) (Fig. 5). In dv hair, at the base and midshaft, the pigment is clumped together into larger aggregates, but the spacing of pigment bands is normal (Fig. 5). The amount of pigment in dv hair is not greatly reduced compared with WT hair as previously reported, except at the tip. In dv hair, the tip is completely devoid of pigment (– score). The results were highly reproducible, and there was little variability within a genotypic class (data not shown). These results indicate that MYO5A is not essential for melanosome transfer to the hair, although it is required for the even incorporation of pigment into the hair. This finding is not surprising given that most melanosomes in dv melanocytes are clumped in the perinuclear region of the cell. The function of MYO5A therefore may be to help ensure that melanosomes are present in the dendritic tips so that pigment can be evenly transferred and incorporated into the developing hair.
Fig. 5.
Pigment distribution in the hair of dv and dsu mice. Base, midshaft, and tips of the underhairs of WT, dsu/dsu, and dv/dv mice as well as dsu/dsu, dv/dv double mutant mice were photographed at X40 after clearing in various alcohols. Shown are representative results for 20 hairs of each genotype.
dsu Loss Alters Pigment Incorporation in the Hair. The molecular and cellular mechanisms involved in melanosome transfer from melanocytes to neighboring keratinocytes and their eventual incorporation into the hair are poorly understood, although a recent study by Seiberg (32) suggest that phagocytosis may be the preferred route. Phagocytosis is a receptor-mediated event that is initiated by the interaction of specialized membrane-bound receptors with specific molecules that are localized on the surface of the particle undergoing transfer. This interaction leads to receptor clustering, which generates a “phagocytic” signal that includes activation of signal transduction pathways. This signaling eventually triggers the local reorganization of the submembrane actin-based cytoskeleton, which is essential for engulfment (33). In skin keratinocytes, the protease-activated receptor 2 controls melanosome ingestion and phagocytosis, but the downstream components of this pathway and the nature of the melanosome ligand are unclear (32). Time-lapse video microscopy indicates that once pigment is internalized, single large melanosomes, or complexes of small melanosomes, are dispersed from the aggregate into the keratinocyte cytoplasm where they are eventually incorporated into the hair.
Given our experiments suggesting that dsu might operate in a MYO5A-independent pathway, we decided to determine whether dsu functions in melanosome transfer to the hair rather than melanosome capture and local transport within the melanocyte dendritic tip. To do this, we examined the distribution of pigment in dsu/dsu and dsu/dsu, dv/dv double mutant hair. As shown in Fig. 5, the dsu mutation rescues most of the pigmentation at the tip in dsu/dsu, dv/dv double-mutant hair (++ score), but it does not rescue clumping in the base or in the midshaft. Rather, dsu contributes its own defect: making smaller spacing between bands in both dsu/dsu and dsu/dsu, dv/dv double-mutant hair (Fig. 5). This characteristic, in combination with increased pigment at the tip, makes the coat look darker in dsu/dsu, dv/dv mice as compared with dv/dv mice. Although the dsu mutation has no effect on coat color in WT mice (to one's eye), it clearly affects band spacing.
Function of dsu in Melanosome Transfer and Pigment Incorporation in the Hair. One way the WT product of dsu might function is in phagocytosis. dsu loss could alter the phagocytic process so that the clumped melanosomes found in dv melanocytes are incorporated more evenly or better dispersed into neighboring keratinocytes and the hair. If this were the case, the chimera experiments would argue that the melanosomes ingested from dv melanocytes must be somehow structurally different from those ingested from WT melanocytes due to the loss of the DSU protein. Neither of these possibilities is consistent, however, with the hair data, which show that pigment is still clumped in the base and midshaft of dsu/dsu, dv/dv hair. dsu loss also might affect the way pigment is incorporated into the developing hair. This hypothesis is consistent with the hair data showing closer spacing between pigment bands and incorporation of pigment into the tips of dsu/dsu, dv/dv double mutant hair.
Conclusions. By using positional cloning, we have identified a unique vertebrate-specific protein that affects the incorporation of pigment into the hair rather than the capture and local transport of melanosomes within the dendritic tips. WT dsu has no domains of its own that would provide clues to its function, and its role in pigmentation would not have been identified by sequence gazing alone. Our studies demonstrate the power of forward genetics for identifying genes that affect pigmentation of the mammalian hair. Additional experiments involving dsu are likely to shed new light on the pigment-transfer process itself, a process that is little studied and little understood.
Acknowledgments
We thank Holly Morris, Rob Koogle, Joanne Dietz, Erika Truffer, Stephanie Springer, and Fran Dorsey for mouse care and maintenance; Kunio Nagashima and Michelle Gignac for generating the Harderian gland thin sections; Richard Frederickson for assistance in figure preparation; and Richard Yip for help in the construction and analysis of the genetic crosses and the BAC rescue experiments. This work was supported by the National Institutes of Health.
Author contributions: X.S.W., J.A.H., N.G.C., and N.A.J. designed research; T.N.O., X.S.W., R.A.R., J.-D.H., D.A.S., L.E.M., and J.A.H. performed research; T.N.O., X.S.W., R.A.R., J.A.H., N.G.C., and N.A.J. analyzed data; and N.G.C. and N.A.J. wrote the paper.
Abbreviations: BAC, bacterial artificial chromosome; YAC, yeast artificial chromosome; Tg, transgenic.
Data deposition: The mouse dsu mRNA sequence described in this paper has been deposited in the GenBank database (accession no. AY628210).
Footnotes
Brauch, L.R. & Russell, W. L. (1946) Genetics 31, 212 (abstr.).
References
- 1.Sweet, H. O. (1983) J. Hered. 74, 305–306. [DOI] [PubMed] [Google Scholar]
- 2.Moore, K. J., Seperack, P. K., Strobel, M. C., Swing, D. A., Copeland, N. G. & Jenkins, N. A. (1988) Proc. Natl. Acad. Sci. USA 85, 8131–8135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilson, S. M., Yip, R., Swing, D. A., O'Sullivan, T. N., Zhang, Y., Novak, E. K., Swank, R. T., Russell, L. B., Copeland, N. G. & Jenkins, N. A. (2000) Proc. Natl. Acad. Sci. USA 97, 7933–7938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matesic, L. E., Yip, R., Reuss, A. E., Swing, D. A., O'Sullivan, T. N., Fletcher, C. F., Copeland, N. G. & Jenkins, N. A. (2001) Proc. Natl. Acad. Sci. USA 98, 10238–10243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hume, A. N., Collinson, L. M., Hopkins, C. R., Strom, M., Barral, D. C., Bossi, G., Griffiths, G. M. & Seabra, M. C. (2002) Traffic 3, 193–202. [DOI] [PubMed] [Google Scholar]
- 6.Fukuda, M., Kuroda, T. S. & Mikoshiba, K. (2002) J. Biol. Chem. 277, 12432–12436. [DOI] [PubMed] [Google Scholar]
- 7.Wu, X. S., Rao, K., Zhang, H., Wang, F., Sellers, J. R., Matesic, L. E., Copeland, N. G., Jenkins, N. A. & Hammer, J. A., III (2002) Nat. Cell Biol. 4, 271–278. [DOI] [PubMed] [Google Scholar]
- 8.Moore, K. J., Swing, D. A., Rinchik, E. M., Mucenski, M. L., Buchberg, A. M., Copeland, N. G. & Jenkins, N. A. (1988) Genetics 119, 933–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moore, K. J., Swing, D. A., Copeland, N. G. & Jenkins, N. A. (1990) Genetics 125, 421–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moore, K. J., Swing, D. A., Copeland, N. G. & Jenkins, N. A. (1994) Genetics 138, 491–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lillie, S. H. & Brown, S. S. (1992) Nature 356, 358–361. [DOI] [PubMed] [Google Scholar]
- 12.Lillie, S. H. & Brown, S. S. (1994) J. Cell Biol. 125, 825–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Catlett, N. L. & Weisman, L. S. (1998) Proc. Natl. Acad. Sci. USA 95, 14799–14804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hill, K. L., Catlett, N. L. & Weisman, L. S. (1996) J. Cell Biol. 135, 1535–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lillie, S. H. & Brown, S. S. (1998) J. Cell Biol. 140, 873–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beningo, K. A., Lillie, S. H. & Brown, S. S. (2000) Mol. Biol. Cell 11, 691–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fletcher, C. F., Lutz, C. M., O'Sullivan, T. N., Shaughnessy, J. D., Jr., Hawkes, R., Frankel, W. N., Copeland, N. G. & Jenkins, N. A. (1996) Cell 87, 607–617. [DOI] [PubMed] [Google Scholar]
- 18.Antoch, M. P., Song, E. J., Chang, A. M., Vitaterna, M. H., Zhao, Y., Wilsbacher, L. D., Sangoram, A. M., King, D. P., Pinto, L. H. & Takahashi, J. S. (1997) Cell 89, 655–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jenkins, N. A., Copeland, N. G., Taylor, B. A., Bedigian, H. G. & Lee, B. K. (1982) J. Virol. 42, 379–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bennett, D. C., Cooper, P. J. & Hart, I. R. (1987) Int. J. Cancer 39, 414–418. [DOI] [PubMed] [Google Scholar]
- 21.Wu, X., Kocher, B., Wei, Q. & Hammer, J. A., III (1998) Cell Motil. Cytoskeleton 40, 286–303. [DOI] [PubMed] [Google Scholar]
- 22.Wu, X., Bowers, B., Wei, Q., Kocher, B. & Hammer, J. A., III (1997) J. Cell Sci. 110, 847–859. [DOI] [PubMed] [Google Scholar]
- 23.Russell, E. S. (1946) Genetics 31, 327–346. [PubMed] [Google Scholar]
- 24.Rinchik, E. M., Russell, L. B., Copeland, N. G. & Jenkins, N. A. (1986) Genetics 112, 321–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dib, C., Faure, S., Fizames, C., Samson, D., Drouot, N., Vignal, A., Millasseau, P., Marc, S., Hazan, J., Seboun, E., et al. (1996) Nature 380, 152–154. [DOI] [PubMed] [Google Scholar]
- 26.Kozak, M. (1999) Gene 234, 187–208. [DOI] [PubMed] [Google Scholar]
- 27.Jung, G., Remmert, K., Wu, X., Volosky, J. M. & Hammer, J. A., III (2001) J. Cell Biol. 153, 1479–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, W., Rusiniak, M. E., Chintala, S., Gautam, R., Novak, E. K. & Swank, R. T. (2004) BioEssays 26, 616–628. [DOI] [PubMed] [Google Scholar]
- 29.Waterston, R. H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J. F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. (2002) Nature 420, 520–562. [DOI] [PubMed] [Google Scholar]
- 30.Russell, E. S. (1948) Genetics 33, 228–236. [DOI] [PubMed] [Google Scholar]
- 31.Russell, E. S. (1949) Genetics 34, 146–166. [DOI] [PubMed] [Google Scholar]
- 32.Seiberg, M. (2001) Pigm. Cell. Res. 14, 236–342. [DOI] [PubMed] [Google Scholar]
- 33.Kwiatkowska, K. & Sobota, A. (1999) BioEssays 21, 422–431. [DOI] [PubMed] [Google Scholar]