Abstract
We developed a quantitative methodology, digital analysis of chromatin structure (DACS), for high-throughput, automated mapping of DNase I-hypersensitive sites and associated cis-regulatory sequences in the human and other complex genomes. We used 19/20-bp genomic DNA tags to localize individual DNase I cutting events in nuclear chromatin and produced ≈257,000 tags from erythroid cells. Tags were mapped to the human genome, and a quantitative algorithm was applied to discriminate statistically significant clusters of independent DNase I cutting events. We show that such clusters identify both known regulatory sequences and previously unrecognized functional elements across the genome. We used in silico simulation to demonstrate that DACS is capable of efficient and accurate localization of the majority of DNase I-hypersensitive sites in the human genome without requiring an independent validation step. A unique feature of DACS is that it permits unbiased evaluation of the chromatin state of regulatory sequences from widely separated genomic loci. We found surprisingly large differences in the accessibility of distant regulatory sequences, suggesting the existence of a hierarchy of nuclear organization that escapes detection by conventional chromatin assays.
Keywords: cis-regulatory elements, DNase I-hypersensitive sites, gene regulation
Comprehensive delineation of functional noncoding sequences in complex genomes is a major goal of modern biology. The activation and function of regulatory sequences is linked to focal alterations in chromatin structure (1, 2), which may be detected experimentally through hypersensitivity to DNase I in the context of nuclear chromatin. DNase I-hypersensitive sites (HSs) are the sine qua non of classical cis-regulatory elements, including promoters, enhancers, silencers, insulators, and locus control regions (3–9). Systematic mapping of DNase I HSs across the genome should therefore yield a comprehensive library of cis-regulatory elements, but is intractable with conventional approaches.
The feasibility of cloning DNase I HSs has recently been demonstrated by using both direct vector-assisted end cloning (10) and a subtractive enrichment approach (11). The former method is limited to quiescent cells and therefore cannot be used in the context of well-studied and widely used cell lines or other proliferating tissues. Moreover, neither method is well suited to efficient recovery of DNase I HSs on a genomewide scale because of the 1:1 mapping between cloning events and sequences; both require large amounts of sequencing and, critically, an independent molecular validation step for each candidate clone.
The application of tag-based or “digital” methodologies has revolutionized the study of transcriptome biology (12–14), enabling both the generation of genomewide data sets and insight into the tremendous dynamic range of gene expression.
Genomewide localization of DNase I HSs and associated cis-regulatory sequences requires development of a high-throughput approach that (i) can efficiently map millions of individual DNase I cutting events, (ii) can be applied to any cell type, and (iii) is self-validating (i.e., it can be applied to automatically map HSs at a high confidence level without requiring a subsequent, independent molecular validation step).
Here we describe an approach that combines molecular and computational methods to achieve these aims. We used 19/20-bp genomic DNA tags to localize individual DNase I cutting events in nuclear chromatin and discovered DNase I HSs by identifying statistically significant tag clustering events by using a quantitative algorithm. This method, digital analysis of chromatin structure (DACS), provides the framework for genome-wide localization of DNase I HSs and associated cis-regulatory sequences in an efficient, quantitative, and automated fashion, opening the door for systematic exposition of the regulatory genome.
Methods
Cell Culture and DNase I Digestion. We cultured K562 (ATCC) cells in RPMI medium 1640 (Invitrogen) supplemented with 10% FBS to a target density of 5 × 105 cells per ml. We performed DNase I (Roche Applied Sciences, Indianapolis) digestions (0.5–2 units per ml) according to the protocol described in ref. 15 and purified DNA by using the Puregene system (Gentra Systems).
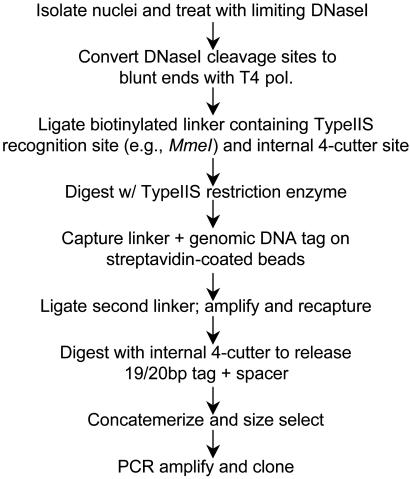
Creation of DACS Libraries. We developed a tagging method for identifying individual DNase I cut sites in nuclear chromatin. Fig. 1 shows a schematic of the protocol, further illustrated in Fig. 6, which is published as supporting information on the PNAS web site. Additional detailed protocol information, including sequences of all oligonucleotides, is provided in the Supporting Text, which is published as supporting information on the PNAS web site. After digestion of isolated nuclei with DNase I under hypersensitive treatment conditions, DNase-cut ends were repaired and ligated to a biotinylated linker adaptor containing dual restriction sites for a type IIs restriction endonuclease (MmeI) and a four-cutter enzyme (MluI), oriented such that the direction of cleavage of MmeI is toward the genomic DNA ligand. After linker ligation, the preparation was digested to completion with MmeI, which released the linker plus 19–20 bp of genomic DNA sequence (owing to a common 1-bp wobble in the MmeI indirect cut site). Linker/genomic DNA tag fragments were then isolated and purified over streptavidin-coated magnetic particles (Dynal, Great Neck, NY). A second linker adaptor containing a BsiWI site was ligated to the exposed genomic DNA end of each fragment bound to the beads. By using primers complementary to each of the linker sequences, the genomic DNA tags were PCR-amplified in situ while attached to the beads. The products of this amplification were collected and separated by electrophoresis through a 10% polyacrylamide gel, purified, and minimally reamplified. The second linker adaptor was released after BsiWI digestion, and the tags were recaptured on streptavidin-coated beads. Tags were cleaved from the beads by digestion with MluI. After purification, tags were ligated to form high-molecular-weight concatemers and size-selected over a 1.5% agarose gel. Fragments >500 bp in length were isolated and cloned into pGEM5z (Promega) in preparation for sequencing. Further detailed protocol information is provided in the Supporting Text.
Fig. 1.
Schematic of DACS library creation. See Methods and Supporting Text for description and Fig. 6 for additional illustration.
DNA Sequencing. All DNA sequencing reactions were carried out on MegaBace 4000 384-capillary sequencers (Amersham Biosciences) by using energy transfer chemistry. Sequences were analyzed with phred, and only those with high-quality base calls were subjected to further analysis.
Mapping of Tags to the Genome. Mapping of short sequence tags to the human genome is inefficient using conventional tools such as blat (16) and blast (17). For example, blat employs an index of nonoverlapping 11-mers and is unable to map sequences smaller than 22 bp. To circumvent these inefficiencies, we used a positional index of all 16-mers (including overlaps and reverse complements) in the human genome (M.H., unpublished work). We then used a multistep procedure to localize tags. First, all tags were examined for an initial exact match within the 16-mer database. Once an initial match was made, the program attempted to extend the match to the full length of the query tag sequence vs. the reference genome. If the initial 16-mer index search did not produce an exact match, subsequent searches of the tag were run and the number of exact subsequent matches to the reference genome, if any, was reported. This process enables 1- to 2-bp mismatch recognition, which is sufficient to account for sequencing errors and genomic variants.
Primer Selection. We designed primers to amplify ≈250-bp genomic segments spanning candidate HS sequences by using primer3 (18).
Analysis of DNase I Hypersensitivity by Hypersensitivity Quantitative PCR (HSqPCR). We used a real-time quantitative PCR-based method to quantify DNase I hypersensitivity in K562 cells as described in refs. 11 and 19.
Conventional DNase I Hypersensitivity Assays. Conventional DNase I hypersensitivity studies were performed by using the indirect end-label technique (20) according to a standard protocol described in ref. 21.
Microarray Expression Analysis. Gene expression analysis was performed on a Human 1A Oligo Microarray (Agilent, Palo Alto, CA). Total RNA was isolated from 5 × 107 K562 cells with an RNeasy total RNA isolation kit (Qiagen, Valencia, CA).
Results
Overview of Digital Analysis of Chromatin Structure. DACS is a hybrid molecular–computational methodology comprising two discrete phases: (i) production of a genomic DNA tag library encompassing individual DNase I-hypersensitive cut sites in nuclear chromatin and (ii) computational analysis of genomic tag distributions to identify statistically significant clusters and derive the underlying HS sequence. A schematic of the process of creating DACS tag libraries from DNase I-treated nuclear chromatin is provided in Fig. 1 and illustrated in Fig. 6. DACS tag libraries provide base-pair resolution of DNase I cutting events and are therefore highly complex compared with RNA-derived libraries. DACS also differs fundamentally from previously described genomic DNA tag-based methods (13, 22, 23) used to study gene expression or copy number in which tags are generated relative to restriction enzyme sites, and subsequent tag localization in the genome relies explicitly on prior knowledge of the restriction site “scaffold.”
Application of DACS to K562 Erythroid Cells. Using the method outlined in Fig. 1, we obtained 257,443 tags from K562 cells. Of these, 237,688 (92.3%) were distinct within the tag population, of which 235,523 (99.1%) could be mapped to the current build of the human genome (Table 1). The number of tags that could be assigned to unique genomic locations was 157,744 (65.1%), and only such tags were used in subsequent analyses. There were 2,290 unmappable tags that were more G+C-rich than the genome as a whole, compatible with derivation from difficult-to-sequence regions such as centromeres. There were 28,215 tags (11.9% of distinct, 13.64% of total) that mapped to 10 or more locations (Table 1). This class encompasses classical repetitive elements, which were thus markedly depleted relative to the genome as a whole, where they account for >40% of the landscape. Although redundant tags may theoretically result from independent DNase I cutting events at identical bases on different alleles, this phenomenon is expected to be rare. Observed tag redundancy is primarily a consequence of PCR amplification, although ≈94% of tags mapping <7 times to the genome were unique within the tag pool (Table 1), indicating amplification did not compromise overall complexity.
Table 1. Mapping DACS tags to the human genome.
| Matches in genome | No. of tags | % of total | No. of distinct tags | % redundant |
|---|---|---|---|---|
| 0 | 2,290 | 0.89 | 2,165 | 5.46 |
| 1 | 163,849 | 63.64 | 154,744 | 5.56 |
| 2 | 27,370 | 10.63 | 25,580 | 6.54 |
| 3 | 10,828 | 4.21 | 10,189 | 5.90 |
| 4 | 5,961 | 2.32 | 5,612 | 5.85 |
| 5 | 3,876 | 1.51 | 3,606 | 6.97 |
| 6 | 2,847 | 1.11 | 2,676 | 6.01 |
| 7 | 2,205 | 0.86 | 2,011 | 8.80 |
| 8 | 1,664 | 0.65 | 1,544 | 7.21 |
| 9 | 1,438 | 0.56 | 1,346 | 6.40 |
| ≥10 | 35,115 | 13.64 | 28,215 | 19.65 |
Of 257,443 tags, 255,153 (99.1%) could be mapped to the current human genome build (National Center for Biotechnology Information build 34/UCSC HG16). Tags are classified according to how many times each matches within the genome. Tags matching ≥10 times are grouped in the bottom row. “No. of distinct tags” refers to tags occurring at least once within each tag subpopulation. “% redundant” refers to the percentage of total tags occurring two or more times within each tag subpopulation (most readily explained as a consequence of PCR amplification during tag preparation). Only nonredundant tags within the singly matching subpopulation were considered in the analyses.
Identification of Statistically Significant Tag Clusters. A priori, DACS libraries are expected to contain three types of tags: (i) tags that derive specifically from HSs, (ii) tags that derive from cutting within DNase-sensitive (although not hypersensitive) domains, and (iii) random tags derived either from nonspecific DNase I cutting or from random fragmentation of genomic DNA occasioned during purification.
To localize DNase I HSs, we developed a two-phase quantitative algorithm to identify statistically significant tag clusters. The algorithm first considers windows of increasing size (100-bp increments) around each DACS tag and identifies cases in which the number of observed tags has exceeded the expected uniform distribution at a threshold of P < 0.001 (see Supporting Text). Next, the algorithm corrects the calculated significance for regions of the genome that have received higher numbers of cuts (e.g., extended open chromatin domains).
DNase I HSs typically comprise a core site-forming domain ≈150–250 bp in size, over which regulatory factor–DNA interactions take place (7, 24); however, the consequent chromatin disruption may give rise to a hypersensitive domain up to several hundred base pairs long including flanking sequences (21, 24). Furthermore, HSs are frequently clustered. As a first approximation of the average size of HS regions over which tag clustering should be evident, we therefore selected a window size of 1,250 bp (250 ± 500 bp). Using this window size, we identified 5,750 statistically significant clusters in the first phase of clustering and 3,714 clusters in the second phase after adjusting for regional differences in tag density. The residual refined set comprised 3,492 two-member clusters (2-clusters), 188 3-clusters, 28 4-clusters, and 10 5-clusters. Further analysis revealed that the algorithm detected no statistically significant 2-clusters where the distance between tags was >650 bp. Higher-order cluster classes were all significant within the full 1,250-bp window. Growth in tag clusters as a function of mapped tags is shown in Fig. 7, which is published as supporting information on the PNAS web site.
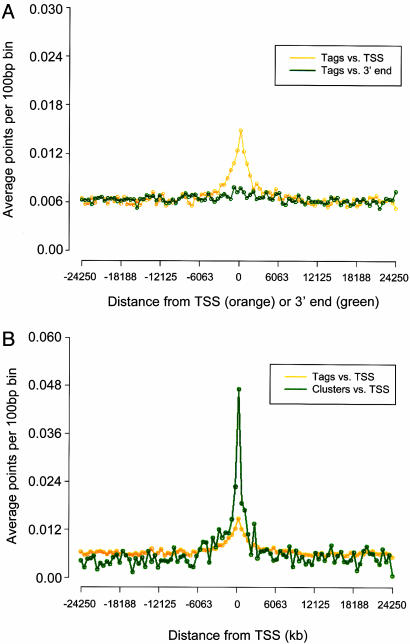
Enrichment at Genomic Landmarks Associated with Functional Elements. We observed marked enrichment of both tags and clusters around transcriptional start sites (TSSs), CpG islands, and evolutionarily conserved noncoding sequences (Fig. 2A and Fig. 8, which is published as supporting information on the PNAS web site).
Fig. 2.
Enrichment of DACS tags and clusters in genomic regions associated with regulation. y axes: average number of individual DACS tags or statistically significant tag clusters per 100-bp bin. x axes: normalized distance (kb) relative to genomic landmark. (A) Distribution of 154,744 distinct, uniquely mapping tags relative to TSSs (orange) and 3′ ends of ≈18,000 RefSeq genes (green). (B) Statistically significant tag clusters (green; n = 3,492) show markedly greater enrichment relative to TSSs vs. individual tags (orange).
DACS clusters displayed substantially greater enrichment around TSSs (Fig. 2B) and CpG islands (data not shown) than did tags alone, compatible with overall enrichment for HSs in these regions. Less marked enrichment was observed over conserved noncoding sequences (Fig. 8). DACS clusters were also preferentially enriched around expressed vs. nonexpressed genes, although the difference was not marked (Fig. 8).
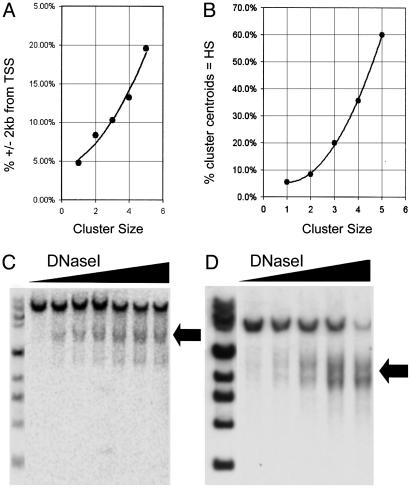
We also determined the proportion of clusters of a given size (i.e., tag number) that fell within ±2 kb of a TSS. This investigation revealed a marked and steady increase as a function of cluster size (Fig. 3A), suggesting a corresponding enrichment in functional elements in higher-density clusters.
Fig. 3.
Genomic localization of 257,443 DACS tags. (A and B) Predictive potential of clusters for HSs increases exponentially. (A) Percentage of cluster centroids within ±2 kb of an annotated RefSeq TSS as a function of cluster size (size = 1 denotes individual tags). (B) Percentage of cluster centroids that coincide precisely with DNase I HSs as a function of cluster size. The ability to correctly predict the location of the HS increases exponentially as a function of cluster size. (C and D) Previously unrecognized elements identified by DACS clusters correspond with classical DNase I HSs. Conventional DNase I hypersensitivity assays were performed to examine previously unrecognized elements identified by DACS. (C) Conventional hypersensitivity assay of the DACS-identified HS in an internal intron of LRBA (see Fig. 4E). (D) The HS element/DACS cluster shown in Fig. 4F (parental bands: 11.1-kb EcoN1 and 6-kb HindIII fragments, respectively).
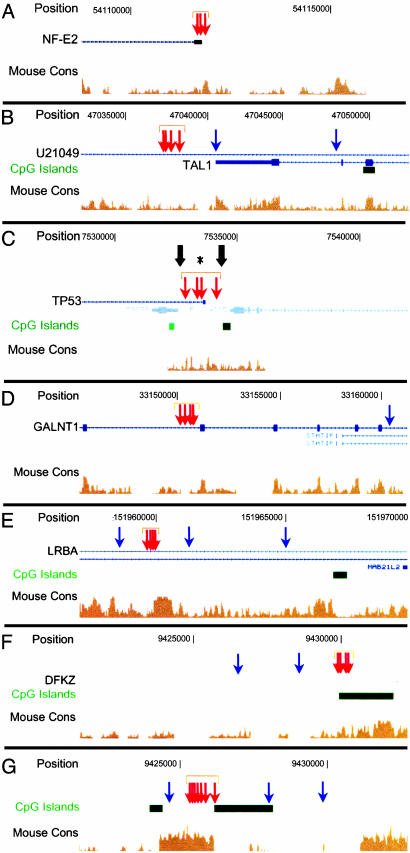
DACS Clusters Identify both Known Regulatory Sequences and Previously Unrecognized Functional Elements. Examples of unbiased identification by DACS clusters of both known and previously unrecognized functional elements are shown in Fig. 4. Examples of known cis-regulatory elements identified included the promoter of the erythroid-specific transcription factor NF-E2 gene (chr12), the major 3′ enhancer of the hematopoietic-specific stem cell leukemia gene (SCL or TAL1) on chr1 (25), and the p53 promoter complex (chr17) (26) (Fig. 4 A–C). Examples of previously unrecognized elements are shown in Fig. 4 D–F. Interestingly, one of these elements lies within a highly conserved sequence block on chr2 located >100 kb from any known gene; however, an adjacent block of equal size and conservation is not an HS. Selected elements identified by DACS and confirmed with HSqPCR were further validated with conventional hypersensitivity assays, demonstrating the correspondence between DACS-identified elements and classical DNase I HSs (Fig. 3 B and C).
Fig. 4.
DACS tag clusters identify known and previously unrecognized functional elements. Tag positions (orange and yellow vertical arrows) are shown relative to chromosomal position, known genes (blue), CpG islands (green), and human–mouse conservation (brown). Statistically significant tag clusters are identified with orange arrows and horizontal brackets. (A–C) Examples of known regulatory elements identified de novo by DACS: promoter of erythroid-specific transcription factor NF-E2 gene (chr12) (A); TAL1/SCL 3′ enhancer (chr1) (26) (B); p53 promoter complex (chr17) (27) (C). Note that the computed cluster centroid (*) falls between two HSs (thick arrows). (D–G) Examples of previously unrecognized elements identified by DACS: element within intron of N-acetylgalactosaminyltransferase gene (GALNT1; chr18) (D); intronic element within lipopolysaccharide-responsive/beige-like anchor protein (LRBA; chr4) (E); cluster over CpG island 12 kb upstream of gene of unknown function on chr3 (F); cluster over highly conserved sequence block on chr2 located >100 kb from any known gene (G). Repeated tagging of specific functional elements in advance of others suggests a discrete hierarchy of nuclear chromatin organization that escapes detection with conventional assays.
Cluster Centroids Localize HS Core Sequences. In principle, the spatial distribution of tags around an HS should permit localization of the HS core domain where hypersensitivity is maximal. In practice, however, the precision is limited by the number of tags available for analysis in any given region.
We hypothesized that, for limiting numbers of tags, the cluster centroid would predict the location of the underlying HS core. On this assumption, the predictive value of the cluster as a whole is determined by two factors: (i) the probability that an HS is contained somewhere within the cluster domain (i.e., that it is a true positive cluster) and (ii) that the HS can be definitively positioned within the cluster bounds on the basis of the tag pattern alone. The latter factor is strictly a function of the density of tags within the cluster window. Both factors are, in turn, a function of the compactness of the cluster, namely, the genomic distance over which the tags are distributed.
To determine the approximate percentage of individual tags (vs. clusters) that overlapped a DNase I HS, we randomly selected 36 tags and assayed them for hypersensitivity in K562 cells by using HSqPCR. Of these, two (5.55%) coincided precisely with an HS. We then tested the centroids of 36 randomly selected clusters comprising two tags, of which three (8.3%) coincided with an HS. To test the correspondence between cluster centroids and HSs, we randomly selected 50% of each pool of clusters containing three, four, and five tags. We identified DNase I HSs at computed centroids of 18/90 (20%) 3-clusters, 5/14 (35.7%) 4-clusters, and 3/5 (60%) 5-clusters (Table 2, which is published as supporting information on the PNAS web site). These numbers reflect an exponential increase in the predictive value of clusters as a function of tag content (Fig. 3B). We also tested one 6-cluster and one 7-cluster, both of which were HSs. Because the 111 clusters tested were widely distributed across the genome, the results confirm a substantial global enrichment for DNase I HSs within DACS clusters.
For the cluster sizes we analyzed, the correspondence between cluster centroids and HSs is not expected to be perfect. The p53 promoter provides a salient example. The p53 gene contains two promoters: one located upstream of the first exon and a second more powerful promoter located within the proximal first intron (26), over which we detected significant tag clustering (Fig. 4C). Testing of the computed cluster centroid with HSqPCR, however, did not initially reveal an HS. To explore this result further, we tiled contiguous 250-bp amplicons across 1,500 bp encompassing the cluster. This tiling revealed the location of two DNase I HSs that coincided with the two previously described promoter elements. However, neither of these abutted the cluster centroid. This further confirmed our prediction that, given the relatively low density of tags in the cluster, the centroid only approximated the HS location. As such, we believe that the predictive values of each cluster size noted previously are likely to be underestimated, perhaps significantly.
Modeling Genome-Scale Discovery of Functional Elements with DACS. We sought to determine the number of tags required to map a given number of DNase I HSs within the human genome with a prespecified positive predictive value (PPV) threshold (i.e., the probability that the cluster centroid accurately predicts the location of an HS). This probability is a function of (i) the number of tags mapped, (ii) the proportion of individual tags in the input tag pool that coincide precisely with HSs (the “enrichment”), and (iii) the relative “intensity” of DNase I HSs.
To quantify the relationship between these variables, we programmed a simulation to determine the number of HSs that would be identified by using DACS at a fixed PPV threshold of 90%, as a function of both the tag pool enrichment and the number of mapped tags. We distributed 50,000 (nonoverlapping) model HSs against the complete human genome sequence, an estimate of the number of HSs that may be active in a particular differentiated cell type. HSs were distributed among noncoding, nonrepetitive regions. Further confinement of the model HSs relative to annotated features (e.g., TSSs) was not necessary because all combinations of nonoverlapping distributions are mathematically equivalent. In silico DACS tag pools of various levels of enrichment (5–20%) were then generated and mapped to the genome in a manner identical to that used to map the experimental tags. Tags that did not derive directly from model HSs were distributed quasirandomly, with a slight bias toward regions in the vicinity (±25 kb) of HSs (to reflect the predicted general accessibility of chromatin in these locales). Only uniquely mapping tags were considered. The simulation then (i) identified statistically significant tag clusters as described above, (ii) computed the centroids, and (iii) determined the proportion of cluster centroids for each cluster size (four tags, five tags, six tags, etc.) that overlapped model DNase I HSs. Finally, it selected the cluster size for which >90% of the centroids overlapped true-positive HSs (i.e., the 90% PPV threshold).
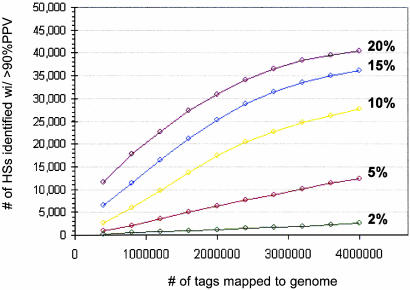
Fig. 5 shows, for a given level of enrichment and number of uniquely mapping tags, the number of DNase I HSs that were localized with a cumulative false-positive rate of <10%. As expected, this number grows rapidly as a function of mapped tags and then levels off as the density of background tags increases. However, the behavior is not asymptotic, because larger numbers of tags will eventually enable identification of most HSs in the population (data not shown). This simulation demonstrates the feasibility of quantitative, automated mapping of HSs in the context of the human genome at a high predictive accuracy threshold (>90%) and, furthermore, that this objective can be achieved with only modest levels of enrichment in the primary tag pool.
Fig. 5.
Modeling genome-scale discovery of HSs with DACS. Shown are results of an in silico simulation of DACS indicating the number of HSs (y axis) that would be identified at a fixed 90% PPV threshold as a function of the number of tags (x axis) mapped for a given input tag-population enrichment for HSs (colored curves; range 2–20%). DACS was simulated against a model genome in which 50,000 model DNase I HSs were distributed against the complete human genome sequence. As expected, the number of HSs predicted with >90% accuracy grows rapidly and then levels off. Larger numbers of tags will eventually enable identification of most HSs in the population (not shown).
Discussion
The development of “digital” or tag-based methodologies to quantify genomic phenomena, including gene expression (12–14) and copy numbers (22, 27), has had a major impact on genome annotation and the analysis of transcriptional regulation and genome dynamics. We have described a digital methodology capable of high-throughput, quantitative, and automatic identification of DNase I HSs and associated cis-regulatory sequences on a genomic scale. Previously described genomic tag-based methods rely on restriction-site scaffolds to simplify radically the genomic mapping process. By contrast, our results demonstrate the practicality of large-scale mapping of 19/20-bp sequences from unrestricted genomic locations, providing the potential for base-pair resolution.
Comprehensive Genomewide Mapping of DNase I HSs. The power of DACS for identification of DNase I HSs is determined principally by the number of tags mapped to the genome, subject to a given level of enrichment for HSs in the primary tag pool. As shown in Fig. 5, high levels of primary enrichment are not required to achieve comprehensive recovery of tens of thousands of DNase I HSs from across the genome. The cells we used (K562) in this pilot study were rapidly dividing, and BrdUrd labeling experiments revealed that ≈30% of cells in logarithmic-phase culture were actively undergoing replication (data not shown). Presumably, this S-phase fraction significantly compromised the observed enrichment (≈5%), because DNase I cutting over chromatin-disassembled S-phase genomes is expected to be largely nonspecific. K562 cells cannot easily be rendered quiescent, nor can they be synchronized, by using conventional methods. To improve enrichment, DACS may be applied to nondividing or synchronized cells, or it may be augmented by a subtractive enrichment step (11).
The precision with which DACS can localize HS core sequences is dictated by the correspondence between the tag pattern within clusters and the actual location of the HS core sequence. However, placed in perspective, even localizing HSs to an ≈1-kb interval would be of considerable value because this size is in the typical range of sequence elements selected for engineering into genetic vectors for further functional studies.
It is clear that analysis of several million tags will be required to produce a comprehensive genomic map of DNase I HSs for a given tissue. However, this figure is comparable to the number of ESTs in the dbEST EST database and is only a small fraction of the number of tag sequences that have been collectively analyzed by other methods, such as serial analysis of gene expression (SAGE). DACS tag pools of this size can be generated readily and economically by using either high-throughput sequencing of tag concatemers or by means of direct “signature” sequencing of DACS tags with massively parallel signature sequencing (MPSS) (28).
Chromatin Organization of Regulatory Sequences: A Quantitative Perspective. A unique feature of DACS is that it provides an unbiased view of the relative chromatin accessibility of functional elements located in widely separated genomic loci. We recovered large numbers of functional elements, including regulatory elements of erythroid- and hematopoietic-specific genes. The fact that the NF-E2 promoter and the SCL/TAL1 enhancer, for example, were repeatedly tagged in advance of other well described regulatory sequences, such as the β-globin locus control region, demonstrates how an unbiased approach can reveal the existence of a discrete hierarchy of chromatin accessibility of specific regulatory sequences. This hierarchy may reflect local structural features, or, more probably, the localization of specific elements in more accessible nuclear compartments. Systematic application of DACS therefore promises to add a new structural dimension to the analysis of cis-regulatory sequences and other functional elements and may provide telling insights into the structural biology of the nucleus.
Digital analysis of chromatin structure will enable systematic cataloging of cis-regulatory sequences in a wide spectrum of normal and diseased tissues and provide quantitative chromatin structural information for each element. Comparisons across tissues will enable classification of tissue-specific, multilineage, and constitutively active HSs. Analysis of differentiating or developing tissue axes should likewise enable identification of elements important in commitment to a given cellular program. Localization of regulatory sequences to small genomic intervals should considerably impact computational studies and the search for functional genetic variation, both of which have been hampered greatly by the vast genomic background.
Supplementary Material
Author contributions: P.J.S., M.M., and J.A.S. designed research; P.J.S., M.H., M.Y., A.S., J.K., R. Hall, J.M., M.O.D., and M.M. performed research; M.H., J.C.W. and R. Humbert contributed new reagents/analytic tools; P.J.S., M.H., J.C.W., R. Humbert, M.M., and J.A.S. analyzed data; and P.J.S., M.H., and J.A.S. wrote the paper.
Abbreviations: DACS, digital analysis of chromatin structure; HS, hypersensitive site; HSqPCR, hypersensitivity quantitative PCR; n-cluster, n-tag cluster; PPV, positive predictive value; TSS, transcriptional start site.
References
- 1.Felsenfeld, G. & Groudine, M. (2003) Nature 421, 448–453. [DOI] [PubMed] [Google Scholar]
- 2.Felsenfeld, G. (1996) Cell 86, 13–19. [DOI] [PubMed] [Google Scholar]
- 3.Tuan, D., Solomon, W., Li, Q. & London, I. M. (1985) Proc. Natl. Acad. Sci. USA 82, 6384–6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Higgs, D. R., Wood, W. G., Jarman, A. P., Sharpe, J., Lida, J., Pretorius, I. M. & Ayyub, H. (1990) Genes Dev. 4, 1588–1601. [DOI] [PubMed] [Google Scholar]
- 5.Burgess-Beusse, B., Farrell, C., Gaszner, M., Litt, M., Mutskov, V., Recillas-Targa, F., Simpson, M., West, A. & Felsenfeld, G. (2002) Proc. Natl. Acad. Sci. USA 99, 16433–16437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Groudine, M., Kohwi-Shigematsu, T., Gelinas, R., Stamatoyannopoulos, G. & Papayannopoulou, T. (1983) Proc. Natl. Acad. Sci. USA 80, 7551–7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gross, D. S. & Garrard, W. T. (1988) Annu. Rev. Biochem. 57, 159–197. [DOI] [PubMed] [Google Scholar]
- 8.Elgin, S. C. (1984) Nature 309, 213–214. [DOI] [PubMed] [Google Scholar]
- 9.Fraser, P. & Grosveld, F. (1998) Curr. Opin. Cell Biol. 10, 361–365. [DOI] [PubMed] [Google Scholar]
- 10.Crawford, G. E., Holt, I. E., Mullikin, J. C., Tai, D., Blakesley, R., Bouffard, G., Young, A., Masiello, C., Green, E. D., Wolfsberg, T. G. & Collins, F. S. (2004) Proc. Natl. Acad. Sci. USA 101, 992–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sabo, P. J., Humbert, R., Hawrylycz, M., Wallace, J. C., Dorschner, M. O., McArthur, M. & Stamatoyannopoulos, J. A. (2004) Proc. Natl. Acad. Sci. USA 101, 4537–4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Velculescu, V. E., Zhang, L., Vogelstein, B. & Kinzler, K. W. (1995) Science 270, 484–487. [DOI] [PubMed] [Google Scholar]
- 13.Saha, S., Sparks, A. B., Rago, C., Akmaev, V., Wang, C. J., Vogelstein, B., Kinzler, K. W. & Velculescu, V. E. (2002) Nat. Biotechnol. 20, 508–512. [DOI] [PubMed] [Google Scholar]
- 14.Shiraki, T., Kondo, S., Katayama, S., Waki, K., Kasukawa, T., Kawaji, H., Kodzius, R., Watahiki, A., Nakamura, M., Arakawa, T., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 15776–15781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reitman, M., Lee, E., Westphal, H. & Felsenfeld, G. (1993) Mol. Cell. Biol. 13, 3990–3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kent, W. J. (2002) Genome Res. 12, 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990) J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 18.Rozen, S. & Skaletsky, H. J. (2000) in Bioinformatics Methods and Protocols, ed. Krawetz, S. A. (Humana, Totowa, NJ), pp. 365–386.
- 19.McArthur, M., Gerum, S. & Stamatoyannopoulos, G. (2001) J. Mol. Biol. 313, 27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu, C. (1980) Nature 286, 854–860. [DOI] [PubMed] [Google Scholar]
- 21.Stamatoyannopoulos, J. A., Goodwin, A., Joyce, T. & Lowrey, C. H. (1995) EMBO J. 14, 106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang, T. L., Maierhofer, C., Speicher, M. R., Lengauer, C., Vogelstein, B., Kinzler, K. W. & Velculescu, V. E. (2002) Proc. Natl. Acad. Sci. USA 99, 16156–16161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dunn, J. J., McCorkle, S. R., Praissman, L. A., Hind, G., Van Der Lelie, D., Bahou, W. F., Gnatenko, D. V. & Krause, M. K. (2002) Genome Res. 12, 1756–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lowrey, C. H., Bodine, D. M. & Nienhuis, A. W. (1992) Proc. Natl. Acad. Sci. USA 89, 1143–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gottgens, B., Nastos, A., Kinston, S., Piltz, S., Delabesse, E. C., Stanley, M., Sanchez, M. J., Ciau-Uitz, A., Patient, R. & Green, A. R. (2002) EMBO J. 21, 3039–3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reisman, D., Greenberg, M. & Rotter, V. (1988) Proc. Natl. Acad. Sci. USA 85, 5146–5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang, T. L., Diaz, L. A., Jr., Romans, K., Bardelli, A., Saha, S., Galizia, G., Choti, M., Donehower, R., Parmigiani, G., Shih I.-M., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 3089–3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brenner, S., Johnson, M., Bridgham, J., Golda, G., Lloyd, D. H., Johnson, D., Luo, S., McCurdy, S., Foy, M., Ewan, M., et al. (2000) Nat. Biotechnol. 18, 630–634. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.