Abstract
Naïve T cells proliferate independently of cognate antigen when introduced into lymphopenic hosts. Lymphopenia-induced proliferation depends on low-affinity MHC/self-peptide complexes and on IL-7. To elucidate the intracellular signals mediating this proliferation, we analyzed changes in gene expression in naïve CD8+ T cells at different times after their transfer into a lymphopenic environment. The genes induced in response to lymphopenia were largely an attenuated subset of those turned up by full antigenic stimulation, including genes related to cell cycling, whereas excluding genes specifically associated with effector activity. After the initial phase of proliferation in an empty compartment, the naïve T cells adopted a stable pattern of gene expression similar to that of antigen-experienced memory cells. Thus, T cells proliferating in lymphopenic hosts do not exhibit a unique gene-expression profile, instead relying on “traditional” signals for this antigen-independent proliferation; this process ultimately results in differentiation to “authentic” memory cells.
Keywords: gene-expression profiling, memory, T cell homeostasis, lymphopenia
The size of the peripheral pool of T lymphocytes is remarkably stable over time. This stability reflects a precise regulation of T cell numbers, balancing their production, proliferation, survival, and death. The size of the T lymphocyte pool appears to be regulated by peripheral mechanisms as the rate of production of new T cells in the thymus is uninfluenced by the number of cells present in secondary lymphoid organs (1). When lymphocyte numbers drop below a certain threshold, mature T cells proliferate, even without exposure to cognate antigen, providing evidence for a mechanism promoting the expansion of T cells in response to lymphopenia (2).
This lymphopenia-induced homeostatic proliferation (HP) requires both T cell receptor (TCR)-transmitted and cytokine-mediated signals. When introduced into a lymphopenic host, naïve CD4+ and CD8+ T cells require interactions with self-MHC-class-II and -class-I molecules, respectively, to undergo homeostatic proliferation (3–6). Additionally, naïve CD4+ and CD8+ T cells both fail to proliferate in an empty compartment in the absence of IL-7 (7, 8). Signals mediated by TCR/MHC:self-peptide and IL-7 receptor (IL-7R):IL-7 interactions are also considered to be essential for long-term survival of naïve T cells in lymphoreplete conditions (2). Furthermore, TCR:MHC/peptide and cytokine receptor/cytokine signals promote the activation, proliferation, and differentiation to effector function when cognate antigen is present. How, then, are these seemingly similar signals translated by the T cell into the appropriate context-dependent response, and how do naïve T cells sense the lymphopenic environment?
During HP, naïve CD8+ T cells enter a phenotypic and functional state with recognizable similarities to memory T cells (9–11). After several weeks in a lymphopenic host, CD8+ T cells, although not directly cytolytic, acquire the ability to make IFN-γ and to lyse targets after short reactivation by antigen, and they express phenotypic memory markers such as CD44 and Ly6C. These cells are distinct from both naïve and effector CD8+ T cells. Effector CD8+ T cells, generated during the active phase of an immune response to cognate antigen, possess direct cytolytic function and are capable of immediate IFN-γ production, whereas truly naïve T cells require an extended exposure to antigen before having these capabilities. It is surprising that, during HP, which occurs in the absence of antigen, naïve T cells would differentiate to memory cells.
We have followed a gene-expression profiling strategy to elucidate the molecular program underlying HP. First, we asked whether T cells placed in a lymphopenic environment display a unique gene-expression signature (a profile different from that induced during conventional activation via the TCR). Second, we investigated whether naïve T cells that have undergone homeostatic proliferation are really “authentic” memory cells. To address these questions, we analyzed the gene-expression profiles of OT-I TCR transgenic CD8+ T cells at different stages of HP.
Materials and Methods
Mice and Adoptive Transfers. All mice were bred and housed under specific pathogen-free conditions. Wild-type female C57/BL6 (B6) mice were obtained from The Jackson Laboratory. OT-I mice expressing a MHC-class-I-restricted TCR transgene (Vα2Vβ5) recognizing a peptide of chicken ovalbumin (357–364 OVAp) presented by H2-Kb were CD90.1 (B6.PLThy1a/Cy) congenic or RAG1-deficient and CD45.1 (Ly5.2) congenic.
OT-I effector and memory cells were generated by i.v. transfer of 1–2 × 106 OT-I T cells into B6 hosts, followed by i.v. infection 1–3 days later with 5 × 106 plaque-forming units of recombinant vaccinia virus encoding OVA cDNA. After infection, CD8+ and CD90.1+ or CD45.1+ cells were sorted after 6 days to obtain effector T cells or after 50 or more days to obtain memory T cells. To generate cells undergoing HP, we sorted naïve OT-I cells (CD8+CD44lo) from pooled spleen and lymph node and transferred them into irradiated (600 rad 1 day before) or untreated B6 or RAG-deficient hosts. At indicated time points after transfer, pooled spleen and lymph node cells were sorted for CD8 and CD90.1 or CD45.1 expression. As a control, naïve T cells were also transferred into unirradiated B6 hosts and sorted after 6 days. All sorted populations were >95% pure. Where indicated, transferred OT-I T cells were labeled with carboxyfluorescein diacetate succinimidyl ester (CFSE) before transfer as described.
Microarrays. RNA preparation and chip hybridizations were performed as described (12). Briefly, RNA was purified from sorted T cell populations (1 × 105 to 1 × 106 cells) by using TRIzol (Invitrogen), and total RNA was amplified by using the MessageAmp aRNA kit (Ambion), followed by biotin labeling with the Bioarray High Yield RNA Transcription Labeling kit (Enzo Life Sciences). The resulting biotin-labeled amplified RNA samples were fragmented and hybridized to Affymetrix Mu74Av2 chips. Replicate samples were as follows: naïve (four samples), effector (three samples), memory (five samples), cells undergoing homeostatic proliferation at day 5 or 6 (HP) (eight samples), 40–44 days after transfer (HP40) (four samples), cells 112–115 days after transfer (HP115) (three samples), and cells 6 days after transfer into unirradiated hosts (no Rad) (four samples).
The raw intensity values for each individual target on the Affymetrix chips (in CEL format) were processed and normalized by using the Robust MultiArray transformation (13) run on the S+ ArrayAnalyzer (Insightful, Seattle). Replicate samples were averaged. Randomized data sets were generated by shuffling the replicate intensities for all of the sample sets; values were then randomly assigned to a pseudosample followed by processing as for the experimental data groups. P values for gene-wise comparisons were calculated with Welch's modified t test, and the significance of the observed distributions was estimated by multiple resampling techniques.
Quantitative PCR. Quantitative real-time PCR was performed by using nonspecific product detection (SYBR green) or specific fluorogenic probes (TaqMan) on cDNA from indicated populations. For each gene, where possible primer pairs were designed to span an intron, they were demonstrated to generate a single product and to amplify in a linear relationship with the housekeeping gene HPRT, which was used in normalization of the samples relative to each other. Probe sets included (5′ to 3′): Prps2 Mm.44199 forward (F), CTGCCTTTGAAGCTGTTGTT, and reverse (R), TCCCCATTGTGGGTTCTT; E2f1 Mm.18036 F, CGCATCTATGACATCACCAA, and R, GCTTACCAATCCCCACCA; Ube2n Mm.30233 F, GTAGCCGAGCAATGGAAGA, and R, CCAACGAAGCCCAAGCCATAGAA; Dck Mm.3446 F, AAAGCCTTGAATTGGATGGA, and R, AGGAGCCAGCTTTCATGTTT; Vcam1 Mm.76649 F, TGAAATGCCTGTGAAGATGG, and R, TCGTTTTGTATTCAGGGGAGA; F2r Mm.24816 F, CGGTCCCTTGCTGTCTTC, and R, TTCTCCTCCTCCTCCTCATC; EST Mm.256588 F, ATCCACCCCAATGATGTTCT, and R, GTACTCCGTGTCCTTCGAGTT; FK506 binding protein 2 Mm.4234 F, ACAGAACCAGCCCTTTGTTT, and R, ATCACCAACTTCCGCTTTTC; hspa1a Mm.6388 F, GGACTTGATTGCAGGACAAA, and R, GGTGCTGGCTAGGAGACAG; and hspa1b F, AATTTAACAGTCAACGCAATTACC, and R, AACAGACTCTTTGCACTTGATAGC.
Results
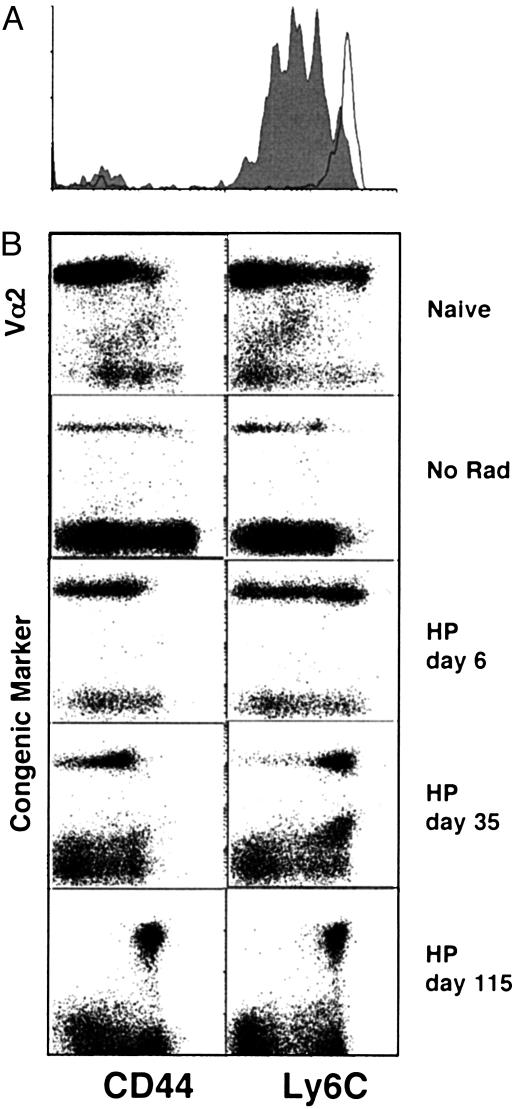
Parallel Induction of Genes During Homeostatic Proliferation and Antigen-Driven Activation. Naïve T cells divide in the absence of cognate antigen when transferred into an empty (irradiated) but not into a full (unirradiated) syngeneic host (2). Naïve OT-I CD8+ T cells were sorted for low expression of CD44, labeled with the intracellular dye CFSE, and transferred into B6 mice, which had or had not been sublethally irradiated (600 cGy). By 6 days after transfer into a lymphopenic mouse, nearly all of the OT-I T cells had proliferated at least once, as indicated by dilution of the CFSE signal, whereas virtually none of the cells transferred into an unirradiated recipient had divided (Fig. 1A). The proliferating naïve T cells remained CD44lo and Ly6Clo 6 days after transfer into both irradiated and unirradiated hosts (Fig. 1B). By 35 days after transfer, all of the naïve cells transferred into the irradiated mice expressed increased levels of CD44 and Ly6C, and they retained high levels of surface expression at 115 days. These data agree with published work showing stable acquisition of memory markers by naïve T cells that have undergone homeostatic proliferation (9, 14), and contrast with our previous report that the expression of memory markers was transient (10). It is likely that the apparent reversion of cells to a naïve phenotype was caused by the transfer of contaminating stem cells as suggested by Ge et al. (14).
Fig. 1.
Proliferation and phenotypic conversion of naïve CD8+ T cells in a lymphopenic host. A total of 2 × 106 naïve OT-I T cells were sorted (CD8+CD44lo) and transferred i.v. to irradiated (600 rad) or unirradiated B6 hosts. (A) Sorted cells were CFSE-labeled before transfer. After 6 days, splenocytes were analyzed by flow cytometry. CFSE levels are shown for CD8+CD45.1+ gated cells transferred to irradiated (filled histogram) or unirradiated (open histogram) hosts. (B) CD8+Thy1.1+ splenocytes recovered from irradiated hosts were analyzed by flow cytometry for expression of CD44 and Ly6C expression.
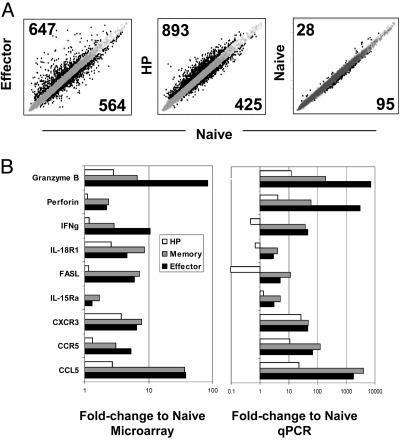
To begin dissecting the signals that trigger and drive HP, we analyzed the gene-expression profiles of cells proliferating in a lymphopenic compartment. OT-I T cells undergoing HP were sorted 6 days after transfer, and RNA was isolated, amplified, biotin-labeled, and hybridized to Affymetrix GeneChips (MU74Av2 arrays). Control data sets were obtained from parallel transfers of OT-I cells into unirradiated hosts. The resulting profiles (compiled from three to eight independent determinations) were compared with data sets obtained in the same manner from naïve, effector, and memory OT-I populations in the context of another study (A.W.G, C.B., D.M., unpublished data). Raw gene-expression data for each sample were processed with the random multiple access (RMA) algorithm for probelevel normalization (13); data from replicate samples were averaged to generate a composite value for each cell. Composite normalized gene-expression values for T cells undergoing HP or an antigen-driven response (effector) were compared with corresponding values for naïve T cells (Fig. 2A Left and Center). First, a false-positive rate was estimated by using two composite data sets derived from naïve cells, generated by subdividing the four naïve cell replicates into two groups (Fig. 2A Right). At a fold-change cutoff of 1.5, the false-positive rate was <8% (123 genes for the “naive cell replicate” comparison versus 1,211 and 1,318 differentially expressed genes for the effector to naïve and HP to naïve cell comparisons, respectively). In both the HP and effector populations, a substantial number of genes were differentially expressed compared with naïve T cells (Fig. 2A Left and Center). Similar numbers were up- and down-regulated in effector and HP cells; however, the degree of change in the values was clearly lower for HP than for the effector population, as judged from the tightness of the gene clouds (Fig. 2A Left and Center; see below).
Fig. 2.
Microarray analysis of effector and HP cells. (A) Log-scale plot of normalized and averaged expression for effector and HP replicates or normalized expression for a subset of the naïve replicates versus normalized and averaged expression of naïve replicates. Genes expressed >1.5-fold higher or lower than the composite naïve sample are highlighted in black. The number of genes up- or down-regulated for each comparison are indicated. (B) Fold-change values obtained from the microarray (Left) or quantitative PCR (qPCR, Right) for the comparison of effector, memory, and HP populations to naïve are shown for a subset of genes.
To provide independent validation of the microarray data, we performed quantitative real-time PCR (qPCR) on cDNA isolated from naïve, effector, memory, and HP populations for a subset of the genes (Fig. 2B). For the nine genes tested, the microarray and qPCR values showed similar relative changes in expression; however, as previously demonstrated, the microarray data generally underestimated the extent of the fold changes, particularly as estimated from the random multiple access (RMA) processing procedure (13, 15, 16).
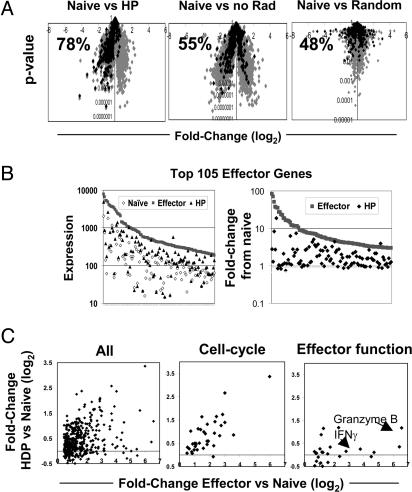
To explore gene-expression profiles in HP cells more profoundly, we focused on their relationship to a set of genes induced at the peak of an antigen-driven response [“effector genes,” a group of 462 genes defined as most significantly induced at day 6 of antigen-driven activation of OT-I cells (vis à vis expression in naïve cells); Table 1, which is published as supporting information on the PNAS web site, and A.W.G., C.B., and D.M., unpublished data]. A parallel increase in expression of these genes was revealed (Fig. 3A and B). For example, when the effector genes were highlighted on a “volcano plot” (P value versus fold-change), the majority of them (78%) showed increased expression in the HP over naïve cell comparison (Fig. 3A Left). This parallel induction contrasted with what was found for the same OT-I cells transferred into a nonlymphopenic host and for a random data set, where minimal skewing of expression was observed, 55% and 48%, respectively (Fig. 3A Center and Right). The correlation between genes expressed by HP and effector cells was very high, because in 100,000 permutations of random data sets, none was skewed >60% (P < 0.0001) (data not shown). Although the majority of genes up-regulated during an antigen-driven response was also induced during HP, the level of induction was, in general, weaker (Fig. 3B); plotting ranked expression (Fig. 3B Left) or fold change values (Fig. 3B Right) for the 105 genes most induced in the “effector gene” set revealed that the HP population had intermediate expression values relative to naïve and effector populations for many of the genes.
Fig. 3.
Genes expressed by effector cells are up-regulated during HP. (A) Fold-change (log2) versus P value plotted for the comparison of HP, no Rad or a random data set to naïve. The subset of genes expressed >1.3-fold or higher in effector compared to naïve with a P value of <0.01 are highlighted in black (462 genes). The percentage of effector genes also up-regulated for the indicated population compared to naïve is given for each plot. (B) The gene-expression values for the 105 genes most induced in effector cells compared to naïve cells are shown for effector, HP, and naïve populations (Left) or fold-change values for effector and HP compared to naïve for the same subset of genes (Right). (C) The fold-change (log2) of HP compared to naïve plotted against the fold-change (log2) of effector compared to naïve for the total effector gene list or a subset of genes involved in the cell-cycle progression or cytolytic effector functions.
Pursuing this issue in more detail, we found that when fold-change values in the comparison to naïve cells were compared for HP and effector cells, once again many, but not all, of the induced genes were common to the two comparisons (Fig. 3C Left). From a combination of database searches, we defined sublists of genes associated with cell-cycle progression or with cytotoxic effector function, and used these lists to focus the comparison of naïve and effector cells. The resulting plots (Fig. 3C Center and Right and Tables 2 and 3, which are published as supporting information on the PNAS web site) reveal a clear difference in the two cell populations. Essentially all of the cell-cycle genes were up-regulated by the population of cells undergoing HP, albeit to a lesser extent than in effector cells (Fig. 3C Center and Table 2). This dampening of the induction makes sense, because the rate of proliferation for cells responding to antigen is faster than that for cells undergoing HP (5). In contrast, the genes known to be associated with cytotoxic T lymphocyte effector functions were generally not up-regulated in the HP population (Fig. 3C Right and Table 3). This observation supports the earlier findings that homeostatically expanding CD8+ T cells did not differentiate into effector cells, as measured by rapid cytoxicity and IFN-γ production upon antigen exposure (5, 6, 9). A similar analysis was completed for genes down-regulated by effector cells compared to naïve; the majority of genes whose expression was diminished during antigen-driven activation were also expressed at lower levels during homeostatic proliferation (data not shown).
HP Leads to a Gene-Expression Profile Like That of Memory T Cells. We used the same approach to reveal the long-term outcome of inducing effector genes. It had been suggested that HP promotes differentiation to memory T cells; however, it has not been clear whether this new phenotype is a stable one (9, 10). A further complication has been the lack of a single marker of memory CD8+ T cells; rather, they have typically been defined by a combination of increased cell-surface expression of phenotypic markers such as CD44 and Ly6C, increased levels of intracellular perforin and granzyme B, and long-term survival after antigen encounter. The assertion that CD8+ T cells become memory-like after HP was based on their expression of a handful of phenotypic markers and their ability to lyse target cells and produce IFN-γ faster than naïve, but slower than effector, T cells.
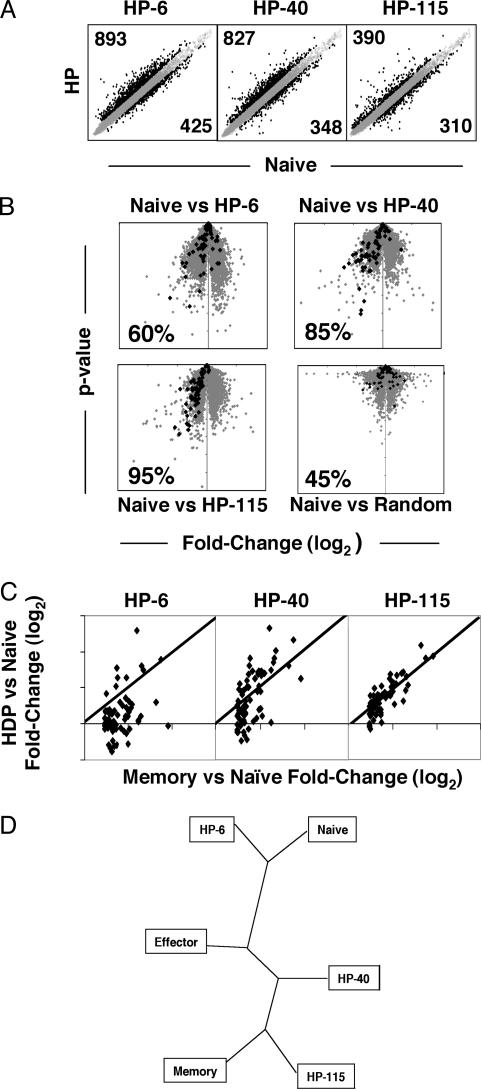
Gene-expression profiles were assessed at different times after transfer of naïve OT-I cells into lymphopenic hosts (the day 6 data sets described above, and comparable sets generated at days 40 and 115 after transfer into lymphopenic hosts). A subset of 74 “memory genes” expressed uniquely in CD8+ memory T cells (compared with effector or naïve cells) was identified as above. By highlighting the designated memory genes on a P value vs. fold change plot that compared HP with naïve T cells, it was apparent that expression of these genes by cells undergoing HP progressively became more similar to that of memory T cells over time (Fig. 4B and Table 4, which is published as supporting information on the PNAS web site). After 6 days in lymphopenic hosts, only 60% of the memory-specific genes were expressed at a higher level in the HP population than in naïve cells (Fig. 4B Upper Left). By 40 days after transfer, 85% of the memory-specific genes were up-regulated to at least some extent in the HP population, and at 115 days, 95% of the genes correlated with memory cell expression (Fig. 4B Upper Right and Lower Left). The comparable value for a random cell dataset was 45% (Fig. 4B Lower Right). When 50,000 randomized data sets were generated, none showed >85% skewing, establishing that the correlation between genes up-regulated in memory T cells and HP cells at days 40 and 115 was highly significant (P < 0.00002) (data not shown). Similarly, when fold-change values (relative to those of naïve cells) for the HP populations at different time points were compared with those for the memory populations, it was clear that the expression of individual genes became more similar to that of memory cells over the HP time course (Fig. 4C). When expression of genes that were down-regulated in the memory population compared to naïve were plotted over the HP time course, a similar correlation of HP115 and memory T cell gene expression was revealed (data not shown).
Fig. 4.
HP leads to expression of memory-specific genes. (A) Log-scale plots of normalized and averaged expression for HP at days 6, 40, and 115 versus normalized and averaged expression of naïve replicates. Genes expressed >1.5-fold higher or lower than naïve are highlighted in black. (B) Fold-change (log2) versus P value plotted for the comparison of HP over a time course compared to naïve. Genes highlighted in black indicate expression relative to naïve for memory-specific genes (genes up-regulated in memory 1.3-fold over naïve with a P value <0.01, but not up-regulated in effector versus naïve). The percentage of memory-specific genes also up-regulated for the indicated population compared to naïve is given for each plot. (C) For the memory-specific gene list, fold-change (log2) values for the indicated HP population compared to naïve are plotted versus the fold-change (log2) values for memory compared to naïve. (D) Representation of the relationship of the indicated populations established by unsupervised hierarchical clustering illustrated as an unrooted phylogenic tree.
Another approach to establishing how gene expression correlates among different cell populations is by arrangement of the experimental populations or individual genes according to similarity of gene expression. To determine which populations were most related, we performed an unsupervised hierarchical clustering analysis by using cluster (http://rana.lbl.gov/EisenSoftware.htm) (17) for the HP populations at different time points and naïve, effector, and memory cells. The results are illustrated as an unrooted phylogenic tree by treeware (http://taxonomy.zoology.gla.ac.uk/rod/treeview). According to this approach, HP-115 and -40 are most related to memory cells, whereas HP-6 maps between naïve and effector cells (Fig. 4C). Thus, over the time course, cells proliferating in a lymphopenic compartment progressively established a gene-expression profile that resembles that of “authentic” memory cells.
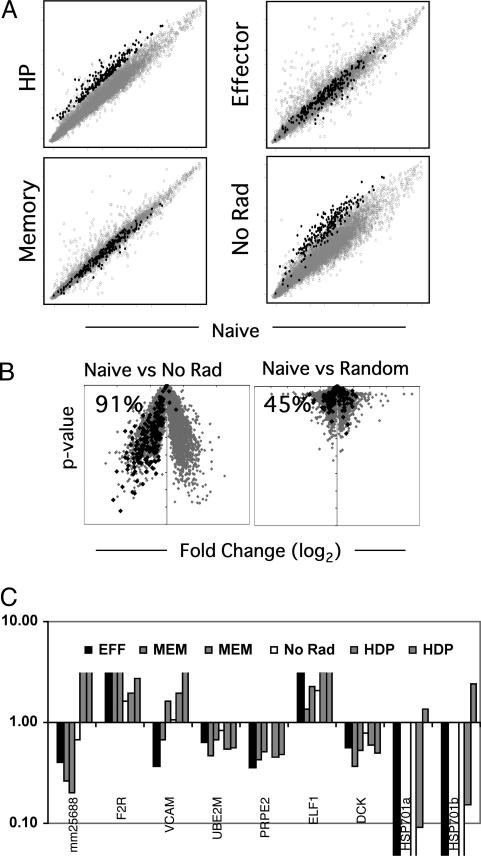
No Evidence of a Unique Gene-Expression Signature Associated with Homeostatic Proliferation. Next, we looked for evidence of a gene-expression profile specific to cells undergoing HP. Although the gene-expression profile of early HP time points included the set of genes also induced during an antigen-driven response, induction of other genes might reflect the operation of additional signaling pathways that control the response to lymphopenia. We reasoned that, if such pathways were present, they should be revealed by analysis of gene expression by T cells 6 days after transfer into lymphopenic hosts because the cells actively proliferate at this time point, in reflection of the host's lymphopenia. To reveal candidates as HP-specific genes, we compiled a list of 219 genes up-regulated in the HP versus naïve cell populations but not in the effector or memory versus naïve populations, termed “HP genes”; expression values for each condition versus expression values for naïve cells, with the HP gene set highlighted, were graphed (Fig. 5A and gene list in Table 5, which is published as supporting information on the PNAS web site). However, one concern was that the purification and transfer of naïve T cells, in and of itself, might induce changes in gene expression like those associated with the HP signal. This issue was addressed by a parallel transfer of the same naïve OT-I CD8+ T cells into an unirradiated host (No Rad). Surprisingly, expression of essentially all of the HP genes was also increased in cells transferred into unirradiated recipients (199 of the 219) (Fig. 5A), with a very strong bias toward up-regulation of the HP genes on the volcano plot (Fig. 5B). Thus, the process of transferring the T cells can result in an unexpectedly marked alteration of gene expression, even several days after transfer and despite highly compatible genetic backgrounds in the donor and host mice.
Fig. 5.
No evidence for a HP-specific gene-expression profile. (A) Log-scale plot of normalized and averaged expression for indicated populations versus naïve expression. Genes up-regulated at day 6 of HP compared to naïve T cells (>1.3-fold over naïve P value < 0.01) and also compared effector and memory cells (>1.3-fold) are highlighted in black (219 genes). (B) Fold-change (log2) versus P value plotted for cells transferred into an unirradiated host (no Rad) compared to naïve or a random data set compared to naïve. Highlighted genes are those up-regulated in HP as described above. (C) Relative expression to naïve T cells by effector (one sample), memory (two samples), no Rad (one sample), and HP (two samples) as determined by quantitative PCR for nine candidate HP-specific genes. Data are representative of at least three unique samples and three different PCR experiments.
After elimination of the genes induced by the adoptive transfer process, very few potentially HP-specific genes remained (Table 6, which is published as supporting information on the PNAS web site). In fact, the presence of only 20 candidate genes for HP-specific induction was well within the range of experimental noise, as estimated by repeated random sampling of the data sets. Nonetheless, we measured by quantitative real-time PCR mRNA levels for nine of the genes (which were estimated to be most induced); only one was found to be consistently increased in the cells undergoing HP vis à vis transcription in naïve, effector or unirradiated cell populations (Fig. 5C). This sequence corresponded to a previously undescribed gene (UniGene Mm.256588) with no clear homology to any known genes. This transcript has an ORF of 421 aa, with no recognizable protein domain (National Center for Biotechnology Information) and no known homologies in nonrodent species. A comparable analysis was completed for genes repressed during HP and, similarly, no HP-specific gene-expression profile emerged.
Thus, when genes normally induced during antigen-driven responses or in memory cells are accounted for, the genes artificially induced by experimental manipulation are discounted, and experimental noise is eliminated, with the exception of a single EST, there does not appear to be a gene-expression signature specific to homeostatic proliferation.
Discussion
Is the difference between proliferation in an empty compartment and survival in a full compartment quantitative or qualitative? The process of HP, the proliferation of lymphocytes in a lymphopenic host, depends on both self-MHC/peptide complexes and IL-7 in the case of naïve T cells (3–8). However, the same elements are required for survival of naïve T cells in a normal host, without resulting in proliferation (2). The explanation may be qualitative: T lymphocytes in a lymphopenic environment may receive a distinct signal to proliferate, possibly generated by stromal cells when they sense a dearth of T cells (through, for example, lack of MHC molecule or CD40 engagement). Or there may be a quantitative explanation: T cells may be able to proliferate in response to abundant IL-7R or TCR ligands, which would normally be in limited supply and the object of competition. Our analysis of gene expression by T cells of different states did not reveal a gene-expression profile specific to cells undergoing HP, which would have been expected if a qualitatively unique signal promotes HP. In fact, the early stages of HP were associated with changes in gene expression that largely resembled those occurring during a weak response to antigen (Fig. 3). This observation argues that, during HP, the TCR interaction with low-affinity MHC/self-peptide ligands in a lymphopenic environment results in a similar but quantitatively weaker signal than that promoted by an agonist ligand. This signal, in coordination with IL-7, appears adequate to promote proliferation when T cell numbers decrease. These findings support the idea that merely increased access to self-MHC molecules and IL-7 are sufficient to induce proliferation in a lymphopenic environment.
Cells undergoing HP do not acquire full effector function (5, 6, 9, 10) despite the fact that their pattern of gene-expression mirrors, at an attenuated level, that of cells responding to cognate antigen (Fig. 3). This incapacity is explained by the fact that transcription of many of the genes known to be associated with cytolysis were not induced during HP (Fig. 3C). The induction of such effector functions might require a full-strength TCR signal, whereas triggering of proliferation and memory formation might occur at a lower threshold. This dichotomy would appear to be an important element of HP because these cells are being stimulated by MHC/self-peptide complexes, and the generation of killing activity against cells expressing the stimulating molecules would be potentially disastrous.
This suboptimal induction of effector genes results in the differentiation to a population of cells with a stable phenotype very similar to that of memory T cells. More than 100 days after transfer into a lymphopenic host, OT-I cells transcribed a set of genes typically expressed by memory cells elicited by cognate antigen. It has been shown that these pseudomemory cells have some of the functional characteristics of memory cells as well, as evidenced by rapid acquisition of the ability to kill targets and make IFN-γ after antigen encounter (9). Thus, T cell memory appears not to be strictly contingent on encounter with cognate antigen or on the acquisition of full effector function, but does seem to progress from a proliferative state with many of the characteristics of antigen-driven activation. This observation is somewhat unexpected, as several studies have provided evidence that the formation of conventional T cell memory resulting from cognate antigen encounter occurs subsequent to the effector stage (18–21). It will be interesting to determine whether the results presented here represent a unique pathway to memory T cell formation or whether they can be generalized, in which case, the expression of effector genes without the full induction of transcripts encoding cytolytic proteins would occur in a subset of cells representing memory precursors during the response to cognate antigen.
Supplementary Material
Acknowledgments
We thank Vincent Butty for running the phylogeny analysis; Vera Bruklich, Tatyana Lipatova, and Quynh-Mai Pham for assistance with mice; Grigoriy Losyev for cell sorting; Jennifer White and Kelly Elder for assistance with the manuscript; and Rob Saccone, Jennifer Johnson, and Jocelyn Yee of the Joslin Genomics Core for microarray processing. This work was supported by Joslin's Diabetes and Endocrinology Research Center Cores (2 P30 DK36836-17), National Institutes of Health Grant AI51530-01 (to C.B. and D.M.), and the W. T. Young Chair for Diabetes Research. A.W.G. was supported by an Irvington Institute–Juvenile Diabetes Research Fellowship and Joslin's National Institutes of Health Training Grant (5 T32 DK007260-24); C.J.L. was funded by Brigham and Women's Training Grant T32 HL07627.
Abbreviations: HP, homeostatic proliferation; TCR, T cell receptor; CFSE, carboxyfluorescein diacetate succinimidyl ester.
Data deposition: The microarray data reported in this paper have been deposited in the National Center for Biotechnology Information GEO Repository (accession no. GSE1921).
References
- 1.Berzins, S. P., Boyd, R. L. & Miller, J. F. (1998) J. Exp. Med. 187, 1839-1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jameson, S. C. (2002) Nat. Rev. Immunol. 2, 547-556. [DOI] [PubMed] [Google Scholar]
- 3.Bender, J., Mitchell, T., Kappler, J. & Marrack, P. (1999) J. Exp. Med. 190, 367-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ernst, B., Lee, D. S., Chang, J. M., Sprent, J. & Surh, C. D. (1999) Immunity 11, 173-181. [DOI] [PubMed] [Google Scholar]
- 5.Goldrath, A. W. & Bevan, M. J. (1999) Immunity 11, 183-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kieper, W. C. & Jameson, S. C. (1999) Proc. Natl. Acad. Sci. USA 96, 13306-13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schluns, K. S., Kieper, W. C., Jameson, S. C. & Lefrancois, L. (2000) Nat. Immunol. 1, 426-432. [DOI] [PubMed] [Google Scholar]
- 8.Tan, J. T., Dudl, E., LeRoy, E., Murray, R., Sprent, J., Weinberg, K. I. & Surh, C. D. (2001) Proc. Natl. Acad. Sci. USA 98, 8732-8737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho, B. K., Rao, V. P., Ge, Q., Eisen, H. N. & Chen, J. (2000) J. Exp. Med. 192, 549-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldrath, A. W., Bogatzki, L. Y. & Bevan, M. J. (2000) J. Exp. Med. 192, 557-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oehen, S. & Brduscha-Riem, K. (1999) Eur. J. Immunol. 29, 608-614. [DOI] [PubMed] [Google Scholar]
- 12.Anderson, M. S., Venanzi, E. S., Klein, L., Chen, Z., Berzins, S., Turley, S. J., von Boehmer, H., Bronson, R., Dierich, A., Benoist, C., et al. (2002) Science 298, 1395-1401. [DOI] [PubMed] [Google Scholar]
- 13.Irizarry, R. A., Hobbs, B., Collin, F., Beazer-Barclay, Y. D., Antonellis, K. J., Scherf, U. & Speed, T. P. (2003) Biostatistics 4, 249-264. [DOI] [PubMed] [Google Scholar]
- 14.Ge, Q., Hu, H., Eisen, H. N. & Chen, J. (2002) Proc. Natl. Acad. Sci. USA 99, 2989-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poirot, L., Benoist, C. & Mathis, D. (2004) Proc. Natl. Acad. Sci. USA 101, 8102-8107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matos, M., Park, R., Mathis, D. & Benoist, C. (2004) Diabetes 53, 2310-2321. [DOI] [PubMed] [Google Scholar]
- 17.Eisen, M. B., Spellman, P. T., Brown, P. O. & Botstein, D. (1998) Proc. Natl. Acad. Sci. USA 95, 14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaech, S. M., Tan, J. T., Wherry, E. J., Konieczny, B. T., Surh, C. D. & Ahmed, R. (2003) Nat. Immunol. 4, 1191-1198. [DOI] [PubMed] [Google Scholar]
- 19.Kaech, S. M., Hemby, S., Kersh, E. & Ahmed, R. (2002) Cell 111, 837-851. [DOI] [PubMed] [Google Scholar]
- 20.Jacob, J. & Baltimore, D. (1999) Nature 399, 593-597. [DOI] [PubMed] [Google Scholar]
- 21.Opferman, J. T., Ober, B. T. & Ashton-Rickardt, P. G. (1999) Science 283, 1745-1748. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.