Summary
Ionotropic neurotransmitter receptors mediate fast synaptic transmission by functioning as ligand-gated ion channels. Fast inhibitory transmission in the brain is mediated mostly by ionotropic GABAA receptors (GABAARs), but their essential components for synaptic localization remain unknown. Here, we identify putative auxiliary subunits of GABAARs, which we term GARLHs, consisting of LH4 and LH3 proteins. LH4 forms a stable tripartite complex with GABAARs and neuroligin-2 in the brain. Moreover, LH4 is required for the synaptic localization of GABAARs and inhibitory synaptic transmission in the hippocampus. Our findings propose GARLHs as the first identified auxiliary subunits for anion channels. These findings provide new insights into the regulation of inhibitory transmission and the molecular constituents of native anion channels in vivo.
Introduction
The major inhibitory transmitter in the brain is γ-aminobutyric acid (GABA), and fast inhibitory transmission is mediated by GABAA receptors (GABAARs), pentameric anion channels consisting of α, β and non- α/β subunits. GABAARs localized postsynaptically regulate membrane potentials to mediate signalling, and the mechanisms for the synaptic localization of GABAARs have been extensively studied. When γ2, the non- α/β GABAAR subunit, is disrupted in primary cortical neurons, synaptic localization of the GABAAR α1 subunit is impaired (Alldred et al., 2005; Essrich et al., 1998). Given that GABAARs likely require protein interactions for their synaptic localization, several proteins have been identified as interacting with GABAAR, including gephyrin and GABAAR-associated protein (Collingridge et al., 2004; Jacob et al., 2008; Kowalczyk et al., 2013; Maric et al., 2011; Mukherjee et al., 2011; Wang et al., 1999). Among these interactors, gephyrin is the most characterized. Elimination of gephyrin by gene targeting or antisense resulted in a reduction in the clustering of some, but not all, GABAAR subunits (Essrich et al., 1998; Kneussel et al., 2001; Kneussel et al., 1999; Levi et al., 2004) and a modest 23% reduction in mIPSC amplitude was observed in gephyrin-deficient primary cultured neurons without changes in mIPSC frequency (Levi et al., 2004). These findings indicate a gephyrin-independent mechanism for GABAAR-mediated transmission. In contrast, neuroligins (NLs) may play a more direct role in the synaptic activity or localization of GABAARs. In Caenorhabditis elegans, there is only one NL isoform and disruption of NL substantially reduced synaptic localization and activity of GABAAR (Maro et al., 2015; Tu et al., 2015). Knocking out three NL isoforms (NL1/2/3) in mice caused a robust reduction in both inhibitory and excitatory transmission in various neurons (Varoqueaux et al., 2006; Zhang et al., 2015). Even though it has been shown that the NL2 isoform preferentially localizes at inhibitory synapses (Chih et al., 2005; Graf et al., 2004; Varoqueaux et al., 2004) and interacts with collybistin and gephyrin (Poulopoulos et al., 2009; Soykan et al., 2014), it remains unclear if and how NLs and GABAARs associate at synapses.
One plausible mechanism for γ2-dependent, gephyrin-independent GABAAR synaptic localization is through an as yet unidentified GABAAR auxiliary subunit. Although ionotropic neurotransmitter receptors were once thought to function independently in the brain, the recent discovery of auxiliary subunits for ionotropic glutamate receptors has changed the understanding of receptor regulation in excitatory transmission. In the brain and neurons, the auxiliary subunits TARP and CNIH determine the localization and properties of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors (AMPARs) (Brockie et al., 2013; Chen et al., 2000; Herring et al., 2013; Jackson and Nicoll, 2011; Kato et al., 2010; Schwenk et al., 2009; Tomita et al., 2005; Yan and Tomita, 2012), whereas Neto auxiliary subunits control the properties of kainate receptors (KARs) (Straub et al., 2011; Tang et al., 2011; Zhang et al., 2009). Disrupting auxiliary subunits impairs mouse behaviour and survival (Hashimoto et al., 1999; Yan et al., 2013). Therefore, it is clear that tetrameric ligand-gated cation channels, such as AMPARs and KARs, function with their auxiliary subunits in vivo. However, it remains unclear whether anion channels likewise have auxiliary subunits that form complexes with the pore-forming subunit of the receptor and that determine their function in the brain.
Here, we identified the GABAAR regulatory Lhfpl (GARLH) family proteins, currently consisting of lipoma HMGIC fusion partner-like 3 and 4 (LH3 and 4), as putative auxiliary subunits of GABAARs. GABAARs in the brain form a tripartite complex with transmembrane GARLH family proteins and NL2. The γ2-containing GABAARs stabilize GARLH expression in the brain. Disrupting GARLH expression reduces the synaptic localization of the γ2-containing GABAARs and GABAAR-mediated synaptic transmission. This identification of the first putative auxiliary subunit of a pentameric ligand-gated anion channel provides a molecular mechanism underlying the synaptic localization of GABAARs to control inhibitory transmission in the brain.
Results
GABAARs form a tripartite complex in the brain
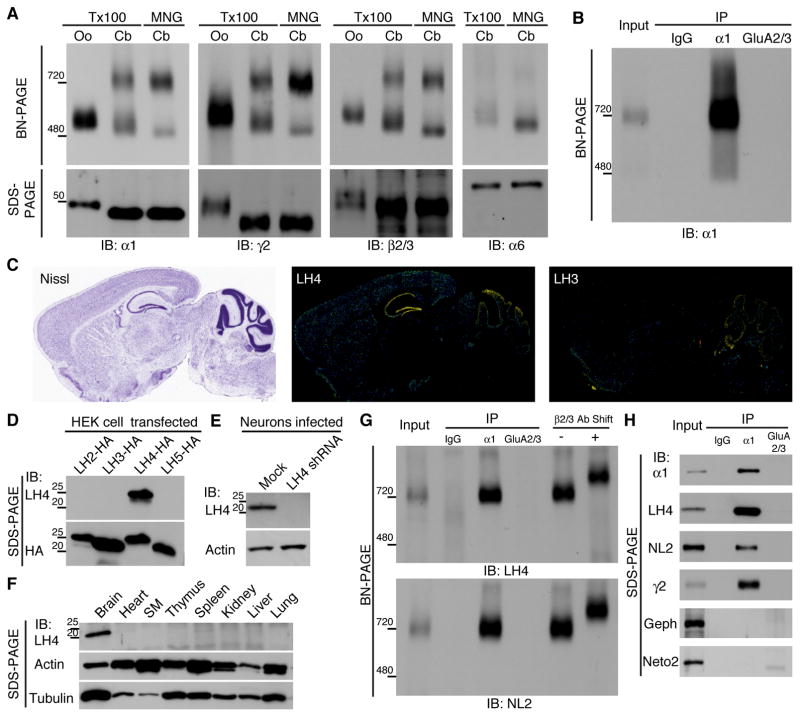
To examine whether native GABAARs form complexes with other proteins, we compared the molecular weights of native and reconstituted GABAARs using blue native polyacrylamide gel electrophoresis (BN-PAGE), a method that preserves protein complexes (Schagger et al., 1994). We expressed GABAARs in Xenopus laevis oocytes by injecting them with cRNAs of three GABAAR subunits (α1, β2 and γ2) (Figure 1A). Using sodium dodecyl sulphate (SDS)-PAGE, the molecular weight of each subunit was found to be approximately 50 kDa, whereas using BN-PAGE, the recombinant GABAAR solubilized with Triton X-100 formed a 520 kDa complex (Fig. 1A), indicating that GABAAR subunits form a hetero-oligomer. The endogenous mouse cerebellar GABAARs containing α1, γ2 or β2/3 subunits formed two distinct complexes, one at 720 kDa and the other at 500 kDa (Figure 1A). When the cerebellum was solubilized with maltose-neopentyl glycol (MNG), native GABAARs migrated mostly to 720 kDa, with a weak band observed at 480 kDa (Figure 1A). The modest differences observed in the migration of proteins from oocytes and cerebellar tissue using BN-PAGE were consistent with those in the molecular weights of the proteins determined using SDS-PAGE (Figure 1A). Similarly, α2- and α3-containing GABAARs migrated to 720 kDa in the mouse hippocampus and cerebral cortex, respectively (Figure S1A). Interestingly, α6-containing GABAARs in the cerebellum migrated mainly to 500 kDa (Figure 1A). The difference in the molecular weights of native GABAARs at 720 kDa and recombinant GABAARs at 500 kDa suggests that the native GABAAR complex contains other protein components.
Figure 1. Native GABAAR complexes contain Lhfpl4 and neuroligin-2.
(A) Recombinant GABAAR expressed in Xenopus laevis oocytes (Oo) by injection of cRNAs of α1, β2 and γ2 GABAAR subunits migrated as a single band of 520 kDa using BN-PAGE when solubilized with Triton X-100 (Tx100). By contrast, the native GABAAR obtained from the cerebellum (Cb) migrated as two bands of 720 kDa and 500 kDa, respectively, with Tx100 solubilization. Using maltose-neopentyl glycol (MNG) solubilization, the native Cb GABAAR migrated as a strong band of 720 kDa and a weak band of 480 kDa. Immunoblots (IB) with antibodies against α1, γ2 and β2/3 subunits showed similar results, whereas the α6-containing Cb GABAAR migrates predominantly to 500 kDa and 480 kDa in Tx100 and MNG, respectively. (B) Immunopurified (IP) native GABAAR complexes obtained using an anti-α1 antibody from MNG-solubilized cerebella migrates to 720 kDa. A normal rabbit IgG and an anti-AMPA receptor GluA2/3 antibody were used as control. (C) The distribution of Lhfpl (LH) 4 and 3 mRNAs in mouse brain according to the Allen Brain Atlas (http://www.brain-map.org/). Nissl staining shows anatomy of the mouse brain (Nissl). LH4 is strongly expressed in hippocampus and cerebellum, whereas LH3 is expressed in cerebellum. (D) The antibody against LH4 recognized LH4 specifically, whereas the anti-HA antibody recognized all HA-tagged LH2/3/4/5 expressed in transfected HEK cells. (E) The anti-LH4 antibody recognized a band in lysate prepared from primary hippocampal neurons, which is eliminated in neurons treated with LH4 shRNA. (F) Among various mouse tissues, the anti-LH4 antibody recognized a 22 kDa band only in the brain. (G) Immunopurified α1-containing cerebellar GABAARs contain LH4 and neuroligin-2 (NL2). Pre-incubation of the immunopurified GABAARs with an anti-β2/3 antibody (Ab) increases the molecular weight. (H) On SDS-PAGE, LH4, NL2 and γ2 co-immunoprecipitated (IP) specifically with α1, but not with the AMPA receptor GluA2/3. Gephyrin (Geph) and Neto2 also failed to co-immunoprecipitate with α1.
To identify these missing components, we immunopurified the GABAAR complex from rat cerebellum using an anti-GABAAR α1 antibody and conducted BN-PAGE (Figure 1B). A rabbit normal IgG and an anti-AMPAR GluA2/3 antibody did not immunoprecipitate the 720 kDa band (Figure 1B). The resulting band at 720 kDa was excised, and its components were analysed using mass spectrometry. Besides GABAAR subunits (α1, β2, γ2, and β3), we identified NL2 and lipoma HMGIC fusion partner-like 4 (LH4) above α6, which mainly formed a complex at 500 kDa (Table and Table S1). LH4 is a four membrane-spanning protein, and its function is currently unknown (Petit et al., 1999). LH4 mRNA is strongly expressed in the hippocampus and cerebellum. In cerebellum, its homologue, LH3, shows complementary expression (Figure 1C). We generated an anti-LH4 antibody that selectively recognized LH4 among three other related proteins (Lhfpl2/3/5) and detected a band at the expected molecular weight of 22 kDa in the brain and primary hippocampal neurons, but not in neurons treated with LH4 shRNA lentivirus (Figure 1D–F and Figure S1B). In addition, with a surface biotinylation assay using acute cerebellar slices, we found that LH4 as well as GluN1 were detected in the biotinylated cell surface fraction, whereas tubulin was detected in the intracellular fraction (non-biotinylated fraction) (Figure S1C). Both anti-LH4 and anti-NL2 antibodies recognized the cerebellar GABAAR complex purified using the anti-α1 antibody, and pre-incubation with an anti-β2/3 antibody increased the molecular weight, indicating that the complex also contained β2/3 subunits (Figure 1G). In addition, the α1 subunit from rat cerebellum co-immunoprecipitated LH4, NL2 and γ2, but not with gephyrin and Neto2, whereas the GluA2/3 AMPARs did not coimmunoprecipitate LH4, NL2 and γ2 (Figure 1H). These results indicate that the cerebellar GABAAR complex observed at 720 kDa contains NL2 and LH4 as well as the GABAAR subunits (α1, β2/3, and γ2).
Table 1. Components of the native GABAAR complex.
The components of the 720 kDa complex were identified using mass spectrometry. Proteins are arranged according to the values of the exclusive spectrum count (Spec.)/exclusive unique peptide count (Pep.). This ratio reflects the relative molar ratio of each peptide. The top seven proteins are listed because the seventh protein, the GABAAR α6 subunit, is a major component of the 500 kDa, but not 720 kDa, GABAAR complex. Detailed peptide information is provided in the Table S1.
LH4-like GARLHs bridge γ2-containing GABAARs and NL2
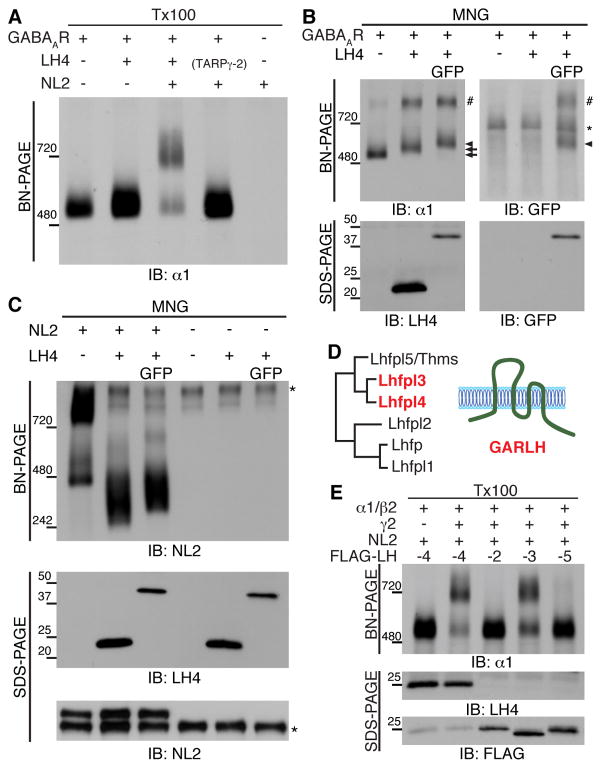
We next investigated the interactions of the GABAAR components using cRNA-injected oocytes. The co-expression of three GABAAR subunits (α1, β2 and γ2) with LH4 and NL2 reconstituted the GABAAR complex at 720 kDa observed in the brain as determined using BN-PAGE with Triton X-100 solubilization (Figure 2A, Lane 3). When either LH4 or NL2 were eliminated, the 720 kDa complex failed to form (Figure 2A, Lane 4 and 2). When membranes from cRNA-injected oocytes were solubilized with MNG, the GABAAR α1 subunit was detected at 500 and 870 kDa (Figure 2B). Because the band detected at 870 kDa was observed in neither the cerebellar lysate nor the Triton X-100-solubilized membranes (Figure 1A and Figure 2A), this band was considered an artifact of the heterologous overexpression system with MNG solubilization. Upon co-expression with LH4 (22 kDa) and LH4 tagged with GFP at its C-terminus (49 kDa), the GABAAR band at 500 kDa shifted to 520 kDa and 550 kDa, respectively (Figure 2B and S2A). Furthermore, anti-GFP antibody recognized the GABAAR band with LH4-GFP at 550 kDa as well as the artificial band around 870 kDa (Figure 2B). These results indicate an interaction of GABAARs with LH4.
Figure 2. LH4-like GARLH family proteins bridge γ2-containing GABAARs and neuroligin-2.
(A) Recombinant GABAARs, consisting of α1, β2 and γ2 subunits in cRNA-injected oocytes, migrated to 520 kDa using BN-PAGE with Triton X-100 (Tx100) solubilization. Co-expression of recombinant GABAARs with Lhfpl4 (LH4) and neuroligin-2 (NL2) increased the molecular weight to 720 kDa, similar to that for cerebellar GABAARs, whereas co-expression of recombinant GABAARs with LH4 alone or NL2/TARPγ-2 failed to similarly increase the molecular weight. (B) Co-expression of LH4 (22 kDa) and LH4 tagged with GFP (LH4-GFP, 49 kDa) shifted recombinant GABAARs at 500 kDa (arrow) to 520 kDa (arrow) and 550 kDa (arrowhead), respectively, in cRNA-injected oocytes solubilized with maltose-neopentyl glycol (MNG). Notably, oocyte membranes solubilized with MNG show a background band (asterisk) and an artificial band of oligomeric GABAARs with LH4 (#). (C) NL2 expressed in oocytes migrates as two bands, a strong band at 800 kDa and weak bands at 460–520 kDa representing various NL2 oligomers. The co-expression of NL2 with LH4 induced the collapse of the NL2 oligomers and emergence of a strong band at 330 kDa, which was shifted up with LH4-GFP co-expression. Asterisk shows non-specific signal. (D) Phylogenetic tree of LH4- homologous proteins. LH4 and 3 proteins are classified here as GABAAR regulatory Lhfpl (GARLH) family proteins. (E) In the cRNA-injected oocyte system, recombinant GABAARs consisting of α1, β2 and extracellularly HA-tagged γ2 with NL2 and FLAG-tagged LH4 reconstituted to 720 kDa, as do native GABAARs, whereas GABAARs containing α1 and β2 without HA-γ2 failed to reconstitute to this molecular weight. Similarly, FLAG-LH3, but not LH2 and LH5, reconstitute cerebellar GABAARs of 720 kDa.
NL2 expressed in cRNA-injected oocytes was detected as two distinct broad bands at 800 kDa and 460 kDa (Figure 2C). Since NL2 is 100 kDa on SDS-PAGE, such large molecular weights on BN-PAGE suggest formation of NL2 oligomers as reported previously (Arac et al., 2007; Dean et al., 2003; Fabrichny et al., 2007; Koehnke et al., 2008). NL2 co-expressed with LH4 and LH4-GFP was detected primarily as a single band at 330 and 360 kDa, respectively (Figure 2C and S2B), suggesting binding of LH4 or LH4-GFP to NL2 disrupts large NL2 oligomers. Because a shift to lower molecular weight upon binding is counterintuitive, we next examined interactions of NL2 with LH4 using a co-immunoprecipitation assay from cRNA-injected oocytes. NL2 co-immunoprecipitated LH4, but not TARPγ-2, in cRNA-injected oocytes (Figure S2C). To determine which NL2 domains are responsible for the LH4 interaction, we generated various deletion construct of HA-tagged NL2 (Figure S2D). All deletion mutants as well as LH4-GFP were co-expressed in oocytes, although the NL2 full length (FL) and the extracellular domain deletion mutant (ΔN) expressed at lower levels (Figure S2E, input). Since expression levels were variable, we used co-immunoprecipitation assay instead of BN-PAGE for evaluating their interaction. We found that NL2 FL, ΔN, and ΔC, but not ΔTM, were co-immunoprecipitated with LH4-GFP using an anti-GFP antibody (Figure S2E). These results indicate the TM domain of NL2 is required for its interaction with LH4.
In order to test which other LH4 homologues assemble with GABAARs and NL2, we evaluated Lhfpl 2, 3 and 5 (amino acid sequence identities to LH4, 26.1%, 78.5%, and 61.8%, respectively) based on phylogenetic analysis (Figure 2D). We tagged the C-terminus of each LH isoform with a FLAG epitope and co-expressed these FLAG-tagged LH isoforms with NL2 and GABAAR subunits in oocytes using cRNA injections. Co-expression of α1 and β2 with FLAG-LH4 and NL2 failed to reconstitute the native GABAAR complex at 720 kDa, but the additional co-expression of extracellularly HA-tagged γ2 succeeded (Figure 2E). Similar to FLAG-LH4, FLAG-LH3 formed a complex at 720 kDa with NL2 and the γ2-containing GABAARs (Figure 2E). Despite expression levels of FLAG-LH2 and -LH5 higher than that of FLAG-LH4 in oocytes injected with the corresponding cRNAs on SDS-PAGE, FLAG-LH2 and -LH5 failed to form the 720 kDa complex (Figure 2E). The Lhfpl family genes were named based on their sequence similarity (Petit et al., 1999); however, our results show that the protein properties of LH3 and LH4 are distinct from those of the other LH genes. To reflect these distinct protein properties we defined a family protein termed GABAAR regulatory Lhfpl (GARLH), comprised of LH3 and LH4 (Figure 2D). Overall, these results indicate that GARLHs form a complex with NL2 and γ2-containing GABAARs.
GABAAR γ2 subunit stabilizes the expressions of LH4 and NL2 proteins
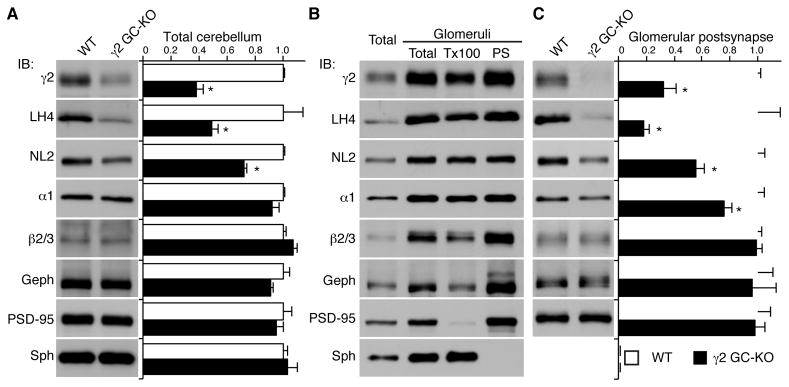
Kainate- and AMPA-type glutamate receptors stabilize the protein expressions of Neto and CNIH auxiliary subunits, respectively (Kato et al., 2010; Zhang et al., 2009); therefore, we next asked whether GABAARs affect LH4 expression in the brain. Because the γ2 subunit is required for formation of the tripartite complex NL2/LH4/GABAAR (Figure 2E), and because most conventional γ2 knockout (KO) mice show postnatal lethality (Gunther et al., 1995), we used cerebellar granule cell (GC)-specific γ2 KO mice generated by crossing the Gabra6 promoter Cre recombinase transgenic mice with conditional γ2 KO mice (Funfschilling and Reichardt, 2002; Schweizer et al., 2003). In cerebella from the γ2 GC-KO mice, γ2 was reduced as expected; the residual γ2 likely originates from cerebellar Purkinje cells, in which γ2 expression was not disrupted. Moreover, the total amounts of both LH4 and NL2 were reduced (Figure 3A). Cerebellar granule cells form GABAergic synapses with axons from cerebellar Golgi cells on cerebellar glomeruli. To further examine whether LH4 and NL2 are reduced at postsynapses in the γ2 KO mice, we purified the mouse glomeruli-enriched fraction and then isolated the Triton X-100-soluble extrasynaptic and insoluble postsynaptic fractions as described previously (Viennot et al., 1991; Wu and Siekevitz, 1988). We found that LH4 and NL2 co-fractionated with γ2, α1 and β2/3 in glomeruli-enriched postsynaptic fractions in WT mice (Figure 3B). Furthermore, in the γ2 GC-KO mice, LH4 and γ2 expression levels in the postsynaptic fraction were reduced to a similar extent as the total glomeruli proteins, and NL2 and α1 expression was significantly, but modestly, reduced (Figure 3C). Taken together, these results suggest that γ2-containing GABAARs stabilize LH4 protein expression in the cerebellum.
Figure 3. The GABAAR γ2 subunit modulates expression of Lhfpl4 and neuroligin-2 in vivo.
(A) Protein levels in the total cerebella from wild-type (WT) and granule cell (GC)-specific γ2 knockout (KO) mice. The amounts of LH4, γ2 and NL2 proteins were significantly reduced in the γ2 GC-KOs without changes in gephyrin (Geph) and PSD-95. Bar graph shows relative expressions to WT mice. (B) Biochemical fractionation of mouse cerebella shows co-segregation of Lhfpl4 (LH4) with GABAAR subunits (α1, γ2 and β2/3) and neuroligin-2 (NL2). The cerebellar homogenate (Total) was fractionated into the cerebellar glomerulus-enriched fraction (Glomeruli, Total) and then separated into the Triton X-100-soluble fraction (Tx100) and the Triton X-100-insoluble postsynaptic fraction (PS). Synaptophysin (Sph) was used as a presynaptic marker. Gephyrin (Geph) and PSD-95 were used as inhibitory and excitatory postsynaptic markers, respectively. (C) Protein levels in the PS fraction of cerebella from WT and GC-γ2 KO mice. The amounts of LH4, γ2, α1 and NL2 proteins were significantly reduced in the GC-γ2 KOs without changes in Geph and PSD-95. Data are mean ± s.e.m.; n = 4 animals; Mann-Whitney U-test; *P < 0.05.
LH4 does not modulate surface expression or agonist/antagonist sensitivity of GABAARs
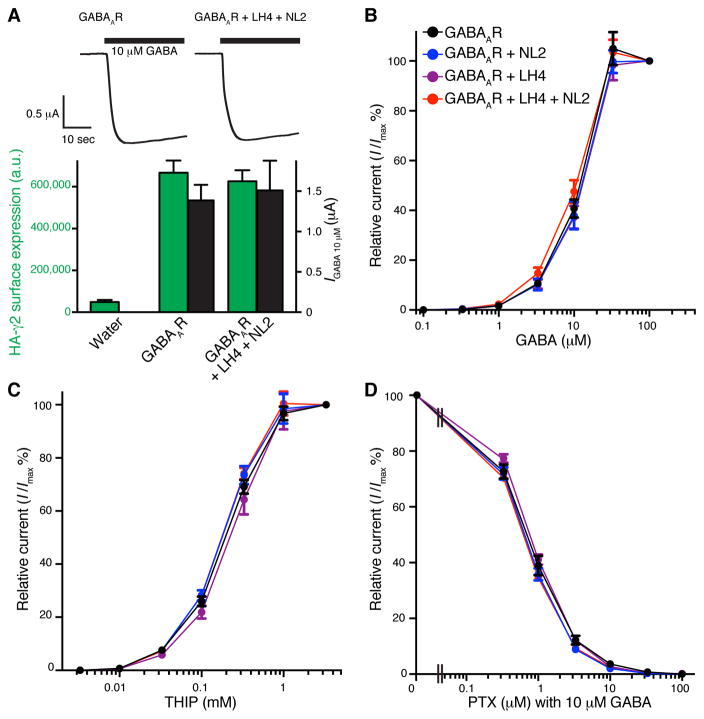
Auxiliary subunits modulate trafficking and/or properties of receptors (Boulin et al., 2012; Yan and Tomita, 2012). Thus, we investigated the potential modulation of surface expression or properties of GABAARs by LH4/NL2 using cRNA-injected oocytes. We expressed α1, β2, and extracellularly HA-tagged γ2 (HA-γ2) with or without LH4 and NL2 in oocytes by corresponding cRNA injections. In this condition these proteins form the 720 kDa complex (Figure 2E). We then measured GABA-evoked currents using two-electrode voltage clamp recording and surface expression of HA-γ2 using a chemiluminescence assay with cell non-permeable labelling of the extracellular HA epitope (Tomita et al., 2004; Zerangue et al., 1999). In these assays, GABA-evoked currents and surface expression of HA-γ2 were not altered by co-expressions of LH4 and NL2 (Figure 4A). To further examine whether LH4 modulates the pharmacology of GABAARs, we measured dose response of the full agonist GABA, the partial agonist THIP, and the antagonist picrotoxin for α1/β2/HA-γ2-containing GABAARs with or without LH4 and/or NL2. We did not see any obvious changes in dose response and estimated EC50/IC50 and Hill coefficient of GABA, THIP and Pictrotoxin (Figure 4B, C, D and Table S2).
Figure 4. LH4 does not modulate GABAAR surface expression or sensitivity to agonists or an antagonist.
(A) Oocytes were injected with cRNAs encoding α1, β2 and extracellularly HA-tagged γ2 with or without Lhfpl4 (LH4) and neuroligin-2 (NL2). GABA (10 μM)-evoked currents and surface expression of HA-γ2 were measured by two electrode voltage clamp (TEVC) recording and chemiluminescence assay, respectively. Co-expression of LH4 and NL2 does not modulate surface expression (green bars) or GABA-evoked currents (black bars) of α1/β2/HA-γ2 containing GABAARs (n = 5 oocytes). (B–D) The cRNAs of α1, β2, and HA-γ2 with combinations of LH4 and NL2 were injected into oocytes, and dose responses were measured with TEVC. Co-expression of LH4 and NL2 does not alter GABAAR sensitivity to the agonists, GABA (B) and 4,5,6,7-tetrahydroisoxazole(5,4-c)pyridin-3-ol (THIP) (C) or an antagonist, picrotoxin (PTX) (D) with 10 μM GABA. Data are mean ± s.e.m. (n = 5 – 6 oocytes).
LH4 regulates synaptic clustering of γ2-containing GABAARs in primary neurons
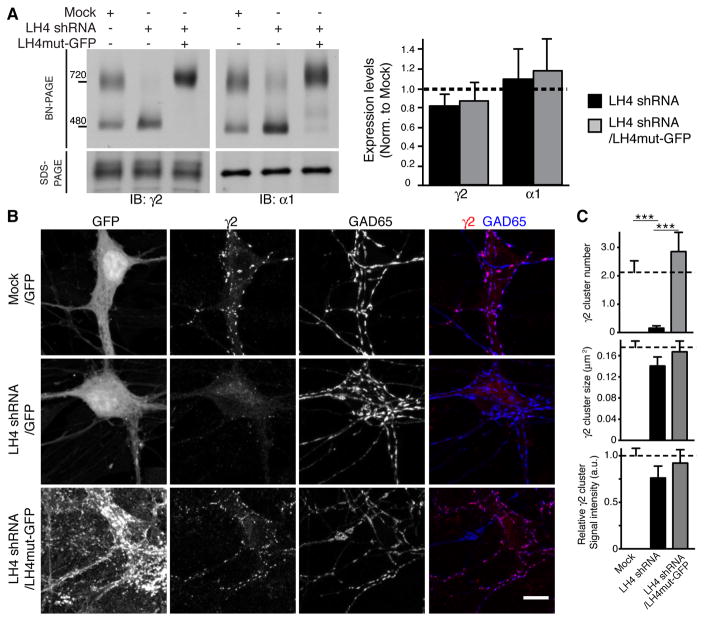
We next examined the role of LH4 associated with GABAARs by reducing LH4 expression using shRNA. To avoid potential confounding redundancy by LH3 (Figure 2D and E), we focused on the hippocampus, which expresses a low level of LH3 (Figure 1C). Similar to the results observed using the cerebellum (Figure 1), both the γ2 and α1 subunits formed two distinct complexes at 480 kDa and 720 kDa in primary hippocampal neurons treated with lentivirus containing GFP alone (Mock, Figure 5A). In neurons treated with lentivirus containing LH4 shRNA, LH4 was undetectable (Figure 1E), the 720 kDa complex was absent, and the intensity of the 480 kDa band was increased (Figure 5A). shRNA may have off target effects. To confirm that loss of the 720 kDa complex in LH4 shRNA-treated neurons was due to loss of LH4, we expressed a C-terminal GFP-tagged LH4 mutant carrying silent mutations in the shRNA target sequence (LH4mut-GFP) in LH4 shRNA-treated neurons. Overexpression of LH4mut-GFP rescued the native GABAAR complex at 740 kDa; this molecular weight was slightly higher than that for the native 720 kDa GABAAR complex due to the addition of GFP (Figure 5A). Moreover, in neurons treated with LH4mut-GFP, the ratio of the 720 to 480 kDa complex was increased (Figure 5A), consistent with overexpression of LH4mut-GFP and suggesting that LH4 limits the complex formation in primary cultured neurons. Total protein expression levels of α1 and γ2 as determined using SDS-PAGE were not altered (Figure 5A).
Figure 5. Lhfpl4 is essential for synaptic GABAAR clustering.
(A) In primary hippocampal neurons, infection of Lhfpl4 (LH4) shRNA lentivirus reduces the molecular weight of the native GABAAR complex from 720 kDa to 480 kDa, as determined using BN-PAGE. Expressing a GFP-tagged LH4 mutant resistant to LH4 shRNA restores the GABAAR complex to 740 kDa. The amounts of γ2 and α1 subunit proteins are not altered, as determined using SDS-PAGE (n = 3 samples from three independent cultures). (B) GABAAR γ2 subunits co-localize with GAD-65 in primary hippocampal neurons infected with a GFP lentivirus. The γ2 subunit signal was decreased in neurons infected with an LH4 shRNA lentivirus and restored by reintroducing LH4mut-GFP in LH4 shRNA-treated neurons. GFP signals indicate infected neurons, and under these conditions, almost all neurons showed GFP signals. (C) Quantification of γ2 subunit immunolabelling shows a significant reduction in the cluster number/10μm dendrite, but not size or intensity, of γ2 clusters in LH4 shRNA-treated neurons. Sample numbers (dendritic segments/clusters) for each condition (Mock, LH4 shRNA, LH4 shRNA/LH4mut-GFP) are as follows; n = 24/168, 22/11, 8/71, respectively, from two independent cultures. Dendritic areas were chosen randomly for analysis of cluster numbers and all detected clusters were subsequently analysed for sizes and signal intensities. Data are mean ± s.e.m.; Kruskal-Wallis test followed by Dunn’s post-test; ***P < 0.001.
We next examined the role of LH4 on the synaptic localization of γ2-containing GABAARs. The γ2 subunit was co-localized with inhibitory presynaptic GAD65 in neurons treated with lentivirus containing only GFP (Mock, Figure 5B). The number of γ2-immunoreactive puncta was markedly reduced in LH4 shRNA-treated neurons (Figure 5B and C and Figure S3A) without changes in the size or signal intensity of the remaining puncta (Figure 5C). γ2 puncta were restored by overexpression of LH4mut-GFP (Figure 5B and C), confirming that loss of γ2 puncta was due specifically to loss of LH4. Moreover, GAD65 and the γ2 subunit partially co-localized with overexpressed LH4mut-GFP that was diffuse along the dendrites, where it could be associated with extrasynaptic γ2-containing receptors, alone or associated with other uncharacterized partners (Figure S3B).
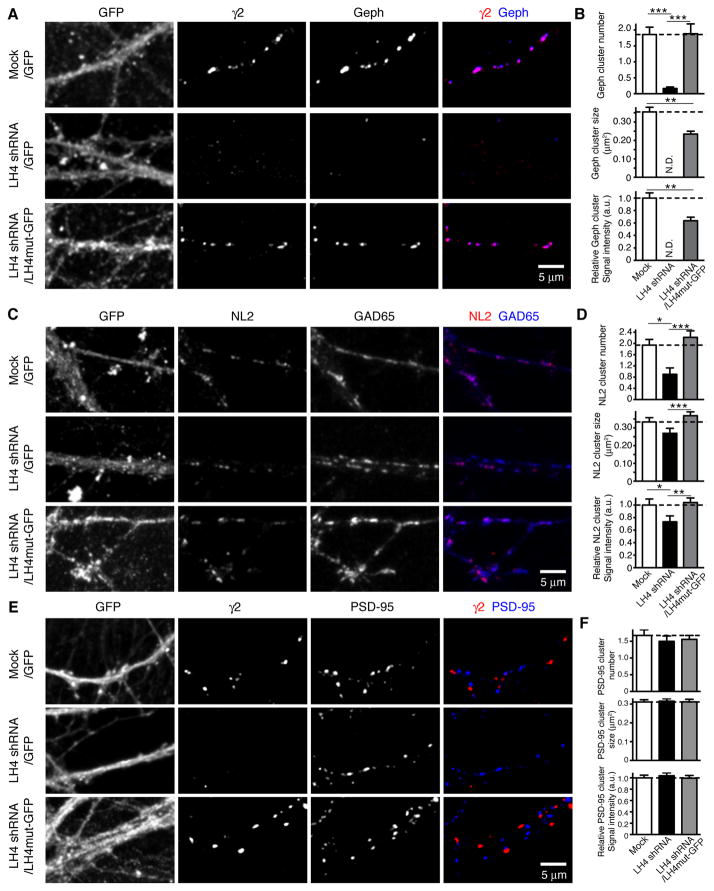
The localization of NL2 and gephyrin was also examined in primary hippocampal neurons with different levels of LH4. We observed a drastic reduction in the number of immunoreactive puncta of gephyrin as well as γ2 in LH4 shRNA-treated neurons (Figure 6A and B). The number of gephyrin-immunoreactive puncta was restored by overexpressing LH4mut-GFP into LH4 shRNA-treated neurons (Figure 6B), indicating that the loss of the gephyrin puncta number was due to a specific loss of LH4. However, the size and signal intensity of gephyrin puncta restored by LH4mut-GFP were partially rescued (Figure 6B). On the other hand, the reduction in the number of NL2-immunoreactive puncta was significant, but modest (Figure 6C and D). Similarly, the size and the intensity of NL2 puncta were reduced modestly, and were restored by expressing LH4mut-GFP (Figure 6D). Thus, a substantial amount of NL2 can localize to synapses without LH4, γ2-containing GABAARs or gephyrin. Notably, excitatory postsynaptic PSD-95 was not obviously altered by changes in LH4 expression (Figure 6E and F).
Figure 6. Lhfpl4 modulates the distribution of gephyrin and Neuroligin-2.
Primary hippocampal neurons at various Lhfpl4 (LH4) expression levels were stained for inhibitory postsynaptic proteins gephyrin (Geph) and neuroligin-2 (NL2). (A) Gephyrin co-localizes with the GABAAR γ2 subunit in primary hippocampal neurons infected with a control GFP lentivirus (Mock). Gephyrin as well as γ2 were reduced in neurons infected with an LH4 shRNA lentivirus, and were restored by overexpressing LH4mut-GFP in LH4 shRNA-treated neurons. (B) The number of gephyrin clusters per 10 μm dendrite was robustly reduced in neurons treated with LH4 shRNA and restored in neurons treated with LH4 shRNA with LH4mut-GFP expression. The substantial reduction in gephyrin puncta in LH4 shRNA-treated neurons precluded the precise evaluation of the size and signal intensity of gephyrin (N.D.). The size and signal intensity of gephyrin puncta were not fully restored by over-expression of LH4mut-GFP in LH4 shRNA-treated neurons. Sample numbers (dendritic segments/clusters) for each condition (Mock, LH4 shRNA, LH4 shRNA/LH4mut-GFP) are as follows; n = 17/148, 18/13, 16/158, respectively, from two independent cultures. Dendritic areas were chosen randomly for analysis of cluster numbers and all detected clusters were subsequently analysed for sizes and signal intensities. (C) NL2 co-localizes with GAD-65 in primary hippocampal neurons infected with a GFP alone lentivirus (Mock). The NL2 signal was reduced in LH4 shRNA-treated neurons and restored by overexpressing LH4mut-GFP in LH4 shRNA-treated neurons. (D) The cluster number and signal intensity of NL2 were modestly, but significantly reduced in LH4 shRNA-treated neurons, and restored by overexpressing LH4mut-GFP. Sample numbers (dendritic segments/clusters) for each condition (Mock, LH4 shRNA, LH4 shRNA/LH4mut-GFP) are as follows; n = 15/130, 18/71, 16/168, respectively, from two independent cultures. (E and F) PSD-95 shows a punctate pattern in primary hippocampal neurons infected with lentivirus carrying GFP alone (Mock), LH4 shRNA or an LH4 mutant resistant to LH4 shRNA (LH4mut-GFP). No significant difference was observed in the cluster number/10 μm dendrite, size and signal intensity of PSD-95. Sample numbers (dendritic segments/clusters) for each condition (Mock, LH4 shRNA, LH4 shRNA/LH4mut-GFP) are as follows; n = 36/373, 36/339, 37/327, respectively, in two independent cultures. Dendritic areas were chosen randomly for analysis of cluster numbers and all detected clusters were subsequently analysed for sizes and signal intensities. Data are mean ± s.e.m.; Kruskal-Wallis test followed by Dunn’s post-test; *P < 0.05; **P < 0.01; *** P < 0.001.
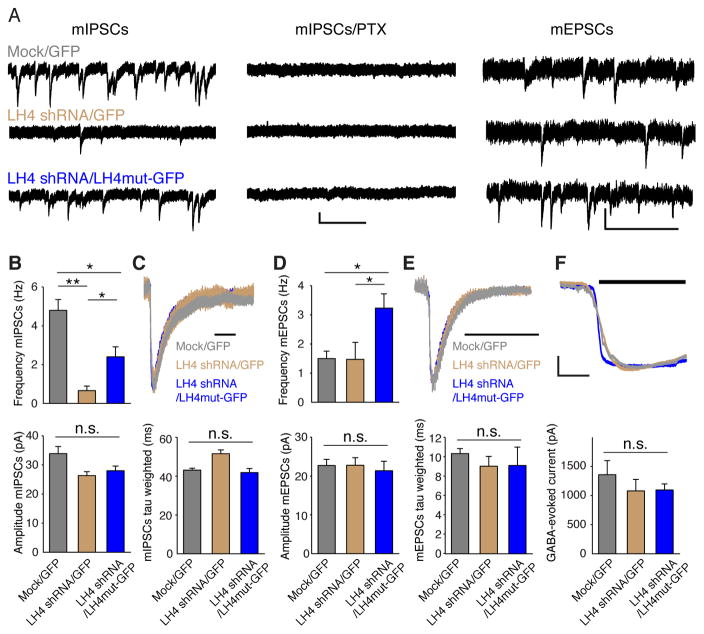
We next examined GABAAR-mediated synaptic transmission in primary hippocampal neurons with various expression levels of LH4 using lentiviruses (Figure 5A) and measured miniature synaptic events (Figure 7A). Synaptic events were measured under whole-cell voltage-clamp configurations (Vh = −70 mV), and miniature inhibitory postsynaptic currents (mIPSCs) were isolated by adding 20 μM CNQX and 100 μM d-APV with 1 μM tetrodotoxin (TTX) (Figure 7A). All mIPSCs were eliminated by further addition of 100 μM picrotoxin (PTX) (Figure 7A). The frequency of mIPSCs was markedly reduced in LH4 shRNA-treated neurons and was partially restored by overexpressing LH4mut-GFP into these LH4 shRNA-treated neurons (Figure 7A and B). No changes in the decay kinetics of the mIPSCs were observed (Figure 7C). The frequency and amplitude of mEPSCs isolated by adding 100 μM PTX and 1 μM TTX were not altered in neurons treated with LH4 shRNA (Figure 7A and D). Unexpectedly, overexpression of LH4mut-GFP in LH4 shRNA-treated neurons increased the mEPSC frequency (Figure 7D). While the reason for this increase is unclear, it is possible that overexpressed LH4mut-GFP induces changes in NLs or other molecules that increase mEPSCs (Chih et al., 2005; Graf et al., 2004). The decay kinetics of the mEPSCs was unaltered in neurons at various LH4 expression levels (Figure 7E). Since the reduction in the mIPSCs upon LH4 knockdown could be due to changes in surface expression or synaptic stabilization of GABAARs, we measured 100 μM GABA-evoked currents in primary cultured neurons treated with shRNA lentivirus and 1 μM tetrodotoxin. No significant differences were observed in GABA-evoked currents in neurons at different LH4 expression levels (Figure 7F). From these, we conclude that the main function of endogenous LH4 is to localize GABAARs to synapses, without affecting the GABAAR surface expression.
Figure 7. Lhfpl4 is required for GABAAR-mediated synaptic transmission.
(A–E) Miniature synaptic events were measured using whole-cell voltage-clamp (Vh = −70 mV) recordings in primary cultured hippocampal neurons treated with GFP lentivirus (Mock), LH4 shRNA lentivirus and lentivirus carrying LH4 shRNA and a GFP-tagged LH4 mutant resistant to LH4 shRNA (LH4mut-GFP). Miniature inhibitory postsynaptic currents (mIPSCs) were isolated by application of TTX (1 μM), CNQX (20 μM) and APV (100 μM), and all events were blocked by further application of picrotoxin (PTX; 100 μM). Scale bar, 20 pA/1 s. Miniature excitatory postsynaptic currents (mEPSCs) were measured by application of TTX (1 μM) and PTX (100 μM). Scale bar, 20 pA/0.5 s. (B) Quantitation of frequency and amplitude of mIPSCs (n=11 – 14 from three independent cultures). (C) Representative traces of mIPSC decay normalized at the peak and quantitation of the mIPSC weighted decay tau estimated by fitting the decay to a biexponential function. Scale bar 50 ms. (D) Quantitation of frequency and amplitude of mEPSCs (n= 8 – 9 neurons from three independent cultures). (E) Representative traces of mEPSC decay normalized at the peak and quantitation of the mEPSC weighted decay tau estimated by fitting the decay to a biexponential function. Scale bar 20 ms. (F) Representative traces and quantitation of peak amplitudes of the GABA-evoked currents upon 5 s applications of 100 μM GABA on primary cultured hippocampal neurons with 1 μM TTX (n = 4 neurons each from two independent cultures). Scale bar, 250 pA/2 s. Data are mean ± s.e.m.; Kruskal-Wallis test followed by Dunn’s post-test; * P < 0.05, ** P < 0.01.
LH4 regulates GABAAR-mediated synaptic transmission in vivo
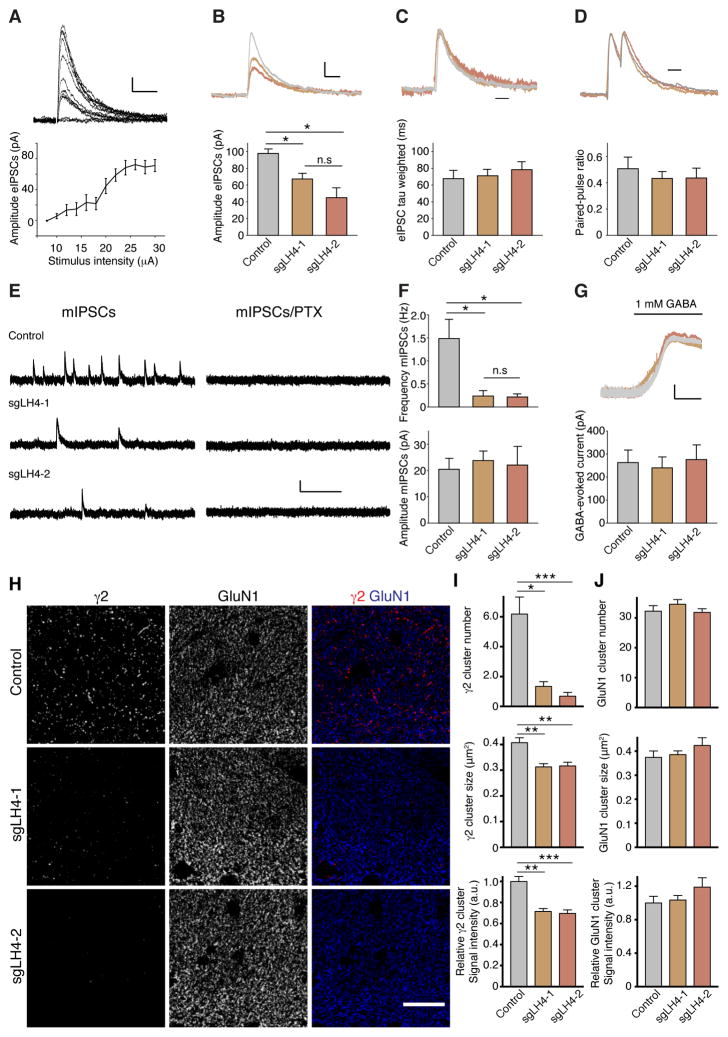
To examine whether LH4 is required for GABAAR transmission and clustering in adult hippocampus, we used stereotaxic injections of adeno-associated virus (AAV) carrying Cre recombinase and single guide RNA (sgRNA) against LH4 into Cre-dependent Cas9 knockin mice as described previously (Platt et al., 2014). AAVs carrying Cre recombinase and sgRNAs (control, sgLH4-1 and sgLH4-2) into Cre-dependent Cas9 knockin mice at 3–4 weeks. Three to nine weeks after AAV injections we prepared acute hippocampal slices and measured under whole-cell voltage-clamp configurations (Vh = 0 mV) electrically evoked IPSCs (eIPSC), mIPSCs, and surface GABAAR activity upon GABA bath application in CA1 pyramidal cells with GFP signal that indicate expressions of both sgRNA and Cas9 (Platt et al., 2014). In this condition, a 90% reduction in LH4 expression was observed in hippocampi expressing two distinct sgRNAs against LH4 (sgLH4-1 and -2) (Figure S4A). To examine eIPSCs, we placed a stimulating pipette in the middle of the stratum pyramidale 150 μm away from the recorded cell (Jurgensen and Castillo, 2015). IPSCs were elicited upon stimulation and the eIPSC amplitude was saturated at 24 μA in CA1 neurons expressing sgLH4-1 (Figure 8A). We stimulated at the saturated level (26 μA) for comparison of eIPSCs, and found that eIPSC amplitudes were reduced 40% and 60% in CA1 neurons expressing sgLH4-1 and sgLH4-2, respectively, compared to those expressing control sgRNA (Figure 8B). The decay kinetics of eIPSCs (Figure 8C) and the paired-pulse ratio with 50 ms inter-stimulus intervals (Figure 8D) were no different. The mIPSCs were measured with 1 μM TTX, and showed a 90% reduction in mIPSC frequency without changes in amplitudes (Figure 8E and F). Furthermore, bath application of 1 mM GABA showed no difference in GABA-evoked currents from neurons expressing each sgRNA compared to controls (Figure 8G). These results are consistent with the results using LH4 shRNA-treated neurons (Figure 7). A robust reduction in mIPSC frequency without changes in mIPSC amplitude and a partial reduction in eIPSC amplitude were also observed in the NL1/2/3 triple knockout mice (Varoqueaux et al., 2006), supporting that LH4 functions with NLs in vivo.
Figure 8. GABAAR-mediated synaptic transmission requires LH4 in adult hippocampus.
Synaptic properties and GABAAR localization were measured in hippocampi from Cas9 knockin mice injected with an AAV virus carrying Cre recombinase and two independent sgRNAs for LH4 (sgLH4-1 and sgLH4-2). (A) Representative traces and quantitation of peak amplitudes of evoked IPSCs with various stimulus intensities in CA1 hippocampal neurons, showing saturation of the eIPSC amplitude beyond 24 μA stimulus intensity (n = 10 neurons from four mice injected with sgLH4-1). Scale bar: 10 pA/100 ms. (B–D) Representative traces and quantitation of peak amplitude (B), weighted decay tau with traces normalized at the peak (C) and the paired-pulse ratio with 50 ms inter-stimulus interval with traces normalized at the peak (D) of IPSCs evoked at 26 μA stimulus intensity from mice injected with each AAV (n = 7 neurons from four animals for control, n = 12 neurons from five animals for sgLH4-1, n = 13 neurons from five animals for sgLH4-2). The amplitude of evoked IPSCs was significantly reduced in neurons expressing sgLH4s compared to control, whereas the decay kinetics and paired-pulse ratio were not. Scale bar: 25 pA/50 ms (B), 50 ms (C), 50 ms (D). (E) Representative traces of miniature IPSCs measured before and after picrotoxin (PTX) (100 μM) application. Scale bar: 20 pA/1.2 sec. (F) Quantitation of frequency and amplitude of mIPSCs from mice injected with each AAV (n = 5 neurons from three animals for control, n = 5 neurons from three animals for sgLH4-1, n = 6 neurons from four animals for sgLH4-2). The frequency, but not the amplitude, of the mIPSCs was significantly reduced in neurons expressing sgLH4s compared to control. (G) Representative traces and quantitation of the 1 mM GABA-evoked currents. Scale bar: 50 pA/1 min. No significant differences were found in the amplitude of the GABA-evoked currents (n = 4 neurons from four animals for control, n = 6 neurons from three animals for sgLH4-1, n = 7 neurons from four animals for sgLH4-2). (H) Punctate localization of γ2 is reduced in hippocampi expressing sgRNAs for LH4. By contrast, GluN1 is not altered. (I) Numbers of γ2-immunolabelled cluster per 100 μm2 are strongly reduced in dendritic area of dentate gyrus from animals carrying sgLH4-1 and sgLH4-2, while slight reductions are observed in γ2-cluster sizes and signal intensities (n = 12 areas from three animals). (J) The cluster number, size and signal intensity of GluN1 were unaltered. Data are mean ± s.e.m.; Kruskal-Wallis test followed by Dunn’s post-test; *P < 0.05; **P < 0.01; ***P < 0.001.
We next examined GABAAR clustering in adult hippocampus, and found that both sgLH4s strongly reduced the number, and slightly reduced size and intensity, of γ2-immunoreactive puncta in the hippocampus 2–5 months after the AAV injection (Figure 8H and I; area quantified is shown in low magnification images in Figure S4B, box). No obvious changes in the signal of NMDA receptor GluN1 subunit (Figure 8H and J), GAD65 (Figure S4C and D) and PSD-95 (Figure S4E and F) were detected. These results strongly suggest that LH4 is required for the synaptic clustering of GABAARs in vivo. Thus, LH4 regulates the synaptic activity and clustering of GABAARs, but not GABAAR activity at the neuronal surface. These results indicate that LH4 controls synaptic stabilization of GABAARs.
Discussion
This study identified the first example of putative auxiliary subunits for pentameric ligand-gated anion channels. We found that the LH4-like GARLH family proteins are putative auxiliary GABAAR subunits that control GABAAR synaptic localization and GABA-mediated synaptic transmission by anchoring the GABAAR to synaptically localized NL2.
GARLH is a putative GABAAR auxiliary subunit
Auxiliary subunits are non-pore-forming subunits that directly and stably interact with a pore-forming subunit, modulate receptor trafficking and/or function, and are necessary for endogenous receptor function in vivo (Yan and Tomita, 2012). We found that most of the γ2 subunit-containing GABAARs form a stable complex with LH4 in the cerebellum and hippocampus, that LH4 is not necessary for pore formation because GABA-evoked currents were detected without LH4 in cRNA-injected oocytes (Fig. 4A), that LH4 interacts with pore-forming subunits of GABAARs and that LH4 is necessary for the synaptic localization of GABAARs in vivo. Therefore, LH4 could be called as an auxiliary subunit of γ2-containing GABAARs. GABAARs also form a complex with LH3. Because LH3 and LH4 display properties distinct from the other four Lhfp-like homologues, we categorized LH3 and LH4 as GABAAR regulatory Lhfpl family proteins (GARLHs).
Auxiliary subunits often modulate channel properties of ion channels. For example, TARP AMPAR auxiliary subunits and Neto KAR auxiliary subunit modulates receptor sensitivity to agonists, antagonists or potentiators (Jackson and Nicoll, 2011; Yan and Tomita, 2012). However, we did not observe obvious difference in the sensitivity to GABA, THIP, and PTX of GABAARs by LH4 co-expression in Xenopus laevis oocytes (Figure 4 and Table S2). This may suggest that GARLHs interact with GABAARs outside of the channel ligand-binding domains or gating domain, although some types of modulation of GABAAR channel properties by GARLHs cannot be ruled out. Modulation of channel properties/functions is one of criteria for auxiliary subunits of ionotropic glutamate receptors (Jackson and Nicoll, 2011; Yan and Tomita, 2012). Here the term “functions” could indicate not only ion channel activity, but also biological functions including their synaptic localization. GARLH is tentatively called as a putative auxiliary subunit of GABAARs here, and could be called as an auxiliay subunit upon characterization of GARLH-modulation of GABAAR channel properties or clarification of definition of auxiliary subunits.
Auxiliary subunits have been identified for tetrameric ligand-gated cation channels (Jackson and Nicoll, 2011; Yan and Tomita, 2012). Here, GARLH could be the first example of auxiliary subunits for pentameric ligand-gated anion channels. This finding supports the proposition that other types of vertebrate pentameric ligand-gated channels, such as the native glycine receptor, serotonin receptor and nicotinic acetylcholine receptor, may contain auxiliary subunits still unidentified in native environments. The levamisole-sensitive acetylcholine receptor in C. elegans functions with single-pass transmembrane proteins LEV-10 and MOLO-1 and the secreted protein LEV-9 (Boulin et al., 2012; Gally et al., 2004; Gendrel et al., 2009), though their mammalian functional homologues have not been identified yet. On the other hand, Neto functions as an auxiliary subunit of the KAR to determine its channel properties in the brain (Zhang et al., 2009), and Neto shares the highest homology with the C. elegans SOL-2, which is indeed an auxiliary subunit for the C. elegans ionotropic glutamate receptor (Wang et al., 2012). Interestingly, GARLHs share similarity with the C. elegans F26D10.11 protein, which may function as a GABAAR auxiliary subunit in C. elegans. Functional preservation of molecular machinery for synaptic transmission during evolution may support similar fundamental mechanisms of synaptic transmission among various species.
The cell biology of inhibitory synapses
We demonstrated that a GARLH isoform, LH4, interacts specifically with γ2-containing GABAARs and NL2, and determines the synaptic localization and activity of GABAARs. Prior work showed that total protein expression levels of cerebellar GABAARs were unchanged in NL KO mice (Zhang et al., 2015). Similarly, here we found no detectable changes in total GABAAR protein levels or activity at the neuronal surface in LH4 shRNA treated neurons and in adult hippocampus disrupting LH4 expressions, despite a loss of synaptic GABAAR clusters. These results suggest that GABAARs diffuse laterally at the neuronal surface and are stabilized at inhibitory synapses by NL2 and LH4.
Our findings open a number of unexpected questions, the answers to which will provide insight in the cell biology of inhibitory synapses. For example, it remains unclear if specific GARLH-NL pairs interact preferentially, potentially defining a code to tune synaptic function. Moreover, we found that cell surface GABAAR activity does not depend on LH4, indicating that LH4 does not play a critical role in the intracellular trafficking of GABAARs. Nonetheless, detailed studies defining the subcellular compartment in which GARLHs and NLs first interact with GABAARs will reveal crucial information about synaptic development. Furthermore, GABAARs are comprised from a pool of 19 subunits, and the rules governing the combinatorial assembly of these subunits into pentamers with distinct functions have been extensively studied (Fritschy et al., 2012; Olsen and Sieghart, 2008; Sigel and Steinmann, 2012). These rules will need to be updated to account for assembly with GARLHs and NLs.
Our study renews focus on NL2 as an upstream factor in determining inhibitory synapse identity and function, and opens many new questions regarding the NL2 function. NL2 overexpressed in neurons localizes preferentially at inhibitory synapses (Chih et al., 2005; Graf et al., 2004), supporting the suggestion that at least NL2 and perhaps other NLs have a signal for localizing at inhibitory synapses, presumably through an interaction with neurexin in vertebrates (Ichtchenko et al., 1995; Missler et al., 2003). Although neurexin was previously reported to interact directly with the GABAARs (Zhang et al., 2010), we did not detect neurexin in the native NL2-LH4-GABAAR complex. In Caenorhabditis elegans, MADD4 interacts with NL to recruit GABAARs at synapses (Maro et al., 2015; Tu et al., 2015). Further identification of NL interactors and reconstitution of native NL-containing complexes at synapses may ultimately provide a unifying model relating synapse development, synaptic adhesion and neurotransmitter receptor localization. During revision of this manuscript, 174 proteins including LH4 were reported in immunoprecipitants of GFP-tagged α2-containing GABAAR complexes (Nakamura et al., 2016). Among these proteins, 7 proteins (Cul1, Ephexin, KCTD12, Mfn2, mGluR5, PAK5/7) were confirmed as potential direct interactors using a GST pulldown experiment. It would be interesting to precisely map the direct physical protein interactions.
Inhibitory presynaptic GAD65 is not changed in LH4 CRISPRed hippocampus even months after AAV injections (Figure S4C), suggesting that presynaptic GAD65 clustering is independent from LH4 clustering, although LH4 deletion in the CRISPRed hippocampus was incomplete (Figure S4A). Surprisingly, we observe a loss of synaptic cluster number of postsynaptic gephyrin as well as GABAAR γ2 subunit in LH4 knockdown neurons (Figure 6), suggesting a molecular signal downstream of GARLH promotes gephyrin clustering. However, the size and signal intensity of gephyrin puncta were partially rescued by LH4-GFP overexpression (Figure 6A and B). These results suggest that gephyrin molecule number/synapse is reduced in LH4-GFP overexpressed neurons. Considering that gephyrin is proposed to form a hexagonal lattice (Kneussel and Betz, 2000), our results may suggest that gephyrin lattice formation might be reduced at synapses in LH4mut-GFP overexpressed neurons. Gephyrin is reported to bind to NLs (Poulopoulos et al., 2009) and we showed that LH4 modulates oligomeric states of NL2 (Figure 2C). Thus, altered oligomeric states of NLs by an excess amount of LH4mut-GFP may modulate machinery that controls gephyrin lattice formation at synapses. LH4mut-GFP overexpression also rescued the mIPSC frequency partially. Because NLs are reported to modulate the expression of presynaptic proteins (Varoqueaux et al., 2006), the change in oligomeric states of NLs may also indirectly modulate presynaptic functions. In summary, our results suggest that formation of inhibitory synapses might occur through GARLH-dependent recruitment of GABAARs, followed by recruitment of gephyrin via direct interactions with GABAAR α and/or β subunits proposed previously (Kowalczyk et al., 2013; Maric et al., 2011; Mukherjee et al., 2011). Perhaps, the NL/GARLH/GABAAR complex initially localizes to inhibitory synapses and recruits gephyrin, followed by formation of a gephyrin lattice with additional GABAARs. Precise mechanisms should be tested in vivo.
STAR Methods
CONTACT FOR REAGENET AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Susumu Tomita (Susumu.Tomita@yale.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
All animal handling was in accordance with protocols approved by the Institutional Animal Care and Use Committee (IACUC) of Yale University. Animal care and housing were provided by the Yale Animal Resource Center (YARC), in compliance with the Guide for the Care and Use of Laboratory Animals (National Academy Press, Washington, D.C., 1996). Animals were maintained in a 12 hr light/dark cycle with ad libitum access to food and water. Wild-type (C57BL/6J, Stock# 000664), the Cre-dependent Cas9 knockin mice (Platt et al., 2014) (Stock # 024857), and the conditional Gabrg2 (Stock# 016830) were obtained from the Jackson Laboratory. The transgenic Cre mouse under the GABAAR α6 promoter (ID# 015966-UCD) was obtained from MMRRC. Sprague Dawley rats were obtained from the Charles River Laboratories. Sample sizes were estimated by previous studies and/or preliminary data. Both male and female animals were used, and were randomly assigned to experimental groups. For analysis of knockout mice, littermates of the same sex were randomly assigned to experimental group. Mice (2–3 month old) and rats (retired breeders) were used for biochemistry. Mice (3–4 week-old) were used for AAV injection. Mice (P0) were used for preparing primary cultured neurons. Female Xenopus laevis frogs were obtained from Nasco, and used for oocyte preparation.
Cell lines
HEK (ATCC, CRL-1573), HEK293FT (Life Technologies), and 293AAV (Cell Biolab, Inc.) were obtained directly from each source listed. Cells were grown at 37 °C, 5% CO2. HEK cells were transfected by using Lipofectamine 2000 (ThermoFisher Scientific) for Fig. 1D.
Primary cultured hippocampal neurons
Primary cultured hippocampal neurons were prepared as described previously (Tomita et al., 2004). Specifically, P0 mice were anesthetized on ice and decapitated. Hippocampi were dissected, treated with papain (1.4 mg/ml at 37 °C for 30 min), triturated with fire-polished glass pipet and cells were plated on poly-D-lysine (PDL) coated glass coverslips and cultured at 37 °C, 5% CO2 in Neurobasal medium containing fetal bovine serum, GlutaMax, B27 supplement and Penicillin Streptomycin. Culture media were replaced to Neurobasal medium containing L-Glutamine, B27 supplement and Penicillin Streptomycin 6 hours later. Neurons were infected with lentivirus at 3 DIV, and half of culture media were refreshed at 7, 11, 15 DIV. Biochemistry, immunocytochemistry (DIV17-21) and electrophysiology (16-21 DIV) were performed.
METHOD DETAILS
Antibodies
The following antibodies were used with indicated dilutions: rabbit polyclonal antibodies to GABAAR α1 (1:3,000 for IB), γ2 (1:2,000 for IB), α6 (1:1,000 for IB), GluA2/3 (1:1,000 for IB) (Millipore), GABAAR γ2 (1:2,000 for ICC, 1:500 for IHC), Neuroligin-2 (1:3,000 for IB, 1:200 for ICC) (Synaptic systems), rabbit normal IgG (Santa Cruz), Neto2 (0.1 μg/mL for IB) (Zhang et al., 2009), α2 (1:500 for IB) (Abcam) and α3 (1:500 for IB) (Novus Biologicals): mouse monoclonal antibodies to α1 (1:2,000 for IB), PSD-95 (1:2,000 for IB, 1:1,000 for ICC and IHC) (NeuroMab), β2/3 (1:1,000 for IB), Actin (1:5,000 for IB) (Millipore), NR1 (1:2,000 for IB, 1:500 for IHC), Gephyrin (1:1,000 for IB), GAD65 (1:1,000 for IHC) (BD Biosciences), Gephyrin (1:1,000 for ICC) (Synaptic Systems), Synaptophysin (1:5,000 for IB), Tubulin (1:5,000 for IB), FLAG (1:1,000 for IB) (Sigma), HA (1:1,000 for IB) (Covance): guinea pig polyclonal antibodies to GFP (0.1 μg/mL for IB, ICC and IHC) (Kim et al., 2010). Polyclonal antisera to Lhfpl4 proteins (GARLH4) were raised by injecting rabbits with a GST-LH4 fusion protein encoding last 52 amino acids of LH4. Antisera were affinity purified on Affi-gel columns (Bio-Rad) containing the His-tagged LH4 fusion proteins (0.1 μg/mL for IB).
Plasmid
GABAAR α1, β2, γ2, neuroligin-2, Lhfpl2, Lhfpl3, Lhfpl4, Lhfpl5 (Open Biosystems), TARPγ-2/stargazin (Tomita et al., 2003), Neto2 (Zhang et al., 2009) cDNAs were cloned into appropriate vectors (pGEM-HE, pcDNA3.1)(Liman et al., 1992). Epitope tags were inserted using the Quick Change mutagenesis (Stratagene). Truncated forms of neuroligin-2 (ΔN, deleted amino acid 30–671; ΔC, deleted amino acid 709–836; ΔTM, deleted amino acid 671–836) were cloned from HA-tagged neuroligin-2 using the Quick Change mutagenesis (Stratagene). Lhfpl4 shRNA (GCAACACTGCTACTGTCTACA) was inserted into FUGW-H1 vector (a gift from Dr. Sally Temple through Addgene, # 25870) (Fasano et al., 2007). The “Lhfpl4 silent mutant” carries six mutations (GCAATACGGCCACGGTTTATA), which do not alter amino acids encoded. The construct was fused with GFP at its C-terminus and replaced with GFP alone in FUGW-H1 vector. For CRISPR, single guide RNAs (sgRNAs) were designed using the CRISPRtool (http://crispr.mit.edu)(Ran et al., 2013) and cloned into AAV:ITR-U6-sgRNA(backbone)-pCBh-Cre-WPRE-hGHpA-ITR (a gift from Dr. Feng Zhang through Addgene #60229) (Platt et al., 2014). sgRNA sequences are:
BN-PAGE
BN-PAGE was performed as described previously (Kim et al., 2010; Schagger et al., 1994). Specifically, membrane fractions of cRNA-injected oocytes, primary neurons and cerebella crude homogenate were solubilized with 1% Triton X-100 or 1% Lauryl Maltose-neopentyl glycol (MNG) (Anatrace, NG310) in 40 mM Tris-Cl (pH 8.0) buffer with Halt inhibitor cocktail (ThermoFisher Scientific). 5 mM EGTA was added to oocyte membrane solubilization. The solubilized proteins were then resolved on BN-PAGE (4–12%), which was followed by Western blot analysis. Molecular weights were determined using the NativeMark Unstained Protein Standard (Life Technologies). For antibody shift, proteins were pre-incubated with anti-β2/3 antibody for 2 hours before BN-PAGE.
Immunoprecipitation and mass spectrometry
Rat cerebella crude homogenate in 0.32 M Sucrose, 10 mM HEPES-Na (pH 7.4) was suspended in 9 volumes of lysis buffer (40 mM Tris-HCl [pH 8.0], 1% MNG) and Halt protease inhibitors (ThermoFisher Scientific), and centrifuged at 100,000 g for 1 hr. Supernatants were then incubated with an anti-α1 antibody (Millipore) and protein A sepharose beads (GE healthcare). Bound proteins were washed with wash buffer containing 0.5% MNG, 150 mM NaCl, 40 mM Tris-HCl (pH 8.0) and eluted by elution buffer containing 1 mM of the α1 antigen peptide QPSQDELKDNTTVFTC synthesized (Genscript), 1% MNG, 40 mM Tris-HCl (pH 8.0), followed by separation on BN-PAGE. The corresponding band was excised and subjected to analysis by mass spectrometry (MS Bioworks LLC, Michigan).
Biochemical fraction
Total cerebellar homogenate and the glomerular postsynapse-enriched fraction were isolated as described with slight modifications (Viennot et al., 1991). Mouse cerebella were homogenized with a Dounce homogenizer, and solubilized with 1% SDS as the total protein fraction. For the glomerular postsynapse-enriched fraction, homogenate was filtered through 150, 125, 80, and 52 μm nylon mesh (Component Supply Company), sequentially, and centrifuged at 0.9 g for 10 min twice. The pellet was resuspended in 20% Ficoll, 0.32 M Sucrose, 1 mM MgCl2, and applied to a five-layer Ficoll Gradient (25%, 20%, 15%, 10% and 5% Ficoll with 0.32 M Sucrose, 1 mM MgCl2). The layered sample was centrifuged at 80,000 g for 2 hrs. The 15–20% and 20–25% interfaces were collected, diluted into homogenization buffer and centrifuged at 100,000 g for 1 hour, and the pellet was recovered. The pellet was treated with 0.5% Triton X-100, followed by centrifugation at 32,600 g for 20 min. The pellet was recovered as the glomerular postsynapse-enriched fraction. Protein concentrations were measured using the BCA protein assay (Pierce). For quantification, protein amounts were adjusted to 5 μg/lane for Fig. 3A, 1 μg/lane for Fig. 3B, C and run on SDS-PAGE. Intensities of signals in western blotting were measured by ImageJ (Schneider et al., 2012).
GABAAR complex using Xenopus laevis oocytes
Oocytes were prepared from Xenopus laevis with collagenase (Tomita et al., 2004). The cRNAs were transcribed in vitro using T7 Message mMachine (Ambion) and injected into Xenopus laevis oocytes. The cRNA amounts per oocyte were as follows; 1.0 ng of α1, β2 and γ2 (Fig. 1A); 0.2 ng of α1, β2 and γ2, 0.1 ng of TARPγ-2 and LH4, 0.5 ng of NL2 (Fig. 2A); 0.2 ng of α1, β2 and HA-γ2, 0.1 ng of LH4 and LH4-GFP (Fig. 2B); 2.0 ng of NL2, 0.4 ng of LH4, 0.8 ng of LH4-GFP (Fig. 2C, S2B); 0.2 ng of α1, β2 and HA-γ2, 0.1 ng of LH4-FLAG, 0.5 ng of LH2-FLAG, 0.33 ng of LH3-FLAG, 1.0 ng of LH5-FLAG, 1.0 ng of NL2 (Fig. 2E); 0.033 ng of α1, β2 and HA-γ2, 0.2 ng of LH4 and 0.5 ng of NL2 (Fig. 4); 2.0 ng of NL2, 0.4 ng of LH4 and TARPγ-2 (Fig. S2C); 2.0 ng of HA-NL2 FL, ΔN and ΔC, 3.0 ng of HA-NL2 ΔTM, 1.0 ng of LH4-GFP and TARPγ-2 (Fig. S2E). After 3–5 days post-injection, 10–14 oocytes were pooled and protein lysates were prepared. Two-electrode voltage clump recording (TEVC) and measurements of surface expression using Xenopus laevis oocytes were performed as described previously (Tomita et al., 2004). After 3–5 days post-injection, TEVC analysis was performed at room temperature in ND96 containing 90 mM NaCl, 2 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2, 5 mM HEPES (pH 7.5). The membrane potential was held at −40 mV using GeneClamp 500B (Axon Instruments). Data were acquired and analyzed by LabChart (ADInstruments). For measurements of surface expression, HA-tagged proteins at the cell surface were labeled with Rat anti-HA antibody (Roche) and horseradish-peroxidase (HRP) conjugated secondary antibody (GE Healthcare). Chemiluminescence assay was performed using SuperSignal ELISA Femto Maximum Sensitivity Substrate (Thermo) and signals were measured by GloMax-Multi+Detection System (Promega).
Stereotaxic injection of AAV
Under sterile conditions, 3–4 week-old animals were anesthetized with isoflurane and secured in a stereotaxic frame, holes the size of the injection needle were drilled into the skull, and injections were done unilaterally with 1 μL of AAV per brain region. Coordinates (in millimeters; caudal to bregma, right to midline, ventral to pial surface) were (2.0, 1.9, 2.2), (2.0, 1.9, 1.9), and (2.0, 1.9, 1.6).
Immunostaining
Primary neurons were fixed and stained as described previously (Tomita et al., 2003). After 2–5 months post-injection of AAV, adult mice were deeply anesthetized with pentobarbitul and perfused transcardially with 4% paraformaldehyde in 0.1 M phosphate buffer pH 7.4. After post-fixation, 40 μm sections were prepared using a vibratome (Leica). Sections were incubated with 1 mg/ml pepsin (Dako) in 0.2 N HCl at 37 °C to enhance signal from synaptic proteins (Fukaya and Watanabe, 2000), followed by staining with appropriate antibodies and imaged using confocal microscopy (Zeiss LSM 710). We acquired data by using Plan-APOCHROMAT 63x/1.40 oil DIC objective lens or Plan-APOCHROMAT 20x/0.8 lens (Zeiss). For quantification, sets of cells and sections were prepared and stained simultaneously. Compared images were acquired at the same time using identical acquisition settings. The numbers of puncta was analyzed using ImageJ (Schneider et al., 2012) in a blinded manner.
Virus production
Lentivirus was prepared as described previously (Zhang et al., 2009). HEK293FT cells (Life Technologies) were transfected with FUGW-H1 with LH4 shRNA, psPAX2 (a gift from Dr. Didier Trono through Addgene, #12260) and pVSVG (Stewart et al., 2003) using the calcium phosphate method. After overnight incubation, medium was changed, and cells were incubated 36–40 hrs. Then supernatant was collected, filtered and concentrated by ultracentrifugation (80,000 g for 2 hours at 4 °C). The viral pellet was resuspended in PBS and stored at −80 °C. Neurons were infected with lentivirus at DIV3.
Adeno-associated virus (AAV) was prepared as described previously (McClure et al., 2011). Briefly, three plasmids of pAAV-DJ, pHelper (Cell Biolabs, Inc.) and AAV-U6-sgRNA-pCBh-Cre encoding LH4 sgRNAs were co-transfected into 293AAV cells (Cell Biolab, Inc.) using the calcium phosphate method. 48–60 hours after transfection, cells were solubilized and AAVs were purified using a HiTrap Heparin column (GE healthcare).
Electrophysiology in primary hippocampal neurons
Cultured neurons at DIV16-21 were transferred to a recording chamber continuously perfused with a bath solution saturated with 95% O2/5% CO2, and containing (in mM): 125 NaCl, 2.4 KCl, 2 CaCl2, 1 MgCl2, 25 NaHCO3, 1.2 NaH2PO4 and 25 Dextrose (301 mOsm, pH 7.4 adjusted with NaOH). Patch pipettes with tip resistances between 10–15 MΩ were filled with internal solution containing (in mM): 145 CsCl, 5 NaCl, 10 HEPES, 5 EGTA, 5 QX-314, 4 Mg-ATP and 0.3 Na-GTP (271 mOsm, pH 7.2 adjusted with CsOH). Miniature postsynaptic currents were monitored at room temperature in whole-cell configuration for 3–15 minutes, using a Multiclamp 700B amplifier (Axon Instruments), at a holding potential of −70 mV. To monitor mIPSCs, APV (100 μM), CNQX (20 μM) and tetrodotoxin (1 μM) were added to the bath solution. In these conditions, mIPSCs manifested as inward currents. To isolate the mEPSCs, picrotoxin (100 μM) and tetrodotoxin (1 μM) were used. Recordings with series resistances > 30 MΩ were rejected. Signals were filtered at 3 kHz and sampled at 25 kHz. Offline analysis was performed using custom Matlab code and Mini Analysis (http://www.synaptosoft.com, Decatur, GA, USA), with a threshold >5pA, a rise time between 0.1–3 ms, decay time >0.5 ms and a half-width of ≥ 0.1 ms. Double exponential equations of the form I(t)=If * exp(−t/τf) + Is * exp(−t/τs), where If and Is are the amplitudes of the fast and slow components, and τf and τs are their respective decay time constants, were used to fit the normalized mIPSCs. A weighted decay tau τw = (τf * If + τs * Is)/(If + Is) was used to compare the decay time between the different groups.
Whole-cell recordings in acute hippocampal slices
Transverse hippocampal slices with a thickness of 300 μm were prepared from P45-P90 mice after 3–9 weeks post AAV injection. Animals were deeply anesthetized with isoflurane and killed by decapitation. Slices were prepared as described (Ting et al., 2014). Briefly, slices were cut with a VT1200S microtome (Leica), and transferred to a chamber containing choline aCSF at room temperature for 10–15 min (in mM: 92 choline chloride, 2.5 KCl, 26 NaHCO3, 1 NaH2PO4, 10 MgSO4, 20 HEPES, 10 dextrose, 0.5 CaCl2, equilibrated with 95% O2/5% CO2), followed by transferring to a chamber containing HEPES aCSF at room temperature (in mM: 92 NaCl, 2.5 KCl, 26 NaHCO3, 1 NaH2PO4, 1.3 MgSO4, 20 HEPES, 10 dextrose, 2 CaCl2, equilibrated with 95% O2/5% CO2). Slices were allowed to recover for at least 1 h before being transferred to a recording chamber containing aCSF at room temperature (in mM: 124 NaCl, 2.5 KCl, 26 NaHCO3, 1 NaH2PO4, 1.3 MgSO4, 10 dextrose, 2.5 CaCl2, equilibrated with 95% O2/5% CO2, 300 mOsm). GFP-positive CA1 hippocampal neurons were identified using an upright epifluorescence microscope. Voltage-clamp recordings (Vhold = 0 mV) were performed using patch pipettes with tip resistances between 7–10 MΩ, filled with internal solution containing (in mM): 123 D-Gluconic acid, 8 NaCl, 10 EGTA, 10 HEPES, 10 dextrose, 1 CaCl2 (270 mOsm, pH 7.2 adjusted with CsOH). The series resistance was continuously monitored, and recordings with series resistances > 40 MΩ were rejected. To isolate GABAAR-mediated currents, the aCSF contained APV (100 μM), CNQX (20 μM), CGP 52432 (5 μM), and strychnine (0.3 μM). To evoke inhibitory postsynaptic currents (IPSCs), square-wave current 200-μs-wide pulses were delivered through a A365 stimulus isolator (WPI) connected to a 1–2 MΩ micropipette filled with aCSF. This stimulating pipette was located in the middle of the stratum pyramidale 150 μm away from the recorded cell. After recording evoked IPSCs, mIPSCs were recorded for 3 min with 1 μM TTX before the subsequent addition of 1 mM GABA and 1 μM TTX to the perfusion system in order to elicit GABA-evoked currents from the same cells. Signals were sampled at 25 kHz, and downsampled at 2.5 kHz.
Surface cell biotinylation of cerebellar slices
Cerebellar slices were prepared from 6–8 week mice as described (Yan et al., 2013). Surface expression of LH4 and GluN1 was quantitated as described (Tomita et al., 2004). Briefly, acute cerebellar slices were labeled for 30 min at 4 °C with 1.5 mg/ml sulfo-NHS-SS biotin (ThermoFisher Scientific). Membranes were prepared and the biotinylated proteins were precipitated with Neutravidin-agarose (ThermoFisher Scientific) and detected by western blotting.
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantification and statistical details of experiments can be found in the figure legends or Method Details section. All data are given as mean ± s.e.m. Statistical significance between means was calculated using Mann-Whitney U-test or Kruskal-willis test followed by Dunn’s post-test; *P < 0.05; **P < 0.01; *** P < 0.001.
Supplementary Material
Highlights.
The γ2-containing GABAARs form a tripartite complex with LH4 and neuroligin-2.
GARLHs are GABAAR putative auxiliary subunits consisting of LH3 and LH4.
The GABAAR γ2 subunit regulates protein expressions of LH4 and neuroligin-2.
GARLH is required for the synaptic GABAAR localization and inhibitory transmission.
Acknowledgments
The authors thank Dr. Satinder Singh for providing her valuable input and the members of the Tomita lab for discussions. We thank Dr. Louis Reichardt for sharing transgenic Cre mice under the GABAAR α6 promoter through the MMRRC, Dr. Bernhard Luscher for conditional γ2 mice, Dr. Feng Zhang for Cre-dependent Cas9 knockin mice through the Jackson laboratory, Addgene for the plasmids listed in the Experimental Procedure and the University of California, Davis/NeuroMab Facility (NIH U24NS050606), for the antibodies. This work was supported by NIH/NIMH MH104984, Yale University (S.T.), and research fellowships from the Japanese Society for the Promotion of Science and the Uehara Memorial Foundation (T.Y.), NIH F30 MH099742 and NIH T32GM007205 (J.S.M.).
Footnotes
Author contributions
S.T. conceived the project. S.T. and T.Y. wrote the manuscript. T.Y. performed all biochemical, histochemical, and oocyte studies. E.H. performed synaptic electrophysiological recordings. J.S.M. produced preliminary results. M.M. generated and maintained gene-targeted mice and antibodies. All authors contributed to the final version of the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alldred MJ, Mulder-Rosi J, Lingenfelter SE, Chen G, Luscher B. Distinct gamma2 subunit domains mediate clustering and synaptic function of postsynaptic GABAA receptors and gephyrin. J Neurosci. 2005;25:594–603. doi: 10.1523/JNEUROSCI.4011-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arac D, Boucard AA, Ozkan E, Strop P, Newell E, Sudhof TC, Brunger AT. Structures of neuroligin-1 and the neuroligin-1/neurexin-1 beta complex reveal specific protein-protein and protein-Ca2+ interactions. Neuron. 2007;56:992–1003. doi: 10.1016/j.neuron.2007.12.002. [DOI] [PubMed] [Google Scholar]
- Boulin T, Rapti G, Briseno-Roa L, Stigloher C, Richmond JE, Paoletti P, Bessereau JL. Positive modulation of a Cys-loop acetylcholine receptor by an auxiliary transmembrane subunit. Nat Neurosci. 2012;15:1374–1381. doi: 10.1038/nn.3197. [DOI] [PubMed] [Google Scholar]
- Brockie PJ, Jensen M, Mellem JE, Jensen E, Yamasaki T, Wang R, Maxfield D, Thacker C, Hoerndli F, Dunn PJ, et al. Cornichons control ER export of AMPA receptors to regulate synaptic excitability. Neuron. 2013;80:129–142. doi: 10.1016/j.neuron.2013.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Chetkovich DM, Petralia RS, Sweeney NT, Kawasaki Y, Wenthold RJ, Bredt DS, Nicoll RA. Stargazin regulates synaptic targeting of AMPA receptors by two distinct mechanisms. Nature. 2000;408:936–943. doi: 10.1038/35050030. [DOI] [PubMed] [Google Scholar]
- Chih B, Engelman H, Scheiffele P. Control of excitatory and inhibitory synapse formation by neuroligins. Science. 2005;307:1324–1328. doi: 10.1126/science.1107470. [DOI] [PubMed] [Google Scholar]
- Collingridge GL, Isaac JT, Wang YT. Receptor trafficking and synaptic plasticity. Nat Rev Neurosci. 2004;5:952–962. doi: 10.1038/nrn1556. [DOI] [PubMed] [Google Scholar]
- Dean C, Scholl FG, Choih J, DeMaria S, Berger J, Isacoff E, Scheiffele P. Neurexin mediates the assembly of presynaptic terminals. Nat Neurosci. 2003;6:708–716. doi: 10.1038/nn1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essrich C, Lorez M, Benson JA, Fritschy JM, Luscher B. Postsynaptic clustering of major GABAA receptor subtypes requires the gamma 2 subunit and gephyrin. Nat Neurosci. 1998;1:563–571. doi: 10.1038/2798. [DOI] [PubMed] [Google Scholar]
- Fabrichny IP, Leone P, Sulzenbacher G, Comoletti D, Miller MT, Taylor P, Bourne Y, Marchot P. Structural analysis of the synaptic protein neuroligin and its beta-neurexin complex: determinants for folding and cell adhesion. Neuron. 2007;56:979–991. doi: 10.1016/j.neuron.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasano CA, Dimos JT, Ivanova NB, Lowry N, Lemischka IR, Temple S. shRNA knockdown of Bmi-1 reveals a critical role for p21-Rb pathway in NSC self-renewal during development. Cell stem cell. 2007;1:87–99. doi: 10.1016/j.stem.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Fritschy JM, Panzanelli P, Tyagarajan SK. Molecular and functional heterogeneity of GABAergic synapses. Cellular and molecular life sciences: CMLS. 2012;69:2485–2499. doi: 10.1007/s00018-012-0926-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaya M, Watanabe M. Improved immunohistochemical detection of postsynaptically located PSD-95/SAP90 protein family by protease section pretreatment: a study in the adult mouse brain. J Comp Neurol. 2000;426:572–586. [PubMed] [Google Scholar]
- Funfschilling U, Reichardt LF. Cre-mediated recombination in rhombic lip derivatives. Genesis. 2002;33:160–169. doi: 10.1002/gene.10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gally C, Eimer S, Richmond JE, Bessereau JL. A transmembrane protein required for acetylcholine receptor clustering in Caenorhabditis elegans. Nature. 2004;431:578–582. doi: 10.1038/nature02893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel M, Rapti G, Richmond JE, Bessereau JL. A secreted complement-control-related protein ensures acetylcholine receptor clustering. Nature. 2009;461:992–996. doi: 10.1038/nature08430. [DOI] [PubMed] [Google Scholar]
- Graf ER, Zhang X, Jin SX, Linhoff MW, Craig AM. Neurexins induce differentiation of GABA and glutamate postsynaptic specializations via neuroligins. Cell. 2004;119:1013–1026. doi: 10.1016/j.cell.2004.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther U, Benson J, Benke D, Fritschy JM, Reyes G, Knoflach F, Crestani F, Aguzzi A, Arigoni M, Lang Y, et al. Benzodiazepine-insensitive mice generated by targeted disruption of the gamma 2 subunit gene of gamma-aminobutyric acid type A receptors. Proc Natl Acad Sci U S A. 1995;92:7749–7753. doi: 10.1073/pnas.92.17.7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto K, Fukaya M, Qiao X, Sakimura K, Watanabe M, Kano M. Impairment of AMPA receptor function in cerebellar granule cells of ataxic mutant mouse stargazer. J Neurosci. 1999;19:6027–6036. doi: 10.1523/JNEUROSCI.19-14-06027.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herring BE, Shi Y, Suh YH, Zheng CY, Blankenship SM, Roche KW, Nicoll RA. Cornichon proteins determine the subunit composition of synaptic AMPA receptors. Neuron. 2013;77:1083–1096. doi: 10.1016/j.neuron.2013.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichtchenko K, Hata Y, Nguyen T, Ullrich B, Missler M, Moomaw C, Sudhof TC. Neuroligin 1: a splice site-specific ligand for beta-neurexins. Cell. 1995;81:435–443. doi: 10.1016/0092-8674(95)90396-8. [DOI] [PubMed] [Google Scholar]
- Jackson AC, Nicoll RA. The Expanding Social Network of Ionotropic Glutamate Receptors: TARPs and Other Transmembrane Auxiliary Subunits. Neuron. 2011;70:178–199. doi: 10.1016/j.neuron.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob TC, Moss SJ, Jurd R. GABA(A) receptor trafficking and its role in the dynamic modulation of neuronal inhibition. Nat Rev Neurosci. 2008;9:331–343. doi: 10.1038/nrn2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurgensen S, Castillo PE. Selective Dysregulation of Hippocampal Inhibition in the Mouse Lacking Autism Candidate Gene CNTNAP2. J Neurosci. 2015;35:14681–14687. doi: 10.1523/JNEUROSCI.1666-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato AS, Gill MB, Ho MT, Yu H, Tu Y, Siuda ER, Wang H, Qian YW, Nisenbaum ES, Tomita S, et al. Hippocampal AMPA receptor gating controlled by both TARP and cornichon proteins. Neuron. 2010;68:1082–1096. doi: 10.1016/j.neuron.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KS, Yan D, Tomita S. Assembly and stoichiometry of the AMPA receptor and transmembrane AMPA receptor regulatory protein complex. J Neurosci. 2010;30:1064–1072. doi: 10.1523/JNEUROSCI.3909-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneussel M, Betz H. Clustering of inhibitory neurotransmitter receptors at developing postsynaptic sites: the membrane activation model. Trends Neurosci. 2000;23:429–435. doi: 10.1016/s0166-2236(00)01627-1. [DOI] [PubMed] [Google Scholar]
- Kneussel M, Brandstatter JH, Gasnier B, Feng G, Sanes JR, Betz H. Gephyrin-independent clustering of postsynaptic GABA(A) receptor subtypes. Mol Cell Neurosci. 2001;17:973–982. doi: 10.1006/mcne.2001.0983. [DOI] [PubMed] [Google Scholar]
- Kneussel M, Brandstatter JH, Laube B, Stahl S, Muller U, Betz H. Loss of postsynaptic GABA(A) receptor clustering in gephyrin-deficient mice. J Neurosci. 1999;19:9289–9297. doi: 10.1523/JNEUROSCI.19-21-09289.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehnke J, Jin X, Trbovic N, Katsamba PS, Brasch J, Ahlsen G, Scheiffele P, Honig B, Palmer AG, 3rd, Shapiro L. Crystal structures of beta-neurexin 1 and betaneurexin 2 ectodomains and dynamics of splice insertion sequence 4. Structure. 2008;16:410–421. doi: 10.1016/j.str.2007.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk S, Winkelmann A, Smolinsky B, Forstera B, Neundorf I, Schwarz G, Meier JC. Direct binding of GABAA receptor beta2 and beta3 subunits to gephyrin. Eur J Neurosci. 2013;37:544–554. doi: 10.1111/ejn.12078. [DOI] [PubMed] [Google Scholar]
- Levi S, Logan SM, Tovar KR, Craig AM. Gephyrin is critical for glycine receptor clustering but not for the formation of functional GABAergic synapses in hippocampal neurons. J Neurosci. 2004;24:207–217. doi: 10.1523/JNEUROSCI.1661-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liman ER, Tytgat J, Hess P. Subunit stoichiometry of a mammalian K+ channel determined by construction of multimeric cDNAs. Neuron. 1992;9:861–871. doi: 10.1016/0896-6273(92)90239-a. [DOI] [PubMed] [Google Scholar]
- Maric HM, Mukherjee J, Tretter V, Moss SJ, Schindelin H. Gephyrin-mediated gamma-aminobutyric acid type A and glycine receptor clustering relies on a common binding site. J Biol Chem. 2011;286:42105–42114. doi: 10.1074/jbc.M111.303412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maro GS, Gao S, Olechwier AM, Hung WL, Liu M, Ozkan E, Zhen M, Shen K. MADD-4/Punctin and Neurexin Organize C. elegans GABAergic Postsynapses through Neuroligin. Neuron. 2015;86:1420–1432. doi: 10.1016/j.neuron.2015.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure C, Cole KL, Wulff P, Klugmann M, Murray AJ. Production and titering of recombinant adeno-associated viral vectors. Journal of visualized experiments: JoVE. 2011:e3348. doi: 10.3791/3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missler M, Zhang W, Rohlmann A, Kattenstroth G, Hammer RE, Gottmann K, Sudhof TC. Alpha-neurexins couple Ca2+ channels to synaptic vesicle exocytosis. Nature. 2003;423:939–948. doi: 10.1038/nature01755. [DOI] [PubMed] [Google Scholar]
- Mukherjee J, Kretschmannova K, Gouzer G, Maric HM, Ramsden S, Tretter V, Harvey K, Davies PA, Triller A, Schindelin H, et al. The residence time of GABA(A)Rs at inhibitory synapses is determined by direct binding of the receptor alpha1 subunit to gephyrin. J Neurosci. 2011;31:14677–14687. doi: 10.1523/JNEUROSCI.2001-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Morrow DH, Modgil A, Huyghe D, Deeb TZ, Lumb MJ, Davies PA, Moss SJ. Proteomic Characterization of Inhibitory Synapses Using a Novel pHluorin-tagged gamma-Aminobutyric Acid Receptor, Type A (GABAA), alpha2 Subunit Knock-in Mouse. J Biol Chem. 2016;291:12394–12407. doi: 10.1074/jbc.M116.724443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen RW, Sieghart W. International Union of Pharmacology. LXX. Subtypes of gamma-aminobutyric acid(A) receptors: classification on the basis of subunit composition, pharmacology, and function. Update. Pharmacol Rev. 2008;60:243–260. doi: 10.1124/pr.108.00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit MM, Schoenmakers EF, Huysmans C, Geurts JM, Mandahl N, Van de Ven WJ. LHFP, a novel translocation partner gene of HMGIC in a lipoma, is a member of a new family of LHFP-like genes. Genomics. 1999;57:438–441. doi: 10.1006/geno.1999.5778. [DOI] [PubMed] [Google Scholar]
- Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M, et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulopoulos A, Aramuni G, Meyer G, Soykan T, Hoon M, Papadopoulos T, Zhang M, Paarmann I, Fuchs C, Harvey K, et al. Neuroligin 2 drives postsynaptic assembly at perisomatic inhibitory synapses through gephyrin and collybistin. Neuron. 2009;63:628–642. doi: 10.1016/j.neuron.2009.08.023. [DOI] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nature protocols. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schagger H, Cramer WA, von Jagow G. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal Biochem. 1994;217:220–230. doi: 10.1006/abio.1994.1112. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer C, Balsiger S, Bluethmann H, Mansuy IM, Fritschy JM, Mohler H, Luscher B. The gamma 2 subunit of GABA(A) receptors is required for maintenance of receptors at mature synapses. Mol Cell Neurosci. 2003;24:442–450. doi: 10.1016/s1044-7431(03)00202-1. [DOI] [PubMed] [Google Scholar]
- Schwenk J, Harmel N, Zolles G, Bildl W, Kulik A, Heimrich B, Chisaka O, Jonas P, Schulte U, Fakler B, et al. Functional proteomics identify cornichon proteins as auxiliary subunits of AMPA receptors. Science. 2009;323:1313–1319. doi: 10.1126/science.1167852. [DOI] [PubMed] [Google Scholar]
- Sigel E, Steinmann ME. Structure, function, and modulation of GABA(A) receptors. J Biol Chem. 2012;287:40224–40231. doi: 10.1074/jbc.R112.386664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soykan T, Schneeberger D, Tria G, Buechner C, Bader N, Svergun D, Tessmer I, Poulopoulos A, Papadopoulos T, Varoqueaux F, et al. A conformational switch in collybistin determines the differentiation of inhibitory postsynapses. EMBO J. 2014;33:2113–2133. doi: 10.15252/embj.201488143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS, Sabatini DM, Chen IS, Hahn WC, Sharp PA, et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. Rna. 2003;9:493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub C, Hunt DL, Yamasaki M, Kim KS, Watanabe M, Castillo PE, Tomita S. Distinct functions of kainate receptors in the brain are determined by the auxiliary subunit Neto1. Nat Neurosci. 2011;14:866–873. doi: 10.1038/nn.2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M, Pelkey KA, Ng D, Ivakine E, McBain CJ, Salter MW, McInnes RR. Neto1 Is an Auxiliary Subunit of Native Synaptic Kainate Receptors. J Neurosci. 2011;31:10009–10018. doi: 10.1523/JNEUROSCI.6617-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting JT, Daigle TL, Chen Q, Feng G. Acute brain slice methods for adult and aging animals: application of targeted patch clamp analysis and optogenetics. Methods Mol Biol. 2014;1183:221–242. doi: 10.1007/978-1-4939-1096-0_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita S, Adesnik H, Sekiguchi M, Zhang W, Wada K, Howe JR, Nicoll RA, Bredt DS. Stargazin modulates AMPA receptor gating and trafficking by distinct domains. Nature. 2005;435:1052–1058. doi: 10.1038/nature03624. [DOI] [PubMed] [Google Scholar]
- Tomita S, Chen L, Kawasaki Y, Petralia RS, Wenthold RJ, Nicoll RA, Bredt DS. Functional studies and distribution define a family of transmembrane AMPA receptor regulatory proteins. J Cell Biol. 2003;161:805–816. doi: 10.1083/jcb.200212116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita S, Fukata M, Nicoll RA, Bredt DS. Dynamic interaction of stargazin-like TARPs with cycling AMPA receptors at synapses. Science. 2004;303:1508–1511. doi: 10.1126/science.1090262. [DOI] [PubMed] [Google Scholar]
- Tu H, Pinan-Lucarre B, Ji T, Jospin M, Bessereau JL. C. elegans Punctin Clusters GABA(A) Receptors via Neuroligin Binding and UNC-40/DCC Recruitment. Neuron. 2015;86:1407–1419. doi: 10.1016/j.neuron.2015.05.013. [DOI] [PubMed] [Google Scholar]
- Varoqueaux F, Aramuni G, Rawson RL, Mohrmann R, Missler M, Gottmann K, Zhang W, Sudhof TC, Brose N. Neuroligins determine synapse maturation and function. Neuron. 2006;51:741–754. doi: 10.1016/j.neuron.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Varoqueaux F, Jamain S, Brose N. Neuroligin 2 is exclusively localized to inhibitory synapses. European journal of cell biology. 2004;83:449–456. doi: 10.1078/0171-9335-00410. [DOI] [PubMed] [Google Scholar]
- Viennot F, Artault JC, Tholey G, De Barry J, Gombos G. An improved method for the preparation of rat cerebellar glomeruli. Journal of neuroscience methods. 1991;38:51–62. doi: 10.1016/0165-0270(91)90154-r. [DOI] [PubMed] [Google Scholar]
- Wang H, Bedford FK, Brandon NJ, Moss SJ, Olsen RW. GABA(A)-receptor-associated protein links GABA(A) receptors and the cytoskeleton. Nature. 1999;397:69–72. doi: 10.1038/16264. [DOI] [PubMed] [Google Scholar]
- Wang R, Mellem JE, Jensen M, Brockie PJ, Walker CS, Hoerndli FJ, Hauth L, Madsen DM, Maricq AV. The SOL-2/Neto auxiliary protein modulates the function of AMPA-subtype ionotropic glutamate receptors. Neuron. 2012;75:838–850. doi: 10.1016/j.neuron.2012.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K, Siekevitz P. Neurochemical characteristics of a postsynaptic density fraction isolated from adult canine hippocampus. Brain Res. 1988;457:98–112. doi: 10.1016/0006-8993(88)90061-3. [DOI] [PubMed] [Google Scholar]
- Yan D, Tomita S. Defined criteria for auxiliary subunits of glutamate receptors. J Physiol. 2012;590:21–31. doi: 10.1113/jphysiol.2011.213868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Yamasaki M, Straub C, Watanabe M, Tomita S. Homeostatic control of synaptic transmission by distinct glutamate receptors. Neuron. 2013;78:687–699. doi: 10.1016/j.neuron.2013.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerangue N, Schwappach B, Jan YN, Jan LY. A new ER trafficking signal regulates the subunit stoichiometry of plasma membrane K(ATP) channels. Neuron. 1999;22:537–548. doi: 10.1016/s0896-6273(00)80708-4. [DOI] [PubMed] [Google Scholar]
- Zhang B, Chen LY, Liu X, Maxeiner S, Lee SJ, Gokce O, Sudhof TC. Neuroligins Sculpt Cerebellar Purkinje-Cell Circuits by Differential Control of Distinct Classes of Synapses. Neuron. 2015;87:781–796. doi: 10.1016/j.neuron.2015.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Atasoy D, Arac D, Yang X, Fucillo MV, Robison AJ, Ko J, Brunger AT, Sudhof TC. Neurexins physically and functionally interact with GABA(A) receptors. Neuron. 2010;66:403–416. doi: 10.1016/j.neuron.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, St-Gelais F, Grabner CP, Trinidad JC, Sumioka A, Morimoto-Tomita M, Kim KS, Straub C, Burlingame AL, Howe JR, et al. A transmembrane accessory subunit that modulates kainate-type glutamate receptors. Neuron. 2009;61:385–396. doi: 10.1016/j.neuron.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.