Abstract
Thermoanaerobacterium sp. strain PSU-2 was isolated from thermophilic hydrogen producing reactor and subjected to draft genome sequencing on 454 pyrosequencing and annotated on RAST. The draft genome sequence of strain PSU-2 contains 2,552,497 bases with an estimated G + C content of 35.2%, 2555 CDS, 8 rRNAs and 57 tRNAs. The strain had a number of genes responsible for carbohydrates metabolic, amino acids and derivatives, and protein metabolism of 17.7%, 14.39% and 9.81%, respectively. Strain PSU-2 also had gene responsible for hydrogen biosynthesis as well as the genes related to Ni-Fe hydrogenase. Comparative genomic analysis indicates strain PSU-2 shares about 94% genome sequence similarity with Thermoanaerobacterium xylanolyticum LX-11. The nucleotide sequence of this draft genome was deposited into DDBJ/ENA/GenBank under the accession MSQD00000000.
Keywords: Whole genome sequencing, Thermoanaerobacterium sp., Hydrogen producing bacteria, Thermophile
| Specifications | |
|---|---|
| Organism/cell line/tissue | Thermoanaerobacterium sp. |
| Strain | PSU-2 |
| Sequencer or array type | 454 pyrosequencing |
| Data format | analyzed |
| Experimental factors | microbial strain |
| Experimental features | draft genome analysis and gene annotation of PSU-2 |
| Consent | N/A |
| Sample source location | Songkhla, Thailand |
1. Direct link to deposited data
The draft genome sequences could be found at the site http://www.ncbi.nlm.nih.gov/nuccore/MSQD00000000
2. Experimental design, materials and methods
The genus Thermoanaerobacterium is a group of anaerobic, gram-positive, rod shaped, reduce thiosulfate to elemental sulfur that belong to Firmicutes as previously described by Lee et al. [1]. Genus Thermoanaerobacterium are thermophilic that specialize in polysaccharide and carbohydrate fermentation, producing primarily L-lactic acid, acetic acid, ethanol, CO2, and H2 [2], [3]. The majority of characterized Thermoanaerobacterium strains have been isolated from hot springs and other thermal environments [4]; however, they have also been isolated from leachate of a waste pile from a canning factory [5], thermophilic bioreactor for biohydrogen production [6], [7] and deep subsurface environments [8]. This genus has been considered for biotechnological applications, such as conversion of lignocellulosic biomass to ethanol [3], biohydrogen and other chemicals [9]. Thermoanaerobacterium strain PSU-2 is a rod shaped, gram-positive, spore-forming and thermophilic hydrogen producing bacteria belonging to Firmicutes that was isolated from a biohydrogen reactor fed with palm oil mill effluent (POME). Phylogenetic analysis based on 16S rRNA genes indicated that strain PSU-2 belonged to the genus Thermoanaerobacterium [6]. This genus had been previously studied for hydrogen production from various carbohydrates, such as starch, sucrose and molasses [10]. Strain PSU-2 has a high hydrogen production capacity within a wide range of pH (4.5–8) and temperature (45–70 °C), with the optimal temperature 60 °C and optimal initial pH about 6.25. The strain performed ethanol–acetate type fermentation in inorganic nitrogen amended medium, while it performed butyrate–acetate type fermentation in organic nitrogen amended medium [6].
The draft genome of strain PSU-2 was sequenced with 454 technology using a GS-FLX pyrosequencer at GATC Biotech, Germany (http://www.gatc-biotech.com). A total of 2,552,497 bases were obtained. Assembly into 44 contigs was done with Newbler version 2.9 accessed through the Lifeportal, University of Oslo (http://www.uio.no/english /services/it/research/hpc/lifeportal) and annotation was conducted on RAST [11]. SEED viewer was used for subsystem functional categorization of the predicted open reading frames (ORFs) and visualization [12]. An average nucleotide identity (ANI) was analysis using the online ANI calculator (http://enve-omics.ce.gatech.edu/ani/index). An In Silico genomic DNA:DNA hybridization was performed by genome-to-genome distance calculator (http://ggdc.dsmz.de).
3. Data description
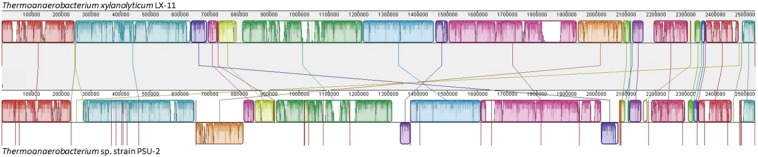
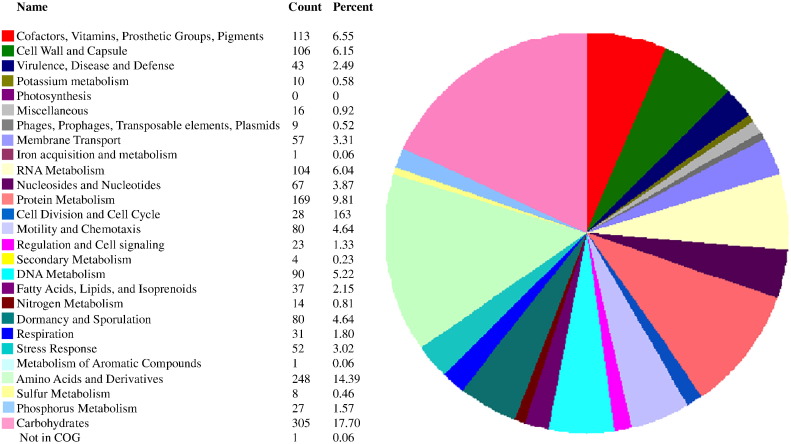
The draft genome of Thermoanaerobacterium sp. strain PSU-2 consisted of single DNA chromosome of 2,552,497 bases, a G + C content of 35.2%. The draft genome was predicted to contain 2555 protein-coding sequence, 8 rRNAs and 57 tRNAs. These genes were annotated and classified into 337 subsystems. Most of the annotated genes were involved in carbohydrates metabolic (17.7%), amino acids (14.39%) and derivatives, and protein metabolism (9.81%) (Fig.1). Strain PSU-2 also had gene responsible for hydrogen biosynthesis as well as the genes related to Ni-Fe hydrogenase. Comparative genomic analysis indicates PSU-2 by an average nucleotide identity (ANI) analysis using the online ANI calculator (http://enve-omics.ce.gatech.edu/ani/index) revealed ANI values of 94, when the PSU-2 draft sequence was compared with complete sequences of Thermoanaerobacterium xylanolyticum LX-11 species, isolated from geothermal areas of Yellowstone National Park, Wyoming, USA (Fig. 2). This indicates that PSU-2 represents a separate species, as this value is lower than the threshold value of 95%, which corresponds to a genomic DNA:DNA hybridization value of 70% and is a common threshold value for distinction between species [13].
Fig. 1.
Draft genome alignment of Thermoanaerobacterium sp. strain PSU-2 with Thermoanaerobacterium xylanolyticum LX-11.
Fig. 2.
Distribution and counts of genes in COG categories for draft genome of Thermoanaerobacterium sp. strain PSU-2.
4. Nucleotide accession number
This whole genome project has been deposited at DDBJ/ENA/ GenBank under accession no. MSQD00000000. The version described in this paper is version MSQD01000000.
Conflict of interest
The authors clarified that this work and writing has no conflict of interest.
Acknowledgements
This work was supported by the Core-to-Core Program, which was financially supported by Japan Society for the Promotion of Science (JSPS), National Research Council of Thailand (NRCT), Vietnam Ministry of Science and Technology (MOST), the National University of Laos, Beuth University of Applied Sciences and Brawijaya University, Research and Development Institute Thaksin University (RDITSU), Agricultural Research Development Agency (ARDA), Research Group for Development of Microbial Hydrogen Production Process from Biomass, Khon Kaen University and Thailand Research Fund through grant number PHD57K0042 and RTA5780002.
References
- 1.Lee Y.E., Jain M.K., Lee C.Y., Lowe S.E., Zeikus J.G. Taxonomic distinction of saccharolytic thermophilic anaerobes description of Thermoanaerobacterium xylanolyticum gen nov, sp nov, and Thermoanaerobacterium saccharolyticum gen nov, sp nov reclassification of Thermoanaerobium brockii, Clostridium thermosulfurogenes, and Clostridium thermohydrosulfuricum E100-69 as Thermoanaerobacter brockii comb nov, Thermoanaerobacterium thermosulfurigenes comb nov, and Thermoanaerobacter thermohydrosulfuricus comb nov, respectively and transfer of Clostridium thermohydrosulfuricum to Thermoanaerobacter ethanolicus. Int. J. Syst. Bacteriol. 1993;34:41–51. [Google Scholar]
- 2.Wiegel J., Mothershed C.P., Puls J. Differences in xylan degradation by various noncellulolytic thermophilic anaerobes and Clostridium thermocellum. Appl. Environ. Microbiol. 1985;49:656–659. doi: 10.1128/aem.49.3.656-659.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lynd L.R., Weimer P.J., van Zyl W.H., Pretorius I.S. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 2002;66:506–577. doi: 10.1128/MMBR.66.3.506-577.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shao W., DeBlois S., Wiegel J. A high-molecular-weight, cellassociated xylanase isolated from exponentially growing Thermoanaerobacterium sp. strain JW/SL-YS485. Appl. Environ. Microbiol. 1995;61:937–940. doi: 10.1128/aem.61.3.937-940.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cann I.K.O., Stroot P.G., Mackie K.R., White B.A., Mackie R.I. Characterization of two novel saccharolytic, anaerobic thermophiles, Thermoanaerobacterium polysaccharolyticum sp nov and Thermoanaerobacterium zeae sp nov., and emendation of the genus Thermoanaerobacterium. Int. J. Syst. Evol. Microbiol. 2001;51:293–302. doi: 10.1099/00207713-51-2-293. [DOI] [PubMed] [Google Scholar]
- 6.O-Thong S., Prasertsan P., Karakashev D., Angelidaki I. Thermophilic fermentative hydrogen production by the newly isolated Thermoanerobacterium thermosaccharolyticum PSU-2. Int. J. Hydrog. Energy. 2008;33:1204–1214. [Google Scholar]
- 7.Ren N., Cao G., Wang A., Lee D.J., Guo W., Zhu Y. Dark fermentation of xylose and glucose mix using isolated Thermoanaerobacterium thermosaccharolyticum W16. Int. J. Hydrog. Energy. 2008;33:6124–6132. [Google Scholar]
- 8.Liu S.Y., Rainey F.A., Morgan H.W., Mayer F., Wiegel J. Thermoanaerobacterium aotearoense sp. nov., a slightly acidophilic, anaerobic thermophile isolated from various hot springs in New Zealand, and emendation of the genus Thermoanaerobacterium. Int. J. Syst. Bacteriol. 1996;46:388–396. [Google Scholar]
- 9.Altaras N.E., Etzel M.R., Cameron D.C. Conversion of sugars to 1,2-propanediol by Thermoanaerobacterium thermosaccharolyticum HG-8. Biotechnol. Prog. 2001;17:52–56. doi: 10.1021/bp000130b. [DOI] [PubMed] [Google Scholar]
- 10.O-Thong S., Prasertsan P., Karakashev D., Angelidaki I. 16S rRNA-targeted probes for specific detection of Thermoanaerobacterium spp., Thermoanaerobacterium thermosaccharolyticum, and Caldicellulosiruptor spp. by fluorescent in situ hybridization in biohydrogen producing systems. Int. J. Hydrog. Energy. 2008;33:6082–6091. [Google Scholar]
- 11.Aziz R.K., Bartels D., Best A.A., DeJongh M., Disz T., Edwards R.A., Formsma K., Gerdes S., Glass E.M., Kubal M. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9(1):75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Overbeek R., Olson R., Pusch G.D., Olsen G.J., Davis J.J., Disz T., Edwards R.A., Gerdes S., Parrello B., Shukla M. The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST) Nucleic Acids Res. 2014;42(D1):D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goris J., Konstantinidis K.T., Klappenbach J.A., Coenye T., Vandamme P., Tiedje J.M. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007;57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]