Significance
Hox genes pattern the anteroposterior axis of all animals that have left and right body sides. In many animals, Hox genes are clustered along the chromosomes and expressed in spatial and temporal order. This coordinated regulation is thought to have preserved the cluster through a developmental constraint. Our study of the genomic organization and the embryonic spatial and temporal expression of Hox genes in sessile marine animals called lampshells (brachiopods) shows that along with having a broken Hox cluster, they lack both temporal and spatial collinearity. Furthermore, we present molecular evidence that the hard tissues (chaetae and shells) of segmented worms, mollusks, and brachiopods share a common origin that dates back to the Early Cambrian.
Keywords: chaetae, shell fields, Lophotrochozoa, Wiwaxia, Hox cluster
Abstract
Temporal collinearity is often considered the main force preserving Hox gene clusters in animal genomes. Studies that combine genomic and gene expression data are scarce, however, particularly in invertebrates like the Lophotrochozoa. As a result, the temporal collinearity hypothesis is currently built on poorly supported foundations. Here we characterize the complement, cluster, and expression of Hox genes in two brachiopod species, Terebratalia transversa and Novocrania anomala. T. transversa has a split cluster with 10 genes (lab, pb, Hox3, Dfd, Scr, Lox5, Antp, Lox4, Post2, and Post1), whereas N. anomala has 9 genes (apparently missing Post1). Our in situ hybridization, real-time quantitative PCR, and stage-specific transcriptomic analyses show that brachiopod Hox genes are neither strictly temporally nor spatially collinear; only pb (in T. transversa), Hox3 (in both brachiopods), and Dfd (in both brachiopods) show staggered mesodermal expression. Thus, our findings support the idea that temporal collinearity might contribute to keeping Hox genes clustered. Remarkably, expression of the Hox genes in both brachiopod species demonstrates cooption of Hox genes in the chaetae and shell fields, two major lophotrochozoan morphological novelties. The shared and specific expression of Hox genes, together with Arx, Zic, and Notch pathway components in chaetae and shell fields in brachiopods, mollusks, and annelids provide molecular evidence supporting the conservation of the molecular basis for these lophotrochozoan hallmarks.
Hox genes are transcription factors that bind to regulatory regions via a helix-turn-helix domain to enhance or suppress gene transcription (1, 2). Hox genes were initially described in the fruit fly Drosophila melanogaster (3, 4) and later reported in vertebrates (5–7) and the nematode Caenorhabditis elegans (8). In all of these organisms, Hox genes were shown to provide a spatial coordinate system for cells along the anteroposterior axis (9). Remarkably, the Hox genes of Drosophila and vertebrates are clustered in their genomes and exhibit a staggered spatial (3) and even temporal—as in mammals (10, 11)—expression during embryogenesis that corresponds to their genomic arrangement (3, 12, 13). These features were used to classify Hox genes into four major orthologous groups—anterior, Hox3, and central and posterior Hox genes—and have been proposed as ancestral attributes of all bilaterally symmetrical animals (1, 13, 14).
Further study of the genomic arrangements and expression patterns of Hox genes in a broader phylogenetic context has revealed multiple deviations from that hypothesized ancestral state. Hox genes are prone to gains (15–17) and losses (18–21), and their arrangement in a cluster can be interrupted, or even completely disintegrated (22–25). Furthermore, the collinear character of the Hox gene expression can fade temporally (24, 26, 27) and/or spatially (28). In addition, Hox genes have diversified their roles during development, extending beyond providing spatial information (29). They also are involved in patterning different tissues (30), and often have been recruited for the evolution and development of novel morphological traits, such as vertebrate limbs (31, 32), cephalopod funnels and arms (28), and beetle horns (33).
Thus, it is not surprising that Hox genes show diverse arrangements regarding their genomic organization and expression profiles in the Spiralia (34), a major animal clade that includes highly disparate developmental strategies and body organizations (35–39). A striking example of this is the bdelloid rotifer Adineta vaga, which belongs to the Gnathifera, the possible sister group to all remaining Spiralia (38, 39). Owing to reduced tetraploidy, the A. vaga Hox complement includes 24 genes, although it lacks posterior Hox genes and a Hox cluster (40). The freshwater flatworms Macrostomum lignano and Schmidtea mediterranea also lack a Hox cluster (41, 42), and parasitic flatworms have undergone extensive Hox gene losses, likely associated with their particular lifestyle (21). Interestingly, the limpet mollusk Lottia gigantea (16) shows a well-organized Hox cluster. Other mollusks (e.g., the Pacific oyster Crassostrea gigas) and the segmented annelid Capitella teleta exhibit split Hox clusters (43, 44). On the other hand, the cephalopod mollusk Octopus bimaculoides has lost several Hox genes and lacks a Hox cluster (22), and the clitellate annelids Helobdella robusta and Eisenia fetida do not have a Hox cluster but have greatly expanded certain Hox classes (16, 17).
Although Hox gene expression is known for a handful of spiralian species (26, 41, 43, 45–54), the relationship between genomic organization and expression domains is known for only three of these species, namely the annelids C. teleta and H. robusta and the planarian S. mediterranea. Consistent with their disintegrated Hox clusters, H. robusta and S. mediterranea show no temporal collinearity and only remnants of spatial collinearity (41, 51, 52). Conversely, C. teleta, which apparently has a split cluster, does exhibit these features (43). In general, these observations suggest that the presence of collinearity—in particular, temporal collinearity—may be associated with the retention of a more or less intact spiralian Hox cluster, as seems to be the case for the vertebrate cluster (14, 23, 55, 56). Nonetheless, more studies combining genomic and expression information, and including the vast spiralian morphological diversity, are essential to draw robust conclusions about Hox gene evolution and regulation in Spiralia and Metazoa (57). These studies also would allow investigators to test whether hypotheses about the correlation between collinearity and cluster organization observed in deuterostomes (23) hold true for protostomes as well.
Here we report a comprehensive study of the genomic arrangement and expression of Hox genes in Brachiopoda, a lineage of Spiralia with origins dating back to the Lower Cambrian (58). We use two brachiopod species—the “articulate” Terebratalia transversa and the “inarticulate” Novocrania anomala—that belong to the two major brachiopod lineages, thereby allowing the reconstruction of putative ancestral characters for Brachiopoda as a whole (Fig. 1A). Our findings demonstrate that the split Hox cluster in the Brachiopoda is not associated with a temporally collinear expression of Hox genes. Furthermore, the spatial expression of Hox genes, together with other transcription factors, such as Zic, Aristaless-related (Arx), and members of the Notch pathway, provide molecular evidence supporting the homology of annelid, brachiopod, and mollusk chaetae and shell fields.
Fig. 1.
Genomic organization of Hox genes in Brachiopoda. (A) Images of adult T. transversa and N. anomala, and phylogenetic position of these species within Brachiopoda and Lophotrochozoa. (B) The 10 Hox genes of T. transversa are ordered along three genomic scaffolds and are flanked by external genes (vertical lines); gene orthology is based on best blast hit. Thus, T. transversa has a split Hox cluster composed of three subclusters. No predicted ORFs were identified between the Hox genes in scaffolds A and B. A colored box represents each Hox gene, and below each box are the direction of transcription and the exon-intron composition. The genomic regions containing Hox genes are represented in scale. (C) The genomic organization of brachiopod Hox genes in a phylogenetic context. Adapted with permission from ref. 22. The genomic order of Hox genes in T. transversa is similar to that observed in other spiralians (e.g., C. teleta, L. gigantea), suggesting that the translocation of the Antp gene upstream to lab is a lineage-specific feature of L. anatina. (In T. transversa and L. anatina, the arrows below the genes show the direction of transcription.) A degenerate-primer screening for Hox genes reported the presence of Lox2 and Lox4 in L. anatina (15). Blastn searches against the sequenced L. anatina genome only confirmed the presence of Lox4, in the same scaffold as Post1 and Post2, although genome annotation pipelines failed to predict this gene (69). The low contiguity of the draft genome assembly of N. anomala hampered the recovery of genomic linkages between the identified Hox genes. Each ortholog group is represented by a specific color.
Results
The Hox Gene Complement of T. transversa and N. anomala.
Transcriptomic and genomic searches resulted in the identification of 10 Hox genes in T. transversa and 7 Hox genes in the transcriptome and two additional fragments corresponding to a Hox homeodomain in the draft genome assembly in N. anomala. Attempts to amplify and extend these two genomic sequences in the embryonic and larval transcriptome of N. anomala failed, suggesting that these 2 Hox genes might be expressed only during metamorphosis and/or in the adult brachiopod. Maximum likelihood orthology analyses resolved the identity of the retrieved Hox genes (Fig. S1 and Table S1). The 10 Hox genes of T. transversa were orthologous to labial (lab), proboscipedia (pb), Hox3, deformed (Dfd), sex combs reduced (Scr), Lox5, antennapedia (Antp), Lox4, Post2, and Post1. The 9 Hox genes identified in N. anomala corresponded to lab, pb, Hox3, Dfd, Scr, Lox5, Antp, Lox4, and Post2.
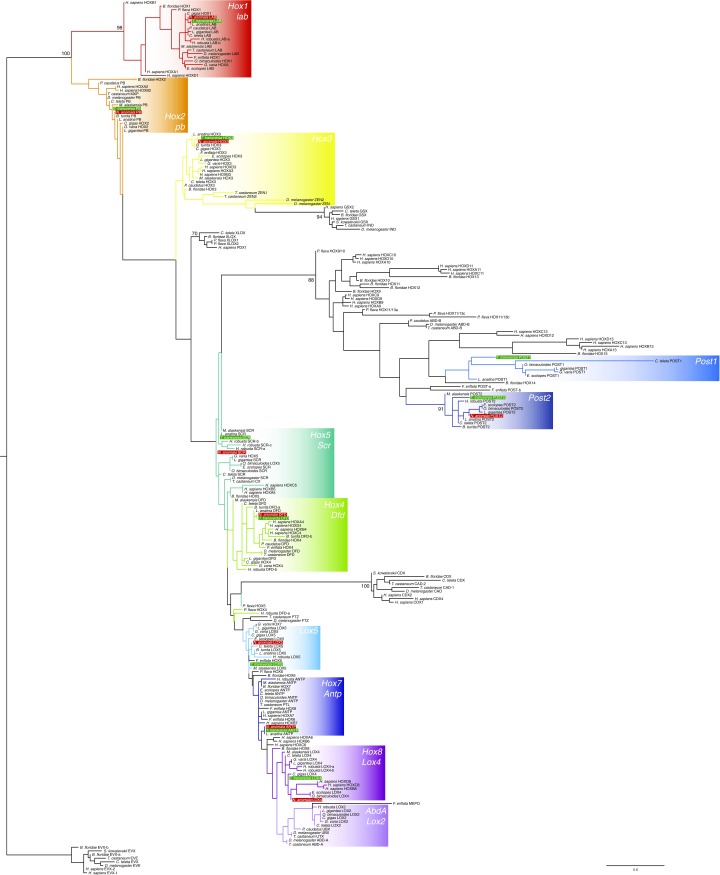
Fig. S1.
Orthology analysis of T. transversa and N. anomala Hox genes. Maximum likelihood phylogenetic analysis of bilaterian Hox and ParaHox genes, using as outgroup the even-skipped (EVX) subfamily. Colored boxes indicate Hox ortholog groups present in spiralian representatives. Hox gene sequences of a representative selection of bilaterian lineages (Table S1) were aligned with MAFFT v.7. The multiple sequence alignment (available on request) was trimmed to include the 60 amino acids of the homeodomain. ProtTest v.3 was used to determine the best-fitting evolutionary model (LG+G+I). Orthology analyses were conducted with RAxML v.8.2.6 using the autoMRE option. T. transversa sequences are highlighted by green boxes; N. anomala sequences, by red boxes. Only bootstrap values of at least 70 at nodes that supposedly represent distinct orthology groups are shown. High support values within orthogroups (e.g., between paralogs) were omitted for the sake of clarity.
Table S1.
Sequences and accession numbers used for Hox orthology assignment
| Organism | Gene | Database | Accession no. |
| Homo sapiens | HoxA1 | GenBank | AAB35423.2 |
| HoxB1 | AAH99633.1 | ||
| HoxD1 | AAG44444.1 | ||
| HoxA2 | NP_006726.1 | ||
| HoxB2 | NP_002136.1 | ||
| HoxA3 | NP_705895.1 | ||
| HoxB3 | AAD10852.1 | ||
| HoxD3 | CAA71102.1 | ||
| HoxA4 | NP_002132.3 | ||
| HoxB4 | AAG45052.1 | ||
| HoxC4 | AAG42145.1 | ||
| HoxD4 | NP_055436.2 | ||
| HoxA5 | CAG47052.1 | ||
| HoxB5 | NP_002138.1 | ||
| HoxC5 | EAW96748.1 | ||
| HoxA6 | NP_076919.1 | ||
| HoxB6 | NP_061825.2 | ||
| HoxC6 | CAG33235.1 | ||
| HoxA7 | CAA06713.1 | ||
| HoxB7 | NP_004493.3 | ||
| HoxB8 | AAG42143.1 | ||
| HoxC8 | AAG42146.1 | ||
| HoxD8 | AAG42152.1 | ||
| HoxA9 | NP_689952.1 | ||
| HoxB9 | AAG42144.1 | ||
| HoxC9 | AAG42151.1 | ||
| HoxD9 | NP_055028.3 | ||
| HoxA10 | AAH07600.1 | ||
| HoxC10 | NP_059105.2 | ||
| HoxD10 | NP_002139.2 | ||
| HoxA11 | NP_005514.1 | ||
| HoxC11 | NP_055027.1 | ||
| HoxD11 | AAF79045.1 | ||
| HoxC12 | AAK16717.1 | ||
| HoxD12 | AAF79044.1 | ||
| HoxA13 | AAC50993.1 | ||
| HoxB13 | AAH70233.1 | ||
| HoxC13 | AAF73439.1 | ||
| HoxD13 | AAC51635.1 | ||
| Gsx1 | NP_663632.1 | ||
| Gsx2 | NP_573574.1 | ||
| Pdx1 | NP_000200.1 | ||
| Cdx-1 | NP_001795.2 | ||
| Cdx-2 | NP_001256.3 | ||
| Cdx-4 | NP_005184.1 | ||
| Evx-1 | NP_001291448.1 | ||
| Evx-2 | NP_001073927.1 | ||
| Branchiostoma floridae | Hox1 | GenBank | BAA78620 |
| Hox2 | BAA78621 | ||
| Hox3 | X68045 | ||
| Hox4 | BAA78622 | ||
| Hox5 | CAA84517 | ||
| Hox6 | CAA84518 | ||
| Hox7 | CAA84519 | ||
| Hox8 | CAA84520 | ||
| Hox9 | CAA84521 | ||
| Hox10 | CAA84522 | ||
| Hox11 | AAF81909 | ||
| Hox12 | AAF81903 | ||
| Hox13 | AAF81904 | ||
| Hox14 | AAF81905 | ||
| Hox15 | ACJ74394.1 | ||
| Gsx | AAC39015.1 | ||
| Xlox | AAC39016.1 | ||
| Cdx | AAC39017 | ||
| Evx-a | AAK58953.1 | ||
| Evx-b | AAK58954.1 | ||
| Ptychodera flava | Hox1 | GenBank | AAR07634.1 |
| Hox4 | AAR07635.1 | ||
| Hox5 | AAR07636.1 | ||
| Hox6 | AAR07637.1 | ||
| Hox9/10 | AAR07638.1 | ||
| Hox11/13a | AAR07639.1 | ||
| Hox11/13b | AAR07640.1 | ||
| Hox11/13c | AAR07641.1 | ||
| Xlox1 | AAR07643.1 | ||
| Xlox2 | AAR07644.1 | ||
| Saccoglossus kowalevskii | Gsx | Uniprot | A0A0U2UDE9 |
| Cdx | GenBank | NP_001158415.1 | |
| Evx | NP_001164694.1 | ||
| Flaccisaggita enflata | Hox1 | GenBank | ABS18809.1 |
| Hox3 | ABS18810.1 | ||
| Hox4 | ABS18811.1 | ||
| Hox5 | ABS18812.1 | ||
| Hox6 | ABS18813.1 | ||
| Hox8 | ABS18814.1 | ||
| MedPost | ABS18817.1 | ||
| Posta | ABS18815.1 | ||
| Postb | ABS18816.1 | ||
| Priapulus caudatus | lab | GenBank | AAD40640.1 |
| pb | AAD40641.1 | ||
| Hox3 | AAD40642.1 | ||
| Dfd | AAD40643.1 | ||
| Ubx | AAD40647.1 | ||
| Abd-B | AAD40649.1 | ||
| Drosophila melanogaster | lab | GenBank | CAB57787 |
| pb | CAA45271 | ||
| Zen | AAF54087.1 | ||
| Zen2 | P09090.2 | ||
| Dfd | P07548 | ||
| Scr | NP_524248 | ||
| ftz | NP_477498 | ||
| Antp | CAA27417 | ||
| Ubx | CAA29194 | ||
| Abd-A | P29555 | ||
| Abd-B | CAB57859 | ||
| Ind | NP_996087.2 | ||
| Cad | AAA28409.1 | ||
| Eve | NP_523670.2 | ||
| Tribolium castaneum | lab | GenBank | EEZ99257.1 |
| Mxp | NP_001107807.1 | ||
| Zen1 | NP_001036813 | ||
| Zen2 | AAK16425.1 | ||
| Dfd | AAK16423.1 | ||
| Cx | NP_001034523.1 | ||
| ftz | AAK16421.1 | ||
| Ptl | NP_001034505.1 | ||
| Utx | EEZ99249.1 | ||
| Abd-A | EEZ99248.1 | ||
| Abd-B | EEZ99247.1 | ||
| Ind | AAW21974.1 | ||
| Cad-1 | NP_001034498.1 | ||
| Cad-2 | XP_008191732.1 | ||
| Eve | NP_001034538.1 | ||
| Capitella teleta | lab | GenBank | ABY67952 |
| pb | ABY67953 | ||
| Hox3 | ABY67954 | ||
| Dfd | ABY67955 | ||
| Scr | ABY67956 | ||
| Lox5 | ABY67957 | ||
| Antp | ABY67962 | ||
| Lox4 | ABY67958 | ||
| Lox2 | ABY67959 | ||
| Post1 | ABY67961 | ||
| Post2 | ABY67960 | ||
| Gsx | AAZ23124.1 | ||
| Cdx | AAZ95508 | ||
| Xlox | AAZ95509.1 | ||
| Evx | ABG82164 | ||
| Hellobdella robusta | lab-a | Simakov et al. (16) | |
| lab-b | Simakov et al. (16) | ||
| Scr-a | Simakov et al. (16) | ||
| Scr-b | Simakov et al. (16) | ||
| Scr-c | Simakov et al. (16) | ||
| Dfd-a | Simakov et al. (16) | ||
| Dfd-b | Simakov et al. (16) | ||
| Lox5 | Simakov et al. (16) | ||
| Antp | Simakov et al. (16) | ||
| Lox4-a | Simakov et al. (16) | ||
| Lox4-b | Simakov et al. (16) | ||
| Lox2 | Simakov et al. (16) | ||
| Post2 | Simakov et al. (16) | ||
| Lingula anatina | lab | ENSEMBL | g10891 |
| pb | g10890 | ||
| Hox3 | g10889 | ||
| Dfd | g10888 | ||
| Scr | g10887 | ||
| Lox5 | g10886 | ||
| Antp | g10892 | ||
| Post1 | g12396 | ||
| Post2 | g12399 | ||
| Crassostrea gigas | Hox1 | ENSEMBL | CGI_10024083 |
| Hox2 | CGI_10024086 | ||
| Hox3 | CGI_10024087 | ||
| Hox4 | CGI_10024091 | ||
| Lox5 | CGI_10026565 | ||
| Lox2 | CGI_10018592 | ||
| Lox4 | CGI_10026562 | ||
| Lottia gigantea | lab | Simakov et al. (16) | |
| pb | Simakov et al. (16) | ||
| Hox3 | Simakov et al. (16) | ||
| Dfd | Simakov et al. (16) | ||
| Scr | Simakov et al. (16) | ||
| Lox5 | Simakov et al. (16) | ||
| Antp | Simakov et al. (16) | ||
| Lox2 | Simakov et al. (16) | ||
| Lox4 | Simakov et al. (16) | ||
| Post1 | Simakov et al. (16) | ||
| Post2 | Simakov et al. (16) | ||
| Octopus bimaculoides | Hox1 | ENSEMBL | Ocbimv22030263 |
| Scr | Ocbimv22018468 | ||
| Lox5 | Ocbimv22010205 | ||
| Antp | Ocbimv22036189 | ||
| Lox2 | Ocbimv22033340 | ||
| Lox4 | Ocbimv22009726 | ||
| Post1 | Ocbimv22015181 | ||
| Post2 | Ocbimv22031197 | ||
| Gibbula varia | HoxA | GenBank | ACX84671.1 |
| Hox2 | ADJ18233.1 | ||
| Hox3 | ADJ18232.1 | ||
| Hox4 | ACX84672.1 | ||
| Hox5 | ADJ18234.1 | ||
| Lox5 | ADJ18235.1 | ||
| Hox7 | ADJ18236.1 | ||
| Lox2 | ADJ18238.1 | ||
| Lox4 | ADJ18237.1 | ||
| Post1 | ACX84673.1 | ||
| Euprymna scolopes | lab | GenBank | AY330184 |
| Hox3 | AY330185 | ||
| Scr | AY330186 | ||
| Lox5 | AY330187 | ||
| Antp | AY330188 | ||
| Lox4 | AY330189 | ||
| Post1 | AY330190 | ||
| Post2 | AY330191 | ||
| Micrura alaskensis | lab | GenBank | KP762174 |
| pb | KP762176 | ||
| Hox3 | KP762173 | ||
| Dfd | KP762180 | ||
| Scr | KP762177 | ||
| Lox5 | KP762179 | ||
| Antp | KP762171 | ||
| Lox4 | KP762175 | ||
| Post2 | KP762178 | ||
| Bugula turrita | pb | GenBank | AAS77225 |
| Hox3 | AAS77226 | ||
| Dfd-a | AAS77227 | ||
| Dfd-b | AAS77228 | ||
| Lox5 | AAS77229 | ||
| Post2 | AAS77230 | ||
| Lepidodermella squamata | Lox4* | This study |
> Lepidodermella_squamata_Lox4.
IITNAVTGANNGSSGKLMGAAHRTAPMYAWMAVVGPNSSQKRRGRQTYTRHQTIELEKEFAFCHYLARKRRIELAAALSLSERQVKIWFQNRRMKLKKEKQQIADMNHISTSTTSTSNSSHSKSNRHDDYNDVNDASSSDEDHLD
Genomic Organization of Hox Genes in T. transversa and N. anomala.
We used the draft assemblies of T. transversa and N. anomala genomes to investigate the genomic arrangement of their Hox genes. In T. transversa, we identified three scaffolds containing Hox genes (Fig. 1B). Scaffold A spanned 81.7 kb and contained lab and pb in a genomic region of 15.4 kb, flanked by other genes with no known linkage to the Hox cluster in other animals. Scaffold B, the longest (284.8 kb) scaffold, included Hox3, Dfd, Scr, Lox5, Antp, Lox4, and Post2, in that order (Fig. 1B), along with the microRNA mir-10 between Dfd and Scr. As in scaffold A, other genes flanked the Hox genes, which occupied a genomic region of 76.2 kb. Finally, Post1 aligned to various short scaffolds. We could not recover any genomic linkage between the identified Hox genes in N. anomala owing to the low contiguity (N50 of 3.5 kb) of the draft genome assembly. Taken together, these data demonstrate that T. transversa has a split Hox cluster broken down into three subclusters, each of them with an organized arrangement. Importantly, the potential genomic disposition of these three subclusters is similar to that observed in other spiralians, such as C. teleta and L. gigantea (Fig. 1C), which suggests that the lineage leading to the brachiopod Lingula anatina experienced genomic rearrangements that modified the order and linkage of the Hox genes.
Hox Gene Expression in T. transversa.
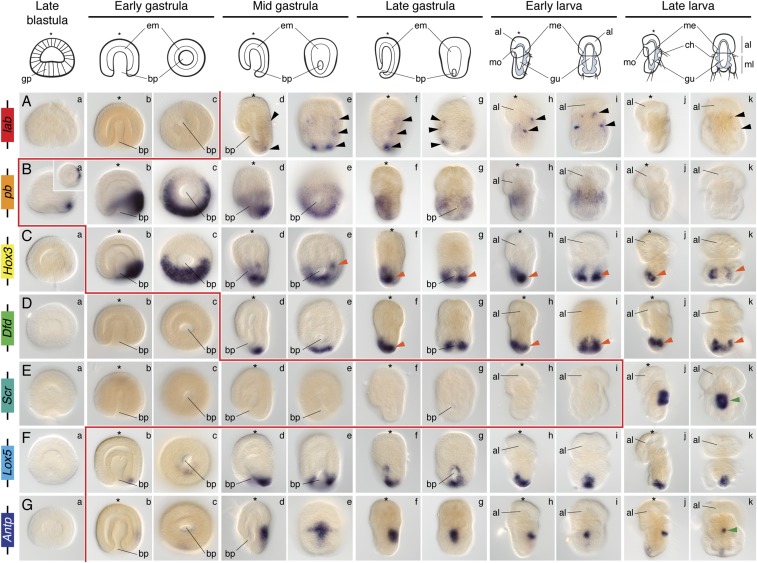
To investigate the presence of temporal and/or spatial collinearity in the expression of the clustered Hox genes in T. transversa, we first performed whole-mount in situ hybridization in embryos from blastula to late, competent larval stages (Fig. 2).
Fig. 2.
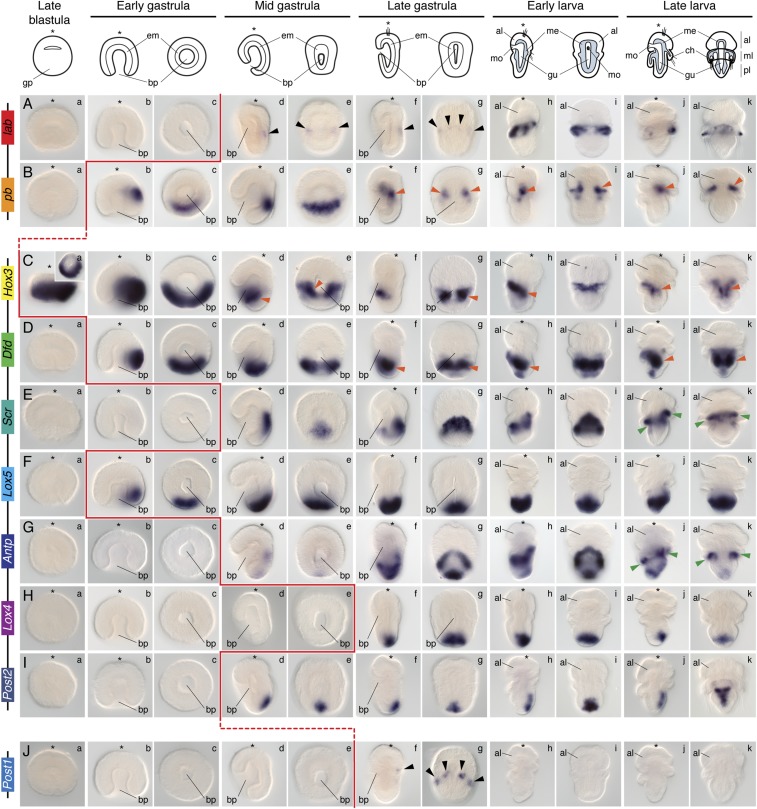
Whole-mount in situ hybridization of each Hox gene during embryonic and larval stages in T. transversa. The Hox genes lab and Post1 are expressed during chaetae formation. The genes pb, Hox3, and Dfd are expressed collinearly along the mantle and pedicle mesoderm. The Hox genes Scr and Antp are expressed in the periostracum, the shell-forming epithelium. Lox5, Lox4, and Post2 are expressed in the posterior ectoderm of the pedicle lobe. These expression patterns are described in detail in the text. Black arrowheads indicate expression in the chaetae sacs. Orange arrowheads highlight mesodermal expression. Green arrowheads indicate expression in the periostracum. The genomic organization of the Hox genes is shown on the left. On top are schematic representations of each analyzed developmental stage on its respective perspective. In these schemes, the blue area represents the mesoderm. Drawings are not to scale. The red line indicates the onset of expression of each Hox gene based on in situ hybridization data. The blastula stage is a lateral view (Inset in Ca is a vegetal view). For each other stage, the left column is a lateral view and the right column is a dorsoventral view. The asterisk demarcates the animal/anterior pole. al, apical lobe; bp, blastopore; ch, chaetae; em, endomesoderm; gp, gastral plate; gu, gut; me, mesoderm; ml, mantle lobe; mo, mouth; pl, pedicle lobe.
Anterior Hox genes.
The anterior Hox gene lab was first detected in the mid gastrula stage in two faint, bilaterally symmetrical dorsal ectodermal domains (Fig. 2A, d and e). In late gastrula stages, lab expression consisted of four dorsal ectodermal clusters corresponding to the position at which the chaetae sacs form (Fig. 2A, f and g). In early larva, lab expression was strong and broad in the mantle lobe (Fig. 2A, h and i), and in late larvae it became restricted to a few mantle cells adjacent to the chaetae sacs (Fig. 2I, j and k). These cells do not colocalize with tropomyosin, which labels the muscular mesoderm of the larva (Fig. 3A). This finding suggests that lab-expressing cells are likely ectodermal, although we cannot exclude the possibility of localization in nonmuscular mesodermal derivates.
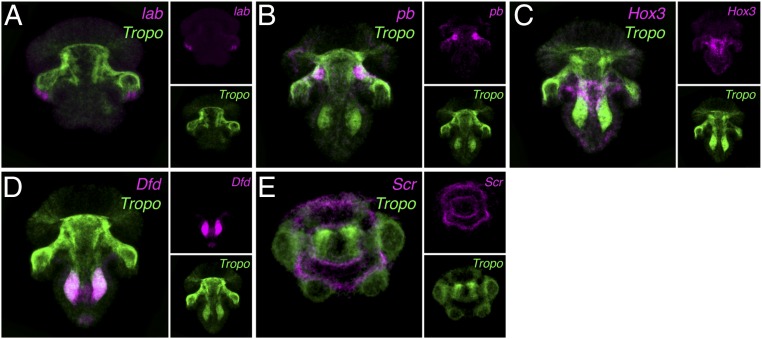
Fig. 3.
Hox expression in mesoderm and periostracum of T. transversa. Double fluorescence in situ hybridization of lab, pb, Hox3, Dfd, and Scr with tropomyosin (Tropo, in green) in late larval stages of T. transversa. (A) The gene lab is expressed in relation to the chaetae sacs, but does not overlap with the tropomyosin-expressing mesoderm. (B–D) The Hox genes pb, Hox3, and Dfd show spatial collinearity along the mantle and pedicle mesoderm. (E) The gene Scr is expressed in the periostracum, the epithelium that forms the shell.
The Hox gene pb was first detected asymmetrically on one side of the ectoderm of the early gastrula (Fig. 2B, b and c). In the mid gastrula, the ectodermal domain was located dorsally and extended as a transversal stripe (Fig. 2B, d and e). Remarkably, this domain disappeared in late gastrula embryos, where pb was detected in the anterior mantle mesoderm (Fig. 2B, f and g). This expression was maintained in early and late larva (Figs. 2 B, h–k and 3B).
Hox3.
The gene Hox3 was detected already in blastula embryos in a circle of asymmetric intensity around the gastral plate (Fig. 2C, a). In early gastrulae, Hox3 was restricted to one half of the vegetal hemisphere, the prospective posterior side (Fig. 2C, b and c). With axial elongation, Hox3 was expressed in the anterior mantle mesoderm and in the ventral ectoderm, limiting the apical and mantle lobe (Fig. 2C, d and e). This expression was maintained in late gastrula stages and in the early larva (Fig. 2C, f–i). In the late larva, Hox3 was detected in part of the ventral internal mantle ectoderm and in the anteriormost part of the pedicle mesoderm (Figs. 2C, j and k and 3C).
Central Hox genes.
The Hox gene Dfd was asymmetrically expressed on one side of the vegetal pole of the early gastrula of T. transversa (Fig. 2D, b and c). This expression was maintained in the mid gastrula and corresponded to the posteriormost region of the embryo (Fig. 2D, d and e). In the late gastrula, Dfd was strongly expressed in the posterior mesoderm (Fig. 2D, f and g). In the early larva, expression remained in the pedicle mesoderm, but new domains in the posterior ectoderm and in the anterior ventral pedicle ectoderm appeared (Fig. 2D, h and i). These expression domains were observed in the late larva as well (Figs. 2D, j and k and 3D).
The central Hox gene Scr was first expressed in the medial dorsal ectoderm of the mid gastrula (Fig. 2E, d and e). In late gastrula stages, Scr expression expanded toward the ventral side, forming a ring (Fig. 2E, f and g). In the early larva, Scr was detected in a ring encircling the anteriormost ectoderm of the pedicle lobe and extending anteriorly on its dorsal side (Fig. 2E, h and i). With the outgrowth of the mantle lobe in the late larva, Scr expression became restricted to the periostracum, the internal ectoderm of the mantle lobe that forms the shell (Fig. 2E, j and k and 3E).
The Hox gene Lox5 was expressed on one side of the early gastrula (Fig. 2F, b and c). During axial elongation, the expression became restricted to the posteriormost ectoderm of the embryo (Fig. 2F, d–g). This domain remained constant in larval stages, where it was expressed in the entire posterior ectoderm of the pedicle lobe (Fig. 2F, h–k).
The Antp gene was weakly detected at the mid gastrula stage, in one posterior ectodermal domain and one dorsal ectodermal patch (Fig. 2G, d and e). In the late gastrula, the posterior expression was maintained and the dorsal domain extended ventrally, encircling the embryo (Fig. 2G, f and g). These two domains remained in the larvae; the ectodermal anteriormost ring-like domain localized to the periostracum, and the posterior domain was limited to the posteriormost tip of the larva (Fig. 2G, h–k).
The Hox gene Lox4 was first detected in the dorsal posterior end of the late gastrula and early larva (Fig. 2H, f–i). In the late larva, Lox4 was expressed dorsally and posteriorly, although it was absent from the posteriormost end (Fig. 2H, j and k).
Posterior Hox genes.
The posterior Hox gene Post2 was first detected in mid gastrula stages at the posterior tip of the embryo (Fig. 2I, d and e). This expression was maintained in late gastrulae (Fig. 2I, f and g). In early larva, Post2 expression extended anteriorly and occupied the dorsoposterior midline of the pedicle lobe (Fig. 2I, h and i). In late, competent larvae, Post2 was detected in a T domain on the dorsal side of the pedicle ectoderm (Fig. 2I, j and k). The Hox gene Post1 was transiently detected in late gastrula stages in the four ectodermal chaetae sacs (Fig. 2J, f and g).
We verified the absence of temporal collinearity in Hox gene expression in T. transversa by real-time quantitative PCR (qPCR) and comparative stage-specific RNA-seq data (Fig. S2).
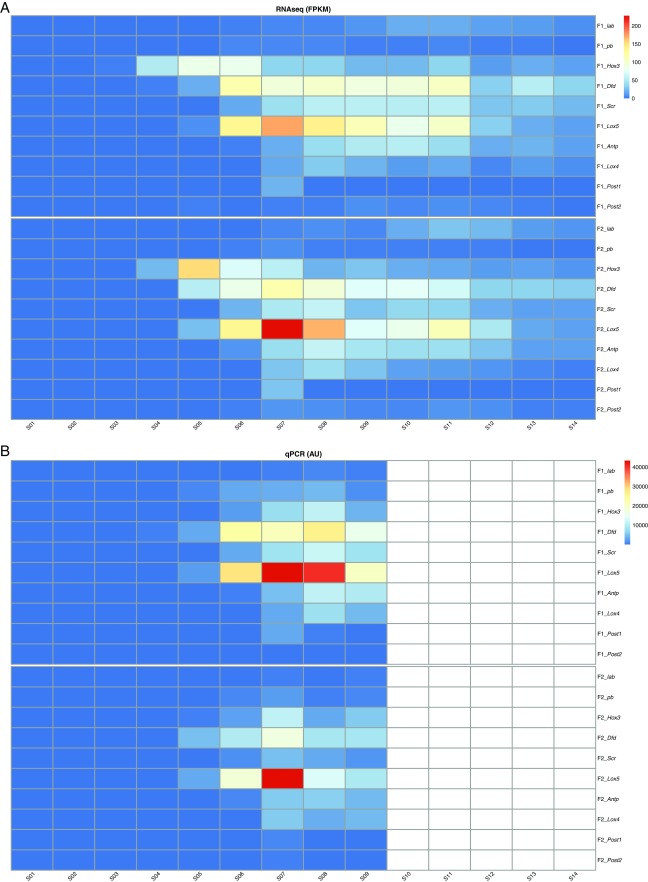
Fig. S2.
Quantitative expression of Hox genes in T. transversa developmental stages. For comparative stage-specific transcriptomic analyses, total RNA was used for constructing Illumina single end libraries and sequenced in four lanes of a HiSeq 2000 platform. Samples were randomized among the lanes. To estimate the abundance of transcripts per stage, we mapped the single end reads to the transcriptome of T. transversa with Bowtie, calculated expression levels with RSEM, and generated a matrix with TMM normalization across samples by running Trinity’s utility scripts. For real-time qPCR, total RNA was DNase-treated and preserved at −80 °C. Gene-specific primers bordering an intron splice site and defining an amplicon of 80–150 bp were designed for each gene (Table S2). Expression levels of two technical replicates performed in two biological replicates were calculated based on absolute quantification units. (A) RNAseq expression levels calculated by fragments per kilobase of exon per million reads mapped (FPKM). As shown by whole-mount in situ hybridization, Hox3 is the first gene up-regulated in the two biological replicates (female 1, F1; female 2, F2). The Hox genes pb and Post2 are detected at low levels compared with other Hox genes, which obscures their temporal variation during development in the heat map. (B) Real-time qPCR expression levels based on absolute quantification units (AU). PCR was not performed for stages 10–14 (white cells). qPCR confirmed the absence of temporal collinearity, although higher levels of Hox3 were not detected at late blastula (S04), as observed by RNAseq and in situ hybridization, likely owing to technical issues with the qPCR. Stages: S01, oocytes; S02, 8 h mid blastula; S03, 19 h late blastula; S04, 24 h moving late blastula; S05, 26 h early gastrula; S06, 37 h mid gastrula; S07, 51 h late gastrula; S08, 59 h bilobed late gastrula; S09, 68 h trilobed late gastrula; S10, 82 h early larva; S11, 98 h late larva; S12, competent larva; S13, 1 d juvenile; S14, 2 d juvenile. Expression levels obtained by real-time qPCR and comparative stage-specific transcriptomics were plotted with R.
Hox Gene Expression in N. anomala.
To infer potential ancestral Hox expression domains for the Brachiopoda, we investigated the expression of the 9 Hox genes of N. anomala during embryogenesis and larval stages (Figs. 4 and 5).
Fig. 4.
Whole-mount in situ hybridization of the Hox genes during embryonic and larval stages in N. anomala. The gene lab is expressed in the chaetae. The Hox genes Hox3 and Dfd are expressed collinearly in the mantle mesoderm. The genes Scr and Antp are expressed in the prospective shell-forming epithelium. The genes pb and Lox5 are detected in the ectoderm of the mantle lobe. The genes Lox4 and Post2 were not detected in transcriptomes and cDNA during embryonic stages. These expression patterns are described in detail in the text. Black arrowheads indicate expression in the chaetae sacs. Orange arrowheads highlight mesodermal expression. Green arrowheads indicate expression in the periostracum. On top are schematic representations of each analyzed developmental stage on its respective perspective. In these schemes, the blue area represents the mesoderm. Drawings are not to scale. The red line indicates the onset of expression of each Hox gene based on in situ hybridization data. The blastula stage is a lateral view (Inset in Ba is a vegetal view). For each other stage, the left column is a lateral view and the right column is a dorsoventral view. The asterisk demarcates the animal/anterior pole. al, apical lobe; bp, blastopore; ch, chaetae; em, endomesoderm; gp, gastral plate; gu, gut; me, mesoderm; ml, mantle lobe; mo, mouth.
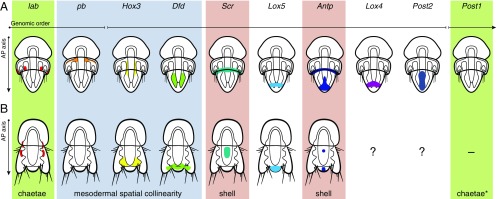
Fig. 5.
Summary of Hox gene expression in T. transversa and N. anomala. Schematic drawings of late larvae of T. transversa (A) and N. anomala (B) depicting the expression of each Hox gene. The Hox genes pb (not in N. anomala), Hox3, and Dfd show staggered expression, at least in one of their domains, associated with the mesoderm (light-blue box). In both brachiopods, the genes Scr and Antp are expressed in the periostracum, or the shell-forming epithelium (red boxes), and lab and Post1 are associated to the developing chaetae (green boxes; asterisk in Post1). Post1 is expressed in the chaetae only during late embryonic stages, not in the mature larva, and only in T. transversa. The expression of Lox4 and Post2 in N. anomala could not be determined in this study. The gene Post1 is missing in N. anomala. Drawings are not to scale.
Anterior Hox genes.
The Hox gene lab was first detected at the mid gastrula stage in three bilaterally symmetrical ectodermal cell clusters that appear to correlate with the presumptive site of chaetae sac formation (Fig. 4A, d and e). The expression in the posteriormost pair was stronger than that in the two anteriormost pairs. This expression level was maintained in the late gastrula stages (Fig. 4A, f and g). In larval stages, lab was detected in the two anteriormost chaetae sacs of the mantle lobe (Fig. 4A, h and i), with expression diminishing in late larvae (Fig. 4A, j and k).
The Hox gene pb was expressed asymmetrically already at blastula stages, in the region that putatively gives rise to the posteriormost body regions (Fig. 4B, a). With the onset of gastrulation, the expression of pb extended around the vegetal pole, almost encircling the entire blastoporal rim (Fig. 4B, b and c). During axial elongation, pb was first broadly expressed in the region that forms the mantle lobe (Fig. 4B, d and e) and later in the ventral mantle ectoderm of the late gastrula (Fig. 4B, f and g). In early larvae, pb was detected in the anterior ventral mantle ectoderm (Fig. 4B, h and i). This domain was not detected in late, competent larvae (Fig. 4B, j and k).
Hox3.
The Hox gene Hox3 was asymmetrically detected around one-half of the vegetal pole of the early gastrulae (Fig. 4C, b and c). In mid gastrulae, the expression almost encircled the entire posterior area and the blastoporal rim (Fig. 4C, d). In addition, a domain in the midposterior mesoderm became evident (Fig. 4C, e). By the end of the axial elongation, Hox3 was strongly expressed in the posterior mesoderm and weakly expressed in the ventral posterior mantle ectoderm (Fig. 4C, f and g). Notably, the posteriormost ectoderm showed no Hox3 expression. This expression pattern was maintained in early and late larval stages (Fig. 4C, h–k).
Central Hox genes.
The central Hox gene Dfd was first detected in the posterior ectodermal tip of mid gastrulae (Fig. 4D, d and e). In late gastrula stages, Dfd was expressed in the posterior ectodermal end (Fig. 4D, f) and in the posterior mesoderm (Fig. 4D, g). Early larvae showed expression of Dfd in the posterior mesoderm and posterior mantle ectoderm (Fig. 4D, h and i). This expression remained in late larvae, although the posteriormost ectodermal end was devoid of expression (Fig. 4D, j and k).
The Hox gene Scr was detected only in late larval stages, in a strong dorsal ectodermal domain (Fig. 4E, j and k). The gene Lox5 was detected asymmetrically around one-half of the blastoporal rim in early gastrula stages (Fig. 4F, b and c). During axial elongation, the expression progressively expanded around the blastoporal rim (Fig. 4F, d and e) and became limited to the ventral midline (Fig. 4F, f and g). In the larvae, Lox5 was expressed in the ventral posteriormost midline (Fig. 4F, h–k).
The Hox gene Antp was first expressed asymmetrically in one lateral side of the early gastrula (Fig. 4G, b and c). In the mid gastrula, Antp was detected in the dorsal ectodermal mantle in a cross-configuration, in the dorsal midline and the mantle cells closer to the apical-mantle lobe boundary (Fig. 4G, d and e). In late gastrulae, Antp was expressed only in a middorsal ectodermal region (Fig. 4G, f and g). This expression pattern also was observed in early larval stages, although the size of the domain was reduced (Fig. 4G, h and i). In late larvae, Antp was detected in a small middorsal patch and a weak ventroposterior ectodermal domain (Fig. 4G, j and k).
We could neither identify nor amplify Lox4 in a transcriptome and cDNA obtained from mixed embryonic and larval stages, suggesting that it is either only very transiently and weakly expressed during embryogenesis or expressed only in later stages (metamorphosis and adulthood).
Posterior Hox genes.
The only posterior Hox gene present in N. anomala, Post2, could not be amplified in cDNA obtained from mixed embryonic and larval stages, suggesting that it is not expressed—or at least expressed only at very low levels—during these stages of the life cycle. The absence of larval expression of Lox4 and Post2 could be related to the lack of the pedicle lobe of craniiform brachiopod larva, which is a characteristic of the lineage (59, 60).
Expression of Arx, Zic, and Notch Components in Brachiopod Chaetal Sacs.
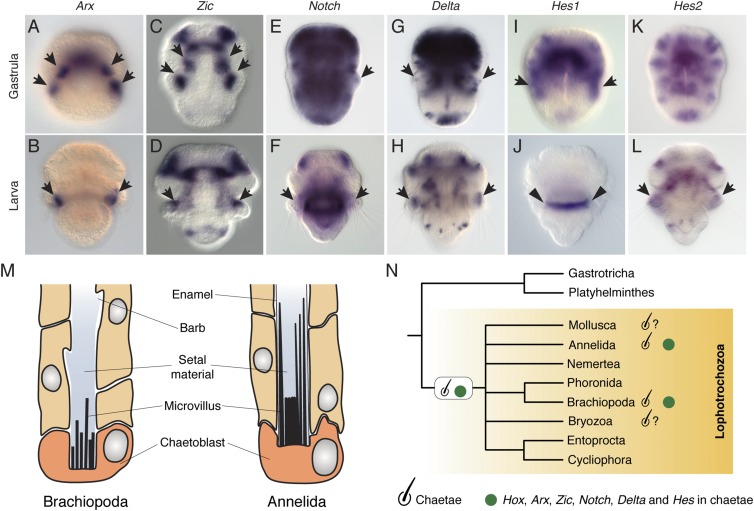
Brachiopods and annelids share the expression of lab and Post1 in the chaetal sacs (26, 43, 61). To further analyze these molecular similarities, we identified and studied the expression of the homeodomain-containing transcription factor Arx, the zinc finger Zic, and components of the Notch signaling pathway, all of which are associated with the development of chaetae in annelids (62–65). Arx was specifically expressed in the developing and mature chaetal sacs (Fig. 6 A and B), whereas Zic was expressed in anterior mesodermal and ectodermal domains as well as in the developing chaetae (Fig. 6 C and D). Similarly, the core components of the Notch pathway, Notch and Delta, were expressed in the developing chaetae, although salt-and-pepper expression in the ectoderm and mesoderm was detected as well (Fig. 6 E–H). The downstream component of the Notch pathway Hes1 was expressed in the mantle lobe ectoderm of the gastrula, where the chaetae sacs form, but was not detected in the larva (Fig. 6 I and J). Finally, the Hes2 ortholog was expressed in the developed chaetae sacs of the larva of the brachiopod T. transversa (Fig. 6 K and L).
Fig. 6.
Expression of chaetae-related genes during T. transversa embryogenesis. (A–L) Whole- mount in situ hybridization of Arx, Zic, Notch, Delta, and two Hes genes in gastrula embryos and larvae of T. transversa. (A) In mid gastrulae, Arx is expressed in the ectoderm of the prospective chaetae sac territories (black arrows) and in a ventral domain. (B) In early larvae, Arx is expressed in the chaetae sacs (black arrows). (C) In late gastrulae, Zic is expressed in the mesoderm of the chaetae sacs (black arrows), apical lobe mesoderm, and anterior ectoderm. (D) In early larvae, Zic is detected in the chaetae sacs (black arrows), in a domain in the pedicle lobe, and in the anterior mesoderm and anterior ectoderm. (E and F) Notch is broadly expressed in the ectoderm and mesoderm of the late gastrula and early larva, particularly in a cluster of a few cells of the developing chaetae (black arrows). (G and H) Delta is strongly expressed in the apical lobe and in a salt and pepper manner in the mantle and pedicle lobe, including the chaetae (black arrows). (I and J) Hes1 is observed in the lateral ectoderm of the gastrula (black arrows), in the area that will subsequently form the chaetae and mantle lobe ectoderm. It is not detected in the larva. (Black arrowheads indicate background expression in J.) (K and L) Hes2 is detected in a salt and pepper manner in the ectoderm and mesoderm of the T. transversa embryo, and in the chaetae of the larva (black arrows). The images are dorsal views (except I and K), with the anterior pole at the top. (M) Morphological similarities between brachiopod and annelid chaetae. Drawing adapted with permission from ref. 84. (N) The shared morphological and molecular characters of chaetae in Brachiopoda and Annelida, together with the presence of chaetae-like structures (chaetae sign with a question mark) in the Mollusca and Bryozoa, support the homology of this lophotrochozoan novelty.
Discussion
The Brachiopod Hox Complement and Evolution of Hox Genes in Spiralia.
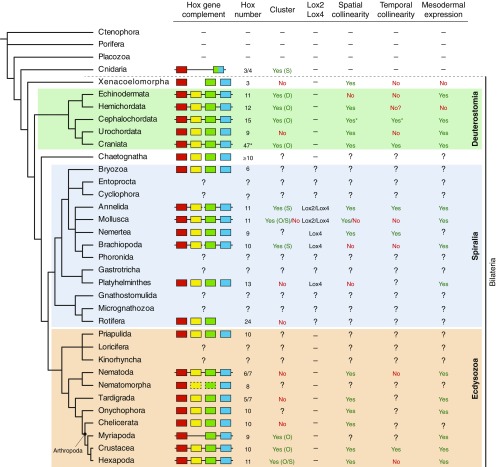
Our findings regarding T. transversa and N. anomala reveal an ancestral brachiopod Hox gene complement consistent with what has been hypothesized to be ancestral for Spiralia and Lophotrochozoa based on degenerate PCR surveys (15, 66–68). This ancient complement comprises ten Hox genes—lab, pb, Hox3, Dfd, Scr, Lox5, Antp, Lox4, Post2, and Post1—and has been confirmed by genomic sequencing of representative annelids and mollusks (16, 22, 44), rotifers and platyhelminthes (21, 40–42), and the linguliform brachiopod L. anatina (69). Whereas T. transversa and L. anatina have retained this ancestral Hox complement, N. anomala has apparently lost Post1 (Fig. 1). Although a previous analysis based on degenerate PCR primers reported the presence of Lox2 in the brachiopod L. anatina (15), current high-throughput sequencing approaches seem to restrict Lox2 to Annelida and Mollusca (Fig. S3). Whether multiple independent losses in diverse spiralian lineages shaped the evolutionary history of this gene, or whether this represents a molecular synapomorphy of annelids and mollusks, requires further study, specifically involving a thorough genomic sequencing of a larger number of lophotrochozoan taxa.
Fig. S3.
Evolution of Hox organization and expression across Metazoa. The table presents the features of the Hox gene complement of each animal lineage in a phylogenetic framework. The Hox complement is summarized by the presence of at least one representative each of the anterior, Hox3, central, and posterior ortholog groups. The Hox number indicates the possible ancestral number, but can vary between species. In Craniata, the number corresponds to the human Hox complement, which consists of four clusters (asterisk). The cluster organization can be of one of three types: organized (O), disorganized (D), or split (S). For species with an atomized cluster, the cluster is described as absent (no presence). Question marks indicate unknown data, and dashes indicate absences. In Cephalochordata, the asterisks indicate that Hox6 and Hox14 do not follow the general temporal and collinear expression in Branchiostoma lanceolatum. See the main text for references.
Our genomic information shows that the Hox cluster of T. transversa is split into three parts, with lab and pb separate from the major cluster and Post1 also on a separate scaffold (Fig. 1B). Overall, this Hox cluster extends for >100 kb, which is significantly shorter than clusters of other lophotrochozoans, such as C. teleta (∼345 kb) (43) and L. gigantea (∼455 kb) (16). Its compact size is related to short intergenic regions and introns, comparable to the situation observed in vertebrate Hox clusters (23). The order and orientation of the Hox genes in T. transversa are preserved and more organized compared with the Hox cluster reported for the brachiopod L. anatina, which exhibits a genomic rearrangement that places a portion of the cluster upstream of lab and in reverse orientation (69). Indeed, the split Hox clusters reported so far in lophotrochozoan taxa exhibit multiple different conformations, indicating that lineage-specific genomic events have shaped Hox gene clusters in Spiralia.
Noncollinearity of Hox Expression in the Split Cluster of T. transversa.
Our analysis of Hox clustering in different animal species together with the temporal and spatial expression patterns of their Hox genes supports the hypotheses that the regulatory elements required for their collinearity—mostly temporal—maintain the clustered organization of the Hox genes in vertebrates and possibly other animals as well (13, 23, 55–57, 70, 71) (Fig. S3). Although there are cases in which spatial collinearity is exhibited in the absence of a cluster, as in the appendicularian chordate Oikopleura dioica (24), all investigated clustered Hox genes show at least one type of collinearity that could account for their genomic organization (23, 57). Given that there are exceptions to the spatial collinearity in vertebrates—e.g., Hoxa2 and Hoxb2 are expressed more anteriorly than Hox1 genes in the vertebrate hindbrain (72)—temporal collinearity is seen as a manifestation of Hox clustering; however, whether temporal collinearity is the agent keeping the cluster together [e.g., through enhancer sharing (73)] remains a matter of debate.
Within Spiralia, this evolutionary scenario appears to be supported by the staggered temporal and spatial expression of the Hox genes in the split cluster of the annelid C. teleta (43). In the other investigated spiralians, there is only either genomic information (e.g., the mollusks L. gigantea and C. gigas) or expression analysis (e.g., the mollusks Gibbula varia and Haliotis asinina) (16, 44, 49, 54). Most of these previous gene expression studies have demonstrated coordinated spatial or temporal expression of Hox genes along the animal’s anteroposterior axis (45, 46, 61) or in organ systems, such the nervous system (49, 54). However, the absence of studies revealing a correlation between Hox gene expression and their genomic organization in these animals hinders reconstruction of the putative mechanisms that preserve Hox clusters in Lophotrochozoa, and thus preclude generalizations about possible scenarios of Hox cluster evolution across all animals.
Our findings robustly demonstrate that overall, the Hox genes of the split Hox cluster of T. transversa exhibit neither strictly spatial nor temporal collinearity (Figs. 2 and 3), and lack quantitative collinearity (57), as has been shown in mice, for example (74). These observations are also supported by the absence of coordinated spatial and temporal expression of the Hox genes in N. anomala (Fig. 4). Although a general trend toward spatial collinearity is present (e.g., the posterior Hox genes are expressed in posterior tissues), the early expression of Hox3 breaks temporal collinearity in T. transversa, whereas it is pb that is first expressed in N. anomala. In both species, Lox5 is also expressed before Scr, as it is also the case in the annelid Nereis virens (61). Ectodermal spatial collinearity is absent in the two brachiopods even when the future location of the larval tissues after metamorphosis is considered (75, 76). The anteriormost class gene lab is expressed exclusively in the chaetae of T. transversa and N. anomala, and thus is not affiliated with anterior neural or foregut tissues as in other lophotrochozoans, such as annelids (43, 77). Similarly, the posteriormost Hox gene, Post1, is very transiently expressed in the chaetae sacs, which occupy a midposition in the larval body. We detected strict spatial collinearity only in the staggered expression of the Hox genes pb, Hox3, and Dfd along the anteroposterior axis of the developing larval mesoderm in both T. transversa and N. anomala (Fig. 5). Thus, the absence of strict temporal and spatial collinearity exhibited by the split Hox cluster in the brachiopod T. transversa supports the idea that temporal collinearity might help keep spiralian Hox genes clustered, as seems to be the case in vertebrates and at least some arthropods (14, 23, 25, 55–57, 78). However, other factors, such as unequal rates of genome rearrangements in different lineages and shared enhancers between genes, also might contribute to the genomic evolution of Hox genes.
Recruitment of Hox Genes for Patterning Lophotrochozoan Chaetae and Shell Fields.
The bristle-like chaetae (or setae) of annelids and brachiopods and shell valves in mollusks and brachiopods are the most prominent hard tissues found in lophotrochozoan spiralians (79) and provide fossilized hallmarks of the Cambrian explosion (80). Chaetae-like structures are also present in the sensory organs of polyplacophoran mollusks (81), Kölliker’s organ of juvenile octopods (82), and the gizzard teeth of some bryozoans (83). The ultrastructural morphology of the brachiopod and annelid chaetae are known to be nearly identical (84–86) (Fig. 6M), and with the placement of brachiopods as close relatives of annelids and mollusks (87), the homology of these structures appears more likely (88). In this context, the anterior Hox gene lab is expressed in the chaetae of Chaetopterus sp. (26) and in the ectoderm around chaetal sacs in N. virens (61), and Post1 is expressed in the chaetae of C. teleta, Platynereis dumerilii, and N. virens (43, 61). Our results show that, similarly, lab and Post1 are expressed specifically in the chaetal sacs of the brachiopods T. transversa and N. anomala (Figs. 2 and 4) and follow the different arrangements of the chaetae in both species. Further evidence of a common, and likely homologous, molecular profile comes from expression of the homeodomain gene Arx, the zinc finger Zic, and components of the Notch signaling pathway. These genes are expressed at each chaetae sac territory in the Platynereis larva (62, 64), in C. teleta (63, 65), and also in the region of the forming chaetae sac territories in T. transversa (Fig. 6 A–L and Fig. S4). Therefore, the expression of the Hox genes lab and Post1 and the homeodomain gene Arx indicate that a similar molecular signature underlays the development of chaetae in annelids and brachiopods. This property, together with the evident and striking morphological similarities shared by brachiopod and annelid chaetae, lend support to considering these two structures homologous and thus common lophotrochozoan innovations (Fig. 6N). This would be consistent with placing the iconic Cambrian fossil Wiwaxia, which contains chaetae, as a stem group lophotrochozoan (89).
Fig. S4.
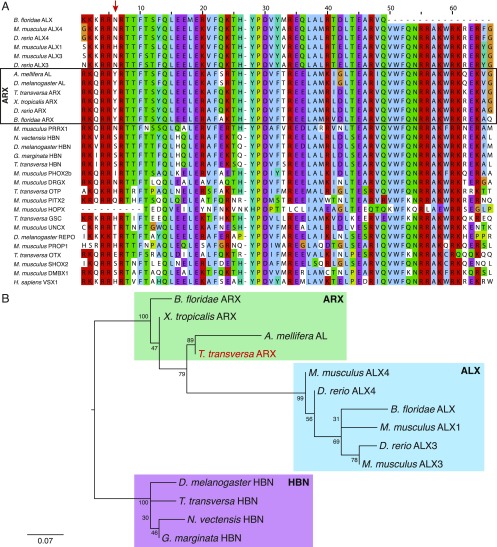
Orthology assignment of the T. transversa Arx gene. (A) Amino acid alignment of the homeodomain region of representative sequences of the PRD gene class. Aristaless-related genes (ARX) are highlighted with a rectangle, and all of them share a unique tyrosine (Y) at position six of the alignment (indicated by a red arrow on top). (B) Maximum likelihood tree of Aristaless-related (ARX), Aristaless-like (ALX), and Homeobrain (HBN; also termed ARX-like) genes. Although the monophyly of ARX genes is dubious, the T. transversa ARX gene is not an ALX or HBN member. Orthology analyses were conducted with RAxML v.8.2.6 using the autoMRE option and LG+G substitution model. Bootstrap values are indicated for each node.
The protective shell is a mineralized tissue present in brachiopods and mollusks. In the gastropod mollusk G. varia, the Hox genes lab, Post1, and Post2 are first expressed in the shell field, and later Dfd is expressed in the shell field as well (53). In H. asinina, lab and Post2 also are related to shell formation (49). In brachiopods, Dfd is associated with the adult shell in L. anatina (69); however, during embryogenesis of T. transversa and N. anomala, only Scr and Antp are expressed in the shell fields, and lab and Post1 are not; here lab and Post1 are expressed in the chaetae sacs. These properties could support the homology of the chitin network formed at the onset of brachiopod and mollusk shell fields. However, the differing deployment of Hox genes in the shell fields of brachiopods and mollusks might indicate that these genes do not have an ancient role in the specification of the shell-forming epithelium. Instead, their consistent deployment during shell development might reflect a more general, conserved role in shaping the shell fields according to their position along the anteroposterior axis.
Conclusion
In this study, we have characterized the Hox gene complement of the brachiopods T. transversa and N. anomala, demonstrating that the last common ancestor to all brachiopods likely had 10 Hox genes (lab, pb, Hox3, Dfd, Scr, Lox5, Antp, Lox4, Post2, and Post1). Noticeably, brachiopod Hox genes do not exhibit global temporal and spatial collinearity, although T. transversa exhibits a split Hox cluster. Only the genes pb (in T. transversa), Hox3 (in both brachiopods), and Dfd (in both brachiopods) show spatial collinearity in the “trunk” mesoderm. The dramatic divergence of the expression of Hox genes from the supposed ancestral state for Hox expression while still retaining a relatively intact Hox cluster might indicate that the loss of constraint on the organization of the Hox cluster in T. transversa is relatively recent. In addition, the Hox genes lab and Post1, as well as the homeobox Arx, are expressed in the developing chaetae, as also has been reported for some annelid species (43, 50, 61). These molecular similarities, together with evident morphological resemblances (85), lend support to considering brachiopod and annelid chaetae homologous structures and reinforce the classification of the fossil Wiwaxia as a stem group lophotrochozoan (89).
Materials and Methods
Animal Cultures.
Gravid adults of T. transversa (Sowerby, 1846) were collected around San Juan Island, WA, and those of N. anomala (Müller, 1776) were collected around Bergen, Norway. Animal husbandry, fertilization, and larval culture were conducted following previously published protocols (90–92).
Hox Cluster Reconstruction in T. transversa and N. anomala.
Male gonads of T. transvesa and N. anomala were preserved in RNAlater (Life Technologies) for further genomic DNA (gDNA) isolation. Paired-end and mate pair libraries of 2 kb and 5 kb insert sizes of T. transversa gDNA were sequenced using the Illumina HiSeq 2000 platform. We first trimmed Illumina adapters with Cutadapt 1.4.2 (93), then assembled the paired-end reads into contigs, scaffolded the assembly with the mate pair reads, and closed the gaps using Platanus 1.21 (94). The genomic scaffolds of T. transversa including Hox genes are available in the GenBank database (accession nos. KX372775 and KX372776). We sequenced the paired-end libraries of N. anomala gDNA using the Illumina HiSeq 2000 platform. We then removed Illumina adapters as above and assembled the paired-end reads with MaSuRCA 2.2.1 (95).
Gene Isolation.
Genes were identified by BLAST searches on public transcriptomes of T. transversa and N. anomala developmental stages (National Center for Biotechnology Information Sequence Read Archive; T. transversa, GenBank accession no. SRX1307070; N. anomala, GenBank accession no. SRX1343816) and their respective draft genomes (see above). All gene sequences have been uploaded to GenBank (accession nos. KX372756–KX372774 and KY124237–KY124242).
Gene Expression Analyses.
Single colorimetric whole-mount in situ hybridization was carried out following an established protocol (detailed protocol available in Protocol Exchange, 2008; www.nature.com/protocolexchange/protocols/480) (96, 97). Double fluorescence in situ hybridization was conducted as described elsewhere (98, 99).
Quantitative Hox Gene Expression in T. transversa.
Thousands of synchronous T. transversa embryos collected at 14 specific stages [oocytes, 8 h mid blastula, 19 h late blastula, 24 h moving late blastula, 26 h early gastrula, 37 h asymmetric gastrula, 51 h bilateral gastrula, 59 h bilobed, 68 h trilobed, 82 h early larva (first chaetae visible), 98 h late larva (long chaetae, eye spots), 131 h competent larva, 1 d juvenile, and 2 d juvenile] were pooled together and preserved in RNAlater (Life Technologies). The same material was used for real-time qPCR and stage-specific transcriptomes (Fig. S2 and Table S2).
Table S2.
Primers used for real-time qPCR experiments
| Gene | Forward | Reverse |
| lab | CAAAGCTCCGTAGCCACTTA | TCGAGCTCTGTCAATTGCTT |
| pb | AACAAATCGGATGGCTCTG | TTCATGGTCTGCTTCCTCTG |
| Hox3 | ACTTCGCGTTAGCCAATCA | TGCAGGAACCCTTCAGAAA |
| Dfd | ATGCCGAGTATAAGCCGTTC | TATACCCGTGGATGAAACGA |
| Scr | ACGTCTGATGCCTGGTGTAG | ATAGCCATGAACAAATGCCA |
| Lox5 | GTGTACGTTTGCCTGGTACG | GCATGTCGCAAGCGTATAGT |
| Antp | TCTCAAGCTCGAGTGTTTGG | GGAGACGCAGATAACGACAG |
| Lox4 | GTTTGTCGACCGCGTCTT | AAATGGATACGGGTCTGCTC |
| Post2 | GCTCCTGTGGCATTGTGTAG | AGCAAGCAAGCCCTGTAGAT |
| Post1 | AACGTTGTCCCATTCTCTCC | CGATATACTATGCGGACCCA |
Acknowledgments
We thank the staff at Friday Harbor Laboratories and the Espeland Marine Station for their invaluable help with animal collection. We also thank Daniel Thiel and Anlaug Boddington; Daniel Chourrout for his valuable comments on early versions of this manuscript; and Kevin Kocot for access to entoproct transcriptomes. This study was funded by a Meltzer Research Fund grant, the Marie Curie International Reintegration Grant FP7-PEOPLE-2009-RG 256450 (to A.H.), and the Sars Centre core budget.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KX372756–KX372774 and KY124237–KY124242). T. transversa Hox profiling is available at 10.6084/m9.figshare.46443.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1614501114/-/DCSupplemental.
References
- 1.McGinnis W, Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68(2):283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- 2.Pearson JC, Lemons D, McGinnis W. Modulating Hox gene functions during animal body patterning. Nat Rev Genet. 2005;6(12):893–904. doi: 10.1038/nrg1726. [DOI] [PubMed] [Google Scholar]
- 3.Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276(5688):565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- 4.McGinnis W, Levine MS, Hafen E, Kuroiwa A, Gehring WJ. A conserved DNA sequence in homoeotic genes of the Drosophila Antennapedia and bithorax complexes. Nature. 1984;308(5958):428–433. doi: 10.1038/308428a0. [DOI] [PubMed] [Google Scholar]
- 5.Carrasco AE, McGinnis W, Gehring WJ, De Robertis EM. Cloning of an X. laevis gene expressed during early embryogenesis coding for a peptide region homologous to Drosophila homeotic genes. Cell. 1984;37(2):409–414. doi: 10.1016/0092-8674(84)90371-4. [DOI] [PubMed] [Google Scholar]
- 6.McGinnis W, Garber RL, Wirz J, Kuroiwa A, Gehring WJ. A homologous protein-coding sequence in Drosophila homeotic genes and its conservation in other metazoans. Cell. 1984;37(2):403–408. doi: 10.1016/0092-8674(84)90370-2. [DOI] [PubMed] [Google Scholar]
- 7.McGinnis W, Hart CP, Gehring WJ, Ruddle FH. Molecular cloning and chromosome mapping of a mouse DNA sequence homologous to homeotic genes of Drosophila. Cell. 1984;38(3):675–680. doi: 10.1016/0092-8674(84)90262-9. [DOI] [PubMed] [Google Scholar]
- 8.Costa M, Weir M, Coulson A, Sulston J, Kenyon C. Posterior pattern formation in C. elegans involves position-specific expression of a gene containing a homeobox. Cell. 1988;55(5):747–756. doi: 10.1016/0092-8674(88)90131-6. [DOI] [PubMed] [Google Scholar]
- 9.Akam M. Hox and HOM: Homologous gene clusters in insects and vertebrates. Cell. 1989;57(3):347–349. doi: 10.1016/0092-8674(89)90909-4. [DOI] [PubMed] [Google Scholar]
- 10.Dollé P, Izpisúa-Belmonte JC, Falkenstein H, Renucci A, Duboule D. Coordinate expression of the murine Hox-5 complex homoeobox-containing genes during limb pattern formation. Nature. 1989;342(6251):767–772. doi: 10.1038/342767a0. [DOI] [PubMed] [Google Scholar]
- 11.Izpisúa-Belmonte JC, Falkenstein H, Dollé P, Renucci A, Duboule D. Murine genes related to the Drosophila AbdB homeotic genes are sequentially expressed during development of the posterior part of the body. EMBO J. 1991;10(8):2279–2289. doi: 10.1002/j.1460-2075.1991.tb07764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duboule D, Morata G. Colinearity and functional hierarchy among genes of the homeotic complexes. Trends Genet. 1994;10(10):358–364. doi: 10.1016/0168-9525(94)90132-5. [DOI] [PubMed] [Google Scholar]
- 13.Lemons D, McGinnis W. Genomic evolution of Hox gene clusters. Science. 2006;313(5795):1918–1922. doi: 10.1126/science.1132040. [DOI] [PubMed] [Google Scholar]
- 14.Garcia-Fernàndez J. The genesis and evolution of homeobox gene clusters. Nat Rev Genet. 2005;6(12):881–892. doi: 10.1038/nrg1723. [DOI] [PubMed] [Google Scholar]
- 15.de Rosa R, et al. Hox genes in brachiopods and priapulids and protostome evolution. Nature. 1999;399(6738):772–776. doi: 10.1038/21631. [DOI] [PubMed] [Google Scholar]
- 16.Simakov O, et al. Insights into bilaterian evolution from three spiralian genomes. Nature. 2013;493(7433):526–531. doi: 10.1038/nature11696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zwarycz AS, Nossa CW, Putnam NH, Ryan JF. Timing and scope of genomic expansion within annelida: evidence from homeoboxes in the genome of the earthworm Eisenia fetida. Genome Biol Evol. 2015;8(1):271–281. doi: 10.1093/gbe/evv243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aboobaker A, Blaxter M. Hox gene evolution in nematodes: Novelty conserved. Curr Opin Genet Dev. 2003;13(6):593–598. doi: 10.1016/j.gde.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 19.Aboobaker AA, Blaxter ML. Hox gene loss during dynamic evolution of the nematode cluster. Curr Biol. 2003;13(1):37–40. doi: 10.1016/s0960-9822(02)01399-4. [DOI] [PubMed] [Google Scholar]
- 20.Smith FW, et al. The compact body plan of tardigrades evolved by the loss of a large body region. Curr Biol. 2016;26(2):224–229. doi: 10.1016/j.cub.2015.11.059. [DOI] [PubMed] [Google Scholar]
- 21.Tsai IJ, et al. Taenia solium Genome Consortium The genomes of four tapeworm species reveal adaptations to parasitism. Nature. 2013;496(7443):57–63. doi: 10.1038/nature12031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Albertin CB, et al. The octopus genome and the evolution of cephalopod neural and morphological novelties. Nature. 2015;524(7564):220–224. doi: 10.1038/nature14668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duboule D. The rise and fall of Hox gene clusters. Development. 2007;134(14):2549–2560. doi: 10.1242/dev.001065. [DOI] [PubMed] [Google Scholar]
- 24.Seo HC, et al. Hox cluster disintegration with persistent anteroposterior order of expression in Oikopleura dioica. Nature. 2004;431(7004):67–71. doi: 10.1038/nature02709. [DOI] [PubMed] [Google Scholar]
- 25.Serano JM, et al. Comprehensive analysis of Hox gene expression in the amphipod crustacean Parhyale hawaiensis. Dev Biol. 2016;409(1):297–309. doi: 10.1016/j.ydbio.2015.10.029. [DOI] [PubMed] [Google Scholar]
- 26.Irvine SQ, Martindale MQ. Expression patterns of anterior Hox genes in the polychaete Chaetopterus: Correlation with morphological boundaries. Dev Biol. 2000;217(2):333–351. doi: 10.1006/dbio.1999.9541. [DOI] [PubMed] [Google Scholar]
- 27.Lowe CJ, Wray GA. Radical alterations in the roles of homeobox genes during echinoderm evolution. Nature. 1997;389(6652):718–721. doi: 10.1038/39580. [DOI] [PubMed] [Google Scholar]
- 28.Lee PN, Callaerts P, De Couet HG, Martindale MQ. Cephalopod Hox genes and the origin of morphological novelties. Nature. 2003;424(6952):1061–1065. doi: 10.1038/nature01872. [DOI] [PubMed] [Google Scholar]
- 29.Godwin AR, Capecchi MR. Hoxc13 mutant mice lack external hair. Genes Dev. 1998;12(1):11–20. doi: 10.1101/gad.12.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chauvet S, et al. Distinct hox protein sequences determine specificity in different tissues. Proc Natl Acad Sci USA. 2000;97(8):4064–4069. doi: 10.1073/pnas.070046997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woltering JM, Duboule D. Tetrapod axial evolution and developmental constraints: Empirical underpinning by a mouse model. Mech Dev. 2015;138(Pt 2):64–72. doi: 10.1016/j.mod.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zakany J, Duboule D. The role of Hox genes during vertebrate limb development. Curr Opin Genet Dev. 2007;17(4):359–366. doi: 10.1016/j.gde.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 33.Wasik BR, Rose DJ, Moczek AP. Beetle horns are regulated by the Hox gene, Sex combs reduced, in a species- and sex-specific manner. Evol Dev. 2010;12(4):353–362. doi: 10.1111/j.1525-142X.2010.00422.x. [DOI] [PubMed] [Google Scholar]
- 34.Barucca M, Canapa A, Biscotti MA. An overview of Hox genes in Lophotrochozoa: Evolution and functionality. J Dev Biol. 2016;4:12. doi: 10.3390/jdb4010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dunn CW, Giribet G, Edgecombe GD, Hejnol A. Animal phylogeny and its evolutionary implications. Annu Rev Ecol Evol Syst. 2014;45:371–395. [Google Scholar]
- 36.Hejnol A. A twist in time: The evolution of spiral cleavage in the light of animal phylogeny. Integr Comp Biol. 2010;50(5):695–706. doi: 10.1093/icb/icq103. [DOI] [PubMed] [Google Scholar]
- 37.Kocot KM. On 20 years of Lophotrochozoa. Org Divers Evol. 2016;16:329. [Google Scholar]
- 38.Laumer CE, et al. Spiralian phylogeny informs the evolution of microscopic lineages. Curr Biol. 2015;25(15):2000–2006. doi: 10.1016/j.cub.2015.06.068. [DOI] [PubMed] [Google Scholar]
- 39.Struck TH, et al. Platyzoan paraphyly based on phylogenomic data supports a noncoelomate ancestry of spiralia. Mol Biol Evol. 2014;31(7):1833–1849. doi: 10.1093/molbev/msu143. [DOI] [PubMed] [Google Scholar]
- 40.Flot JF, et al. Genomic evidence for ameiotic evolution in the bdelloid rotifer Adineta vaga. Nature. 2013;500(7463):453–457. doi: 10.1038/nature12326. [DOI] [PubMed] [Google Scholar]
- 41.Currie KW, et al. HOX gene complement and expression in the planarian Schmidtea mediterranea. Evodevo. 2016;7:7. doi: 10.1186/s13227-016-0044-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wasik K, et al. Genome and transcriptome of the regeneration-competent flatworm, Macrostomum lignano. Proc Natl Acad Sci USA. 2015;112(40):12462–12467. doi: 10.1073/pnas.1516718112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fröbius AC, Matus DQ, Seaver EC. Genomic organization and expression demonstrate spatial and temporal Hox gene colinearity in the lophotrochozoan Capitella sp. I. PLoS One. 2008;3(12):e4004. doi: 10.1371/journal.pone.0004004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang G, et al. The oyster genome reveals stress adaptation and complexity of shell formation. Nature. 2012;490(7418):49–54. doi: 10.1038/nature11413. [DOI] [PubMed] [Google Scholar]
- 45.Fritsch M, Wollesen T, de Oliveira AL, Wanninger A. Unexpected co-linearity of Hox gene expression in an aculiferan mollusk. BMC Evol Biol. 2015;15:151. doi: 10.1186/s12862-015-0414-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fritsch M, Wollesen T, Wanninger A. Hox and ParaHox gene expression in early body plan patterning of polyplacophoran mollusks. J Exp Zoolog B Mol Dev Evol. 2016;326(2):89–104. doi: 10.1002/jez.b.22671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hiebert LS, Maslakova SA. Expression of Hox, Cdx, and Six3/6 genes in the hoplonemertean Pantinonemertes californiensis offers insight into the evolution of maximally indirect development in the phylum Nemertea. Evodevo. 2015;6:26. doi: 10.1186/s13227-015-0021-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hiebert LS, Maslakova SA. Hox genes pattern the anterior-posterior axis of the juvenile but not the larva in a maximally indirect developing invertebrate, Micrura alaskensis (Nemertea) BMC Biol. 2015;13:23. doi: 10.1186/s12915-015-0133-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hinman VF, O’Brien EK, Richards GS, Degnan BM. Expression of anterior Hox genes during larval development of the gastropod Haliotis asinina. Evol Dev. 2003;5(5):508–521. doi: 10.1046/j.1525-142x.2003.03056.x. [DOI] [PubMed] [Google Scholar]
- 50.Irvine SQ, Martindale MQ. Comparative analysis of Hox gene expression in the polychaete Chaetopterus: Implications for the evolution of body plan regionalization. Am Zool. 2001;41(3):640–651. [Google Scholar]
- 51.Kourakis MJ, Martindale MQ. Hox gene duplication and deployment in the annelid leech Helobdella. Evol Dev. 2001;3(3):145–153. doi: 10.1046/j.1525-142x.2001.003003145.x. [DOI] [PubMed] [Google Scholar]
- 52.Kourakis MJ, et al. Conserved anterior boundaries of Hox gene expression in the central nervous system of the leech Helobdella. Dev Biol. 1997;190(2):284–300. doi: 10.1006/dbio.1997.8689. [DOI] [PubMed] [Google Scholar]
- 53.Samadi L, Steiner G. Involvement of Hox genes in shell morphogenesis in the encapsulated development of a top shell gastropod (Gibbula varia L.) Dev Genes Evol. 2009;219(9-10):523–530. doi: 10.1007/s00427-009-0308-6. [DOI] [PubMed] [Google Scholar]
- 54.Samadi L, Steiner G. Expression of Hox genes during the larval development of the snail, Gibbula varia (L.): Further evidence of non-colinearity in molluscs. Dev Genes Evol. 2010;220(5-6):161–172. doi: 10.1007/s00427-010-0338-0. [DOI] [PubMed] [Google Scholar]
- 55.Duboule D. Temporal colinearity and the phylotypic progression: A basis for the stability of a vertebrate Bauplan and the evolution of morphologies through heterochrony. Dev Suppl. 1994;1994:135–142. [PubMed] [Google Scholar]
- 56.Ferrier DEK, Minguillón C. Evolution of the Hox/ParaHox gene clusters. Int J Dev Biol. 2003;47(7-8):605–611. [PubMed] [Google Scholar]
- 57.Monteiro AS, Ferrier DEK. Hox genes are not always colinear. Int J Biol Sci. 2006;2(3):95–103. doi: 10.7150/ijbs.2.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rudwick MJS. Living and Fossil Brachiopods. Hutchinson; London: 1970. [Google Scholar]
- 59.Freeman G. A developmental basis for the Cambrian radiation. Zoolog Sci. 2007;24(2):113–122. doi: 10.2108/zsj.24.113. [DOI] [PubMed] [Google Scholar]
- 60.Vellutini BC, Hejnol A. Expression of segment polarity genes in brachiopods supports a non-segmental ancestral role of engrailed for bilaterians. Sci Rep. 2016;6:32387. doi: 10.1038/srep32387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kulakova M, et al. Hox gene expression in larval development of the polychaetes Nereis virens and Platynereis dumerilii (Annelida, Lophotrochozoa) Dev Genes Evol. 2007;217(1):39–54. doi: 10.1007/s00427-006-0119-y. [DOI] [PubMed] [Google Scholar]
- 62.Gazave E, Guillou A, Balavoine G. History of a prolific family: The Hes/Hey-related genes of the annelid Platynereis. Evodevo. 2014;5:29. doi: 10.1186/2041-9139-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thamm K, Seaver EC. Notch signaling during larval and juvenile development in the polychaete annelid Capitella sp. I. Dev Biol. 2008;320(1):304–318. doi: 10.1016/j.ydbio.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 64.Fischer A. 2010. Mesoderm formation and muscle development of Platynereis dumerilii (Nereididae, Annelida). PhD dissertation. (Freie Universität Berlin, Berlin)
- 65.Layden MJ, Meyer NP, Pang K, Seaver EC, Martindale MQ. Expression and phylogenetic analysis of the zic gene family in the evolution and development of metazoans. Evodevo. 2010;1(1):12. doi: 10.1186/2041-9139-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Balavoine G, de Rosa R, Adoutte A. Hox clusters and bilaterian phylogeny. Mol Phylogenet Evol. 2002;24(3):366–373. doi: 10.1016/s1055-7903(02)00237-3. [DOI] [PubMed] [Google Scholar]
- 67.Halanych KM, Passamaneck Y. A brief review of metazoan phylogeny and future prospects in Hox research. Am Zool. 2001;41:629–639. [Google Scholar]
- 68.Passamaneck YJ, Halanych KM. Evidence from Hox genes that bryozoans are lophotrochozoans. Evol Dev. 2004;6(4):275–281. doi: 10.1111/j.1525-142X.2004.04032.x. [DOI] [PubMed] [Google Scholar]
- 69.Luo YJ, et al. The Lingula genome provides insights into brachiopod evolution and the origin of phosphate biomineralization. Nat Commun. 2015;6:8301. doi: 10.1038/ncomms9301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ferrier DEK, Holland PWH. Ciona intestinalis ParaHox genes: Evolution of Hox/ParaHox cluster integrity, developmental mode, and temporal colinearity. Mol Phylogenet Evol. 2002;24(3):412–417. doi: 10.1016/s1055-7903(02)00204-x. [DOI] [PubMed] [Google Scholar]
- 71.Patel NH. Evolutionary biology: Time, space and genomes. Nature. 2004;431(7004):28–29. doi: 10.1038/431028a. [DOI] [PubMed] [Google Scholar]
- 72.Tümpel S, Wiedemann LM, Krumlauf R. Hox genes and segmentation of the vertebrate hindbrain. Curr Top Dev Biol. 2009;88:103–137. doi: 10.1016/S0070-2153(09)88004-6. [DOI] [PubMed] [Google Scholar]
- 73.Sharpe J, Nonchev S, Gould A, Whiting J, Krumlauf R. Selectivity, sharing and competitive interactions in the regulation of Hoxb genes. EMBO J. 1998;17(6):1788–1798. doi: 10.1093/emboj/17.6.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Spitz F, Gonzalez F, Duboule D. A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell. 2003;113(3):405–417. doi: 10.1016/s0092-8674(03)00310-6. [DOI] [PubMed] [Google Scholar]
- 75.Nielsen C. The development of the brachiopod Crania (Neocrania) anomala (O. F. Müller) and its phylogenetic significance. Acta Zoologica. 1991;72:7–28. [Google Scholar]
- 76.Freeman G. Metamorphosis in the brachiopod Terebratalia: Evidence for a role of calcium channel function and the dissociation of shell formation from settlement. Biol Bull. 1993;184:15–24. doi: 10.2307/1542376. [DOI] [PubMed] [Google Scholar]
- 77.Steinmetz PRH, Kostyuchenko RP, Fischer A, Arendt D. The segmental pattern of otx, gbx, and Hox genes in the annelid Platynereis dumerilii. Evol Dev. 2011;13(1):72–79. doi: 10.1111/j.1525-142X.2010.00457.x. [DOI] [PubMed] [Google Scholar]
- 78.Kao D, et al. The genome of the crustacean Parhyale hawaiensis, a model for animal development, regeneration, immunity and lignocellulose digestion. eLife. 2016;5:e20062. doi: 10.7554/eLife.20062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brusca RC, Moore W, Shuster SM. Invertebrates. Sinauer Associates; Sunderland, MA: 2016. [Google Scholar]
- 80.Budd GE, Jensen S. A critical reappraisal of the fossil record of the bilaterian phyla. Biol Rev Camb Philos Soc. 2000;75(2):253–295. doi: 10.1017/s000632310000548x. [DOI] [PubMed] [Google Scholar]
- 81.Leise EM, Cloney RA. Chiton integument: Ultrastructure of the sensory hairs of Mopalia muscosa (Mollusca: Polyplacophora) Cell Tissue Res. 1982;223(1):43–59. doi: 10.1007/BF00221498. [DOI] [PubMed] [Google Scholar]
- 82.Brocco SL, O’Clair RM, Cloney RA. Cephalopod integument: The ultrastructure of Kölliker’s organs and their relationship to setae. Cell Tissue Res. 1974;151(3):293–308. doi: 10.1007/BF00224540. [DOI] [PubMed] [Google Scholar]
- 83.Gordon DP. The resemblance of bryozoan gizzard teeth to “annelid-like” setae. Acta Zoologica. 1975;56:283–289. [Google Scholar]
- 84.Gustus RM, Cloney RA. Ultrastructural similarities between setae of brachiopods and polychaetes. Acta Zoologica. 1972;53(2):229–233. [Google Scholar]
- 85.Lüter C. Ultrastructure of larval and adult setae of Brachiopoda. Zool Anz. 2000;239:75–90. [Google Scholar]
- 86.Orrhage L. Light and electron microscope studies of some brachiopod and pogonophoran setae. Z. Morph. Tiere. 1973;74(4):253–270. [Google Scholar]
- 87.Halanych KM, et al. Evidence from 18S ribosomal DNA that the lophophorates are protostome animals. Science. 1995;267(5204):1641–1643. doi: 10.1126/science.7886451. [DOI] [PubMed] [Google Scholar]
- 88.Lüter C, Bartolomaeus T. The phylogenetic position of Brachiopoda: A comparison of morphological and molecular data. Zool Scr. 1997;26:245–253. [Google Scholar]
- 89.Smith MR. Ontogeny, morphology and taxonomy of the soft-bodied cambrian “mollusc” Wiwaxia. Palaeontology. 2014;57:215–229. [Google Scholar]
- 90.Freeman G. Regional specification during embryogenesis in the articulate brachiopod Terebratalia. Dev Biol. 1993;160(1):196–213. doi: 10.1006/dbio.1993.1298. [DOI] [PubMed] [Google Scholar]
- 91.Freeman G. Regional specification during embryogenesis in the craniiform brachiopod Crania anomala. Dev Biol. 2000;227(1):219–238. doi: 10.1006/dbio.2000.9857. [DOI] [PubMed] [Google Scholar]
- 92.Reed C. Phylum Brachiopoda. In: Strathmann MF, editor. Reproduction and Development of the Marine Invertebrates of the Northern Pacific Coast. Univ of Washington Press; Seattle: 1987. pp. 486–493. [Google Scholar]
- 93.Martin M. 2011 Cutadapt removes adapter sequences from high-throughput sequencing reads. journal.embnet.org/index.php/embnetjournal/article/view/200/479. Accessed January 22, 2017.
- 94.Kajitani R, et al. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 2014;24(8):1384–1395. doi: 10.1101/gr.170720.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zimin AV, et al. The MaSuRCA genome assembler. Bioinformatics. 2013;29(21):2669–2677. doi: 10.1093/bioinformatics/btt476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hejnol A, Martindale MQ. Acoel development indicates the independent evolution of the bilaterian mouth and anus. Nature. 2008;456(7220):382–386. doi: 10.1038/nature07309. [DOI] [PubMed] [Google Scholar]
- 97.Santagata S, Resh C, Hejnol A, Martindale MQ, Passamaneck YJ. Development of the larval anterior neurogenic domains of Terebratalia transversa (Brachiopoda) provides insights into the diversification of larval apical organs and the spiralian nervous system. Evodevo. 2012;3:3. doi: 10.1186/2041-9139-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Martin-Duran JM, Passamaneck YJ, Martindale MQ, Hejnol A. The developmental basis for the recurrent evolution of deuterostomy and protostomy. Nat Ecol Evol. 2016;1:0005. doi: 10.1038/s41559-016-0005. [DOI] [PubMed] [Google Scholar]
- 99.Grande C, Martín-Durán JM, Kenny NJ, Truchado-García M, Hejnol A. Evolution, divergence and loss of the Nodal signalling pathway: New data and a synthesis across the Bilateria. Int J Dev Biol. 2014;58(6-8):521–532. doi: 10.1387/ijdb.140133cg. [DOI] [PubMed] [Google Scholar]