Significance
PEMapper and PECaller are paired software programs that simplify mapping and variant calling for whole-genome datasets. Whole-genome sequencing data are fast becoming the most natural dataset for all genetic studies. Analysis tools for data at this scale are essential. This manuscript describes tools, which solve the challenges of data analysis at whole-genome scale, using an approach involving 16-mer mapping and SNP calling based on a Pólya–Eggenberger distribution for SNP genotypes. We show that our software package is faster (cheaper to run), uses much less disk space (cheaper to store results), requires no previous knowledge of existing genetic variation (easier to deploy to nonhuman species), and achieves calling results that are as good as Genome Analysis Toolkit best practices.
Keywords: genome sequencing, GATK, sequence mapping, SNP calling, software
Abstract
The analysis of human whole-genome sequencing data presents significant computational challenges. The sheer size of datasets places an enormous burden on computational, disk array, and network resources. Here, we present an integrated computational package, PEMapper/PECaller, that was designed specifically to minimize the burden on networks and disk arrays, create output files that are minimal in size, and run in a highly computationally efficient way, with the single goal of enabling whole-genome sequencing at scale. In addition to improved computational efficiency, we implement a statistical framework that allows for a base by base error model, allowing this package to perform as well or better than the widely used Genome Analysis Toolkit (GATK) in all key measures of performance on human whole-genome sequences.
Whole-genome sequencing (WGS) using short reads on the Illumina platform is an increasingly cost-effective approach for identifying genetic variation, with growing potential for both research and clinical applications (1–4). A critical challenge is in the development of efficient algorithms capable of rapidly and accurately identifying variable sites from among the enormous collection of sequence reads (5). Given the large size of eukaryotic genomes, even modest false-positive or -negative error rates can act as barriers to the success of genetic studies and would inhibit the utility of such studies for both research and clinical applications.
The de facto standard methodology for mapping and calling variants is the so-called Burrows--Wheeler aligner (BWA)/Genome Analysis Toolkit (GATK) best practices pipeline (6), which was devised and validated for whole-exome experiments and has greatly facilitated whole-exome studies for identification of disease-causing variants (7–9). Although this pipeline can be used successfully at whole-genome scales, there are barriers to its use, particularly as the number of samples increases. BWA (10), Bowtie (11), and most other commonly used read mapping software packages are designed to run in low-memory footprints [i.e., less than 8 or 16 gigabytes (GB) random-access memory (RAM)]. Because whole-genome datasets are large (necessarily greater than 100 GB uncompressed for 30× coverage), these read mappers must continuously read and write large quantities of data to and from the disk. Sorting reads, in particular, is highly disk input/output (I/O)-intensive. Although a high-performance disk array can provide the needed I/O performance for a single instance of BWA/GATK processing (6), no disk array can possibly accommodate the I/O performance required to run multiple GATK instances simultaneously on parallel processors. Moreover, even if the disk array itself could meet the demand, the network/fiber interconnects between the array and the computational nodes quickly become saturated. Simply put, although BWA/GATK best practices do an excellent job in a nonclustered environment, the “network cost” in a clustered environment significantly limits its performance for large WGS datasets.
GATK best practices have additional limitations. First, output files can be quite large. Binary sequence alignment/map (BAM) files, required to store sequence alignment data, are almost always larger than the initial fastq files of nucleotide sequences, and Haplotype Caller (HC) output can be nearly one-half the size of the BAM files. Thus, total storage requirements to run the pipeline can approach 300 GB compressed per sample for WGS data. Second, variant calling begins with individual samples (not collections of samples; i.e., joint calling), and as a result, the distinction between sites called as homozygous reference genotypes and those called as missing (insufficient evidence to make a call) is not always maintained. Third, the GATK best practices joint genotyping caller, required to generate the highest-quality genotype calls, does not scale well to whole-genome data. As currently implemented, the joint caller simply will not run on whole-genome size files in sample collections larger than 10–20 human genomes, even on computers with 512 GB RAM. Crashing with more than 20 samples seriously limits the utility of GATK for large-scale sequencing. Finally, the entire GATK best practices pipeline relies on and uses enormous quantities of “previous knowledge” about the position and frequency of SNPs and indels (insertion/deletion variants). Using known positions of variants is both a strength, in that it leverages outside knowledge to improve performance, and a weakness, in the sense that it makes its application to nonhuman systems difficult and may also create biases in variant calling.
Here, we describe two software programs intended to overcome the limitations of GATK best practices called PEMapper and PECaller. PEMapper solves the inherent limitations of the BWA/GATK pipeline by performing all of the necessary read sorting, storing, and mapping procedures in RAM. Human genome indices are preloaded, and final output is written only once (never reloaded, resorted, etc.). These technical changes lead to substantial performance gains as detailed below. PEMapper requires a large RAM allocation (typically nearly 200 GB for the sequence of a whole human genome) but in exchange, does not overburden the network or disk subsystems. Modern computational clusters, such as those found at many universities or available from cloud providers [i.e., Amazon Web Services (AWS)], are well-equipped to run many simultaneous instances of PEMapper in parallel to expedite experiments. Additionally, output from PEMapper/PECaller comes in much smaller files, decreasing the long-term storage requirements for WGS data (Table 1).
Table 1.
Data storage requirements for a single sample using each pipeline
| File type | GB |
| GATK | |
| FASTQ files | 78.9 |
| BAM file | 115 |
| Individual VCF file | 53 |
| Combined VCF file (per sample) | 0.561 |
| Total | ∼247.5 |
| PEMapper/PECaller | |
| FASTQ files | 78.9 |
| Pileup file | 7.8 |
| Mapping files | 4 |
| SNP file (per sample) | 0.035 |
| Indel file | 0.0001 |
| Total | ∼91 |
Unlike PEMapper, with innovations that are strictly in implementation, PECaller represents an intellectual departure from several other genotype-calling models (6, 12) First, variant detection occurs simultaneously (joint calling in the initial stage) in all samples from the same experiment. Joint calling from the outset is important, because it ensures that the distinction between missing data (data with insufficient evidence for any genotype) and homozygous reference data is recognized from the inception. In addition, it allows the imposition of a population genetics-inspired prior on the data and the ability to fit sophisticated models of read error to help distinguish bases with high error rates from those that actually harbor variants. The population genetics prior accounts for the fact that most sites are expected to be invariant but conditional on the site containing a variant; the variant is expected to be in Hardy–Weinberg equilibrium. Second, another innovation of the PECaller method involves the underlying statistical model used to describe the data. Formally, we assume that read depths are drawn from a Pólya–Eggenberger (PE; Dirichlet multinomial) distribution, not the more conventional multinomial assumption. Using a PE distribution allows us to model a nucleotide base as having both a relatively high “error rate” but also, importantly, a large variance in that rate. Use of the PE distribution helps us reduce false-positive variant calling, while at the same time, enabling us to call true heterozygotes, even when the relative fraction of the two alleles is highly uneven (another common occurrence). The fact that PEMapper/PECaller does not use any information about “known” SNPs or indels makes it far better suited for nonhuman systems or human diseases, such as cancer, with large numbers of de novo mutations. We show that PEMapper/PECaller, despite not using any such information, performs as well or better than GATK best practices in all aspects of variant discovery and calling.
Results
Performance of PEMapper/PECaller.
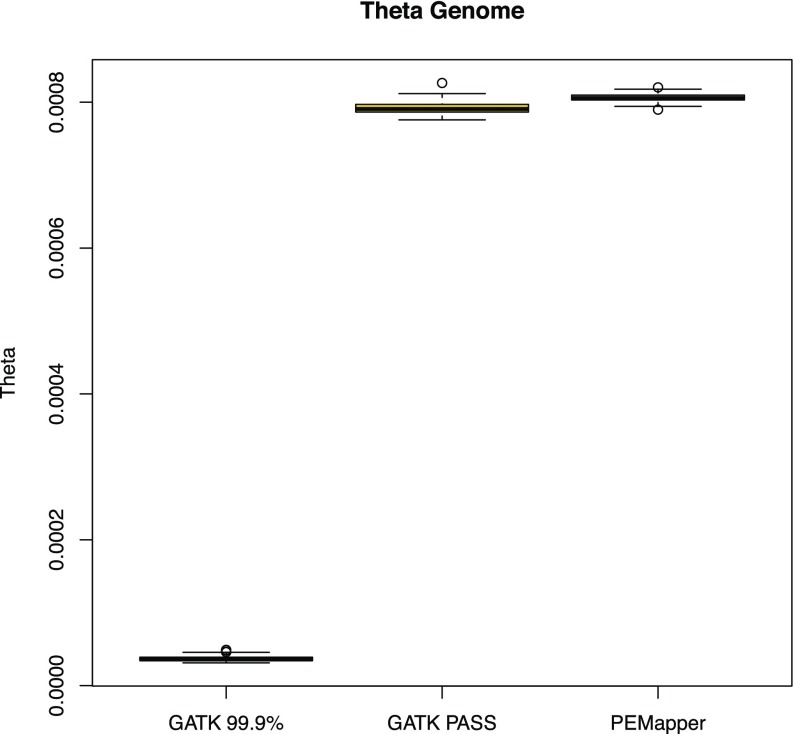
The simplest measure of variation, named theta (13), counts the number of heterozygotes called per sample per base. Theta is estimated to be somewhere between 0.0008 and 0.001 in humans (14, 15). Fig. 1 shows theta for each of 97 sequenced human genome samples that passed quality control (QC) (Methods). Most remarkable is the extremely consistent levels of variation called between samples, with individuals ranging from 0.0008351 to 0.0008624. The overall variation levels are consistent with previous estimates.
Fig. 1.
Theta across all 97 samples based on the calls from PEMapper/PECaller, GATK PASS, and GATK Tranche99.9. PEMapper/PECaller and GATK PASS samples sit between 0.00075 and 0.0009 variants per base as expected. Tranche99.9 calls are much lower.
False-Positive Calling.
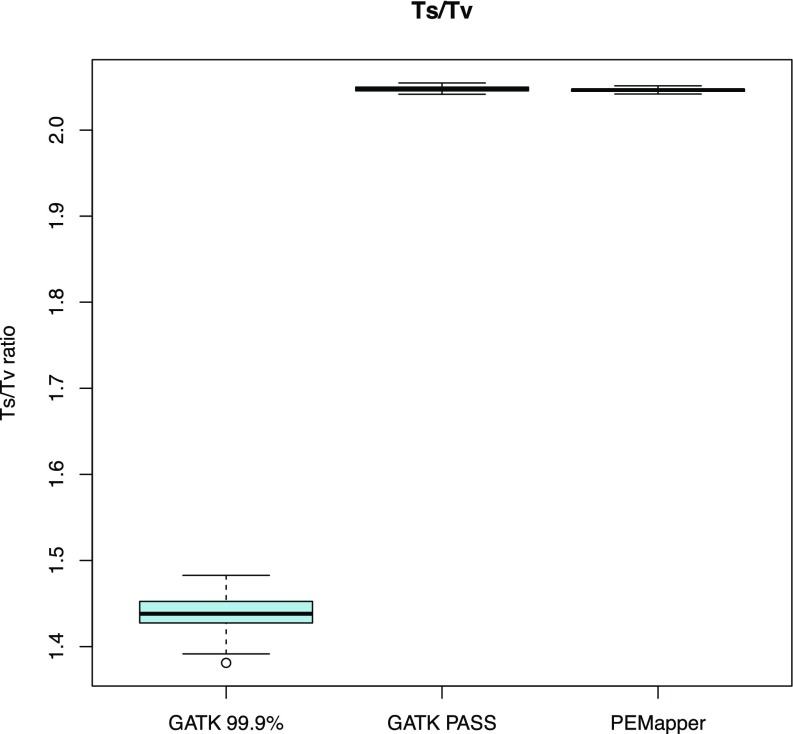
Our analysis provides ample evidence that this called variation contains very few false-positive findings (nonvariant sites called variant in error). Sequence changes from A->G, G->A, C->T, and T->C are called “transitions.” All other changes are called “transversions.” There are twice as many transversions possible as transitions. Many mutational mechanisms favor transitions over transversions (oxidative deamination, etc.). Selection also likely favors transitions over transversions (much more likely to be silent in exons; similar binding for transcription factors; e.g., wobble binding). However, random genotype calling error likely results in increased transversions (because there are twice as many ways to get a transversion as a transition when you make an error). Thus, real data ought to be enriched for transitions over transversions, and false data ought to be enriched for transversions. Picking nucleotides at random would give a 0.5:1 transition to transversion (Ts/Tv) ratio. It is widely believed that the overall Ts/Tv ratio is ∼2.0 in humans (genome.sph.umich.edu/wiki/SNP_Call_Set_Properties). For every sample in this study, the Ts/Tv ratio was between 2.042:1 and 2.051:1 (Fig. 2). Looking at the entire collection of variants, the ratio was 2.073:1. This overall ratio can be used to estimate the fraction of false-positive variant calls. If we assume that the “true” ratio is 2.12:1, a value determined from all variants called by both PEMapper/PECaller and GATK (see below), and we assume that false-positive variant calls have a ratio of 0.5:1 (as expected by chance), then an observed ratio of 2.073:1 implies that, over all 97 samples, ∼3% of the variants were false positives. On a per sample basis, less than 1 in 3,000–5,000 called variants per sample were false positives. The data quality from PEMapper/PECaller compares favorably with that of other next-generation sequencing (NGS) analytical tools (16).
Fig. 2.
Comparison of Ts/Tv ratios for PEMapper/Caller, GATK PASS, and GATK Tranche99.9 called variants. PEMapper/PECaller and GATK PASS are virtually identical at near 2.04 and 2.05 per sample, respectively, indicating excellent quality calls. GATK Tranche99.9 is much lower, between 1.3 and 1.5 per sample, indicating much lower-quality calls.
Exonic Variation.
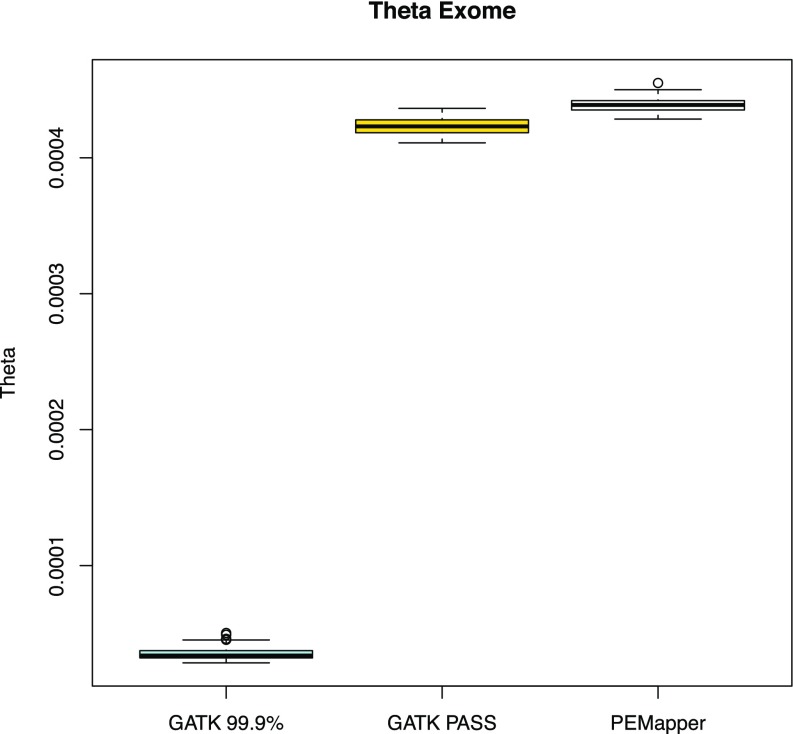
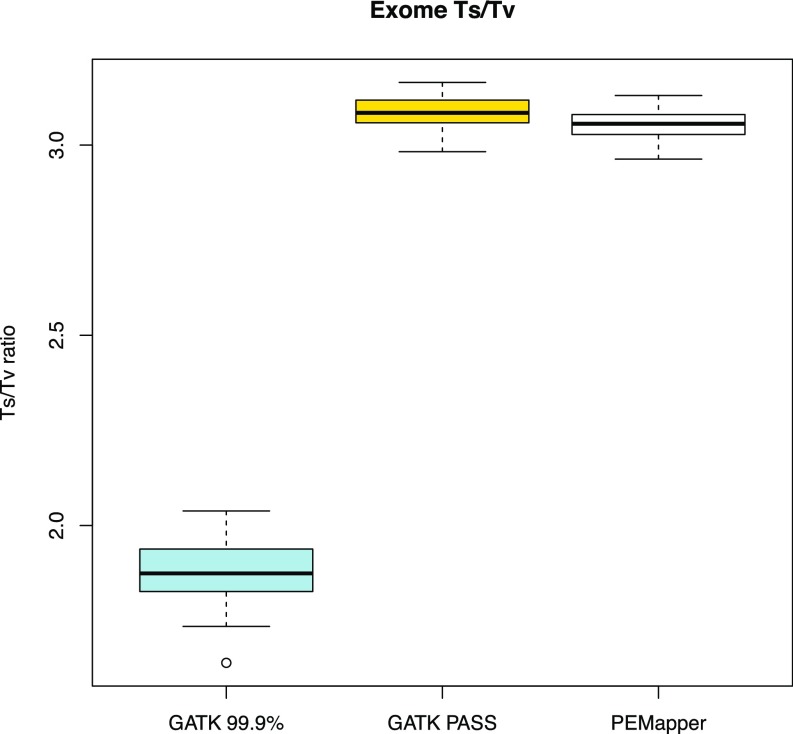
In general, there ought to be far less variation in exons than in the genome as a whole. In these samples, we saw theta in exons to be between 0.0004284 and 0.0004550 per sample (Fig. 3) (i.e., slightly more than one-half its value for the genome as a whole). We also found a much higher Ts/Tv ratio (2.963:1–3.130:1) (Fig. 4), consistent with selection for transitions in exons. Of the variants in exons, one expects approximately one-half to be “silent” (making no change to the amino acid sequence) and one-half to be replacement (changing the amino acid sequence). The average silent to replacement ratio (17) per sample was 1.101:1, with a range from 1.074:1 to 1.127:1 (Fig. 5). On average, there were ∼20,000 variants in the CCDS-defined (Consensus Coding Sequence Project; https://www.ncbi.nlm.nih.gov/projects/CCDS/CcdsBrowse.cgi) exome of each individual. Over the entire collection of sites, 44.54% of all exonic variants were silent. This number is very similar to published estimates from 100× whole-exome sequencing (18). Of note, Tennessen et al. (18) restricted themselves to ∼16,000 well-covered genes, where here, we use the whole-CCDS exome.
Fig. 3.
Theta in all sample exomes based on PEMapper/PECaller, GATK PASS, and Tranche99.9 calls. GATK PASS and PEMapper/PECaller samples are near 0.00045 as expected, with PEMapper/PECaller calling slightly more variants.
Fig. 4.
Ts/Tv ratio across all sample exomes based on PEMapper/PECaller, GATK PASS, and Tranche99.9 calls. All samples called by PEMapper/PECaller and GATK PASS are near three as expected. Tranche99.9 calls are much lower again.
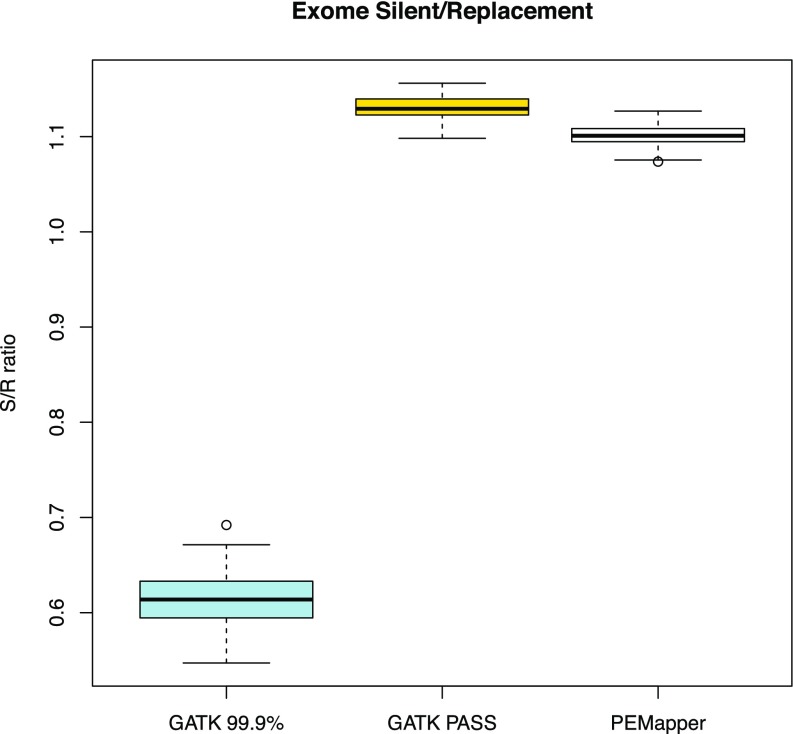
Fig. 5.
Silent to replacement (S/R) ratio across all sample exomes based on PEMapper/PECaller, GATK PASS, and Tranche99.9 calls. All samples called by PEMapper/PECaller and GATK PASS are between 1.05 and 1.15 as expected. Again, Tranche99.9 calls are significantly lower.
Calling Rare Variation.
Naively, we might imagine most false positives to be in the “singleton” category (i.e., variants seen only once in our sample set). Here, singletons have a Ts/Tv ratio of 2.105–1, better than the PEMapper/PECaller average of 2.073–1 and very close to Ts/Tv ratio of the overlap set between GATK and PEMapper/PECaller. Therefore, singletons, despite the additional potential for false-positive calls, actually seem to be as reliable or more reliable than the set of all sites.
dbSNP 146 contains all variants currently reported in the ExAC (7) dataset as well as all variants discovered by 1000 Genomes (19). An exonic variant not found in dbSNP 146 is almost surely either a false-positive call or a variant that is exceedingly rare in the general population. Exonic sites that change the amino acid (replacement sites) and are not found in dbSNP should be the category of variation most enriched for false-positive calls. For the entire set of replacement SNPs, the Ts/Tv ratio is 2.173. Replacement SNPs in dbSNP are 2.254, whereas those not in dbSNP are 1.762. For singleton replacement SNPs, the Ts/Tv ratio is 2.328. Singleton replacement SNPs in dbSNP are 2.562, whereas those not in dbSNP are 1.846. This set of singleton replacement sites that are not found in dbSNP is the set that ought to be most enriched for false positives. Despite this enrichment, replacement sites that are not in dbSNP have a Ts/Tv ratio only ∼10% lower than SNPs overall, suggestive that, although this set may be the most enriched for false positives of any possible set, it is still comprised largely of true-positive calls.
Completeness and Accuracy.
Overall, more than 98.4% of the nonrepeat-masked genome had high-quality calls. As expected, more than 99% of these sites were called homozygous reference in all 97 samples. At sites called variant in at least one sample, our overall data completeness was 99%. Most of these samples (93) were also genotyped on Illumina 2.5M arrays. These arrays provide over 140 million genotypes that can be compared with the sequence-called genotype. Over these 140 million genotype calls, PECaller data were 99.85% complete and agreed with array call 99.76% of the time. Partitioning these numbers by array-called genotype, we note that, if the genotyping array called “homozygote reference,” the sequencing call was 99.95% complete and agreed 99.94% of the time. If the array called a “heterozygote,” the sequencing was 99.81% complete and agreed 99.23% of the time. Finally, if the array called a “homozygote nonreference,” the sequencing was 99.88% complete and agreed with the array 99.56% of the time.
Lack of agreement between sequencing- and array-based calls can be caused by errors in either the array or the sequencing call. One can show that, if the arrays are 99.8% accurate, regardless of true genotype, the agreement level above is consistent with sequencing being 99.9% accurate overall (i.e., if arrays are only 99.8% accurate, most of the disagreements between array and sequencing are caused by array errors).
Rare Variant False Negatives.
Although the overall completeness and accuracy at high-frequency sites are excellent (99.85% complete and more than 99.76% accurate), it is possible that data completeness and accuracy at low-frequency variants might be considerably worse. This worsening could occur, because joint calling of samples can increase one’s confidence for high-frequency variants, while providing comparatively little benefit for rare variant calling. To assess the probability of “missing” rare variants, we look at variants called by the Illumina 2.5M array where the variant allele was observed in only one of our samples. In this collection of ∼40,000 singleton variants, we do not see evidence for increased missing data rates in singleton variants, with only 0.24% missing data. We also do not find any substantial genotyping error in these variants, assuming the array is less than 99.991% accurate at sites where all samples are homozygote.
Performance of GATK.
We have run the complete “best practices” pipeline, including the latest version (3.6) of the HC and complete joint calling with variant recalibration and filtering on 97 samples (6, 12). PEMapper seems to perform as well or better than GATK in all measurable ways. GATK tends to conflate missing data with error. Variant call files (VCFs) do not report sites that do not have high-quality variant sites in at least one sample. Thus, if a site is not in the VCF file, it is not immediately clear whether the site is missing (insufficient evidence) or “error” (falsely believed to be high quality and reference). To try to disentangle the two in a way that displays GATK in the best possible light, we imposed the following rules. If a site was not in the VCF file and the array called homozygous reference at the site in the sample, those sites were scored as “complete” and “agree” with the array. If a site was called variant by the array in at least one sample but missing from the VCF file, this site was called missing in individuals who are not homozygous reference.
GATK calls two classes of SNPs: PASS (their highest-quality calls) and Tranche99.9to100 (their second highest quality; called Tranche99.9 hereafter). Using this paradigm, GATK finds theta in these samples to be 0.000829 (0.000792 coming from PASS and 0.0000371 coming from Tranche99.9). GATK finds the Ts/Tv ratio to be 2.09 for PASS and 1.439 for Tranche99.9, indicating that variants in Tranche99.9 are not especially trustworthy and are quite likely to be false positives.
GATK Exonic Variation.
GATK finds the value of theta in the exomes of these samples to be between 0.00041 and 0.00043, averaging 0.00423 in PASS variants. Using both PASS and Tranche99.9, theta in exomes averages 0.000458. The Ts/Tv ratio in exons averages 3.086 in PASS variants and 1.88 in Tranche99.9 variants. The silent to replacement site ratio averages 1.131 in PASS sites and 0.613 in Tranche99.9 sites, again suggesting that Tranche99.9 variants are not high quality. The individual samples averaged ∼19,000 exonic variants identified by GATK PASS.
GATK Vs. PEMapper/PECaller.
To a great extent, PEMapper/PECaller and GATK generally make the same genotype calls at variant sites in the same samples. This level of overlap is a remarkable achievement for PEMapper/PECaller given the impressive accuracy and extensive use of training set data for GATK (20, 21). Over all 97 samples, PEMapper called 6,588,872 SNPs (SNPs with exactly two alleles) (Fig. 1), with an overall Ts/Tv ratio of 2.07–1. In category PASS, there are 6,338,222 SNPs, with a Ts/Tv ratio of 2.09–1; of these SNPs, 6,241,660 (98.4%) were also called by PECaller. In Tranche99.9, there were 424,564 SNPs, with a Ts/Tv ratio of 1.25–1. Of those SNPs, “only” 145,373 variants were called in common with PECaller, and those SNPs had a much better Ts/Tv ratio than Tranche99.9 overall (1.72–1). The PASS GATK calls not made by PECaller (96,562) had a Ts/Tv ratio of 1.25–1. The Tranche99.9 GATK calls not made by PECaller had a Ts/Tv (266,521) ratio of 1.06–1. Finally, PECaller SNPs not called by GATK (197,660) had a Ts/Tv ratio of 1.31–1 (Fig. 2 and Table 2). Overall, these results mean that PEMapper/PECaller calls slightly more variants than GATK PASS and slightly fewer than GATK total (PASS + Tranche99.9). SNPs called by GATK but not called by PEMapper/PECaller look to be of worse quality than SNPs called by PEMapper/PECaller but not called by GATK. The performance of Tranche99.9 SNPs in all ways suggests that they should probably not be used for analysis, because they are likely to have significant numbers of false positives.
Table 2.
Comparison of the number of variants called and the Ts/Tv ratio for those variants between PEMapper/PECaller and GATK
| Category | No. of variants called | Ts/Tv ratio |
| PEMapper/PECaller | 6,588,872 | 2.07:1 |
| GATK PASS | 6,338,222 | 2.09:1 |
| GATK 99.9 | 424,564 | 1.21:1 |
| PEMapper/PECaller and GATK 99.9 | 145,373 | 1.72:1 |
| GATK PASS but not PEMapper/PECaller | 96,562 | 1.25:1 |
| GATK 99.9 but not PEMapper/PECaller | 266,521 | 1.06:1 |
| PEMapper/PECaller but not GATK | 197,660 | 1.31:1 |
Variants not called by PEMapper/PECaller (but called by GATK) are of worse quality than those not called by GATK (but called by PEMapper/PECaller).
Using the Illumina 2.5M Array as the gold standard, we were able to compare the completeness and accuracy of both PEMapper/PECaller and the GATK pipeline. Across the board, PEMapper/PECaller outperformed GATK, albeit only slightly (Table 3). If the array called homozygous reference, PEMapper/PECaller was 99.95% complete and 99.94% agreement with the array compared with GATK, with 98.98% complete and 99.83% agreement. If the array called heterozygous, PEMapper/PECaller was 99.81% complete and 99.23% agreement with the array compared with GATK, with 99.31% complete and 99.78% agreement. If the array called homozygous nonreference, PEMapper/PECaller was 99.88% complete and 99.56% agreement with the array compared with GATK, with 99.68% complete and 99.15% agreement. Overall, PEMapper/PECaller was 99.85% complete and 99.76% agreement with the array compared with GATK, with 99.82% complete and 99.74% agreement with array.
Table 3.
Comparison of calling completeness and accuracy compared with the Illumina 2.5M array gold standard for PEMapper/PECaller and GATK
| Variant call type | PEMapper/PECaller, % | GATK, % | ||
| Completeness | Accuracy | Completeness | Accuracy | |
| Ref/ref | 99.95 | 99.94 | 99.98 | 99.83 |
| Ref/alt | 99.81 | 99.23 | 99.31 | 99.78 |
| Alt/alt | 99.88 | 99.56 | 99.68 | 99.15 |
| Overall | 99.85 | 99.76 | 99.82 | 99.74 |
PEMapper/PECaller performs slightly better than GATK. Alt/alt, two alternate alleles; Ref/alt, one reference and one alternate allele; Ref/ref, two reference alleles.
Essentially, both callers are primarily “limited” by microarray-based errors. This limitation means that it may be that both callers are nearly always getting the right answer when the array is correct and that, when the array is in error, they differ in differing ways. To a first approximation, the difference between the two can be summarized, because GATK is slightly more likely than PEMapper to fail to report a site called variant by the array. The sites that GATK excludes but PEMapper calls are slightly more likely than average to disagree between PEMapper and the array. There is certainly no evidence that GATK is doing a substantially better job than PEMapper. We also point out that all of this is despite the fact that GATK is using knowledge about the position of high-frequency variants to help align sequences and set thresholds for calling. PEMapper/PECaller uses none of this information, and it is mapping and calling variants “naively” and yet, achieves the same overall results.
In a slightly different comparison experiment, we know that, with the Illumina arrays, GATK, and PECaller, we have three separate sets of calls. Dropping any call that is missing in the array, GATK, or PECaller, there are ∼140 million genotypes called in common between the arrays and either GATK or PECaller and over 633 million variant calls that can be compared between GATK and PECaller. For each of three, we can assume that one of three is the “gold standard” for accuracy and ask what the error rate is at variant sites relative to this gold standard. These results are shown in Table 4. Several conclusions can be drawn. First, all three are excellent and in close agreement. Second, GATK looks to be a slight outlier. If GATK is set as the gold standard, both the array and PECaller seem to have approximately a 1% error rate at heterozygous sites and very low error rates at homozygous sites. Conversely, comparing GATK with the array gold standard, heterozygotes seem to have an excellent error rate, but homozygous nonreference calls have an abnormally high error rate. The simplest explanation of both of these observations is that GATK is slightly “overcalling” heterozygotes at the expense of homozygous calls but only very slightly, because overall calling is truly excellent.
Table 4.
Comparison of error rates using three possible gold standards (Illumina array with 140 million calls, 630 million PECaller calls, and 630 million GATK calls)
| Variant type | Illumina array as gold standard | PECaller as gold standard | GATK as gold standard | |||
| PECaller | GATK | Array | GATK | Array | PECaller | |
| Ref/ref | 0.00061 | 0.00174 | 0.00224 | 0.00157 | 0.00080 | 0.00136 |
| Ref/alt | 0.00766 | 0.00217 | 0.00351 | 0.00712 | 0.01032 | 0.01132 |
| Alt/alt | 0.00439 | 0.00849 | 0.00123 | 0.00739 | 0.00107 | 0.00240 |
| All | 0.00235 | 0.00261 | 0.00235 | 0.00300 | 0.00261 | 0.00300 |
When Illumina array calls are the gold standard, PECaller has much less error in homozygous reference and homozygous alternate calls, while having more in heterozygous calls. Overall, PECaller has slightly less error. Using all three, it is possible to discern that GATK is overcalling heterozygotes at the expense of homozygous calls. Alt/alt, two alternate alleles; Ref/alt, one reference and one alternate allele; Ref/ref, two reference alleles.
Insertion and Deletion Comparisons.
Calling of small insertions and deletions remains a very difficult category of variation to evaluate, largely because, unlike SNPs, there is no obvious external gold standard (such as genotyping arrays) with which to compare. Various investigators have attempted to create artificial gold standards [e.g., Chong et al. (22) use sets of “artificial” indels created by the investigators at known locations], but given that GATK calling is highly dependent on using known training set data to improve its accuracy, it is not clear whether such testing paints an appropriate picture of GATK accuracy. It is certain that GATK does a very poor job (less than 20% accurately detected) at determining the exact position and presence of small indels when those indels have not previously been seen in its training set, but given that GATK uses such extensive training sets, it surely is better than 20% accuracy overall. What is absolutely clear is that GATK and PEMapper/PECaller often call the same small indels. When they do, they usually agree on genotype (homozygous reference vs. heterozygous indel vs. homozygous indel) for individual samples. When the two calling algorithms disagree concerning the presence or absence of an indel, it is very difficult to interpret. Precise calling of copy number variants (CNVs) requires specialized software and is not part of either the PEMapper/PECaller or GATK package (22, 23).
PECaller called 406,015 small deletions, of which 84% (342,094) were called in exactly the same position by GATK. PECaller also called 212,272 insertions, of which 84% (178,478) were called by GATK. Therefore, a large percentage of the indels called by PEMapper/PECaller overlap perfectly with GATK. In the other direction, GATK called many more indels than PECaller. A total of 37% of deletions called by GATK and 57% of GATK-called insertions were not called by PECaller. This difference was not primarily because of the fact that the Smith–Waterman mapping parameters in PEMapper were set to drop any read with a large (larger than ∼10 bp) indel, because even for the smallest of indels, there was often considerable disagreement (30% of the one-base deletions called by GATK were not called by PEMapper, and 55% of the one-base insertions were not called). It should be noted that this comparison required the indel to be called in exactly the same position (i.e., not even one base different from one another). In even slightly repetitive sequence, precise indel position is often unknowable, and it is hardly surprising that indels called by one algorithm are sometimes given slightly different positions by another; even allowing for such, there is a considerable number of small indels called by each algorithm not called by the other, with vastly more indels called by GATK than called by PEMapper/PECaller.
However, when both algorithms called an indel at the same position, there is great agreement between them in individual genotype calls. Among individuals called homozygous reference by PECaller at sites containing a deletion, GATK called those same individuals homozygous reference 99% of the time; individuals called heterozygous deletions by PECaller 99% of the time were also called heterozygous deletions by GATK. Individuals called as homozygous deletions by PECaller were called homozygous deletions by GATK 97% of the time. Insertions were slightly less consistent with 96, 93, and 94% consistency in the calls for homozygous reference, heterozygous insertion, and homozygous insertions, respectively, in individuals at sites containing an insertion. PEMapper/PECaller and GATK are in great agreement when they both call an indel at the same site.
Exome Comparison.
Given that Tranche99.9 variants are of poor quality, we look at only the comparison between PEMapper/PECaller and GATK PASS variants in the exome. Overall, PEMapper/PECaller calls ∼1,000 more variants per exome than GATK PASS (Fig. 3). The statistics for these variants are nearly identical, with PEMapper/PECaller producing a Ts/Tv ratio of 3.06 compared with 3.09 for GATK (Fig. 4). PEMapper/PECaller produced a silent to replacement ratio of 1.11 compared with 1.13 for GATK (Fig. 5). Essentially, GATK seems to use its prior knowledge of variant locations to find slightly more silent sites but may call slightly fewer potentially novel exonic replacement variants, because it is limited by the existing variant lists.
Computational Time.
The PEMapper and PECaller pipeline is dramatically faster than the GATK pipeline. In both cases, the first one-half of the pipeline was run offsite using AWS resources; the best practices require read sorting that cannot be run in parallel on our local cluster, because our cluster (like many others) uses a shared disk array environment. Total central processing unit (CPU) time will scale similarly, because all AWS instances use the same number of processors. Likewise, in both cases, the second one-half of the pipeline was run locally using the Emory Libraries and Information Technology’s “Tardis” resource. This computing cluster offers 12 nodes, each with 64 cores and 512 GB RAM. We report wall clock time for these tasks as well, resulting in a fair comparison. The time to map and call 97 genomes is ∼1.2 d using the PEMapper/PECaller pipeline and ∼3 d using the GATK BWA/HC pipeline per genome analyzed (Table S1). Thus, PEMapper/PECaller is more than twice as fast, even when all disk operations occurred in an isolated disk environment. In a shared disk environment, we could only run PEMapper. It should further be noted that PECaller jointly called the entire batch of 97 samples, something the GATK Unified Caller was incapable of doing, even on a node with 512 GB RAM. Some of the time saved using AWS is because of the fact that the GATK output is significantly larger than the output from PEMapper (∼150 GB per sample); therefore, the data transfer time is longer, but given that it averaged over approximately 30 megabytes per second (MB/s) of transfer, this additional download time added only ∼1.5 h per genome. Additionally, PECaller output requires less than 10th of the data storage space as GATK (Table 1). Including the raw sequencing data, PEMapper/PECaller requires only 40% of the storage space that GATK requires for the same sample. Finally, it should be noted that PECaller called all samples in a single batch, which allowed missing data vs. homozygous reference allele calls to be distinct for all samples.
Table S1.
Comparison of time to run PEMapper/PECaller and GATK best practices
| Program | Time to run mapping and calling |
| GATK (BWA and HC in parallel on AWS) | 70.6 ± 9 h per genome |
| GATK Unified Caller (batches of 10 genomes) | 0.21 h per genome |
| GATK Recalibration Tool (batches of 10 genomes) | 0.11 h per genome |
| GATK total | ∼72 h (3 d) per genome |
| PEMapper (in parallel on AWS) | 29 ± 5.8 h per genome |
| PECaller (batch of 97 genomes) | 0.34 h per genome |
| PEMapper/PECaller total | ∼29.34 h (1.2 d) per genome |
PEMapper is much faster than BWA/HaplotypeCaller, whereas PECaller and the GATK Unified Caller and Recalibration Tool take about the same time to run. When combined in a pipeline comparison, PEMapper/PECaller is nearly twice as fast as GATK for 97 samples.
All of these comparisons mean that it is both faster and easier to run PEMapper/PECaller than the GATK pipeline for studies with more than even a handful of samples. It is also less expensive because of the reduced use of computational resources. Taken together, PEMapper/PECaller enables more genomes to be analyzed, allowing for larger study sizes.
Discussion
The future of genomics is WGS on greater than thousands of genomes. Analyzing that many genomes at once, both efficiently and accurately, is a tremendous computational challenge. The GATK best practices pipeline is the de facto standard for analysis of sequencing data because it does an excellent job and has proven its utility in vast numbers of exome studies. Although a user may be well-advised to continue using the GATK pipeline for exome analysis or small numbers of whole genomes (24), we show here that PEMapper/PECaller is the decidedly better option for large-scale mapping and calling of genomes (25). PEMapper/PECaller is significantly more efficient than GATK, requiring fewer computational resources and less storage space and thus, costing less (5). PEMapper/PECaller manages to do this while providing nearly identical (or better) calling quality than GATK. PEMapper/PECaller also does not rely on any more outside information than a reference genome, making it applicable to both human and nonhuman sequencing studies.
PEMapper/PECaller completely overcomes the technical challenges of GATK best practices. It runs well in a shared disk environment. Batch calling can occur in batches of hundreds to thousands of whole genomes easily [although computation time scales as Nlog(N) of batch size]. All sites are output together with a confidence score, so that the missing vs. homozygous reference distinction is always maintained trivially. This distinction is important, because it allows straightforward implementation of genome-wide association study (GWAS) style QC procedures—e.g., sites can be filtered on call rate and Hardy–Weinberg. The most natural way to handle these data is simply to convert them to PLINK format, QC, and analyze them like any other GWAS, except that these data just happen to include all of the rare and common sites from the onset.
Overall, GATK best practices and PEMapper/PECaller make identical calls at almost every site. When they differ from one another, there is evidence that neither is very reliable. GATK best practices achieves its excellent results in large part by incorporating preexisting knowledge into the pipeline. Reads are realigned based on preexisting knowledge of SNPs and indels. Variants are classified, filtered, or dropped based on extensive training sets of known human variants. PEMapper/PECaller achieves essentially the same result based on no specific prior knowledge but an intelligent genotyping model that uses nothing more than the observed data at hand. In principle, the PECaller variants could be similarly filtered/tranched/etc., but we show there is no obvious need. By not using any preexisting knowledge, PEMapper/PECaller is far easier to use in nonhuman systems.
PEMapper/PECaller is not only much simpler to use than GATK best practices, but also, it produces data that are of the same or very slightly higher quality. It is clear that either calling platform is more than adequate to support modern genetic studies (26), but PEMapper/PECaller is far easier to run, uses less computational time and storage, and behaves far better in a shared disk environment. These benefits will enable researchers to analyze large numbers of whole-genome sequences both faster and more efficiently. Using PEMapper/PECaller to map and call large-scale genome sequencing will also further precision medicine efforts (27). Large studies using whole-genome sequences are now much easier to complete computationally using PEMapper/PECaller by reducing the currently most challenging bottleneck from experiments of this type.
Methods
The PEMapper/PECaller assumes that a reference target sequence is available, but no other information is needed. All mapping and genotype calling occur relative to this reference sequence. The PEMapper pipeline is composed of a series of three interconnected programs. The first of three prepares a hashed index of the target sequence. The remaining two programs form a pipeline, with the output of PEMapper forming the input of PECaller. PEMapper is computationally intensive but extremely gentle on disk and network subsystem. To make this possible, the underlying philosophy behind the PEMapper is that memory use should be killed for speed and limited I/O. As a result, PEMapper uses ∼45 bytes of memory per base in the reference sequence plus approximately 1 GB of memory per computational core. Therefore, a whole-human genome sequence on a 64-core workstation typically uses ∼200 GB RAM. The source code is freely available at https://github.com/wingolab-org/pecaller.
The first of three programs in the PEMapper/PECaller is called index_target. Following BLAT (28), Maq, and several other published algorithms, the target region is decomposed into 16-nt reads. The positions of all overlapping 16-mers in the target are stored. This program needs to be run only once for each target region examined. Unlike GATK best practices, no additional information on “known SNPs,” “indels,” or training sets is required or used.
The next stage called PEMapper, which also builds on approaches similar to BWA, contains a small innovation to help enable indel mapping. Reminiscent of several other algorithms, the 16-mers are allowed to have up to one sequence mismatch from the target. Thus, when mapping a 100-base read with a 16-base index, an individual read could have up to six errors and still be properly mapped as long as those errors are evenly distributed along the read. However, the algorithm also allows the 16-mers some “wobble” room, so that, relative to each other, they can map a few bases away from their expected location (up to eight bases for a 16-mer). Finally, only one-half of the 16-mers need to map in the correct order, orientation, and distance apart from one another. Positions that satisfy these requirements are taken as “potential mapping” positions.
PEMapper takes this list of putative mapping locations for each read and performs a Smith–Waterman alignment in each potential location to determine the optimal position and alignment score. At this stage, reads are rejected if the final Smith–Waterman alignment score is less than a user-defined percentage of the maximum score possible for the given read length (29). For results described below, we required 90% of the maximum alignment score and used the following alignment penalties: match = 1, mismatch = −1/3, gap open = −2, and gap extend = −1/36. The primary output of PEMapper is the “pileup” statistic for each base in the target. PEMapper pileup output files include the number of reads where an A, C, G, or T nucleotide was seen together with the number of times that base appeared deleted or there was an insertion immediately after the base. Thus, each base appears to have six “channels” of data: the numbers of A, C, G, T, deletion, and insertion reads.
The PE Distribution.
The PE distribution is a multidimensional extension of the beta-binomial distribution. Although it arises in numerous contexts and was initially described in connection with an urn sampling model (30), for our purposes, we view the PE distribution as the result of multinomial sampling when the underlying multinomial coefficients are themselves drawn from a Dirichlet distribution (31) in the same way that the 1D analog, the beta-binomial distribution, can be thought of as binomial sampling with beta-distributed probability of success. Intuitively, we envision six channels of data (numbers of A, C, G, T, deletion, and insertion reads) as being multinomially sampled with some probability of drawing a read from each of the channels, but that the probability varies from experiment to experiment and is itself drawn from a Dirichlet distribution. The coupling of the Dirichlet distribution with the multinomial distribution is common in Bayesian inference, because the former distribution is often used as a conjugate prior for parameters modeled in the latter distribution (31). Here, our purpose is subtly different. In Bayesian estimation, the assumption is that the observations are fundamentally multinomial but that the parameters of that multinomial are unknown, and the Dirichlet is used to measure the degree of that uncertainty in the parameter estimates. In the Bayesian estimation case, as the data size gets sufficiently large, convergence to a multinomial occurs. Here, however, we assume that the observations are fundamentally overdispersed relative to a multinomial, and there is not necessarily a multinomial convergence.
At any given base, a diploid sample could be 1 of 21 possible genotypes (a homozygote of A, C, G, T, deletion, or insertion and all 15 possible heterozygotes). We assume that the number of reads seen in each of six possible channels (A, C, G, T, deletion, and insertion) of data for an individual with genotype j is drawn from a PE distribution in six dimensions. We further assume that each of 21 possible genotypes is characterized by its own PE distribution and that these 21 distributions vary from base to base but are shared by all samples at a given base. A 6D PE distribution is characterized by six parameters, so let aj be a 6D vector corresponding to the parameters for genotype j. If ni is a 6D vector containing six channels of data observed in individual i at a given base and if individual i has genotype j, then the probability of those observations is , where Ni is the total number of reads observed (Ni = ni,1 + ni,2 + ni,3 + ni,4 + ni,5 + ni,6) for individual i, Aj is the corresponding sum of the parameters for genotype j (Aj = aj,1 + aj,2 + aj,3 + aj.4 + aj,5 + aj,6), and Γ is the usual gamma function (32). Note that the expected proportion of reads coming from channel k is given simply by aj,k/Aj.
Genotype-Calling Overview.
Genotype calling occurs across all samples simultaneously in a fundamentally Bayesian but iterative manner. First, the PE parameters for all 21 genotypes are set to “default values” and assumed to be known. Second, the genotypes of all of the samples are called in a Bayesian manner, conditional on the known PE parameters. Third, the PE parameters are estimated, conditional on the genotypes called in step 2. The process then iterates, with the genotypes recalled and parameters reestimated. The iteration continues until either calls no longer change or a maximum number of iterations is reached. For all of the results described here, the maximum was set at five iterations, which was seldom reached.
PE Parameter Initialization.
For all 21 genotype models, Aj is set to either the average read depth across samples or 100, whichever is larger. For homozygote base calls (A, C, G, and T but not indels), the expected proportion of reads coming from channels different from the channel associated with the homozygote allele (i.e., the expected proportion of error reads) is set at 1/Aj or 0.3%, whichever is larger for each channel; thus, at initialization, we assume between 0.3 and 1% error reads in every channel. The remainder of the reads is expected to come from the “correct” channel. For heterozygote genotype calls, the error channels are set similarly, except for the “deletion” channel, which is expected to have 5% of the reads, indicating a prior assumption that ∼5% of true heterozygous reads will map incorrectly as deletions. If the heterozygote genotype does not involve the reference allele, the remaining reads are expected to come equally to both of the appropriate channels. However, if the heterozygote includes the reference allele, we assume that 52% of the remaining reads map to the reference allele and that 48% map to the nonreference allele. This assumption incorporates our notion that, some portion of the time, nonreference alleles will not map or will map incorrectly as indels.
To meet the challenge of mapping indel variation, we made the following assumptions: for deletion homozygotes, we again assume a 0.3–1% read proportion in all of the channels that do not involve the reference allele or the deletion; however, we expect the remaining reads to divide 80% deletion and 20% reference, indicating our assumption that a substantial fraction of deletion reads mismap as reference, even when the deletion is homozygous. When the deletion is heterozygous, we assume the nonerror channels to divide 60 and 40% between the reference channel and the deletion channel, respectively. Insertions after the current base are again assumed to have 0.3–1% reads in the error channels. For homozygotes, 80% of the remaining reads are expected to include the reference base and have an insertion afterward, whereas 20% of the reads will only include the reference allele. For heterozygous insertions, 40% of the remaining reads are expected to include both the reference allele and an insertion, and 60% are expected to include only the reference allele.
Bayesian Genotype Calling with a Population Genetics Prior.
We assume that m samples have been sequenced. Each of those m samples can be any of 21 possible genotypes. Thus, there are a total of (21)m possible genotype configurations of those m samples. Let ck be one such configuration; ck is an m-dimensional vector, where element ck,i is an integer between 1 and 21, and indicates the genotype of sample i. Genotypes of all of the samples are assumed to be independent, and therefore, the likelihood of configuration ck is
Most sites will not be segregating, and all m samples will be identical to the reference allele. Let c0 be the configuration where all samples are the reference allele. By assumption, the prior probability that this configuration is
where θ is a user-supplied parameter corresponding to 4Neμ, Ne is the effective size of the population from which the samples were drawn, and μ is the per site and per generation mutation rate (13). For humans, it is generally assumed to be ∼0.001 (14). All other configurations have at least one sample with at least one allele different from the reference allele. Let f(ck,r) be the number of nonreference alleles of type r, 0 < r < 6, found in configuration ck. The prior probability of configuration ck is assumed to be
where HW(ck) is the Hardy–Weinberg exact P value (33) associated with configuration ck, and the sum in the denominator is taken over all (21)m − 1 genotype configurations (Methods, Computational Efficiencies).
Overall, this prior can be summarized as follows. The population from which these samples are drawn is assumed to be of constant size and neutral, and the reference allele is assumed to be the ancestral allele at every site. The prior probability that a site is segregating is the one derived by Watterson (13) for an infinite site neutral model. Conditional on the site segregating, the assumption is that the site is in Hardy–Weinberg equilibrium, and the derived allele frequency was drawn from an infinite site neutral model. Thus, the prior probability is a combination of two terms, one of which derives from the Hardy–Weinberg P value and the other that derives from the number of different alleles seen to be segregating. Finally, we should note that we have tacitly assumed that all of the sequenced samples are randomly drawn from the underlying population (i.e., not intentionally picked to be relatives of one another). Alternatively, the user may provide a standard linkage/ped (PLINK pedigree format) (34) to specify the relationship between samples. When this option is invoked, Hardy–Weinberg is calculated only among unrelated individuals (i.e., founders), and for every configuration, ck, the minimum number of de novo mutations, Dn(ck), is calculated for the configuration. Each de novo mutation is assumed to occur with user specified probability μ, and the prior probability of the configuration is modified to
The posterior probability of configuration ck is
where the sum is taken over all possible genotype configurations (but see below). If 0 < gi < 22 is the genotype of individual i, then
where I(cs,i = j) is an indicator function that equals one whenever element i of configuration cs is equal to j and zero otherwise. Thus, we take the probability that the genotype of individual i is j to be the sum of the posterior probabilities of the genotype configurations in which we call sample i genotype j. The PECaller calls sample i genotype j whenever Pr(gi = j) is greater than some user-defined threshold, and otherwise, the genotype is called “N” for undetermined. For all of the results presented here, the threshold was set at 0.95.
Estimating PE Parameters and Repeating.
Because of local sequence context, the repetitive nature of many organisms’ sequence, and specific issues with sequencing chemistry as a function of base composition, not all bases have the same error characteristics. Some bases may appear to have a very high fraction of reads containing errors, whereas other bases have almost none. Some heterozygotes may exhibit nearly 50:50 ratios of the two alleles; others can be highly asymmetrical. To account for this fact, we wish to estimate the PE parameters independently at every base. There are three technical challenges. First and most importantly, the genotypes of the samples are not known with certainty; hence, we do not know with certainty which observations are associated with which underlying PE distribution. Second, for technical reasons (one lane “worked better” than another, etc.), some samples may have many more reads than other samples, and we do not want these high-read samples to dominate our estimates disproportionately. Third, because it is necessary to estimate parameters repeatedly, the algorithm must be computationally efficient. With this requirement in mind, we chose moment-based estimators of our parameters (35).
In principle, we would like to estimate the PE coefficients for genotype j, aj, by averaging over the observed number of reads seen in every sample that has genotype j; however, we do not know this with certainty. Therefore, let fi be a 6D vector, where element fi,k = ni,k/Ni contains the fraction of individual i’s reads that were observed in channel k. Let
and
Thus, Mj,k and Vj,k are the “weighted” mean and variance, respectively, in read fraction from channel k among individuals with genotype j, where both moments are weighted by our confidence that the individual truly is genotype j. Usually, most genotypes will have little weight (i.e., few if any samples are called that genotype), and even when samples are called that genotype, sometimes there is little to no variation seen in read fractions (i.e., 100% of the reads come from one channel in all of the samples called that genotype). Let Yj be the number of channels for genotype j that have nonzero observed variance in read fraction. Thus,
where I(Vj,k > 0) is an indicator that genotype j has nonzero variance in channel k. For any genotype with Wj < 1.5 (i.e., less than two samples called that genotype) or Yj < 2 (i.e., less than two channels with variance in read fraction), all PE parameters are returned to their initialization values. Otherwise, let channel z be the channel with nonzero variance (Vj,z > 0) but minimal mean (Mj,z < Mj,k for all other k with nonzero variance) estimate
and
Sj can be thought of as a “leave one out” moment estimate of the “precision” of the PE distribution, and Mj,k is a first-moment estimate of the mean read fraction in each channel (35). Notice that all channels with a small expected read fraction are rounded up to one (see below). After the PE parameters for all of the genotype models are estimated, the process repeats, and genotypes are recalled until genotype calls no longer change or a maximum of five iterations is reached.
Computational Efficiencies.
The sample space of configurations is impossibly large. For anything other than a trivially small number of samples, the sums over the configuration sample space cannot be done. Nevertheless, the prior distribution is remarkably “flat,” and this property can be used to great advantage. If two configurations, cu and cv, differ by only a single sample’s genotype, then we know that the ratio of their prior probabilities is bounded by
To see this idea, note that the largest difference in prior probabilities occurs when configuration cu has a single homozygote of an allele not seen in configuration cv. The difference in Hardy–Weinberg P values associated with this is less than 1/2m (33), and the difference caused by the number of alleles segregating is θ/2. Thus, if
then
The immediate implication of this is that dropping configuration cv from the sum will have little effect on the posterior probabilities of any of the likely configurations of the genotypes, and a simple, nearly linear time algorithm to enumerate all of the likely configurations and ignore the unlikely ones is suggested.
We build the list of likely configurations by moving through the samples one at a time. Initially, we start with a set of 21 configurations that correspond to all of the possible genotypes for sample 1. We calculate the likelihood of all 21 one-sample configurations and then, remove any configuration with likelihood less than 10−6 times the largest likelihood. Additionally, we always save the configuration associated with all samples being homozygote reference, because a priori, this configuration is the most likely configuration of samples. Next, to each of the remaining configurations, we add all 21 possible genotypes for the second sample, thereby increasing the number of sample configurations by a factor of 21. However, we again immediately remove all configurations with likelihood less than 10−6 times the largest likelihood. We repeat until we have gone through all m samples. In principle, each step could increase the number of likely configurations by a factor of 21, but in practice, it almost never increases the number by more than a factor of 2 (i.e., there are almost never more than two likely genotypes for one sample); most of the time, it does not increase the number of configurations at all (i.e., most of the time, there is only one likely genotype for a sample). Even when m is in the hundreds, most bases have only a handful of likely configurations, and seldom is the total number of likely configurations more than a few thousand.
PECaller takes advantage of two other computational efficiency tricks. First, HW exact probabilities are fundamentally discrete and a simple function of the numbers of heterozygous and homozygous genotypes. Those values can be calculated ahead of time and stored in lookup tables, greatly aiding that computation. Second, PE distributions contain several gamma functions, and although gamma functions can be computationally expensive to calculate, in a special case, they are cheap. If x is an integer, Γ(x) is equal to (x − 1) factorial, and therefore, we round all PE coefficients to their nearest integer ≥1. PE distributions can be calculated strictly in terms of factorials, and it is easy to precalculate and store all factorial values less than, say, 10,000. It should be noted as well that all likelihood calculations occur computationally as natural logs and are raised to an exponential only when necessary for posterior probability determinations. Thus, as a practical matter, the natural log of factorials is computed and stored.
Finally, both PEMapper and PECaller can be set to disregard highly repetitive sequences. By default, during the initial placement of reads, PEMapper ignores any 16-mer that maps to over 100 different locations in the genome. Thus, to even attempt Smith–Waterman alignment, at least one-half of the 16-mers in a read must map to less than 100 places in the genome. Any read more repetitive than this is dropped. Similarly, PECaller can be given a file in bed format that constitutes the “target” region to be called. This file can be used, for example, to specify the exome only for exome studies or the nonrepeat-masked regions of the human genome for WGS studies. Because variation in repeat-masked regions is both extremely difficult to interpret and highly prone to error/mismapping, all of the results described will be for the unique portion of the genome (i.e., nonrepeat masked).
Bisulfite Sequencing and Other User Options.
A possible application of next-generation sequencing is to determine the pattern of methylation in a given region sequenced. One way of doing this is to first treat the DNA with bisulfite, which converts Cs to Ts, unless the C has been methylated. Bisulfite treatment can pose unique challenges for mapping short sequence reads. The PEMapper/PECaller contains a user option to gracefully handle bisulfite-treated DNA. When the user selects this option, all mapping is initially done in a “three-base genome,” where Cs and Ts are treated as if they are the same nucleotide. Indexing of the genome is done in this three-base system as is initial mapping. Final placement of reads with Smith–Waterman alignment is done in a four-base system, but C–T mismatches are scored as if they are perfect matches. The methylation status of any C allele can then be immediately calculated from the pileup files, which gives the numbers of C and T alleles mapping at any base.
Many second generation sequencing technologies can create both single-ended and pair-ended reads, with either single files per sample or multiple files per sample. The PEMapper can take all of these forms of data, and for pair-ended data, the user specifies the minimum and maximum expected distances between the mate-pair reads. For mate-pair data, the PEMapper will first attempt to place the reads in a manner consistent with the library construction rules, but if no such placement can be made, it will place one or both reads if they individually map uniquely with sufficiently high score.
Throughout Methods, Bayesian Genotype Calling with a Population Genetics Prior, we assumed that every sample was diploid and therefore, that there were 21 possible genotypes for any sample at a given base. If the user specifies that this is haploid data, only six possible genotypes are assumed (homozygotes for any of six alleles), and the Hardy–Weinberg P value is removed from the prior.
WGS.
We tested the performance of GATK and PEMapper on 97 WGS samples sequenced as part of the International Consortium on Brain and Behavior in 22q11.2 Deletion Syndrome (www.22q11-ibbc.org). The collaboration, an initiative supported by the National Institute of Mental Health, combines genomic with neuropsychiatric and neurobehavioral paradigms to advance the understanding of the pathogenesis of schizophrenia and related disorders given the high risk for these conditions (more than one in four) in individuals with the 22q11.2 deletion (36). Rigorous approaches are applied across the International Consortium on Brain and Behavior in 22q11.2 Deletion Syndrome to characterize the phenotypes, the 22q11.2 deletion, and the remaining genome. DNA samples from 97 participants (37, 38) each had a typical 2.5-Mb hemizygous 22q11.2 deletion. Eight of these participants have previously published WGS data using different methods (38).
All samples were sequenced at the Hudson–Alpha Institute of Biotechnology (Birmingham, AL) on Illumina HiSeq-2500 machines using their published protocols. Briefly, the concentration of each DNA sample was measured by fluorometric means (typically PicoGreen reagent from Invitrogen) followed by agarose gel electrophoresis to verify the integrity of DNA. After sample quality control, all samples with passing metrics were processed to create a sequencing library. For each sample, 2 µg blood-extracted genomic DNA was sheared with a Covaris sonicator, the fragmented DNA was purified, and paired-end libraries were generated using standard reagents. Yields were monitored after sonication and ligation and at the complete library stage with additional PicoGreen quantitation steps. Every library in the project was tagged with a 2D barcode that leverages the Illumina sequencer’s ability to perform four sequencing reads per run (two data reads and two index reads). Two types of quality control were performed on each library before sequencing. First, the size distribution of the library was determined with a Perkin–Elmer/Caliper LabChip GX to verify a correctly formed and appropriately sized library. To avoid overlapping reads, a physical size of 500–600 bp was verified on the Caliper or Agilent instrument. This observed physical size corresponds to an alignment-based insert size of slightly over 300 bp. Second, the next step in the quality control process was a real-time quantitative PCR assay with universal primers to precisely quantity the fragments that are able to be sequenced in the library. The real-time PCR results in combination with the size data were used to normalize all libraries to a 10-nM final concentration. After quality control, each plate of 96 libraries was pooled into a single complex pool. The final library pool was sequenced on a test run using the Illumina MiSeq instrument and a paired-end 150-nt sequencing condition with indexing reads. The data from the MiSeq served as a final quality control step for both samples and the libraries. Libraries that passed QC were subjected to full sequencing on the Illumina HiSeq 2500 instruments according to current Illumina protocols, essentially as described in the work by Bentley et al. (39). The unique barcoding features of the described library construction allow up to 96 samples to be pooled and sequenced simultaneously. Of these samples, 93 were also run on Illumina Omni 2.5 genotyping arrays (https://www.illumina.com/techniques/popular-applications/genotyping.html), which served as an additional sequencing quality control.
PEMapper/PECaller Methods.
PEMapper was run on AWS r3.8xlarge instances with 32 CPUs with 244 GB RAM for each sample. Globus Genomics (https://www.globus.org) was contracted to facilitate the running of PEMapper on AWS. A PEMapper workflow is available through Globus Genomics, which leverages batch submission, such that multiple samples can be submitted for mapping simultaneously. The sequencing files (fastq format) were uploaded to AWS via Globus Genomics, and the PEMapper output is subsequently returned to the user’s local machine. PEMapper was run with all default parameters and a 90% threshold for Smith–Waterman alignment. PECaller was run with a default theta value of 0.001 (Results) and a 95% posterior probability for a genotype to be considered called (less than 95% is reported as missing or N). Sites with less than 90% complete data were dropped. All mapping and genotyping occurred relative to the human HG38 reference as reported by the University of California at Santa Cruz (UCSC) Genome Browser on July 1, 2015. We report results only for the nonrepeat-masked portion of the genome.
End User Instructions.
Running the PEMapper/PECaller pipeline is very straightforward for an end user. One begins with fastq files from WGS (the number does not matter; however, many represent the complete sequencing of the sample of interest). If the end user has opted to use the Globus Genomics pipeline on AWS, the fastq files are uploaded to the PEMapper workflow, and the user receives three important files in return: a pileup file, a summary file, and an indel file. If the end user is running PEMapper locally, he or she must have a copy of the reference genome and load that into memory before running PEMapper with the map_directory_array.pl script. In either case, the user will run PECaller locally. To do so, one gathers the pileup and indel files for each sample to be processed in a single folder. The script call_directory.pl is used to launch PECaller. That script generates an .snp file (containing all SNPs but no indels in an unsorted list) as output. Then, merge_indel_snp.pl is run to merge the indels into the list of SNPs. This script produces a merged .snp file (containing SNPs and indels in a sorted list). This file can be converted simply to a PLINK pedigree format and represents the primary output of the pipeline. Several additional scripts permit easy quality control assessments of the data. The first script, snp_tran_counter.pl, generates a file with Ts/Tv ratio information about the samples. At this point, the web-based annotation program, SeqAnt (https://seqant.genetics.emory.edu/) (17), can be used to annotate the merged .snp file. Finally, a second script, snp_tran_silent_rep.pl, takes the output from SeqAnt and generates a file with silent/replacement information about the samples.
GATK Methods.
The initial steps of GATK, BWA and HC, were similarly run on AWS r3.8xlarge instances with 32 CPUs with 244 GB RAM for each sample. Globus Genomics (https://www.globus.org) was also contracted to facilitate the running of GATK. A GATK workflow is available through them that runs, in order, BWA v0.7.12-r1039, sambaba v0.5.4, and GATK v3.5–0-g36282e4. The reference genome used was hg38 downloaded from the Broad Institute. This workflow leverages batch submission, such that multiple samples can be submitted for mapping simultaneously. The sequencing files (fastq format) were uploaded to AWS through Globus Genomics, and the GATK output (BAM and VCF files) was subsequently returned to the user’s local machine.
Joint genotyping and variant recalibration were done in GATK v3.6 locally in batches of 10 samples because of the intensive computational load. The joint genotyping and variant recalibration tools were run on nodes with 64 cores and 512 GB RAM. All mapping and genotype calling were relative to same reference hg38 genome in PEMapper/PECaller, with SNP sets, etc. taken from the hg38 resource bundle provided by GATK. All repeat-masked regions of the genome were dropped. The Unified caller would not run on the entire 97 sample dataset, even on compute nodes with 512 GB RAM free (it always eventually reported an “out of heap space” error whether run on the whole genome or each chromosome separately). We attempted to run the unified caller on subsequently smaller batches of data: it would complete in a batch size of 10 genomes but failed at a batch size of 20. Results below are from nine batches of 10 samples each and one batch of 7 samples.
Acknowledgments
We thank the members of the laboratories of M.E.Z. and D.J.C. for comments on the manuscript, Cheryl T. Strauss for editing, and the Emory–Georgia Research Alliance Genome Center supported in part by Public Heath Service Grant UL1 RR025008 from the Clinical and Translational Science Award Program, the NIH, and the National Center for Research Resources for performing the Illumina sequencing runs. The TARDIS Emory High Performance Computing Cluster was used for this project. This work was supported by NIH/National Institute of Mental Health Grants U54 HD082015 and U01 MH101720, which are part of the International Consortium on Brain and Behavior in 22q11.2 Deletion Syndrome, and the Simons Foundation Autism Research Initiative (M.E.Z.).
Footnotes
The authors declare no conflict of interest.
1A complete list of the International Consortium on Brain and Behavior in 22q11.2 Deletion Syndrome can be found in SI Appendix.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1618065114/-/DCSupplemental.
Contributor Information
Collaborators: Bernice Morrow, Beverly Emanuel, Donna M. McDonald-McGinn, Steve Scherer, Anne Bassett, Eva Chow, Joris Vermeesch, Ann Swillen, Raquel Gur, Carrie Bearden, Wendy Kates, Vandana Shashi, Tony Simon, Pankj Chopra, Joseph Cubells, David J. Cutler, Michael P. Epstein, H. Richard Johnston, Jennifer Mulle, Viren Patel, Stephen T. Warren, Thomas S. Wingo, Michael E. Zwick, Linda Campbell, Gabriela Repetto, Jacob Vorstman, Therese Van Amelsvoort, Stephen Eliez, Nicole Philip, Doron Gothelf, Marianne Van Den Bree, Michael Owen, Clodagh Murphy, Declan Murphy, Sixto Garcia-Minaur, Damian Neine-Suner, Kieran Murphy, Marco Armando, and Stefano Vicari
References
- 1.Bainbridge MN, et al. Whole-genome sequencing for optimized patient management. Sci Trans Med. 2011;3(87):87re83. doi: 10.1126/scitranslmed.3002243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saunders CJ, et al. Rapid whole-genome sequencing for genetic disease diagnosis in neonatal intensive care units. Sci Trans Med. 2012;4(154):154ra135. doi: 10.1126/scitranslmed.3004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levy SE, Myers RM. Advancements in next-generation sequencing. Annu Rev Genomics Hum Genet. 2016;17:95–115. doi: 10.1146/annurev-genom-083115-022413. [DOI] [PubMed] [Google Scholar]
- 4.Stavropoulos DJ, et al. Whole-genome sequencing expands diagnostic utility and improves clinical management in paediatric medicine. NPJ Genomic Med. 2016;1:15012. doi: 10.1038/npjgenmed.2015.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muir P, et al. The real cost of sequencing: Scaling computation to keep pace with data generation. Genome Biol. 2016;17(1):53. doi: 10.1186/s13059-016-0917-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lek M, et al. Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Rubeis S, et al. DDD Study; Homozygosity Mapping Collaborative for Autism; UK10K Consortium Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515(7526):209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Purcell SM, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506(7487):185–190. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKenna A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watterson GA. On the number of segregating sites in genetical models without recombination. Theor Popul Biol. 1975;7(2):256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- 14.Consortium TIH. International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437(7063):1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cutler DJ, et al. High-throughput variation detection and genotyping using microarrays. Genome Res. 2001;11(11):1913–1925. doi: 10.1101/gr.197201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yi M, et al. Performance comparison of SNP detection tools with illumina exome sequencing data--an assessment using both family pedigree information and sample-matched SNP array data. Nucleic Acids Res. 2014;42(12):e101. doi: 10.1093/nar/gku392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shetty AC, et al. SeqAnt: A web service to rapidly identify and annotate DNA sequence variations. BMC Bioinformatics. 2010;11:471. doi: 10.1186/1471-2105-11-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tennessen JA, et al. Broad GO; Seattle GO; NHLBI Exome Sequencing Project Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337(6090):64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abecasis GR, et al. 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fonseca NA, Rung J, Brazma A, Marioni JC. Tools for mapping high-throughput sequencing data. Bioinformatics. 2012;28(24):3169–3177. doi: 10.1093/bioinformatics/bts605. [DOI] [PubMed] [Google Scholar]
- 21.Hwang S, Kim E, Lee I, Marcotte EM. Systematic comparison of variant calling pipelines using gold standard personal exome variants. Sci Rep. 2015;5:17875. doi: 10.1038/srep17875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chong Z, et al. novoBreak: Local assembly for breakpoint detection in cancer genomes. Nat Methods. 2017;14(1):65–67. doi: 10.1038/nmeth.4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Albers CA, et al. Dindel: Accurate indel calls from short-read data. Genome Res. 2011;21(6):961–973. doi: 10.1101/gr.112326.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zook JM, et al. Integrating human sequence data sets provides a resource of benchmark SNP and indel genotype calls. Nat Biotechnol. 2014;32(3):246–251. doi: 10.1038/nbt.2835. [DOI] [PubMed] [Google Scholar]
- 25.Belkadi A, et al. Whole-genome sequencing is more powerful than whole-exome sequencing for detecting exome variants. Proc Natl Acad Sci USA. 2015;112(17):5473–5478. doi: 10.1073/pnas.1418631112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnston HR, Hu Y, Cutler DJ. Population genetics identifies challenges in analyzing rare variants. Genet Epidemiol. 2015;39(3):145–148. doi: 10.1002/gepi.21881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ashley EA. Towards precision medicine. Nat Rev Genet. 2016;17(9):507–522. doi: 10.1038/nrg.2016.86. [DOI] [PubMed] [Google Scholar]
- 28.Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12(4):656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith TF, Waterman MS. Identification of common molecular subsequences. J Mol Biol. 1981;147(1):195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 30.Eggenberger F, Pólya G. Über die Statistik verketteter Vorgänge. Z Angew Math Mech. 1923;3(4):279–289. [Google Scholar]
- 31.Johnson NL, Kotz S, Balakrishnan N. Discrete Multivariate Distributions. Wiley; New York: 1997. [Google Scholar]
- 32.Abramowitz M, Stegun IA. 1964. Handbook of Mathematical Functions: With Formulas, Graphs, and Mathematical Tables (National Bureau of Standards, Washington, DC)
- 33.Wigginton JE, Cutler DJ, Abecasis GR. A note on exact tests of Hardy-Weinberg equilibrium. Am J Hum Genet. 2005;76(5):887–893. doi: 10.1086/429864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ng KW, Tian G-L, Tang M-L. Dirichlet and Related Distributions. Wiley; New York: 2011. [Google Scholar]
- 36.Schneider M, et al. International Consortium on Brain and Behavior in 22q11.2 Deletion Syndrome Psychiatric disorders from childhood to adulthood in 22q11.2 deletion syndrome: Results from the International Consortium on Brain and Behavior in 22q11.2 Deletion Syndrome. Am J Psychiatry. 2014;171(6):627–639. doi: 10.1176/appi.ajp.2013.13070864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bassett AS, Marshall CR, Lionel AC, Chow EWC, Scherer SW. Copy number variations and risk for schizophrenia in 22q11.2 deletion syndrome. Hum Mol Genet. 2008;17(24):4045–4053. doi: 10.1093/hmg/ddn307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Merico D, et al. Whole-genome sequencing suggests schizophrenia risk mechanisms in humans with 22q11.2 deletion syndrome. G3 (Bethesda) 2015;5(11):2453–2461. doi: 10.1534/g3.115.021345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456(7218):53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]