Significance
Antibiotic resistance is a rising health threat worldwide, against which novel strategies are urgently needed. We have taken a systems approach to examine a potent and underexplored class of broad-spectrum antibiotics, the dithiolopyrrolones (DTPs). Our results indicate that DTPs disrupt cellular processes by high-affinity chelation of essential metal ions and inhibition of a subset of metalloenzymes. This mode of action is unique amongst antibiotics and may be further explored for treatment of multidrug-resistant infections. Our study also highlights chemical genomics as a powerful approach for the identification of antimicrobial mechanisms of action.
Keywords: antibiotics, mode of action, natural products, zinc chelation, metallophore
Abstract
Natural products harbor unique and complex structures that provide valuable antibiotic scaffolds. With an increase in antibiotic resistance, natural products once again hold promise for new antimicrobial therapies, especially those with unique scaffolds that have been overlooked due to a lack of understanding of how they function. Dithiolopyrrolones (DTPs) are an underexplored class of disulfide-containing natural products, which exhibit potent antimicrobial activities against multidrug-resistant pathogens. DTPs were thought to target RNA polymerase, but conflicting observations leave the mechanisms elusive. Using a chemical genomics screen in Escherichia coli, we uncover a mode of action for DTPs—the disruption of metal homeostasis. We show that holomycin, a prototypical DTP, is reductively activated, and reduced holomycin chelates zinc with high affinity. Examination of reduced holomycin against zinc-dependent metalloenzymes revealed that it inhibits E. coli class II fructose bisphosphate aldolase, but not RNA polymerase. Reduced holomycin also strongly inhibits metallo–β-lactamases in vitro, major contributors to clinical carbapenem resistance, by removing active site zinc. These results indicate that holomycin is an intracellular metal-chelating antibiotic that inhibits a subset of metalloenzymes and that RNA polymerase is unlikely to be the primary target. Our work establishes a link between the chemical structures of DTPs and their antimicrobial action; the ene-dithiol group of DTPs enables high-affinity metal binding as a central mechanism to inhibit metabolic processes. Our study also validates the use of chemical genomics in characterizing modes of actions of antibiotics and emphasizes the potential of metal-chelating natural products in antimicrobial therapy.
Studies on the mode of action of natural products have uncovered valuable antimicrobial targets and provided starting points for the design of new antibiotics (1). Although considerable effort is directed toward discovering novel natural products as next-generation antibiotics, many “old” classes of natural products remain underexplored and unused. As we show here, a more complete understanding of the mode of action of these neglected natural products could provide novel antimicrobial targets and revive the potential of these compounds as therapeutics.
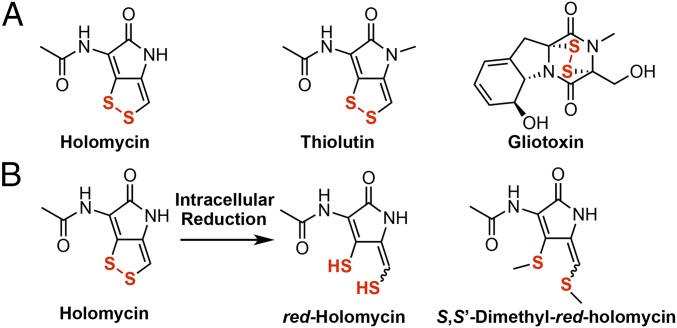
Dithiolopyrrolone (DTP) natural products display broad-spectrum inhibitory activity against bacteria, fungi, and human cell lines (2). DTPs are active against both Gram-negative and Gram-positive pathogens, with high potency against methicillin-resistant Staphylococcus aureus (MRSA), Klebsiella pneumoniae, and Pseudomonas aeruginosa. Members of the DTP family share a unique bicyclic core consisting of a dihydropyrrolone ring fused to an ene-disulfide, with a variety of N-acyl groups (2, 3); holomycin and thiolutin contain an N-acetyl group (Fig. 1A), whereas thiomarinol has the polyketide antibiotic marinolic acid as the N-acyl group linked to the DTP (4, 5).
Fig. 1.
Structures of disulfide-containing natural products. (A) Holomycin, thiolutin, and gliotoxin are investigated using the chemical genomics screen. (B) Our prior work suggests that holomycin is intracellularly converted to reduced holomycin (red-holomycin) as the active form. The holomycin-producing bacterium Streptomyces clavuligerus generates S,S’-dimethyl–red-holomycin as a product of a backup protection mechanism, when the gene encoding for dithiol oxidase HlmI is deleted.
The promising antimicrobial activity of DTPs has prompted several studies of their mode of action. Holomycin and thiolutin have been examined against Escherichia coli and Saccharomyces cerevisiae, with conflicting results. Thiolutin reversibly inhibits RNA and protein synthesis in both E. coli and S. cerevisiae in whole-cell assays and inactivates yeast RNA transcription in vitro; thus, RNA polymerase was proposed as the primary target in yeast (6, 7). Oliva et al. confirmed inhibition of growth and macromolecule synthesis in E. coli (8). However, they found holomycin only weakly inhibited E. coli RNA polymerase in vitro, shedding doubts on RNA polymerase being the primary target in E. coli (8). Glucose utilization and respiration in E. coli have also been shown to be inhibited by thiolutin (9). Given this conflicting evidence, the mode of action for this intriguing class of antibiotics is unclear.
We wondered whether the DTP scaffold might impact multiple pathways that contribute to its overall antimicrobial effect. Earlier work showed that the ene-disulfide of holomycin is redox-active and integral to its antimicrobial action (Fig. 1B) (10, 11). The producing bacterium encodes a dithiol oxidase HlmI in the holomycin biosynthetic pathway to keep the ene-disulfide closed for self-protection against the ene-dithiol form (11). The hlmI deletion mutant generates S,S’-dimethylated reduced holomycin with no antimicrobial activity (Fig. 1B) as a backup self-protection mechanism (10). These results suggest that holomycin is a prodrug whose activation involves intracellular reduction of the ene-disulfide to the active form ene-dithiol (Fig. 1B). The mechanism by which reduced holomycin (red-holomycin) inhibits bacterial growth and the role of the ene-dithiol group remain unknown.
To identify the pathways that DTPs may inhibit, we used a powerful chemical genomics platform in E. coli that profiles global drug–gene interactions (12). In the original screen, the Keio collection of 3,900 nonessential E. coli K-12 gene knockouts was screened against varying dosages of 114 antibiotics and chemical and physical stressors, yielding 324 conditions (12, 13). Here, we applied this platform to gain a global view of cellular pathways affected by the antibiotics holomycin and thiolutin as well as gliotoxin, a fungal toxin that is also thought to function via a redox-active disulfide (Fig. 1A) (14). Our screen identified a role for all three compounds in disrupting metal homeostasis. Our mode of action studies on holomycin as a prototypical DTP reveal that holomycin is an intracellular metallophore with high affinity for metal ions, especially zinc, and can inhibit a subset of zinc-dependent metalloenzymes. Our work provides a model for the function of holomycin and related DTPs as antibiotics.
Results and Discussion
A Unique Sensitivity Profile for Holomycin, Thiolutin, and Gliotoxin.
To gain insights into the mode of action for holomycin, thiolutin, and gliotoxin, we investigated these compounds in a chemical genomics screen in E. coli K-12 with other neglected antibiotics for a total of 51 unique conditions (15). Mutants from the Keio deletion library were grown as individual colonies on agar plates with subinhibitory concentrations of each compound, colony opacity was quantified using image analysis, and a quantitative fitness score was assigned to each mutant based on differences in colony opacity from the strain average (13). Positive fitness scores reflect the degree of resistance of a mutant to an environmental stress, and negative scores reflect the sensitivity to a stress. We use “sensitivity profile” to refer to the set of fitness scores across all mutants in the library for a particular stress condition.
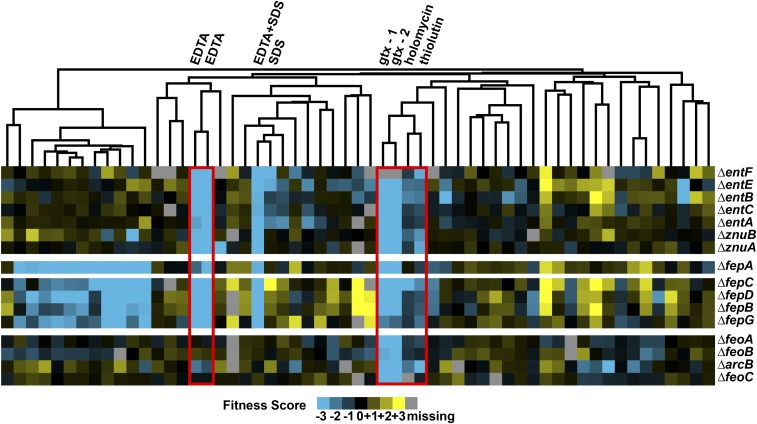
The sensitivity profiles of holomycin, thiolutin, and gliotoxin were correlated (R > 0.40, Pearson correlation coefficient with a nonparametric cutoff yielding P < 0.05), reflecting a similar set of genetic susceptibilities to these compounds in E. coli. Interestingly, the sensitivity profiles of these three compounds were correlated to neither rifampicin nor actinomycin D (R < 0.05), two antibiotics known to directly inhibit bacterial transcription. Instead, hierarchical clustering of the dataset revealed that gene deletion mutants in the acquisition of extracellular iron and zinc were sensitized to all three compounds relative to the wild type (hypersensitive mutants) (Fig. 2 and SI Appendix, Table S2).
Fig. 2.
Chemical genomics screen identifies iron and zinc uptake mutants with increased sensitivity toward dithiolopyrrolones. Fitness scores of the chemical genomics dataset were visualized as a heat map with 57 stress conditions (51 unique conditions) on the abscissa and Keio mutants on the ordinate. Stress conditions described in this work are boxed in red including EDTA, gliotoxin, holomycin, and thiolutin. The dendrogram above the heat map represents clustering of stress conditions with similar sensitivity profiles, which shows that holomycin and thiolutin cluster with two gliotoxin conditions (gtx-1 and gtx-2). The rest of the conditions can be found in SI Appendix, Table S1, and their analysis is described in ref. 15. The first 12 conditions on the Left of the heat map are conducted on iron-limited M9 minimal media, whereas the rest are on LB.
The deleted genes in these hypersensitive mutants are primarily located in two loci. The ent-fep locus encodes enzymes responsible for both the biosynthesis of the iron siderophore enterobactin (ent genes) and the transport machinery responsible for uptake of extracellular ferric enterobactin (fep genes). The second locus is at the znu operon that encodes an inner membrane zinc uptake complex. All three groups of genes were sensitized to holomycin, thiolutin, and gliotoxin but not rifampicin or actinomycin D. Instead, the sensitivity profiles of these compounds were more similar to the metal chelator EDTA (Fig. 2), and are unique among the ∼300 stress conditions tested in the previous and present screens (12, 15). Given these shared sensitivity profiles in metal uptake, we hypothesized that holomycin, thiolutin, and gliotoxin exert antimicrobial activity by interfering with cellular metal homeostasis.
Disrupting Metal Uptake Systems Sensitizes E. coli to Holomycin.
We validated the sensitivities of the E. coli mutants discovered in the screen by an independent assay. At least one representative deletion from each uptake pathway [zinc(II) and iron(III)] was constructed in the E. coli MG1655 background using P1 transduction and flippase recombinase (FLP)-mediated recombination to excise the kanamycin resistance cassette (13). Holomycin was chosen to represent the DTPs because it has a higher efficacy against E. coli K-12 than thiolutin, and we have developed an efficient synthetic route for holomycin. MOPS minimal medium (MOPS MM) was used as a growth medium to control extracellular metal content. Further, holomycin exhibits an order of magnitude lower minimal inhibitory concentration (MIC) in MOPS MM (0.2 µg/mL) liquid culture than in LB (2 µg/mL).
We used serial spot dilution assays to examine the sensitivities of each deletion to holomycin. E. coli was initially grown in the presence of 0.2 µg/mL holomycin; however, because MG1655 was less sensitive on solid media than in liquid culture, the sensitivities of MG1655 and mutants were compared on concentrations from 0.1 to 0.4 µg/mL. The deletion mutants ∆entF, ∆fepD, and ∆znuB were sensitive to 0.2 µg/mL of holomycin (SI Appendix, Fig. S1A). Interestingly, the ∆fepA mutant has roughly equal sensitivity to holomycin as MG1655 (SI Appendix, Fig. S1A), potentially because E. coli harbor secondary receptors that translocate iron-bound enterobactin degradation products (catechols), which complement FepA function (16). Notably, growth of the ∆znuB mutant is strongly inhibited even at a lower holomycin concentration (0.1 µg/mL) (SI Appendix, Fig. S1A). In contrast to the zinc import mutants, those in zinc export (∆zntA, ∆zntR) and regulation (∆zur) exhibit no change in sensitivity relative to MG1655 (SI Appendix, Fig. S1A). Importantly, none of the mutants showed growth defects in the absence of antibiotic (SI Appendix, Fig. S1B). The hypersensitivity of mutants defective in iron and zinc import indicates that holomycin alters intracellular metal availability, which the cell is unable to rectify due to the mutations of these genes.
We chose to investigate the effect of holomycin on zinc homeostasis because zinc uptake mutants were the most sensitive to holomycin treatment among those identified from the chemical genomics screen (SI Appendix, Fig. S1A). If cellular zinc sequestration is the primary effect of holomycin, complementation of deletion strains with native levels of zinc import proteins should rescue sensitivity to holomycin, and overexpression of entire zinc transport complexes should enable resistance. Complementation of ∆znuB by overexpression of znuB rescues the sensitivity of ∆znuB without interfering with normal growth (SI Appendix, Fig. S1 C–G). Furthermore, expression of the entire operon in both wild type and ∆znuB allows E. coli MG1655 to resist a higher concentration of holomycin (0.4 µg/mL) (SI Appendix, Fig. S1 F and G). In this case the znuABC operon was introduced on a multiple copy vector, which leads to enhanced expression of the transporter complex, allowing E. coli to partly alleviate the zinc homeostatic stress. To evaluate the effect of holomycin treatment on the cellular zinc content, we conducted inductively coupled plasma–mass spectrometry (ICP-MS) analysis on E. coli cells. Cells treated with twice the MIC of holomycin retain a similar cellular zinc quota within error (13 ± 2 ppb Zn/mg dry cells) as those that are treated with DMSO vehicle alone (11.8 ± 0.3 ppb Zn/mg dry cells) (SI Appendix, Fig. S2B). This result demonstrates holomycin does not affect the total zinc content and suggests that it may instead affect the cellular zinc availability.
Reduced Holomycin Chelates Zinc and Other Divalent Metals.
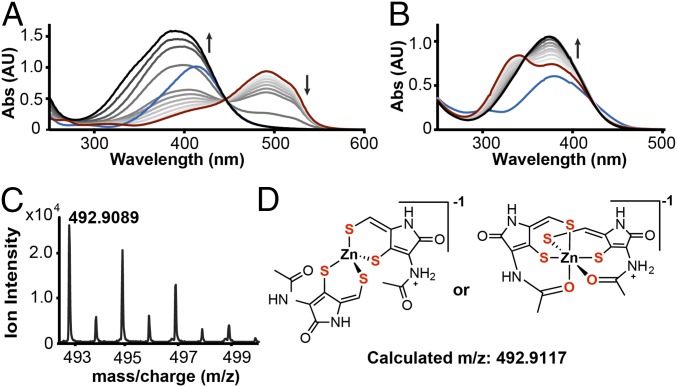
We first investigated the ability of holomycin to chelate essential metals, focusing on zinc. Spectrophotometric and mass spectrometric methods were used to examine the affinity of both the disulfide and the reduced dithiol forms of holomycin (red-holomycin) for several metal ions. We used 4-(2-pyridylazo)-resorcinol (PAR) as a spectrophotometric binding competitor for zinc. Titration of holomycin into the Zn(PAR)2 complex solution in the presence of the reducing agents tris(2-carboxyethyl)phosphine (TCEP) or 1,4-dithiothreitol (DTT) resulted in the loss of the characteristic Zn(PAR)2 absorbance at 493 nm, indicating red-holomycin competes with PAR for zinc binding (Fig. 3A and SI Appendix, Fig. S3). No loss in the 493-nm peak is observed in the absence of reducing agent, showing reduction of the DTP disulfide is required for zinc chelation (SI Appendix, Fig. S4). DTT or TCEP alone could only weakly dissociate the Zn(PAR)2 complex (SI Appendix, Fig. S3).
Fig. 3.
Reduced holomycin chelates zinc with high affinity. (A) Competition experiment of reduced holomycin against Zn(PAR)2. PAR (blue) and Zn (II) forms Zn(PAR)2 complex (red); titrations of 0–5 equivalents of reduced holomycin (a gradient from gray to black) yield the Zn[red–holo-dithiolate]2 complex (black). (B) Direct titration of zinc into reduced holomycin leads to formation of the Zn[red–holo-dithiolate]2 complex (λmax 375 nm). Holomycin (blue) is converted to reduced holomycin (red) using TCEP; titrations of 0–1 equivalents of Zn(II) (a gradient from gray to black) yield the new complex (black). The arrows in A and B indicate the direction of absorbance changes. (C) Mass spectrum of the [2M+H+Zn]−1 complex with distinct isotopic distribution for Zn(II). (D) Two possible coordinations for the Zn[red–holo-dithiolate]2 complex.
We characterized the formation of a potential Zn–red-holomycin complex. In the PAR competition experiments, the decrease in absorbance at 493 nm of the Zn(PAR)2 complex is accompanied by an increase in a new absorbance band centered between 370 and 400 nm. Direct titration of zinc into red-holomycin resolves this peak, which represents the newly formed complex with a λmax at 375 nm, distinct from the λmax of red-holomycin at 340 nm, with an extinction coefficient of 30,840 M–1cm−1 (Fig. 3B). This titration also shows an isosbestic point at 350 nm, suggesting only one species of Zn–red-holomycin complex is formed, and it is related linearly to red-holomycin. The absorbance of this complex reaches its maximum at 0.5 equivalents of zinc, indicating a stoichiometry of 2:1 ligand:metal (L:M) (Fig. 3B), which has also been reported for the reduced form of gliotoxin (17). A pH screen indicates that the complex forms in a wide pH range (pH 4–10.5) based on the characteristic 375-nm peak (SI Appendix, Fig. S5). High-resolution mass spectrometry confirmed the stoichiometry and the observed mass indicated that both ene-thiols exist in the completely deprotonated thiolate form in the complex. Masses corresponding to both [2M+Zn+H]–1 and [2M+Zn+Na]–1 (M = red-holomycindithiolate or red–holo-dithiolate) were detected, with distinct isotopic distributions for zinc (Fig. 3C). The exact coordination chemistry is unknown and is depicted as tetrahedral or octahedral as two possibilities (Fig. 3D).
Based on the PAR competition data and the stoichiometry obtained from mass spectrometry, we determined a stability constant for the formation of the Zn[red–holo-dithiolate]2 complex. The average pKD’ (logβ’2) and SEM for complex formation is 14.8 ± 0.1 based on three observations (stability constant, 1014.8 ± 0.1 M−2) and corresponds to a pZn value of 11 (SI Appendix). The pZn value for red-holomycin is in a similar range as the pZn for EDTA we calculated (pZn = 15, SI Appendix). These values demonstrate that red-holomycin binds zinc with high affinity. red-Holomycin exhibits stronger chelating ability compared with simple dithiol-containing compounds such as DTT (SI Appendix, Fig. S3) or dihydrolipoic acid (SI Appendix, Fig. S6), which may be due to lower pKa values or more rigid geometry of the ene-dithiols than DTT and dihydrolipoic acid. The aforementioned S,S’-dimethyl–red-holomycin, produced by a holomycin biosynthetic mutant in S. clavuligerus, is unable to chelate zinc, indicating that the free ene-dithiol is essential for zinc binding (SI Appendix, Fig. S7). This result is consistent with the lack of antimicrobial activity of this compound against Streptomyces spp. and further supports that S-methylation is a general mechanism for self-protection by microbial producers of disulfide-containing natural products such as DTPs and gliotoxin (10, 18). Holomycin also forms complexes with Cu(I) and Fe(II). By direct titration of Fe(II) or Cu(I) into red-holomycin, we observed a λmax centered around 379 nm and 386 nm for red-holomycin complexed with Fe(II) and Cu(I), respectively (SI Appendix, Figs. S8 and S9). We focus on zinc chelation in the rest of this study because the E. coli mutants deficient in zinc uptake are most sensitive to holomycin, as described above.
Prodrug Mechanism of Holomycin.
We have previously proposed that holomycin is a prodrug that is intracellularly reduced to its active form (10). To validate this mechanism, we examined whether the reduced and metallated forms of holomycin can inhibit bacterial growth. Preformed red-holomycin or Zn[red-holo]2 complex added extracellularly display much weaker antibacterial activity than the disulfide form of holomycin as shown by a disk diffusion assay (SI Appendix, Fig. S2A), suggesting that both the reduced and metallated forms of holomycin are insufficiently taken up by the bacteria. In addition, the Zn[red-holo]2 complex, once formed, is stable in the presence of physiologically relevant ligands, such as glutathione (both reduced and oxidized), citric acid, and cysteine (SI Appendix, Fig. S10). Together these results strongly indicate that holomycin enters the cell in the disulfide form as a prodrug, and the resulting reduced form acts as an intracellular chelator and yields a stable complex inside the cell. This proposed prodrug mechanism is reminiscent of the disulfide-containing natural product FK228, which undergoes intracellular reduction in a glutathione-dependent manner, unmasking an alkyl thiolate that ligands to zinc in histone deacetylases (19). Holomycin joins FK228 as a redox-controlled zinc-liganding natural product.
Holomycin Inhibits FbaA but not E. coli RNA Polymerase in Vitro.
RNA polymerase contains structural zinc ions, the chelation of which has been proposed as the antimicrobial mechanism for the natural product lomofungin (20, 21). To examine whether holomycin inhibits RNA polymerase via a similar chelation mechanism and resolve the conflicting reports regarding RNA polymerases as targets for DTPs, we reevaluated the activity of red-holomycin against purified E. coli RNA polymerase in vitro using a [α-32P] CTP transcription assay. We observed less than 20% inhibition of E. coli RNA polymerase by up to 100 μM (∼20 μg/mL) red-holomycin but complete inhibition by 50 μM rifampicin, a RNA polymerase inhibitor (SI Appendix, Fig. S2C). This result is in agreement with the prior report by Oliva et al. (8) on the weak inhibition of RNA polymerase by holomycin under reducing conditions containing DTT and demonstrates that RNA polymerase is unlikely the primary antimicrobial target of holomycin in E. coli. Inhibition of yeast RNA polymerase reported for thiolutin may result from inhibiting zinc-dependent transcription factors in the eukaryotic preinitiation complex, which is absent in prokaryotes, rather than RNA polymerase itself.
To explore the ability of holomycin to inhibit metalloproteins, we evaluated the class II zinc-dependent fructose bisphosphate aldolase (FbaA) as a holomycin target because it is essential for E. coli growth and has already been explored as an antimicrobial target due to its general absence from plants and animals (22). Using a coupled spectrophotometric assay that measures NADH consumption, we showed that red-holomycin inhibits 85–90% of FbaA activity at 25 µM (SI Appendix, Fig. S11). This result suggests that inhibition of FbaA contributes to the overall inhibitory effect of holomycin. Inhibition of FbaA also explains the previously observed phenotypes for thiolutin treatment of E. coli: thiolutin prevents glucose utilization while permitting growth on glycerol, lactate, succinate, or fumarate, all of which enter metabolism downstream of FbaA (9). Furthermore, inhibition of glycolysis would result in ppGpp accumulation and a decrease in initiating nucleotide triphosphate (iNTP). This situation can lead to the stringent response and inhibition of RNA synthesis, as has been observed for holomycin (8, 9, 23). The inhibition of FbaA but not RNA polymerase suggests that holomycin, despite its small size, selectively targets a subset of metalloproteins. Metal chelation in metalloproteins by holomycin provides a unified mechanism for the inhibition of the multiple pathways previously reported; transcription is unlikely the primary target of holomycin and may only represent a component of the downstream effects.
Holomycin Inhibits Metallo–β-Lactamases in Vitro.
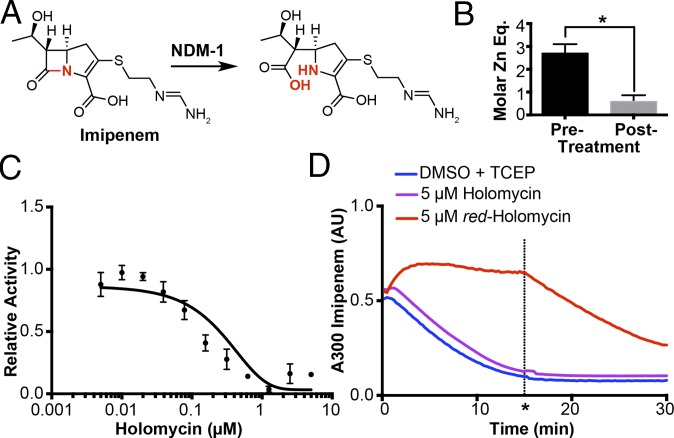
Due to the high affinity of red-holomycin for zinc and its inhibition of FbaA, we investigated the utility of holomycin against other therapeutically relevant metalloproteins. As a proof of concept, we examined the inhibitory activity of holomycin against a metallo–β-lactamase (MBL), New Delhi MBL-1 (NDM-1). MBLs are a significant clinical health threat due to their ability to confer antimicrobial resistance by hydrolyzing all types of β-lactams, including “last-resort” carbapenems (24). Importantly, MBLs use a zinc-dependent catalytic mechanism that is distinct from serine hydrolase β-lactamases and are therefore resistant to the clinically used β-lactamase inhibitor clavulanic acid. We monitored the activity of reconstituted NDM-1 by the hydrolysis of imipenem and nitrocefin, two well-established substrates for NDM-1 (Fig. 4 A and C and SI Appendix, Fig. S12) (24). Preincubation of red-holomycin with NDM-1 inhibited hydrolysis of both imipenem and nitrocefin (Fig. 4C and SI Appendix, Fig. S12). The dose–response curve of red-holomycin against NDM-1 yielded an IC50 of 110 nM for imipenem and 153 nM for nitrocefin, demonstrating its high potency.
Fig. 4.
Inhibition of NDM-1 by reduced holomycin. (A) Hydrolysis reaction of imipenem carried out by NDM-1. (B) Treatment of NDM-1 with red-holomycin decreases the number of zinc equivalents (Eq) in NDM-1 as detected by ICP-MS, error bars denote SEM of biological quadruplicate data. (C) Dose–response curve of 250 pM NDM-1 treated with varying concentrations of holomycin. Error bars represent SEM of three replicates. (D) The hydrolysis of imipenem by NDM-1 is inhibited by 5 µM reduced holomycin (red); NDM-1 activity can be restored upon addition of 4x molar excess Zn(II) (relative to holomycin) at 15 min (*). Neither the disulfide form of holomycin (purple) nor TCEP and DMSO vehicle (blue) inhibits imipenem hydrolysis by NDM-1.
We next investigated the mechanism of NDM-1 inhibition by red-holomycin. NDM-1 contains two active site zincs and requires at least one for imipenem hydrolysis (25). We reconstituted NDM-1 with zinc and removed unbound zinc by buffer exchange. ICP-MS analysis of reconstituted NDM-1 confirmed that 2.7 ± 0.4 equivalents of zinc are bound to NDM-1. After red-holomycin treatment, only 0.6 ± 0.3 equivalents of zinc remain bound to NDM-1, indicating that red-holomycin has removed 2.1 ± 0.5 equivalents of zinc from NDM-1 (Fig. 4B and SI Appendix, Table S3). In addition, inhibition of NDM-1 by red-holomycin is reversible by the addition of four times excess ZnSO4 relative to holomycin (Fig. 4D). These results indicate that red-holomycin inhibits NDM-1 reversibly by removing active site zinc. Previous reports show that EDTA removes both zinc ions, and PAR only removes one zinc from NDM-1 (25). The zinc-binding affinity of red-holomycin is lower than EDTA but higher than PAR, thus corroborating its ability to remove zinc from NDM-1. A similar mechanism of zinc metalloenzyme inhibition has also been reported for gliotoxin, which ejects zinc from p300 and blocks the HIF-1α-mediated hypoxic response (26). As expected, the disulfide form of holomycin does not inhibit NDM-1 because it lacks the ability to chelate zinc (Fig. 4D). These results demonstrate that red-holomycin is a potent inhibitor of NDM-1 through a zinc chelation mechanism.
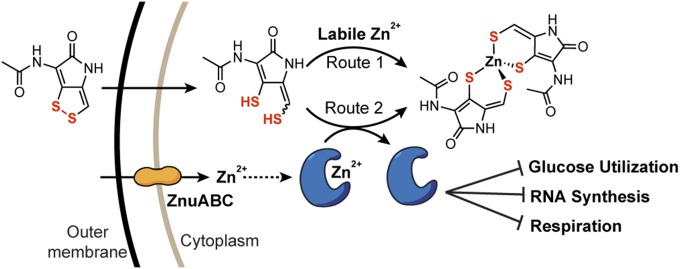
A Model for Holomycin Action.
We propose a model for the antimicrobial action of holomycin: holomycin enters target bacteria as the disulfide form and is reductively activated in the cytoplasm; reduced holomycin likely redistributes the metal pool by sequestering free metal ions (route 1) and removing metals from a subset of metalloproteins (route 2); these two actions enable holomycin to inhibit multiple essential pathways simultaneously (Fig. 5). Metal ions are essential for bacterial growth, and the disruption of metal homeostasis has profound impacts on viability (27). Route 1 is based on the high-affinity chelating activity of holomycin and the increased bacterial resistance to holomycin when zinc import genes are overexpressed (SI Appendix, Fig. S1 C–G). Although bacteria maintain zinc at a relatively high total concentration (between 0.1 and 0.2 mM), little intracellular iron or zinc exists as free metal ions (free zinc concentration in E. coli between 2.0 and 11.5 × 10−16 M) (28, 29). Sequestration of this labile zinc pool by holomycin can have dramatic impacts on zinc homeostasis. Route 2 is in accordance with our in vitro findings on the direct inhibition of essential metalloproteins such as FbaA. Rather than directly impacting proteins involved in the maintenance of metal homeostasis, such as the zinc uptake machinery ZnuABC that we identified by chemical genomics (Fig. 5), holomycin likely targets essential proteins that depend on metal for activity. This route may be more prominent in the mode of action, as supplementation with higher concentrations of zinc does not alleviate growth inhibition by holomycin (SI Appendix, Fig. S2 and Table S4), in contrast to the deactivation of extracellular chelators like EDTA by exogenous metals. Although a significant portion of cellular zinc is thought to bind RNA polymerase and ribosomal proteins, the most abundant proteins in the cell (28, 30), a great deal remains unknown about zinc homeostasis, such as zinc distribution in the cell and mechanisms of zinc trafficking and insertion into proteins (31). These observations lay the foundation for future efforts to understand the specificity of holomycin and other DTPs against metalloproteins.
Fig. 5.
A model for the mechanism of action of holomycin. Holomycin is internalized as a prodrug by a target bacterium such as E. coli, and reductive activation in the cytoplasm generates red-holomycin, which disrupts zinc homeostasis by two possible routes: (1) red-holomycin chelates labile zinc and limits zinc availability; (2) red-holomycin chelates and removes zinc from a subset of zinc-dependent enzymes, inactivation of which have a multitude of impacts such as inhibition of glucose utilization, RNA synthesis, and respiration. Although both routes contribute to the inhibitory effect of holomycin, route 2 is likely more prominent. Disruption of the zinc import machinery ZnuABC restricts zinc uptake and further sensitizes E. coli to holomycin.
Implications for Antimicrobial Therapy.
The classic function of metallophores and siderophores is the acquisition of essential metals; however, additional functions for natural product metallophores have emerged in microbe–microbe and microbe–host interactions in recent years (32), including protection from host defenses (33), and cell–cell communication and signaling (34). Although many natural products bind metals, only a few have been reported to exhibit antimicrobial activity, including the aforementioned lomofungin (20) and actinonin, which has been explored as an inhibitor of polypeptide deformylase (35). Our studies of holomycin suggest that DTPs are a unique class of intracellular metallophores that function as potent antibiotics. The structural and activity distinction of holomycin from lomofungin, actinonin, and gliotoxin (SI Appendix, Fig. S13) indicates that nature has evolved diverse chemical scaffolds all with chelating ability but diverse targets.
Although metal homeostasis has been explored as an antimicrobial target, prior efforts have primarily focused on the inhibition of metal uptake, specifically, the biosynthesis of siderophores (36). Many pathogens, however, produce multiple siderophores, diminishing the efficacy of any single inhibitor (37). Our work further supports that direct chelation of metal ions can simultaneously alter metal availability and interfere with essential metalloprotein-dependent cellular processes; such effects have been reported for metal chelators in targeting bacterial infections, cancer, and neurodegenerative disorders (38, 39). This multifaceted strategy decreases the likelihood of bacterial resistance. Consistent with this hypothesis, only mutants with two- to fourfold increases in resistance were obtained in attempts to raise spontaneous suppressor mutants against holomycin in E. coli in LB (SI Appendix). These mutants contain loss-of-function mutations in glutathione biosynthesis, consistent with the prodrug mechanism of holomycin; however, glutathione alone cannot reduce holomycin in vitro (SI Appendix, Fig. S14), suggesting additional mechanisms are involved in the intracellular activation of holomycin. The cytotoxicity of DTPs against human cell lines may complicate the use of DTPs in antimicrobial therapies, but it also suggests the medicinal applications of DTPs may extend beyond antimicrobial therapy (2).
The potent and reversible inhibition of MBLs by red-holomycin identifies DTP as a potential scaffold to inhibit carbapenem resistance by MBLs. Holomycin joins the natural product aspergillomarasmine A (AMA) (40) and the synthetic spiro-indoline–thiadiazoles (41) and the bisthiazolidines (42) as MBL inhibitors via a zinc-chelation mechanism. Both AMA and SIT-Z5 resensitized NDM-1 positive K. pneumoniae to carbapenem antibiotics (40, 41), revealing the promise of zinc chelators as carbapenem potentiators. Although red-holomycin potently inhibits NDM-1 in vitro, the effects of holomycin and meropenem are additive in whole-cell assays against NDM-1 positive K. pneumoniae (SI Appendix, Fig. S15). This effect is likely due to the cellular localization of NDM-1 in the oxidizing periplasm of Gram-negative bacteria that does not allow reductive activation of holomycin. Consequently, holomycin cannot inhibit periplasmic NDM-1 before entry to the cytoplasm. This result further supports that holomycin is selective for cytoplasmic metal chelation. Further, holomycin differs from other zinc-chelating inhibitors in two major ways: holomycin is a potent antibiotic, whereas the other inhibitors harbor little antimicrobial activity alone; holomycin also has a distinctive zinc coordination and binding mode: two red-holomycin molecules likely supply four thiolates and potentially two carbonyl ligands, whereas the other inhibitors use a combination of thiol, carboxylic acid, and nitrogen ligands. These unique chelating properties promote DTP natural products as a starting point for developing scaffolds to inhibit MBLs and other therapeutically relevant zinc-dependent enzymes.
Conclusion
We have connected the chemical activities of DTPs with their mechanisms of action. Holomycin acts as a prodrug, whose intracellular reduction facilitates high-affinity chelation with metal ions, particularly zinc. Chelation allows holomycin to disrupt metal homeostasis and inhibit bacterial growth by inhibiting a subset of metalloenzymes. As an application and exploitation of this activity, we show that holomycin potently inhibits the β-lactam resistance enzyme NDM-1 in vitro. Our work highlights that systematic mode-of-action studies are critical to understand how antibiotics function and to revive the therapeutic potential of previously neglected antibiotics.
Supplementary Material
Acknowledgments
B.L. thanks Dr. Chris T. Walsh for his encouragement and support and Drs. Carol Gross, Kathy Franz, and Roberto Kolter for useful insights. Purified E. coli RNA polymerase was a gift from Dr. Dorothy Erie, University of North Carolina at Chapel Hill, Chapel Hill, NC, and the authors thank her and Matt Satusky for their assistance with RNA polymerase inhibition assays. The authors thank Drs. Jim Imlay, Alex Miller, Jason Peters, Anthony Richardson, Nicholas Vitko, and Adam Palmer for helpful discussions. The authors also thank Dr. Brandie Ehrmann and Peter Cable for assistance and advice with inductively-coupled plasma mass spectrometry analyses; Dr. Albert Bowers for assistance with holomycin synthesis; Dr. Sidi Chen for assistance with sequence analysis; and Drs. Albert Bowers, Linda Spremulli, Gary Pielak, and Dorothy Erie for insightful comments about the manuscript. This work was supported by National Institutes of Health Grant GM099904-04 (to B.L.). S.Z.A.R is supported by the National Science Foundation Graduate Research Fellowship Program (Distinguished Graduate Fellowship Award 1106401).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1612810114/-/DCSupplemental.
References
- 1.Brown ED, Wright GD. Antibacterial drug discovery in the resistance era. Nature. 2016;529(7586):336–343. doi: 10.1038/nature17042. [DOI] [PubMed] [Google Scholar]
- 2.Li B, Wever WJ, Walsh CT, Bowers AA. Dithiolopyrrolones: Biosynthesis, synthesis, and activity of a unique class of disulfide-containing antibiotics. Nat Prod Rep. 2014;31(7):905–923. doi: 10.1039/c3np70106a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kenig M, Reading C. Holomycin and an antibiotic (MM 19290) related to tunicamycin, metabolites of Streptomyces clavuligerus. J Antibiot (Tokyo) 1979;32(6):549–554. doi: 10.7164/antibiotics.32.549. [DOI] [PubMed] [Google Scholar]
- 4.Shiozawa H, et al. Thiomarinol, a new hybrid antimicrobial antibiotic produced by a marine bacterium. Fermentation, isolation, structure, and antimicrobial activity. J Antibiot (Tokyo) 1993;46(12):1834–1842. doi: 10.7164/antibiotics.46.1834. [DOI] [PubMed] [Google Scholar]
- 5.Dunn ZD, Wever WJ, Economou NJ, Bowers AA, Li B. Enzymatic basis of “hybridity” in thiomarinol biosynthesis. Angew Chem Int Ed Engl. 2015;54(17):5137–5141. doi: 10.1002/anie.201411667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jimenez A, Tipper DJ, Davies J. Mode of action of thiolutin, an inhibitor of macromolecular synthesis in Saccharomyces cerevisiae. Antimicrob Agents Chemother. 1973;3(6):729–738. doi: 10.1128/aac.3.6.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khachatourians GG, Tipper DJ. In vivo effect of thiolutin on cell growth and macromolecular synthesis in Escherichia coli. Antimicrob Agents Chemother. 1974;6(3):304–310. doi: 10.1128/aac.6.3.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oliva B, O’Neill A, Wilson JM, O’Hanlon PJ, Chopra I. Antimicrobial properties and mode of action of the pyrrothine holomycin. Antimicrob Agents Chemother. 2001;45(2):532–539. doi: 10.1128/AAC.45.2.532-539.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bergmann R. Thiolutin inhibits utilization of glucose and other carbon sources in cells of Escherichia coli. Antonie van Leeuwenhoek. 1989;55(2):143–152. doi: 10.1007/BF00404754. [DOI] [PubMed] [Google Scholar]
- 10.Li B, Forseth RR, Bowers AA, Schroeder FC, Walsh CT. A backup plan for self-protection: S-methylation of holomycin biosynthetic intermediates in Streptomyces clavuligerus. ChemBioChem. 2012;13(17):2521–2526. doi: 10.1002/cbic.201200536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li B, Walsh CT. Streptomyces clavuligerus HlmI is an intramolecular disulfide-forming dithiol oxidase in holomycin biosynthesis. Biochemistry. 2011;50(21):4615–4622. doi: 10.1021/bi200321c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nichols RJ, et al. Phenotypic landscape of a bacterial cell. Cell. 2011;144(1):143–156. doi: 10.1016/j.cell.2010.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol Syst Biol. 2006;2(1):2006 0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scharf DH, et al. Biosynthesis and function of gliotoxin in Aspergillus fumigatus. Appl Microbiol Biotechnol. 2012;93(2):467–472. doi: 10.1007/s00253-011-3689-1. [DOI] [PubMed] [Google Scholar]
- 15.Shiver AL, et al. A chemical-genomic screen of neglected antibiotics reveals illicit transport of kasugamycin and blasticidin S. PLoS Genet. 2016;12(6):e1006124. doi: 10.1371/journal.pgen.1006124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nikaido H, Rosenberg EY. Cir and Fiu proteins in the outer membrane of Escherichia coli catalyze transport of monomeric catechols: Study with beta-lactam antibiotics containing catechol and analogous groups. J Bacteriol. 1990;172(3):1361–1367. doi: 10.1128/jb.172.3.1361-1367.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woodcock JC, Henderson W, Miles CO. Metal complexes of the mycotoxins sporidesmin A and gliotoxin, investigated by electrospray ionisation mass spectrometry. J Inorg Biochem. 2001;85(2-3):187–199. doi: 10.1016/s0162-0134(01)00193-3. [DOI] [PubMed] [Google Scholar]
- 18.Duell ER, et al. Sequential inactivation of gliotoxin by the S-methyltransferase TmtA. ACS Chem Biol. 2016;11(4):1082–1089. doi: 10.1021/acschembio.5b00905. [DOI] [PubMed] [Google Scholar]
- 19.Furumai R, et al. FK228 (depsipeptide) as a natural prodrug that inhibits class I histone deacetylases. Cancer Res. 2002;62(17):4916–4921. [PubMed] [Google Scholar]
- 20.Fraser RS, Creanor J. The mechanism of inhibition of ribonucleic acid synthesis by 8-hydroxyquinoline and the antibiotic lomofungin. Biochem J. 1975;147(3):401–410. doi: 10.1042/bj1470401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruet A, Bouhet JC, Buhler JM, Sentenac A. On the mode of action of lomofungin, an inhibitor of RNA synthesis in yeast. Biochemistry. 1975;14(21):4651–4658. doi: 10.1021/bi00692a014. [DOI] [PubMed] [Google Scholar]
- 22.Capodagli GC, Sedhom WG, Jackson M, Ahrendt KA, Pegan SD. A noncompetitive inhibitor for Mycobacterium tuberculosis’s class IIa fructose 1,6-bisphosphate aldolase. Biochemistry. 2014;53(1):202–213. doi: 10.1021/bi401022b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schneider DA, Gourse RL. Changes in the concentrations of guanosine 5′-diphosphate 3′-diphosphate and the initiating nucleoside triphosphate account for inhibition of rRNA transcription in fructose-1,6-diphosphate aldolase (fda) mutants. J Bacteriol. 2003;185(20):6192–6194. doi: 10.1128/JB.185.20.6192-6194.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Berkel SS, et al. Assay platform for clinically relevant metallo-β-lactamases. J Med Chem. 2013;56(17):6945–6953. doi: 10.1021/jm400769b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomas PW, et al. Characterization of purified New Delhi metallo-β-lactamase-1. Biochemistry. 2011;50(46):10102–10113. doi: 10.1021/bi201449r. [DOI] [PubMed] [Google Scholar]
- 26.Cook KM, et al. Epidithiodiketopiperazines block the interaction between hypoxia-inducible factor-1α (HIF-1α) and p300 by a zinc ejection mechanism. J Biol Chem. 2009;284(39):26831–26838. doi: 10.1074/jbc.M109.009498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cisar JS, Tan DS. Small molecule inhibition of microbial natural product biosynthesis—an emerging antibiotic strategy. Chem Soc Rev. 2008;37(7):1320–1329. doi: 10.1039/b702780j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Finney LA, O’Halloran TV. Transition metal speciation in the cell: Insights from the chemistry of metal ion receptors. Science. 2003;300(5621):931–936. doi: 10.1126/science.1085049. [DOI] [PubMed] [Google Scholar]
- 29.Outten CE, O’Halloran TV. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science. 2001;292(5526):2488–2492. doi: 10.1126/science.1060331. [DOI] [PubMed] [Google Scholar]
- 30.Hensley MP, Tierney DL, Crowder MW. Zn(II) binding to Escherichia coli 70S ribosomes. Biochemistry. 2011;50(46):9937–9939. doi: 10.1021/bi200619w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma Z, Jacobsen FE, Giedroc DP. Coordination chemistry of bacterial metal transport and sensing. Chem Rev. 2009;109(10):4644–4681. doi: 10.1021/cr900077w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnstone TC, Nolan EM. Beyond iron: Non-classical biological functions of bacterial siderophores. Dalton Trans. 2015;44(14):6320–6339. doi: 10.1039/c4dt03559c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chaturvedi KS, Hung CS, Crowley JR, Stapleton AE, Henderson JP. The siderophore yersiniabactin binds copper to protect pathogens during infection. Nat Chem Biol. 2012;8(8):731–736. doi: 10.1038/nchembio.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chaturvedi KS, Henderson JP. Pathogenic adaptations to host-derived antibacterial copper. Front Cell Infect Microbiol. 2014;4:3. doi: 10.3389/fcimb.2014.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen DZ, et al. Actinonin, a naturally occurring antibacterial agent, is a potent deformylase inhibitor. Biochemistry. 2000;39(6):1256–1262. doi: 10.1021/bi992245y. [DOI] [PubMed] [Google Scholar]
- 36.Ferreras JA, Ryu J-S, Di Lello F, Tan DS, Quadri LEN. Small-molecule inhibition of siderophore biosynthesis in Mycobacterium tuberculosis and Yersinia pestis. Nat Chem Biol. 2005;1(1):29–32. doi: 10.1038/nchembio706. [DOI] [PubMed] [Google Scholar]
- 37.Shapiro JA, Wencewicz TA. Acinetobactin isomerization enables adaptive iron acquisition in Acinetobacter baumannii through pH-triggered siderophore swapping. ACS Infect Dis. 2016;2(2):157–168. doi: 10.1021/acsinfecdis.5b00145. [DOI] [PubMed] [Google Scholar]
- 38.Weekley CM, He C. Developing drugs targeting transition metal homeostasis. Curr Opin Chem Biol. 2016;37:26–32. doi: 10.1016/j.cbpa.2016.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prachayasittikul V, Prachayasittikul S, Ruchirawat S, Prachayasittikul V. 8-Hydroxyquinolines: A review of their metal chelating properties and medicinal applications. Drug Des Devel Ther. 2013;7:1157–1178. doi: 10.2147/DDDT.S49763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.King AM, et al. Aspergillomarasmine A overcomes metallo-β-lactamase antibiotic resistance. Nature. 2014;510(7506):503–506. doi: 10.1038/nature13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Falconer SB, et al. Zinc chelation by a small-molecule adjuvant potentiates meropenem activity in vivo against NDM-1-producing Klebsiella pneumoniae. ACS Infect Dis. 2015;1(11):533–543. doi: 10.1021/acsinfecdis.5b00033. [DOI] [PubMed] [Google Scholar]
- 42.Hinchliffe P, et al. Cross-class metallo-β-lactamase inhibition by bisthiazolidines reveals multiple binding modes. Proc Natl Acad Sci USA. 2016;113(26):E3745–E3754. doi: 10.1073/pnas.1601368113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.