Significance
Comprehensive clinical, neurological, and genetic examinations characterized a generalized myoclonic epilepsy syndrome with photosensitivity in young Rhodesian Ridgeback dogs. The average age of onset of seizures was 6 mo. Genetic analyses revealed a defective DIRAS family GTPase 1 (DIRAS1) gene and protein. DIRAS1 is widely expressed in the brain and has been suggested to regulate acetylcholine release and play a role in neurodevelopment. This study reveals a candidate gene for human myoclonic epilepsies, and a translational model to further elucidate the role of DIRAS1 in neurotransmission and neurodevelopment, and its modulation as a therapeutic option in common epilepsy.
Keywords: seizure, juvenile, canine, photosensitivity, Ras
Abstract
The clinical and electroencephalographic features of a canine generalized myoclonic epilepsy with photosensitivity and onset in young Rhodesian Ridgeback dogs (6 wk to 18 mo) are described. A fully penetrant recessive 4-bp deletion was identified in the DIRAS family GTPase 1 (DIRAS1) gene with an altered expression pattern of DIRAS1 protein in the affected brain. This neuronal DIRAS1 gene with a proposed role in cholinergic transmission provides not only a candidate for human myoclonic epilepsy but also insights into the disease etiology, while establishing a spontaneous model for future intervention studies and functional characterization.
Dogs provide physiologically relevant models of human disease. Aggressive breeding has resulted in a unique genetic architecture that facilitates gene discovery (1). Many breeds originate from a limited number of founder animals and the use of popular sires is a common practice. As a consequence, each breed represents an isolated population with high levels of phenotypic homogeneity, reduced genetic diversity, and enrichment of breed-specific disorders (2). Hundreds of naturally occurring canine conditions are analogous to human diseases, such as diabetes, cancers, epilepsies, eye diseases, autoimmune diseases, and monogenic diseases.
Epilepsy is the most common chronic neurological disease in dogs (3). A strong genetic background is suspected in many dog breeds with a high prevalence (4) and several genes have been discovered in both symptomatic and idiopathic epilepsy. Most of these genes represent orthologs to the corresponding human epilepsy genes, such the canine models for progressive myoclonic epilepsy, including NHLRC1 in Lafora disease (5, 6) and CLN1, CLN2, ATP13A2, CLN5, CLN6, CLN8, and MFSD8 in different types of neuronal ceroid lipofuscinosis (1, 7). Only two genes have been associated with idiopathic epilepsy in dogs, ADAM23 and LGI2 (8, 9).
In this study, we describe a unique model of genetic generalized epilepsy in Rhodesian Ridgeback (RR) dogs characterized by a young age of onset. The RR is an African dog breed, originating from Rhodesia, now Zimbabwe. The breed-defining characteristic is a dorsal ridge, caused by a lateral instead of caudal orientation of the hair in this region (10). The RR reflects a mixture of several European dog breeds and the local ridged Hottentot Khoi dog and was initially bred for lion hunting (10, 11). The presence of multiple affected dogs with a distinct phenotype in many litters proposed an inherited condition, which warranted us to embark a comprehensive study to describe the clinical features and find the genetic cause.
Results
Generalized Myoclonic Epilepsy with Photosensitivity in Young RR Dogs.
Altogether, we studied 95 RR dogs, of which 24 (15 males, 9 females) shared a unique epilepsy phenotype of frequent myoclonic jerks/twitches, with an onset in young dogs (mean 6 mo; median 3.5 mo; range 6 wk–18 mo) as the outstanding feature. Eleven dogs were 5- to 18-mo-old (juvenile, adolescence) at age of onset, in 12 dogs onset was between 2 and 4 mo of age (corresponding to 2–10 y in humans, childhood), and in one dog it was at 6 wk (infantile). Photosensitivity was reported in eight dogs. The disease progressed to generalized tonic-clonic seizures (GTCS) in 38% of dogs within 6 mo (median; 1.5–29 mo) after onset of myoclonic seizures. Owners of three dogs (>8 y of age) reported that dogs retained normal cognition throughout life.
Myoclonic jerks were described by the owners as severe startling or even resembling an electric shock. Preceding alterations in behavior were not observed. Myoclonic twitches mainly occurred when the animals were in a recumbent position and relaxed, drowsy, or in the first stages of sleep, and with the eyes either closed or open. Occasionally twitches occurred also when the dogs were sitting, standing, or walking (Movie S1). No autonomic signs occurred during the myoclonic seizures. Based on video review, myoclonic jerks were predominantly confined to the trunk, proximal limb musculature (especially the thoracic limbs), cervical musculature producing nodding movements of the head (Movie S2), and the face (masticatory muscles resulting in chewing movements, eyelid and ear twitches). Myoclonic jerks would often be limited to or start at one side of the body; however, a consistent side predilection could not be detected. Intensity varied between events and individual dogs. Some muscle contractions were rather subtle, with just a small range of motion, whereas others were very vigorous and at times made the dogs jump into the air or dash against the floor, wall, or furniture. Although a single event lasted less than 1 s, twitches often occurred in series as repetitive myoclonic muscle contractions. GTCS were also frequently preceded by a series of myoclonic twitches. Some dogs appeared confused or scared following the episodes and seemed to be very agitated after the events, rising up and wandering around restlessly. Hence, sleep appeared impaired in these dogs. Dogs were normal between events. Owners reported daily (87%) or almost daily (13%; every second to third day) occurrence of myoclonic twitches with a frequency of up to 150 twitches per day. Up to 50 jerks per hour were recorded with EEG in some dogs. Two dogs showed increased myoclonic jerks during heat (cases 2 and 11). In three siblings (cases 8, 9, and 10) and another dog (case 6) onset of myoclonus was observed 2 d after vaccination. Onset of GTCS appeared to be temporally related to vaccination in another two dogs (cases 6 and 7).
Diagnostic investigations (Table S1) failed to identify any consistent structural abnormalities. A few dogs had potential brain abnormalities on neuroimaging evaluations that may be incidental findings, such as ventricular asymmetry (12) (Table S2). Twenty-one RRs with myoclonic epilepsy were treated with a variety of antiepileptic drugs (AEDs: phenobarbital, potassium bromide, primidone, levetiracetam, clonazepam, imepitoin; monotherapy or combination) in adequate dosages and with serum concentrations (phenobarbital: mean 28.6 mg/L; potassium bromide: mean 1,353 mg/L) within therapeutic range (13). Levetiracetam, which is also an effective drug for juvenile myoclonic epilepsy (JME) in humans (14), and potassium bromide seemed to be the most effective based on response of dog owners.
Table S1.
Clinical protocols
| RR dog | On site observation of myoclonic seizures | Video documentation | Completed questionnaire | Pedigree available | Neuroimaging | CSF | Neurometabolic screening* | EEG | EEG with ILS |
| Case 1 | Yes | Yes | No | No | MRI | Yes | Yes | Yes | No |
| Case 2 | Yes | Yes | No | Yes | MRI | Yes | Yes | Yes† | No |
| Case 3 | Yes | Yes | No | No | CT | Yes | Yes | No | No |
| Case 4 | Yes | Yes | Yes | No | CT | Yes | Yes | Yes | No |
| Case 5 | Yes | No | Yes | No | CT, MRI | Yes | No | No | No |
| Case 6 | Yes | Yes | Yes | Yes | — | Yes | Yes | Yes | No |
| Case 7 | Yes | No | No | No | MRI | Yes | No | No | No |
| Case 8 | Yes | No | No | Yes | MRI | Yes | Yes | No | No |
| Case 9 | Yes | Yes | Yes | Yes | — | No | No | Yes | No |
| Case 10 | Yes | Yes | Yes | Yes | — | No | No | Yes | No |
| Case 11 | Yes | Yes | Yes | Yes | — | No | Yes | Yes | No |
| Case 12 | Yes | Yes | Yes | Yes | — | No | Yes | Yes | No |
| Case 13 | Yes | Yes | Yes | Yes | MRI | Yes | Yes | Yes‡ | Yes |
| Case 14 | No§ | No§ | Yes | Yes | CT | Yes | No | No | No |
| Case 15 | Yes | Yes | Yes | No | MRI | Yes | No | Yes‡ | No |
| Case 16 | Yes | Yes | Yes | Yes | CT | Yes | No | No | No |
| Case 17 | Yes | Yes | Yes | No | MRI | Yes | No | Yes‡ | No |
| Case 18 | Yes | Yes | Yes | No | MRI | Yes | Yes | No | No |
| Case 19 | Yes | Yes | Yes | Yes | MRI | Yes | No | Yes | No |
| Case 20 | Yes | Yes | Yes | Yes | — | No | No | Yes | Yes |
| Case 21 | Yes | Yes | No | Yes | — | No | No | Yes | Yes |
| Case 22 | No | Yes | No | Yes | — | No | No | Yes | Yes |
| Case 23 | Yes | Yes | Yes | Yes | CT, MRI | Yes | No | Yes | Yes |
| Case 24 | Yes | Yes | Yes | Yes | CT, MRI | Yes | No | Yes | Yes |
| Control 1 | — | — | — | No | — | — | No | Yes | No |
| Control 2 | — | — | — | No | — | — | Yes | Yes‡ | No |
| Control 3 | — | — | — | No | — | — | No | Yes | No |
| Control 4 | — | — | — | No | — | — | No | Yes | No |
| Control 5 | — | — | — | No | — | — | No | Yes | No |
| Control 6 | — | — | — | No | — | — | No | Yes | No |
| Control 7 | — | — | — | No | — | — | No | Yes | Yes |
| Control 8 | — | — | — | Yes | — | — | No | Yes | Yes |
| Control 9 | — | — | — | Yes | — | — | No | Yes | Yes |
| Control 10 | — | — | — | Yes | — | — | No | Yes | Yes |
| Control 11 | — | No | Yes | Yes | MRI | Yes | No | Yes† | No |
Summary of the used diagnostic investigation protocols. ILS, intermittent light stimulation.
Neurometabolic screening: urinary amino acids and urinary organic acids; cases 2, 6, and 13 also serum amino acids.
Recording upon awakening from general anesthesia (isoflurane, propofol).
Sedation for electrode placement with 0.015 mg/kg dexmedetomidine, reversal with 0.15 mg/kg atipamezole.
This dog was included despite lack of on-site observation or video documentation of myoclonic twitches, as one sibling (dog 13) was affected as well and those two dogs were possessed by the same owner.
Table S2.
Clinical and EEG data
| RR | Sex | Genotype | Onset myoclonic seizures | Onset GTCS | Clinical photosensitivity | Neurologic examination | Brain imaging | CSF | Neurometabolic screen | Ictal EEG findings | Interictal EEG findings | EEG with ILS | Outcome (observation time) |
| Case 1 | F | dd | 18 mo | — | No | Spinal pain | T1 and T2 hypointensity in the transition area of the frontal lobe and the olfactory bulb (MRI) | WNL | WNL | No myoclonic jerks during recording* | WNL | — | Alive |
| Case 2 | F | dd | 17 mo | 1 y 11 mo | No | PL proprioceptive deficits, mild lumbosacral pain | WNL (MRI) | WNL† | Serum alanine elevated | Generalized 4–5 Hz SWC, at times epileptogenic discharges precede the myoclonic jerks | Generalized 4–5 Hz SWC | — | Dead (killed because of poor seizure control; nercropsy WNL) (7 mo) |
| Case 3 | M | — | 7 mo | — | — | WNL | WNL (CT) | WNL | Results not available, reported WNL | —‡ | — | — | Dead (unknown reason) (3 mo) |
| Case 4 | M | dd | 10 wk | — | No | Abraded claws of all limbs | Asymmetric lateral ventricles§ (CT) | WNL | WNL | Generalized 4 Hz SWC with anterior maximum, at times preceded by 5 Hz slowing, at times polyspikes | Generalized 4 Hz SWC with anterior maximum | — | Alive (6 y 11 mo) |
| Case 5 | M | — | 3 mo | 1 y 5 mo | No | Inconsistent menace OS | Asymmetric lateral ventricles§ (CT, MRI) | WNL | — | —‡ | — | — | Dead (ana-plasmosis, AIHA, ARF) (2 y 5 mo) |
| Case 6 | M | dd | 5 mo | 8 mo | No | WNL | — | WNL | WNL | Myoclonic jerks without clear EEG correlate¶ | WNL | — | Dead (ruptured splenic tumor) (4 y) |
| Case 7 | F | — | 11 mo | 1 y 5 mo | No | WNL | WNL (MRI) | WNL | — | —‡ | — | — | Dead (foreign body, renal tumor) (5 y 5 mo) |
| Case 8 | F | — | 10 wk | — | Yes | WNL | WNL (MRI) | WNL | WNL | —‡ | — | — | Dead (killed due to poor seizure control) (5 y) |
| Case 9 | F | dd | 10 wk | — | Yes | WNL | — | — | — | Right side dominant (C4, T4, F8, Cz, C3) 5–6 Hz biphasic spikes that approach a 4–5 Hz SWC morphology at points; events generalize, occasionally with a time lag, and often with independent spikes occurring at different channels | Right side dominant spikes and 4–5 Hz SWC (C4, T4, F8, Cz, C3), at times spikes in C4/T4 and C3 do not generalize | — | Alive (8 y 9 mo) |
| Case 10 | M | dd | 10 wk | — | Yes | WNL | — | — | — | Single spikes or polyspikes | Single spikes with anterior maximum | — | Alive (8 y 9 mo) |
| Case 11 | F | dd | 14 mo | — | Yes | WNL | — | — | WNL | 4–5 Hz (occasionally 3 Hz) generalized SWC with anterior maximum, at times preceded by 4 Hz slowing, bursts of 7–8 Hz polyspikes | Generalized 4 Hz SWC with anterior maximum | — | Alive (4 y 4 mo) |
| Case 12 | M | dd | 9 wk | 2 y 7 m | No | WNL | — | — | Smear in urinary organic acids (qualitative analysis) | Singlet or douplet SW | WNL | — | Alive (5 y 11 mo) |
| Case 13 | M | dd | 3 mo | 4.5 m | Yes | WNL | Lateral ventricle asymmetry§ and mild meningeal contrast enhancement (MRI) | WNL | WNL | Generalized 4–5 Hz SWC with anterior maximum, at times SW or slowing precede the myoclonic jerks | Generalized 4 Hz SWC with anterior maximum | Photoconvulsive responses with 4 Hz generalized SWC (stimulation frequencies 8–14 Hz) | Alive (1 y 8 mo) |
| Case 14 | M | — | 6 mo | — | No | WNL | WNL (CT) | WNL | — | —‡ | — | — | Dead (killed because of poor seizure control) (3 mo) |
| Case 15 | M | dd | 8 mo | — | No | WNL | WNL (MRI) | WNL | — | Generalized 4 Hz SWC or single spikes, central maximum, at times switching side, at times jerks are preceded by a crescendo of epileptogenic discharges, occasionally myoclonic jerks without clear EEG correlated | Generalized 4 Hz SWC or single spikes, central maximum, at times switching side | — | Alive (1 y 4 mo) |
| Case 16 | M | dd | 11 mo | — | No | WNL | WNL (CT) | WNL | — | —‡ | — | — | Alive (9 y 2 mo) |
| Case 17 | M | dd | 6 mo | — | No | WNL | WNL (MRI) | WNL | — | Single spikes, occasionally myoclonic jerks without clear EEG correlate‖ | Single spikes | — | Alive (1 y 2 mo) |
| Case 18 | F | dd | 6 mo | 8 mo | no | WNL | Slight reduction of brain volume (MRI)** | WNL | WNL†† | —‡‡ | — | — | Alive (1 y 4 mo) |
| Case 19 | M | dd | 6 wk | 7 mo | No | WNL | WNL (MRI) | WNL | — | Irregular generalized 4 Hz SWC, generalized 4 Hz slowing with anterior maximum switching between different leads, at times emerging into SWC, occasionally myoclonic jerks without clear EEG correlated | Irregular generalized 4 Hz SWC with anterior maximum switching between different leads, generalized 4 Hz slowing, at times emerging into SWC | — | Alive (1 y 2 mo) |
| Case 20 | M | dd | 8 wk | 8 mo | Yes | WNL | — | — | — | Single spikes and SW | Single spikes and SW | Photoconvulsive responses with 4 Hz generalized SWC with occipital maximum (stimulation frequencies 6–17 Hz) | Alive (6 mo) |
| Case 21 | F | dd | 4 mo | — | No | WNL | — | — | — | Single spikes | WNL | No PPR | Alive (4 mo) |
| Case 22 | F | dd | 10 wk | — | No | WNL | — | — | — | No myoclonic jerks during recordingi | WNL | No PPR | Alive (5 mo) |
| Case 23 | M | dd | 10 wk | — | Yes | WNL | WNL (CT, MRI) | WNL | — | Polyspikes and SW | WNL | Photoconvulsive responses with 4 Hz generalized SWC with occipital maximum (stimulation frequencies 3–4 Hz) | Alive (5 mo) |
| Case 24 | M | dd | 8 wk | — | Yes | WNL | WNL (CT, MRI) | WNL | — | No myoclonic jerks during recording without activation procedures* | No epilepto-genic discharges during recording without activation procedures | Photoconvulsive responses with 4 Hz generalized SWC with occipital maximum (stimulation frequencies 6–10 Hz) | Alive (10 mo) |
| Control 1 | F | DD | — | — | — | WNL | — | — | — | — | WNL | — | Alive (5 y 1 mo) |
| Control 2 | M | Dd | — | — | — | WNL | — | — | WNL | — | WNL | — | Alive (9 y 2 mo) |
| Control 3 | M | DD | — | — | — | WNL | — | — | — | — | WNL | — | Alive (5 y) |
| Control 4 | M | DD | — | — | — | WNL | — | — | — | — | WNL | — | Alive (9 y 9 mo) |
| Control 5 | F | DD | — | — | — | WNL | — | — | — | — | WNL | — | Alive (7 y 3 mo) |
| Control 6 | F | — | — | — | — | WNL | — | — | — | — | WNL | — | Alive (8 y 10 mo) |
| Control 7 | M | — | — | — | — | WNL | — | — | — | — | WNL | No PPR | Alive (4 y 4 mo) |
| Control 8 | F | Dd | — | — | — | WNL | — | — | — | — | WNL | No PPR | Alive (3 y 10 mo) |
| Control 9 | F | Dd | — | — | — | WNL | — | — | — | — | WNL | No PPR | Alive (3 y 10 mo) |
| Control 10 | F | Dd | — | — | — | WNL | — | — | — | — | WNL | No PPR | Alive (2 y 1 mo) |
| Control 11 | M | DD | — | 1 y 10 m | No | WNL | WNL (MRI) | WNL | — | — | WNL | — | Alive (2 y 10 mo) |
Summary of all clinical and EEG data of generalized myoclonic epilepsy in 24 RR dogs (cohort used for clinical characterization). AIHA, autoimmune hemolytic anemia; ARF, acute renal failure; CT, computed tomographic imaging; DD, wild-type genotype; Dd, heterozygous DIRAS1 mutation (carrier); dd, homozygous DIRAS1 mutation; GTCS, generalized tonic-clonic seizures; ILS, intermittent light stimulation; OS, left eye; PPR, photoparoxysmal response; SW, spike-and-wave; SWC, spike-and-wave complexes; WNL, within normal limits.
Treated with AED.
Mild increase in monocytes and banded neutrophils in cytospin preparations, normal total nucleated cell count, normal protein.
Unavailable for EEG.
May be clinically irrelevant as ventricle asymmetry is seen in 38% of healthy dogs and 44% of dogs with idiopathic epilepsy (15).
EEG correlate was obscured by muscle artifact.
EEG correlate was occasionally obscured by muscle artifact.
Unremarkable on repeat 3T MRI.
Mild elevation in amino acids cystathionine and homocysteine; organic acids WNL.
Nondiagnostic EEG (panting).
By the time of submission, three dogs were euthanized at 9 mo, 2 y, and 5 y of age because of poor seizure control; three dogs died from causes unrelated to the epilepsy and one died for unknown reasons (Table S2). One dog was available for postmortem examination (case 2) that ruled out extracranial pathologies. In this single brain, histology showed postictal changes only including mild clustered neuronal hypereosinophilia in lateral geniculate nucleus and pyramidal cell layers of the neocortex. Histoarchitectural changes, dysmorphic neurons, and reactive gliosis were not evident. The remaining dogs were alive without any evidence of mental or cognitive decline.
Ambulatory Wireless Video EEG Defines the Electroclinical Syndrome.
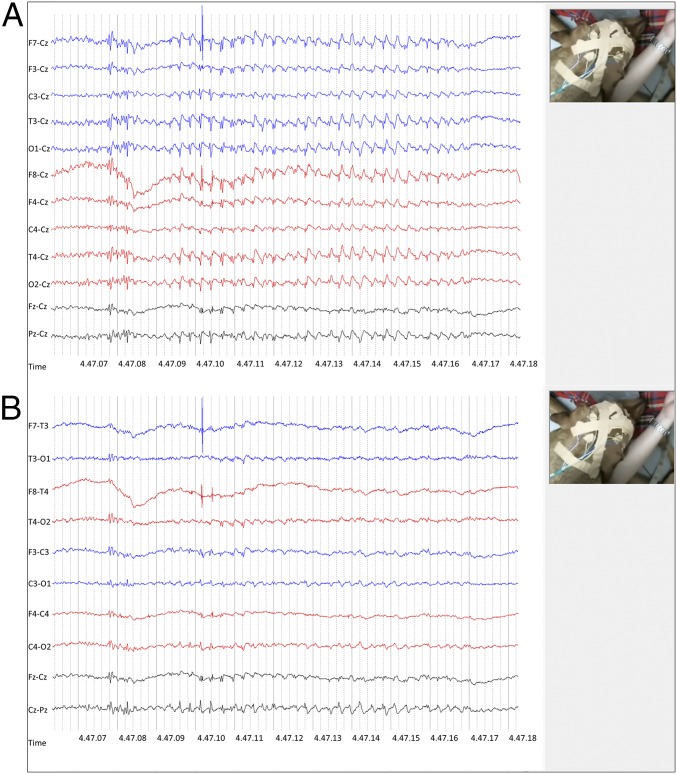
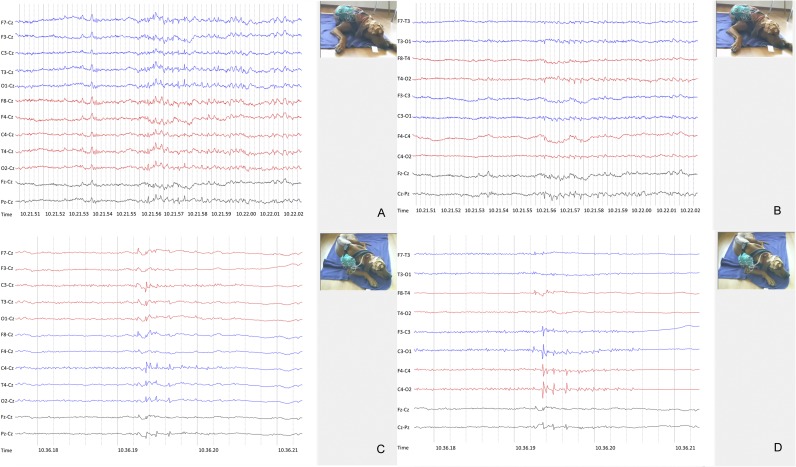
Simultaneous video and EEG recordings documented the epileptic origin of the events in 82% of examined cases (Table S2). EEG was recorded for prolonged times (>1 h, 13 recording leads) in 17 affected RRs displaying myoclonic twitches, and 11 breed-matched controls (10 healthy RRs, 1 RR with idiopathic epilepsy with GTCS). Background activity was appropriate to state in all dogs (15). Myoclonic twitches of variable intensity occurred in all but two cases during EEG recording. The characteristic ictal pattern was generalized 4–5 Hz spike-and-wave complexes (SWC) (Fig. 1 A and B) or polyspike-wave complexes (PSWC) during the initial phase, with a predominantly fronto-central maximum that often switched between different leads over both hemispheres, and occasionally generalized with a time lag. Another ictal pattern comprised biphasic spikes and paroxysmal bursts consisting of 7- to 8-Hz spikes that at times again were followed by SWC and an occasional occurrence of focal activity (Fig. S1). In some dogs, myoclonic activity was consistent with onset of ictal discharges, whereas in others myoclonic twitches were preceded by a crescendo of EEG paroxysms. Not all motor activity was accompanied by EEG paroxysms, but myoclonic jerks appeared identical and muscle artifact may have obscured the EEG correlate on some occasions. Affected RRs displayed also epileptiform discharges comprising ictal spikes or interictal 4- to 5-Hz SWC (Fig. S1). Furthermore, some dogs intermittently displayed rhythmical 4- to 5-Hz slowing that at times morphed into SWC accompanied by myoclonic jerks. During EEG recording, myoclonic twitches emerged predominantly during quiet rest, drowsiness, or slow-wave sleep. In some dogs, single episodes were recorded while awake and even less often while standing. Similarly, EEG paroxysms emerged with higher frequency when the dogs were less alert. The effect of sleep deprivation was not assessed. For the EEG, instead of sleep deprivation, we encouraged the dogs to nap. Unremarkable EEG recordings were obtained from control dogs.
Fig. 1.
Ictal EEG. LFF: 1 s; HFF: 70 Hz. (A and B) Minor head and eyelid twitches were accompanied by 4-Hz spike-and-wave complexes with a central maximum (A: Cz referential montage; B: bipolar montage). EEG is also presented as online supporting material (Fig. S1 and Movie S4).
Fig. S1.
Video-EEG with interictal SWC and ictal spikes. LFF: 1 s; HFF: 70 Hz. (A and B) These 4-Hz spike-and-wave complexes were not accompanied by any visible movements (A: Cz referential montage; B: bipolar montage). (C and D) Ictal spikes (C: Cz referential montage; D: bipolar montage).
Photosensitivity Is a Feature of Generalized Myoclonic Epilepsy in RR Dogs.
Visually induced seizures were reported in 8 of 23 (35%; confidence interval 95%: 18.7–55.2%) RRs with generalized myoclonic epilepsy (Table S2). These were described as myoclonic seizures triggered by visual stimuli, such as light flashes, sudden incidence of light when opening the shutters in the morning, or sunlight interrupted by trees while walking through the forest. A videotape was provided where each photic stimulus (produced by photoflashes) was followed by myoclonic jerks (Movie S3). Upon video-EEG recording with photic stimulation in six affected RRs, four dogs (66%) displayed photoconvulsive responses time-locked with the onset of the photic stimulus (Table S2 and Movie S4). Video-EEG with photic stimulation did not reveal any abnormalities in clinically healthy RR controls, including three heterozygous carriers of the gene deletion. Besides light, noise was also a triggering factor in three siblings (cases 8–10).
Genetic Analyses Reveal a 4-bp Deletion Mutation in DIRAS1.
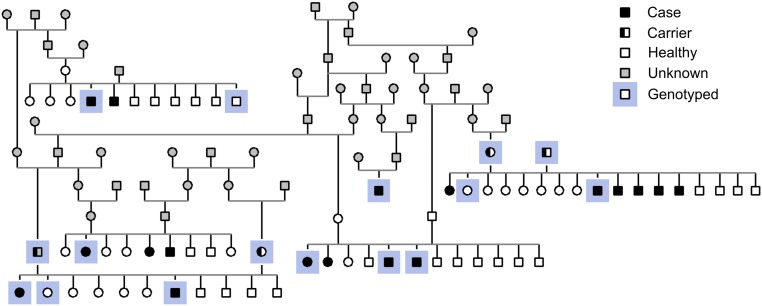
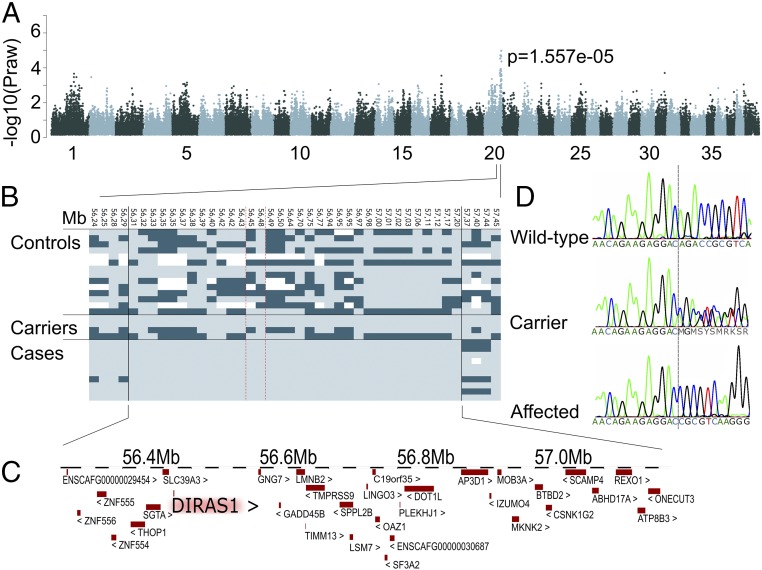
The pedigree established around the affected dogs suggested an autosomal recessive inheritance (Fig. S2). To identify the genetic cause of the generalized myoclonic epilepsy in RRs, we combined a genome wide association study (GWAS) and next-generation sequencing analyses using whole-exome (WES) and whole-genome (WGS) resequencing. Assuming a recessive mode of inheritance, the WES analysis of two unrelated cases against 169 exomes from nonepileptic dogs (Table S3) resulted in a group of 10 variants, of which 6 were in the predicted coding regions (Table S4). Only one nonsynonymous variant was found, a 4-bp deletion in the exon 2 of the DIRAS1 gene (c.564_567delAGAC; gene structure according to the Broad Institute CanFam3 Improved Annotation Data v1) (Fig. 2D), resulting in a frameshift and a stop loss (Fig. S3). A GWAS and haplotype analysis in 10 RR cases and 18 RR controls supported the WES study by identifying the best-associated region (P = 0.977 × 10−5) (Table S5) in a 1.6-Mb region (55,597,243–57,195,857) at chromosome 20 (Fig. 2A), including the DIRAS1 gene (Fig. 2C). The critical region was further split into a 300-kb and an 890-kb region (Fig. 2B) by a 400-kb recombination in one of the cases. WGS of one epileptic dog also identified the c.564_567delAGAC deletion in DIRAS1, but it was absent from 99 control whole genomes (Table S3). Structural variation analysis in the WGS data were performed within the original 1.6-Mb associated region of the epileptic RR dog. Only one 35-kb duplication (56,210,949–56,246,523) was found in the region; however, it resided outside of the 890-kb disease-associated haploblock, which starts at 56.3 Mb (Fig. 2B). In addition, the duplication was present in several nonepileptic control dogs within our 99 dogs WGS data (Table S3), excluding it as an epilepsy candidate in RRs.
Fig. S2.
Pedigree. An example of a RR pedigree with generalized myoclonic epilepsy, suggesting a recessive mode of inheritance.
Table S3.
Breeds with NGS data
| Breed | Whole exome | Whole genome |
| Akita | 1 | — |
| Alaskan Malamute | 5 | 1 |
| American Hairless Terrier | 4 | — |
| American Staffordshire Terrier | — | 1 |
| Australian Cattle dog | — | 2 |
| Australian Kelpie | 11 | — |
| Australian Terrier | 13 | — |
| Beagle | — | 1 |
| Bearded Collie | — | 7 |
| Belgian Shepherd | 16 | — |
| Berger Blanc Suisse | — | 1 |
| Bichon frisé | 4 | — |
| Border Collie | 8 | 25 |
| Boston Terrier | 1 | — |
| Boxer | 4 | — |
| Central Asian Shepherd dog | — | 1 |
| Chihuahua | 2 | — |
| Coton de Tulear | 2 | — |
| Dachshund | — | 1 |
| Dalmatian dog | — | 1 |
| Dandie Dinmont Terrier | — | 1 |
| Doberman Pinscher | 9 | 3 |
| Elo | — | 1 |
| English Bulldog | 1 | — |
| Entlebucher Sennenhund | — | 8 |
| Eurasier | — | 2 |
| Finnish Hound | 3 | — |
| Finnish Lapphund | 6 | 1 |
| Finnish Spitz | 6 | — |
| Fox Terrier | 4 | — |
| French Bulldog | 1 | 1 |
| German Pointer | 3 | — |
| German Shepherd | 1 | 1 |
| German Wirehaired | — | 1 |
| Great Dane | 16 | — |
| Irish Soft-Coated Wheaten Terrier | 4 | 1 |
| Irish Terrier | — | 1 |
| Italian Greyhound | 2 | 1 |
| Karelian Beardog | 6 | 1 |
| King Charles Spaniel | 2 | — |
| Kromfohrländer | — | 1 |
| Kuvasz | 3 | — |
| Labrador Retriever | — | 3 |
| Lagotto Romagnolo | 1 | 4 |
| Lancashire heeler | 1 | — |
| Landseer | — | 2 |
| Leonberger | — | 1 |
| Malinois | — | 2 |
| Miniature Schnautzer | 2 | — |
| Mixed breed | — | 1 |
| Newfoundland Dog | 4 | — |
| Norwegian Lundehund | 1 | — |
| Parson russel terrier | 1 | — |
| Perro de Agua Español | — | 1 |
| Pinscher | 1 | — |
| Pomeranian | — | 1 |
| Pyrenean Shepherd | 1 | — |
| Rhodesian Ridgeback (cases) | 2 | 1 |
| Rhodesian Ridgeback | — | 2 |
| Rottweiler | 2 | 2 |
| Saluki | 2 | 1 |
| Samoyeed | 1 | — |
| Schnautzer | 5 | — |
| Shetland Sheepdog | 3 | — |
| Siberian Husky (not purebred) | — | 3 |
| Sloughi | — | 3 |
| Swedish Vallhund | 2 | 2 |
| Welsh Springer Spaniel | — | 2 |
| Westhighland White Terrier | — | 1 |
| Whippet | 4 | 1 |
| White Shepherd | — | 1 |
| Yorkshire Terrier | — | 1 |
| Total | 171 | 100 |
A summary of the breeds with whole exome and genome data.
Table S4.
Candidate mutations
| Chr | Position (bp) | Reference | Alternative | Gene name | Function (Ensembl) | Exonic function (Ensembl) | Effect (snpEff) | Impact (snpEff) |
| 16 | 1855004 | G | A | Y_RNA | Downstream | — | Intergenic | Modifier |
| X | 41819512 | G | A | WDR13 | Splicing | — | Downstream | Modifier |
| 16 | 2006387 | G | A | CUL1 | Splicing | — | Intron | Modifier |
| 3 | 85098858 | C | CTTTAAATGTAACATTTTGTTTTTTTGTTTTTTTTTTTT | SEPSECS | Intronic | — | Intron | Modifier |
| 20 | 56474664 | GGACAGAC | GGAC | DIRAS1 | Exonic | Frameshift deletion | Frameshift | High |
| 2 | 77117099 | C | T | WNT4 | Exonic | Synonymous SNV | Synonymous coding | Low |
| X | 57124177 | C | T | CDX4 | Exonic | Synonymous SNV | Synonymous coding | Low |
| X | 60408672 | G | T | TAF9B | Exonic | Synonymous SNV | Synonymous coding | Low |
| 16 | 2023357 | G | A | CUL1 | Exonic | Synonymous SNV | Synonymous coding | Low |
| X | 55708156 | C | T | ENSCAFG00000017103_11 | Exonic | Synonymous SNV | Synonymous coding | Low |
A summary of 10 case-specific variants identified in the whole exome sequencing analysis after filtering against 169 control exomes.
Fig. 2.
GWAS. (A) Manhattan plot indicates best P values at chromosome 20. (B) An 890-kb haplotype is shared by cases. (C) The associated region contains 33 genes including DIRAS1. (D) Chromatograms of an affected, carrier, and wild-type dog indicate the c.564_567delAGAC variant.
Fig. S3.
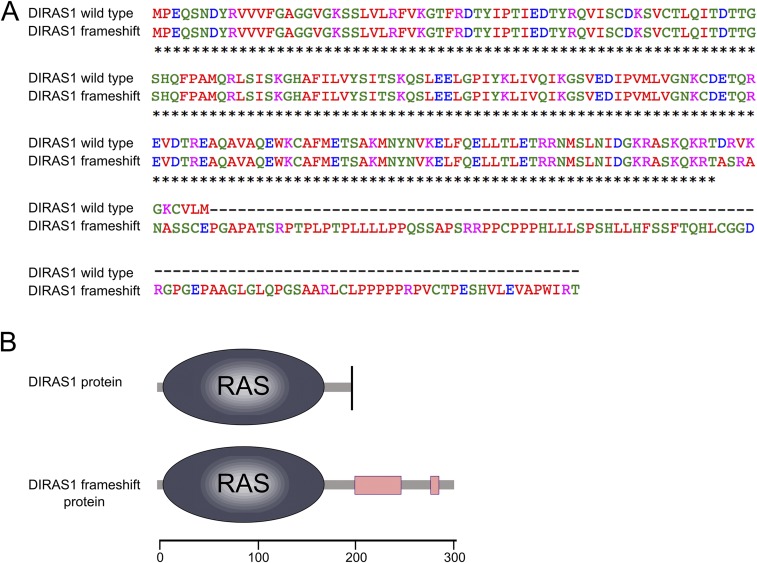
Sequence comparison. (A) Sequence alignment of the normal and mutated DIRAS1 protein was constructed to assess the effect of the c.564_567delAGAC frameshift. The variant changes the last 10 amino acids of the normally 198 amino acids-long protein and produces a stop loss leading to a protein 104 amino acids longer than the wild-type. The Ras domain remains intact. (B) Schematic overview.
Table S5.
GWAS hits
| Chr | Predictor name | Position (bp) | P value | SNP model or adjusted P value |
| Ped-GWAS | Marginal P value | SNP model | ||
| 20 | BICF2G630446654 | 53461931 | 0.97687E-05 | Add |
| 20 | BICF2P1269879 | 52411334 | 0.97687E-05 | Add |
| 20 | TIGRP2P277323_RS | 53534730 | 0.10225E-04 | Add |
| 5 | BICF2S23234898 | 44292103 | 0.21872E-04 | Add |
| 5 | BICF2P419745 | 44230649 | 0.21872E-04 | Add |
| 5 | BICF2G630187408 | 44061256 | 0.21872E-04 | Add |
| 20 | TIGRP2P278055_RS | 55666391 | 0.37939E-04 | Add |
| 20 | TIGRP2P278052_RS | 55661675 | 0.37939E-04 | Add |
| 20 | BICF2S23636929 | 55653830 | 0.37939E-04 | Add |
| 20 | BICF2P1342156 | 55615324 | 0.45206E-04 | Add |
| PLINK | Unadjusted P value | Adjusted P value | ||
| 20 | BICF2S23636929 | 55653830 | 1.557e-05 | 1.557e-05 |
| 20 | TIGRP2P278052_RS8588937 | 55661675 | 1.557e-05 | 1.557e-05 |
| 20 | TIGRP2P278055_RS8819835 | 55666391 | 1.557e-05 | 1.557e-05 |
| 20 | BICF2G630445927 | 54625801 | 2.834e-05 | 2.834e-05 |
| 20 | BICF2G630447057 | 52856293 | 2.834e-05 | 2.834e-05 |
| 20 | BICF2G630445375 | 55637322 | 3.219e-05 | 3.219e-05 |
| 20 | BICF2G630445416 | 55621611 | 3.219e-05 | 3.219e-05 |
| 20 | BICF2P1342156 | 55615324 | 3.219e-05 | 3.219e-05 |
| 20 | BICF2G630446654 | 53461931 | 4.32e-05 | 4.32e-05 |
| 20 | BICF2P1269879 | 52411334 | 4.32e-05 | 4.32e-05 |
Top 10 SNPs identified by Ped-GWAS and PLINK in the GWAS.
The genotyping of the DIRAS1 deletion in 14 clinically verified RR cases and 26 controls revealed a homozygous mutant genotype in all cases, a heterozygous genotype in the obligate carriers, and the homozygous wild-type genotype in controls (Fig. 2D), indicating a complete segregation of the deletion allele with the disease. Genotyping additional 498 RRs from 13 countries indicated a carrier frequency of ∼15% (Table S6). To investigate the breed and epilepsy specificity of the deletion, we genotyped an additional 965 epileptic dogs in 12 breeds, but did not find any carriers, indicating that the mutation is specific to generalized myoclonic epilepsy in RRs. Collectively, these results strongly suggest that the deletion in the coding region of DIRAS1 causes the generalized myoclonic epilepsy in the breed.
Table S6.
Genotyped dogs: Epidemiological investigation
| Breed | Country | Wild-type | Carrier | Mutation homozygote | Total |
| Rhodesian Ridgeback | Czech Republic | 31 | 3 | — | 34 |
| Rhodesian Ridgeback | Denmark | 5 | — | — | 5 |
| Rhodesian Ridgeback | Finland | 189 | 32 | — | 221 |
| Rhodesian Ridgeback | Germany | 46 | 15 | 18 | 79 |
| Rhodesian Ridgeback | Hungary | 2 | 1 | — | 3 |
| Rhodesian Ridgeback | Latvia | 6 | 2 | — | 8 |
| Rhodesian Ridgeback | Poland | 4 | — | — | 4 |
| Rhodesian Ridgeback | Russia | 24 | 11 | — | 35 |
| Rhodesian Ridgeback | Slovakia | 11 | — | — | 11 |
| Rhodesian Ridgeback | South-Africa | 3 | — | — | 3 |
| Rhodesian Ridgeback | Spain | — | — | 1 | 1 |
| Rhodesian Ridgeback | Sweden | 2 | — | 1 | 3 |
| Rhodesian Ridgeback | Switzerland | 111 | 19 | — | 130 |
| Rhodesian Ridgeback | Unknown | 1 | — | — | 1 |
| Total | 435 | 83 | 20 | 538 | |
| Percent | 80,9 | 15,4 | 3,7 | 100 | |
| Percent (Germany excluded) | 84,7 | 14,8 | 0,4 | 100 | |
| Australian Shepherd | 45 | — | — | 45 | |
| Belgian Shepherd | 175 | — | — | 175 | |
| Border Collie | 88 | — | — | 88 | |
| Border Terrier | 92 | — | — | 92 | |
| Finnish Spitz | 158 | — | — | 158 | |
| Kromfohrländer | 44 | — | — | 44 | |
| Lagotto Romagnolo | 46 | — | — | 46 | |
| Miniature Pinscher | 45 | — | — | 45 | |
| Pyrenean Shepherd | 22 | — | — | 22 | |
| Saluki | 78 | — | — | 78 | |
| Schipperke | 120 | — | — | 120 | |
| Whippet | 52 | — | — | 52 | |
| Total | 965 | 965 |
A summary of the breeds and dogs genotyped for the DIRAS1 mutation.
Altered Intracellular Expression Pattern of Mutant DIRAS1.
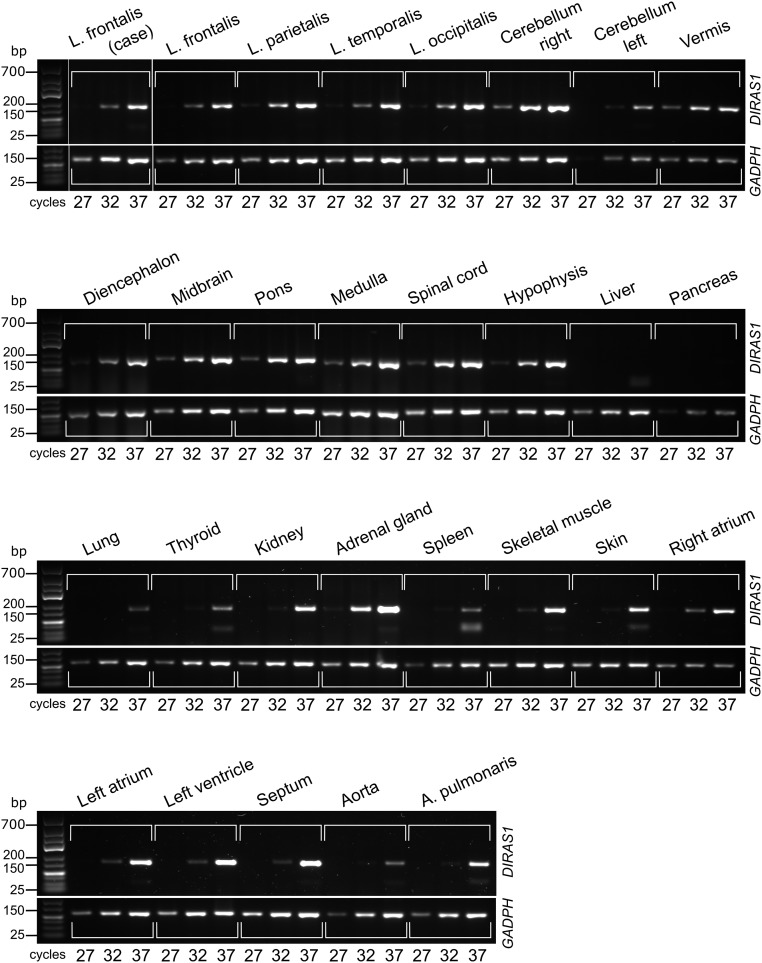
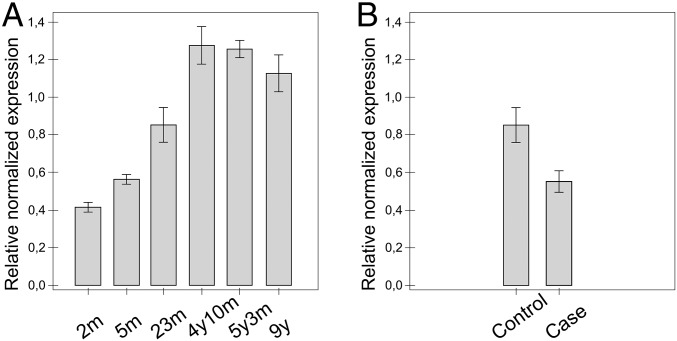
The expression pattern of the DIRAS1 transcript is poorly characterized and suggested to be limited to the brain and heart (16). We amplified the transcript in 28 canine tissues, including 12 brain regions, the spinal cord, and 15 peripheral tissues, and found abundant expression in all brain regions, whereas the pattern was more limited and variable in extra neural tissues (Fig. S4). The possible developmental expression pattern of DIRAS1 was also investigated in the frontal cortices at six different time points: 2, 5, and 23 mo and 4, 5, and 9 y. The results indicate increased expression until adulthood (Fig. 3A).
Fig. S4.
DIRAS1 expression pattern. Expression profiling of the DIRAS1 transcript across 28 tissues indicates abundant pattern in the different regions of the brain but more limited pattern is found in nonneural tissues. The stability of the DIRAS1 transcript is not markedly altered in the frontal lobes of a case (RR) and a control (Belgian Shepherd) dog by a semiquantitative PCR using GADPH as a loading control. A, arteria; L, lobus.
Fig. 3.
DIRAS1 expression. (A) The increase in the expression of the DIRAS1 transcript by age was observed when comparing six different age points in the frontal cortex (the ages of 2 mo, 5 mo, 23 mo, 4 y and 10 mo, 5 y and 3 mo, and 9 y, n = 1 in each). (B) The stability of the DIRAS1 transcript was studied in the frontal cortices of age-matched (24- and 23-mo-old) case (RR, n = 1) and control (Great Dane, n = 1) dogs by a quantitative PCR. The result suggests a modest decrease in the stability of the mutant transcript. The error bars refer to variance in experimental triplicates (SD < 0.13 in each). YWHAZ and GADPH were used as loading controls in quantitative PCR.
The 4-bp deletion resides at the end of the DIRAS1 coding region, resulting in a frameshift at the C-terminal end of the predicted protein (Fig. S3). The last 10 amino acids of DIRAS1 change causing a stop loss, which is followed by 104 extra amino acids. The only functional domain, RAS, remains intact, but the protein has additional low complexity regions toward its C terminus (Fig. S3), likely rendering the mutated protein functionally altered. The effect of the deletion mutation on the stability of the DIRAS1 transcript was investigated by quantitative PCR in the frontal cortices between the age-matched 2-y-old case and control dogs. The result suggested only a modest decrease in the case (Fig. 3B), which also agrees with an unremarkable change in the semiquantitative PCR (Fig. S4).
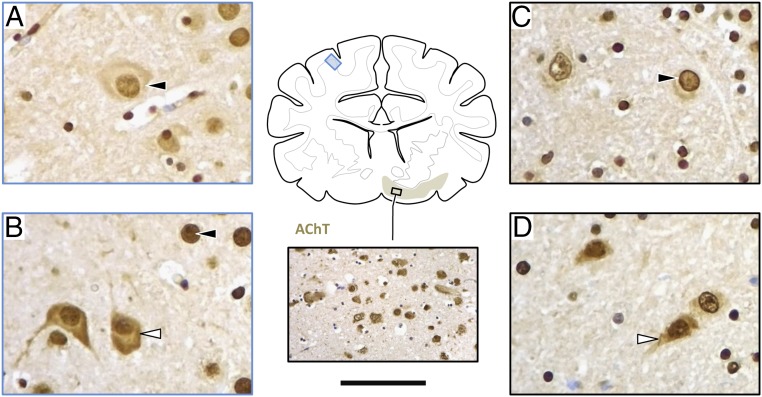
Immunolabeling revealed abundant expression of DIRAS1 antigen throughout the brain (brainstem, cerebellum, and prosencephalon), including the cholinergic basal forebrain nuclei, depicted in Fig. 4. The intracellular expression pattern of wild-type and mutant DIRAS1 somewhat differed between the single affected dog and control dogs. Distinctive nuclear and membranous pattern observed in the control dogs (RRs and non-RRs) (Fig. 4 A and C) had changed to advanced diffuse staining of nerve cell somata in the affected RR (Fig. 4 B and D). These results suggest that there is a persistent expression of the mutated DIRAS1 protein with an altered intracellular localization.
Fig. 4.
Immunohistochemical DIRAS1 expression. Wild-type RRs show predominantly nuclear staining (black arrowhead) as seen in the parietal cortex (A; blue frame) and cholinergic forebrain nuclei (C; black frame). With DIRAS1 mutation (B and D) protein expression is abundant and there is a more diffuse staining of nerve cell perikarya (white arrowhead) in all brain regions, including the brainstem. Figure shows expression in parietal cortex (B; blue frame), and forebrain nuclei (D; black frame). Cholinergic target areas were confirmed by staining for the vesicular acetylcholine transporter (AChT), as demonstrated in the Inset. (Scale bar: A–D, 35 μm; inlet AChT, 150 μm.)
Discussion
This study characterizes a breed-specific generalized myoclonic epilepsy with an early onset. Results from genetic and functional studies suggest that the epilepsy is caused by a 4-bp deletion in the coding region of the DIRAS1 gene, resulting in a frameshift and a stop loss. We found abundant expression of DIRAS1 throughout the canine brain with a difference in subcellular expression patterns between wild-type and mutant proteins. Although mammalian functions are unknown, previous studies suggest that DIRAS1 is needed for acetylcholine transmission at neuromuscular junctions in Caenorhabditis elegans (17) and neuronal development in zebrafish (18). Therefore, this canine DIRAS1 defect provides not only a candidate gene for generalized myoclonic epilepsies but also insights to the disease etiology, while establishing a spontaneous model for preclinical studies and functional characterization.
Common human idiopathic generalized epilepsies recognized by the International League Against Epilepsy include childhood absence epilepsy, epilepsy with myoclonic absences, epilepsy with myoclonic atonic seizures, epilepsy with GTCS alone, juvenile absence epilepsy, myoclonic epilepsy in infancy, and JME (19). Generalized myoclonic epilepsy in RR dogs reveals important parallels to JME, which is one of the most common forms of epilepsy in humans (14, 20–23). As in humans, jerks are bilateral, arrhythmic, at times asymmetric, and predominate upon the upper limbs and trunk (20, 21), whereby additional nodding movements of the head were present in some RRs. EEG recordings revealed a pattern found in human JME patients: SW or PSW discharges with a fronto-central accentuation and a normal background activity with an occasional occurrence of focal activity, EEG asymmetries switching sides, and diffuse or intermittent slowing (22, 24, 25). An important characteristic shared by human JME and generalized myoclonic epilepsy in RRs is the manifestation with photosensitivity, particularly as JME has one of the strongest associations with photosensitivity among all epilepsies (26, 27). Phenotypic heterogeneity was apparent because not all dogs were photosensitive, which may reflect influences of age, sex, or individual genetic background of the dogs. In humans, photosensitivity is an age-dependent phenomenon and is more prevalent in children with a peak age of onset about 12 y (27). There is also strong evidence for a genetic component of photoparoxysmal response (PPR) in humans and many loci have been identified (27–30). Thus, affected RRs provide another spontaneous large animal model to investigate the neural mechanisms of photosensitivity (31, 32).
However, there are also a number of differentiating characteristics and phenotypic heterogeneity: the low prevalence of GTCS (JME 80–95%; RRs 38%), the absence of absence seizures (although this seizure type might be difficult to recognize in dogs), and a variable age of onset, with several dogs showing a relatively early onset (6–10 wk) in the socialization period (6–12 wk) and others during the juvenile period (starting at 12 wk) and adolescence up to 18 mo, when behavioral maturation tends to reach adult values in the dogs (33–35). Differences between dog breeds exist and differences in the order of development of social and motor skills between dogs and humans have been encountered (34). Thus, early age of onset may still be in line with human genetic generalized epilepsy syndromes, such as JME, in which 25% may have absence—and not myoclonic—seizures in childhood. People with JME also have 2- to 3-Hz and 4- to 6-Hz interictal epileptic discharges, and most have polyspikes (22). EEG failed to demonstrate clear ictal discharges in association with myoclonus on some occasions. Although it was considered that EEG was obscured by muscle artifact of the myoclonus, myoclonic behavior needs to be monitored with telemetry for further investigations. There is also a possibility that the generator for myoclonic seizures is not superficial, rather subcortical. EEG will not be able to detect deep neuronal function if the generator is at the brainstem level. Although photosensitivity was observed in 35% of dogs, the prevalence of photosensitivity based upon the EEG studies appears to be higher (66%) than in humans and, in humans, the photoparoxysmal and photoconvulsive responses are maximal fronto-centrally and not occipitally. There is a high prevalence of MRI findings, which is not typical for human JME. We acknowledge that the presence of MRI findings point toward a symptomatic etiology; however, there were no consistent findings. Ventricle asymmetry is also frequently present in dogs without epileptic seizures, and thus may be clinically not relevant (12). Similarly, a small amount of meningeal enhancement is consistently demonstrated in normal dogs (36). However, we cannot exclude that some of the structural abnormalities interacted with the phenotype: for example, lowered seizure threshold on both hemispheres. The EEG phenotype is consistent with generalized myoclonic epilepsy, and certainly not focal epilepsy. Finally, human JME is characterized by strong chronodependency, with myoclonic jerks and GTCS in the morning after awakening or during relaxation periods in the evening (37). Although generalized myoclonic epilepsy in RRs also shows a strong association with the sleep–wake cycle, myoclonic twitches and EEG discharges appeared predominantly in the relaxed state, at rest, or during the first stages of sleep, mirroring subtypes of JME (38). The observed differences may reflect species-specific differences in biorhythmicity and sleep regulation, or may indicate parallels to other genetic sleep-associated epilepsies, such as myoclonic epilepsy in infancy, which sometimes progresses to JME (39), or autosomal dominant nocturnal frontal lobe epilepsy (NFLE) (40).
DIRAS1 is a novel epilepsy gene with a robust expression pattern in the CNS tissues. It is part of the Ras family of small GTPases, which have been linked to many cellular signaling pathways in cell growth and differentiation, synaptic plasticity, learning, and memory (41–43). DIRAS1 and DIRAS2 form a biochemically and functionally distinct branch of Ras GTPases, which are characterized by a fast guanidine–nucleotide exchange rate (16). The biological function of DIRAS1 in mammals is poorly characterized. DIRAS1 has been suggested to function as a tumor suppressor in glioblastoma and other tumor cell lines through the inhibition of Ras-mediated transformation, altered NF-κB transcription activity, diminished ERK1/2 and MAPK signaling, and antagonization of pro-oncogenic small Ras GTPases (44). Studies in C. elegans have demonstrated that the DIRAS1 and exchange protein directly activated by cAMP (EPAC) orthologs colocalize at the presynaptic membranes and are needed for the maintenance of normal presynaptic acetylcholine release at neuromuscular junctions (17). DIRAS1 was also suggested to play a role in cell migration, neurite outgrowth, and dendrite architecture in the developing nervous system of a zebrafish model (18).
Understanding the role and mechanisms of DIRAS1 in cholinergic neurotransmission and epilepsy remains an important task. Nicotinergic cholinergic activity influences brain excitability and cognition, regulates the excitatory/inhibitory switch of GABA during neuronal development (45), stimulates glutamate release from thalamocortical terminals, controls GABA release onto pyramidal neurons, and maintains nonrapid eye movment sleep by low levels of acetylcholine, whereby cholinergic stimulation is associated with microarousals in this sleep stage (46). Mutations in nicotinergic acetylcholine receptor (nAChR) subunits CHRNA4, CHRNA2, and CHRNB2 are associated with autosomal dominant NFLE and sporadic NFLE (47). CHRNA7 coding for the α7 subunit of the nAChR is also a potential candidate gene for JME in humans (48). Abnormal DIRAS1 function could alter cholinergic neurotransmission or formation of neuronal circuits and network assembly in the developing brain resulting in myoclonic epilepsy and photosensitivity. This canine model establishes a prime resource to address these questions and mechanisms in future experiments, including mutation-specific–induced neuronal cultures.
In summary, careful clinical and genetic studies identified a candidate gene for one of the most common forms of human epilepsy with a postulated function in cholinergic neurotransmission. While inspecting the gene in human myoclonic and epilepsy cohorts for risk variants, future functional studies should identify the DIRAS1-mediated mechanisms in neurotransmission and provide drug targets for common epilepsies.
Materials and Methods
Study Cohorts.
Twenty-four RR cases were identified (Table S2). Inclusion criteria were clinical observation of myoclonic jerks on video recordings or observation at one of the study sites and completion of an online questionnaire or an interview. Altogether, 538 EDTA-blood and tissue samples were collected from privately owned RRs in Germany, Finland, and 11 other countries (Table S6). A cohort of 965 epileptic dogs from 12 other breeds from Finland was included (Table S6). Sample collection was ethically approved by the Animal Ethics Committee of State Provincial Office of Southern Finland, Hämeenlinna, Finland (ESAVI/6054/04.10.03/ 2012), “Cantonal Committee for Animal Experiments” (Canton of Bern; permit 23/10), and the German Animal Welfare Act. Further details are provided in SI Materials and Methods.
Neurodiagnostic Investigation.
All RR cases underwent a clinical, neurological, and laboratory examination. Structural epilepsy was excluded by imaging through MRI in 12 RR cases and postmortem examination of 1 dog. Additional investigations comprising cerebrospinal fluid (CSF) analysis, neurometabolic screening, imaging through CT, skin biopsy, and AED serum concentration measurements were performed for a number of studied dogs. Further details are provided in SI Materials and Methods.
EEG.
Awake ambulatory wireless video-EEG was conducted in 17 RR cases and 11 RR control dogs. Recordings were performed in a quiet environment, with dogs encouraged to lie down. EEG was recorded routinely using 15 (7 in one dog) subdermal needle electrodes. In six cases and four controls an additional video-EEG with photic stimulation was conducted at the end of the EEG study. Further details are provided in SI Materials and Methods.
Postmortem Examination.
Postmortem examination was conducted on one affected RR. The animal underwent routine autopsy in which the brain was removed in toto and trimmed according to standardized algorithms (49). Relevant brain areas (prosencephalon, cerebellum, brainstem) were sampled and histologically evaluated using neurohistological standard stains on paraffin sections.
GWAS.
Genotyping of 10 affected RRs from the initial study cohort and 18 unaffected RRs was performed. The genotype data were filtered and frequency and genotyping pruned. A case-control association test was performed by PLINK (50) and by Mendel software's Ped-GWAS (51). Further details are provided in SI Materials and Methods.
Resequencing.
Dog exome libraries for two German RR cases were generated. The sequencing data were analyzed and filtered under a recessive model against 169 additional exomes (Table S3). The pathogenicity of the coding variants was predicted in the CanFam 3.1 annotation. One RR case was whole-genome sequenced and filtered against 99 additional whole genomes (Table S3) and the presence of the candidate mutation was inspected visually. Further details are provided in SI Materials and Methods.
Sanger Sequencing and TaqMan Genotyping.
The identified candidate variant was validated by a standard PCR followed by Sanger sequencing in 33 German RR samples, including 12 cases from the initial study cohort. For a larger mutation screening in additional samples (Table S6), a TaqMan assay was run. Further details are provided in SI Materials and Methods.
Gene Expression.
Fresh postmortem samples were collected (for the full list, Fig. 3 and Fig. S4) from one case and six control dogs. RNA was extracted and reverse-transcribed into cDNA. The canine DIRAS1 transcript was amplified and sequenced. Semiquantitative and quantitative PCRs were performed. Further details are provided in SI Materials and Methods.
Immunohistochemistry.
Tissue studies were conducted on the brains (prosencephalon, cerebellum, brainstem) of one RR case and three control RRs (LMU Munich neuropathology brain archive). Primary antibodies (pAB) were directed at DIRAS1 and the vesicular acetylcholine transporter. The slides were antigen-demasked, incubated with pAB, and stained using polymer technology and a diaminobenzidine tetrahydrochloride. Further details are provided in SI Materials and Methods.
SI Materials and Methods
Study Cohorts.
Twenty-four RR cases were identified. Four study sites (Veterinary Hospital Trier, Trier, Germany; Veterinary Practice Bathen-Noethen, Cologne, Germany; Department of Small Animal Medicine and Surgery, University of Veterinary Medicine, Hannover, Germany; Clinic of Small Animal Medicine, LMU Munich, Germany) participated in recruitment of the initial cohort. Additional cases were recruited via call for study participants on the homepage of LMU Munich and referral by diplomates of the European College of Veterinary Neurology. Inclusion criteria were clinical observation of myoclonic jerks on video recordings or observation at one of the study sites and completion of an extensive online questionnaire (semse.vetmed.uni-muenchen.de/umfragen/index.php?sid=%2018469&lang=de) or personal interview. Dogs with other types of muscle contractions (e.g., tremor, fasciculations) or GTCS or focal seizures as the only manifestation of epilepsy were excluded. Dogs without video documentation of myoclonic jerks with insufficiently completed questionnaires were denied study participation.
Altogether, 538 EDTA-blood and tissue samples were collected from privately owned RRs in Germany, Finland, and 11 other countries (Table S6). The German RR study cohort included blood samples of 19 affected RRs displaying the generalized myoclonic epilepsy phenotype, 11 unaffected relatives (dam and sire of two litters, four littermates, one additional dam and her sister, one cousin) and 18 controls (13 healthy RRs, 4 RRs with idiopathic GTCS, 2 RRs with other types of muscle twitches). Genotyping analysis included also a cohort of 965 epileptic dogs from 12 other breeds from Finland (Table S6). The samples were stored at −20 °C until genomic DNA was extracted using a semiautomated Chemagen extraction robot (PerkinElmer Chemagen Technologie) or the Nucleon Bacc2 kit (GE Healthcare). DNA concentration was determined either with the NanoDrop ND-1000 UV/Vis Spectrophotometer or Qubit 3.0 Fluorometer (Thermo Fisher Scientific). Sample collection was ethically approved by the Animal Ethics Committee of State Provincial Office of Southern Finland, Hämeenlinna, Finland (ESAVI/6054/04.10.03/ 2012), “Cantonal Committee For Animal Experiments” (Canton of Bern; permit 23/10), and the “Institutional Review Board” of the Clinic of Small Animal Medicine (LMU Munich, Germany 28.05.13).
Neurodiagnostic Investigation.
All dogs underwent a clinical and neurological examination by a veterinary neurologist. Laboratory investigations comprised complete blood cell count and serum biochemistry profile (alanine aminotransferase, alkaline phosphatase, total bilirubin, urea, creatinine, total protein, albumin, glucose, sodium, potassium, chloride, calcium, phosphate), fasted venous ammonia (14 cases), fasted and postprandial bile acids (7 cases), creatine kinase activity (8 cases), ionized calcium (4 cases), coagulation profile (PT, PTT, TT; 1 case), and thyroid panel (total T4, free T4, thyroid stimulating hormone; 7 cases). AED serum concentration measurements (IDEXX Laboratories) were accomplished in six dogs treated with phenobarbital and four dogs treated with potassium bromide. Neurometabolic screening comprised urinary organic acids, oligosaccharides, mucopolysaccharides, and urinary and serum amino acids in 10 cases and 1 related control RR in specialized laboratories (Children’s Hospital Frankfurt, Germany; Biocontrol, Labor Ingelheim, Ingelheim, Germany; Comparative Biochemical Genetics Laboratory Veterinary Unit, University of California, San Diego, CA; Metabolic Genetics Screening Laboratory, School of Veterinary Medicine, University of Philadelphia, PA). Skin biopsy from one affected dog was immersed in 2.5% glutaraldehyde and was screened for ultrastructural evidence of metabolic disorders by electron microscopy (ZM 906, Zeiss). Advanced imaging through MRI (LMU Munich: 1.5 T Magnetom Symphony Syngo MR Siemens; University of Leipzig: Philips MR Ingenia 3T with Omega HP Gradient; Philips Medical Systems; various other high- and low-field MR machines) or CT was undertaken in 12 and 7 dogs, respectively. CSF for analysis was obtained following sterile preparation by atlantooccipital puncture with the dogs under general anesthesia in 17 cases.
EEG.
Awake ambulatory wireless video-EEG was conducted in 17 RR cases and 11 RR control dogs (10 healthy RRs and 1 RR with idiopathic epilepsy with GTCS) using a Trackit MK3 EEG/Polygraphy Recorder with video (Lifelines Neurodiagnostic Systems) and Persyst 12 software (Persyst Development Corporation and Micromed Morpheus Home LTM ambulatory EEG recorder (Micromed Neurodiagnostic). Because the episodes in question emerged primarily when the dogs were relaxed, recordings were performed in a quiet environment where the dogs had the possibility to lie down in a comfortable position. EEG was recorded routinely using 15 subdermal stainless steel needle electrodes (F3, Fz, F4, F7, F8, C3, Cz, C4, O1, Pz, O2, T1, T2, Ref, Neut; Natus Europe, product number 019-475800). Electrode placement was as described by Pellegrino and Sica (52) for mesencephalic dogs, but modified for our demands: we placed subdermal needles at the anterior canthus of the base of the ear for the temporal loci (T3, T4) and the reference more caudally between the medial canthi of the eyes both as proposed by James et al. (53). We did not use the frontopolar leads, but added two additional electrodes 1- to 1.5-cm dorsolateral of the lateral canthi of the eyes (F7, F8). After positioning, all electrodes were fixed in place with medical tape (Leukoplast). Subsequently, the recorder—as well as the remaining wires—were attached to a harness on the dog’s back via cohesive bandages (Co-Flex). In one dog, seven subdermal electrodes were used, including two frontal (F3, F4), one central (V), and two occipital electrodes (O1, O2), as well as a reference at the dorsum of the nose (Ref) and a ground in the neck (Neut). Three cases and one control required sedation for electrode placement (0.015 mg/kg dexmedetomidine, reversal with 0.15 mg/kg atipamezole). In one case and one control, video-EEG recording was undertaken upon awakening from general anesthesia. Impedance was kept under 10 kΩ in most cases (up to 20 kΩ in rare cases). In six cases and four controls, an additional video-EEG with photic stimulation was conducted at the end of the EEG study (Micromed Morpheus Home LTM ambulatory EEG recorder, Micromed Neurodiagnostic). A lamp with circular reflector and a viewing distance of 30 cm was used. Stimulation was conducted for 10 s followed by a rest of 5 s per each flash frequency. The following flash frequencies (in Hz) were used in this order: 1–6–11–18–7–12–16–4–25–10–17–9–14–3.
GWAS.
Genotyping of 10 affected and 18 unaffected RRs from the German study cohort (Fig. S2) was performed using Illumina’s Canine HD array (173 k). The genotype data were filtered with a SNP call rate of >95%, array call rate of >95%, minor allele frequency of >5% and by using a Hardy–Weinberg equilibrium of P ≤ 0.00005. After frequency and genotyping pruning (with a window size of 50, a step of five SNPs, and an r2 threshold of 0.5) no individual dogs were removed and 110,492 SNPs remained for analysis. A case-control association test (GWAS) was performed by PLINK (50) and by Ped-GWAS, using Mendel software (51). Two cases and four controls were excluded in PLINK analysis because of relatedness. After Ped-GWAS quality control analysis, 107,852 SNPs were included in the analysis.
Resequencing.
Dog exome libraries for two German RR cases were generated from standard indexed Illumina libraries using a custom Roche/Nimblegen solution-based capture library (120705_CF3_Uppsala_Broad_EZ_HX1) according to the protocol described previously by Elvers et al. (54). The sequencing data were analyzed using our in-house scripts as previously described (55). The average sequencing coverage varied between 25–35× per sample and revealed on average ∼108,909 variants per sample. The two affected dogs shared 30,506 homozygous variants; however, filtering under a recessive model against 169 additional exomes from other nonaffected breeds available (Table S3) resulted in a total of 10 case-specific homozygous variants (Table S4), of which 6 variants were in the predicted coding regions (1 indel, 5 synonymous). SnpEff (56) was used to predict the pathogenicity of the coding variants in the CanFam 3.1 annotation. One RR case was whole genome-sequenced in the Science for Life Laboratory in Stockholm, Sweden, as described previously (57). The RR whole genome (∼25×) was filtered against 99 additional whole genomes from other nonaffected breeds available (Table S3). Variants in the 890-kb critical region at chromosome 20 were investigated, revealing the previously identified mutation in the case, although none of the other dogs carried it. The presence of the candidate mutation was inspected visually in the BAM file using the Integrative Genomics Viewer. Structural variation analysis in the associated region was performed using DELLY v0.6.7 to detect possible case-specific deletions or duplications in the WGS data of an epileptic RR against 99 nonepileptic control genomes including two RRs (Table S3). The detected sites were merged and regenotyped across all samples using DELLY v0.7.5. Genotyped copy number variants were required to have both split-read (SR) and paired-end (PE) support (SR > 1 and PE > 5 or SR > 5 and PE > 1) to retain a high-confidence call set.
Sanger Sequencing and TaqMan Genotyping.
The identified candidate variant (chr20:g.56’474’668_56’474’671delAGAC) was first validated by a standard PCR followed by Sanger sequencing in 33 German samples, including 12 cases from the initial study cohort. Forward (ACAATGTCAAGGAGCTCTTCC) and reverse (GCTCACATGAGGACGCATT) primers were designed to amplify the mutation region using AmpliTaq Gold 360 Mastermix (Applied Biosystems, Life Technologies) or Biotools’ DNA Polymerase. The sequencing reactions were then performed on an ABI 3730 capillary sequencer (Applied Biosystems, Life Technologies), after treatment with exonuclease I and shrimp alkaline phosphatase. The Sanger sequence data were analyzed using Sequencher 5.1 (GeneCodes). For a larger mutation screening in additional samples (Table S6), a TaqMan assay (Applied Biosystems) was developed (probes: GGAACATGAGCCTCAACATCGAT and GGACGCATTTGCCCTTGAC) and reactions were run by the Bio-Rad’s CFX96 Touch Real-Time PCR Detection System instrumentation according to the manufacturer’s instructions.
Sequence Alignment.
The CanFam 3.1 assembly was used for all analyses. All numbering within the canine DIRAS1 gene correspond to the accessions XM_005633100.2 and XP_005633157.1. For the annotation of the gene and the identified risk variant, we have used Broad Institute CanFam3 Improved Annotation Data v1. The sequence alignments of the normal and mutated DIRAS1 proteins were constructed by using the Clustal Omega tool (www.ebi.ac.uk/Tools/msa/clustalo/). The aligned protein sequences were derived from the Entrez Protein database (https://www.ncbi.nlm.nih.gov/protein). The domain structures were investigated by SMART (smart.embl-heidelberg.de/).
Gene Expression.
Fresh postmortem samples were collected from seven dogs. Twenty-eight different tissues were collected from one Finnish epilepsy-free Belgian Shepherd dog (for the full list, Fig. S4) killed at the age of 4 y and 10 mo. The frontal cortex was collected from five other epilepsy-free control dogs (one 2-mo-old Wire Fox Terrier, one 5-mo-old Labrador Retriever, two Great Danes aged 23 mo and 5 y and 3 mo, one 9-y-old Saluki) and one case RR. The 24-mo-old female RR case was killed because of severe seizures nonresponsive to treatment with phenobarbital and the control dogs because of nonepilepsy-related disorders or behavior by owners’ consent. Samples were harvested immediately after killing and snap-frozen in liquid nitrogen before storage in −80 °C. Total RNA was extracted from the canine tissues by using the RNeasy Mini Kit (Qiagen), and sample concentrations were measured by using a Qubit 3.0 Fluorometer (Thermo Fisher Scientific). The High Capacity RNA-to-cDNA Kit (Applied Biosystems, Life Technologies) was then used to reverse-transcribe equal amounts of total RNA into cDNA. In both semiquantitative and quantitative PCR, the canine DIRAS1 transcript was amplified and sequenced by using a primer pair (TGAAGATGGACGTCATACTGCTT and GAACACCACCACTCGGTAGTC) that was designed to span over exon–intron boundaries to control for genomic DNA contamination. Semiquantitative PCR was performed in three different cycles (27, 32, and 37 cycles) before visualization of the amplicons on a 2.5% (wt/vol) agarose gel (100 V, 2 h). GAPDH was used as a loading control. Quantitative PCR was performed for triplicates of each sample using Bio-Rad’s CFX96 Touch Real-Time PCR Detection System instrumentation according to the manufacturer’s instructions. YWHAZ and GAPDH were used as loading controls. The relative normalized expression pattern was calculated with Bio-Rad’s CFX Manager Software. The SE between triplicates was below 0.13 in each studied sample.
Immunohistochemistry.
Tissue studies were conducted on the brain of one affected RR (case 2) donated by the owner for postmortem examination. The animal underwent routine autopsy on which the brain was removed in toto and trimmed according to standardized algorithms (49). Relevant brain areas (prosencephalon, cerebellum, brainstem) were sampled and histologically evaluated using neurohistological standard stains on paraffin sections. Identical sections were obtained and examined from three unaffected (two dogs without seizures and one with seizures secondary to acute hemorrhagic stroke) RRs admitted for diagnostic reasons unrelated to the purpose of this study. pAB were directed at DIRAS1 protein (rabbit polyclonal, PA5-26409, Fisher Scientific) and Vesicular Acetylcholine Transporter (VAChT, rabbit polyclonal, Ab68984, Abcam). Anti-DIRAS1 polyclonal antibody was developed against synthetic peptide between 109 and 136 amino acids from the central region of human DIRAS1. This epitope has 100% amino acid identity with the corresponding canine epitope. Therefore, sequence homology scale renders specific cross-reaction of the antibody very likely. Before staining, the slides underwent microwave-based antigen demasking in citrate buffer. Slides were incubated with pAB (DIRAS1 1:500; VAChT 1:100) for 18 h at 4 °C. Subsequent staining used polymer technology (IMPRESS Linaris) and a diaminobenzidine tetrahydrochloride staining kit.
Supplementary Material
Acknowledgments
We thank Martin J. Schmidt, Tanja A. Steinberg, Verena Butz, Andreas Brühschwein, Martin Wrzosek, and Anja Waselau for their support of this study; Tom Pieper, Stephan Arnold, and Silke Link for training in EEG; Adrian Sewell for discussing results of metabolic screening; Sini Karjalainen and Riccardo Solda for excellent technical assistance; the Dog Biomedical Variant Database Consortium (Gus Aguirre, Catherine André, Danika Bannasch, Doreen Becker, Cord Drögemüller, Eva Furrow, Urs Giger, Christophe Hitte, Marjo Hytönen, Vidhya Jagannathan, Tosso Leeb, Hannes Lohi, Jim Mickelson, Anita Oberbauer, Jeffrey Schoenebeck, Claire Wade) for providing access to whole-genome variants from control dogs; and all the dog owners for contributing samples and particularly those who participated in clinical studies. The study was supported by the Academy of Finland (1268091), the Sigrid Juselius Foundation, the Jane and Aatos Erkko Foundation, ERCStG (260997), Epilepsiatutkimussäätiö, Biocentrum Helsinki, the Munich University Society, the Canine Health Foundation (CHF Grant 02248), the Albert Heim Foundation (Project 105), and Canada Foundation for Innovation and Ontario Ministry of Research and Innovation (30953).
Footnotes
Conflict of interest statement: H.L. is one of the owners of the Genoscoper Ltd., which provides the gene test developed based on the study.
This article is a PNAS Direct Submission.
Data deposition: Whole-genome and exome sequences can be found in the BioSample database, https://www.ncbi.nlm.nih.gov/biosample (accession nos. SAMN06161402, SAMN06161403, SAMN06161404).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1614478114/-/DCSupplemental.
References
- 1.Potschka H, Fischer A, von Rüden E-L, Hülsmeyer V, Baumgärtner W. Canine epilepsy as a translational model? Epilepsia. 2013;54(4):571–579. doi: 10.1111/epi.12138. [DOI] [PubMed] [Google Scholar]
- 2.Lindblad-Toh K, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438(7069):803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- 3.Volk HA. International Veterinary Epilepsy Task Force consensus reports on epilepsy definition, classification and terminology, affected dog breeds, diagnosis, treatment, outcome measures of therapeutic trials, neuroimaging and neuropathology in companion animals. BMC Vet Res. 2015;11(1):174. doi: 10.1186/s12917-015-0460-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hülsmeyer VI, et al. International Veterinary Epilepsy Task Force’s current understanding of idiopathic epilepsy of genetic or suspected genetic origin in purebred dogs. BMC Vet Res. 2015;11(1):175. doi: 10.1186/s12917-015-0463-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lohi H, et al. Expanded repeat in canine epilepsy. Science. 2005;307(5706):81. doi: 10.1126/science.1102832. [DOI] [PubMed] [Google Scholar]
- 6.Hajek I, et al. NHLRC1 repeat expansion in two beagles with Lafora disease. J Small Anim Pract. 2016;57(11):650–652. doi: 10.1111/jsap.12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashwini A, et al. Neuronal ceroid lipofuscinosis associated with an MFSD8 mutation in Chihuahuas. Mol Genet Metab. 2016;118(4):326–332. doi: 10.1016/j.ymgme.2016.05.008. [DOI] [PubMed] [Google Scholar]
- 8.Seppälä EH, et al. LGI2 truncation causes a remitting focal epilepsy in dogs. PLoS Genet. 2011;7(7):e1002194. doi: 10.1371/journal.pgen.1002194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koskinen LLE, et al. Identification of a common risk haplotype for canine idiopathic epilepsy in the ADAM23 gene. BMC Genomics. 2015;16(1):465. doi: 10.1186/s12864-015-1651-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salmon Hillbertz NHC, et al. Duplication of FGF3, FGF4, FGF19 and ORAOV1 causes hair ridge and predisposition to dermoid sinus in Ridgeback dogs. Nat Genet. 2007;39(11):1318–1320. doi: 10.1038/ng.2007.4. [DOI] [PubMed] [Google Scholar]
- 11.Hillbertz NH, Andersson G. Autosomal dominant mutation causing the dorsal ridge predisposes for dermoid sinus in Rhodesian ridgeback dogs. J Small Anim Pract. 2006;47(4):184–188. doi: 10.1111/j.1748-5827.2006.00016.x. [DOI] [PubMed] [Google Scholar]
- 12.Pivetta M, De Risio L, Newton R, Dennis R. Prevalence of lateral ventricle asymmetry in brain MRI studies of neurologically normal dogs and dogs with idiopathic epilepsy. Vet Radiol Ultrasound. 2013;54(5):516–521. doi: 10.1111/vru.12063. [DOI] [PubMed] [Google Scholar]
- 13.Bhatti SFM, et al. International Veterinary Epilepsy Task Force consensus proposal: Medical treatment of canine epilepsy in Europe. BMC Vet Res. 2015;11(1):176. doi: 10.1186/s12917-015-0464-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noachtar S, et al. N166 Levetiracetam Study Group Levetiracetam for the treatment of idiopathic generalized epilepsy with myoclonic seizures. Neurology. 2008;70(8):607–616. doi: 10.1212/01.wnl.0000297512.18364.40. [DOI] [PubMed] [Google Scholar]
- 15.Holliday TA, Williams C. Clinical electroencephalography in dogs. Vet Neurol Neurosurg J. 1999;1(1):1. [Google Scholar]
- 16.Kontani K, et al. Di-Ras, a distinct subgroup of ras family GTPases with unique biochemical properties. J Biol Chem. 2002;277(43):41070–41078. doi: 10.1074/jbc.M202150200. [DOI] [PubMed] [Google Scholar]
- 17.Tada M, et al. Neuronally expressed Ras-family GTPase Di-Ras modulates synaptic activity in Caenorhabditis elegans. Genes Cells. 2012;17(9):778–789. doi: 10.1111/j.1365-2443.2012.01627.x. [DOI] [PubMed] [Google Scholar]
- 18.Yeh C-W, Hsu L-S. Zebrafish diras1 promoted neurite outgrowth in neuro-2a cells and maintained trigeminal ganglion neurons in vivo via Rac1-dependent pathway. Mol Neurobiol. 2016;53(10):6594–6607. doi: 10.1007/s12035-015-9550-2. [DOI] [PubMed] [Google Scholar]
- 19.Berg AT, et al. Revised terminology and concepts for organization of seizures and epilepsies: Report of the ILAE Commission on Classification and Terminology, 2005-2009. Epilepsia. 2010;51(4):676–685. doi: 10.1111/j.1528-1167.2010.02522.x. [DOI] [PubMed] [Google Scholar]
- 20.Welty TE. Juvenile myoclonic epilepsy: Epidemiology, pathophysiology, and management. Paediatr Drugs. 2006;8(5):303–310. doi: 10.2165/00148581-200608050-00003. [DOI] [PubMed] [Google Scholar]
- 21.Genton P, Thomas P, Kasteleijn-Nolst Trenité DGA, Medina MT, Salas-Puig J. Clinical aspects of juvenile myoclonic epilepsy. Epilepsy Behav. 2013;28(1) Suppl 1:S8–S14. doi: 10.1016/j.yebeh.2012.10.034. [DOI] [PubMed] [Google Scholar]
- 22.Serafini A, Rubboli G, Gigli GL, Koutroumanidis M, Gelisse P. Neurophysiology of juvenile myoclonic epilepsy. Epilepsy Behav. 2013;28(1) Suppl 1:S30–S39. doi: 10.1016/j.yebeh.2012.11.042. [DOI] [PubMed] [Google Scholar]
- 23.Jallon P, Latour P. Epidemiology of idiopathic generalized epilepsies. Epilepsia. 2005;46(Suppl 9):10–14. doi: 10.1111/j.1528-1167.2005.00309.x. [DOI] [PubMed] [Google Scholar]
- 24.Anderson J, Hamandi K. Understanding juvenile myoclonic epilepsy: Contributions from neuroimaging. Epilepsy Res. 2011;94(3):127–137. doi: 10.1016/j.eplepsyres.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 25.Wolf P, et al. Juvenile myoclonic epilepsy: A system disorder of the brain. Epilepsy Res. 2015;114:2–12. doi: 10.1016/j.eplepsyres.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 26.Guerrini R, Genton P. Epileptic syndromes and visually induced seizures. Epilepsia. 2004;45(Suppl 1):14–18. doi: 10.1111/j.0013-9580.2004.451011.x. [DOI] [PubMed] [Google Scholar]
- 27.Verrotti A, Beccaria F, Fiori F, Montagnini A, Capovilla G. Photosensitivity: Epidemiology, genetics, clinical manifestations, assessment, and management. Epileptic Disord. 2012;14(4):349–362. doi: 10.1684/epd.2012.0539. [DOI] [PubMed] [Google Scholar]
- 28.Galizia EC, et al. EuroEPINOMICS CoGIE Consortium CHD2 variants are a risk factor for photosensitivity in epilepsy. Brain. 2015;138(Pt 5):1198–1207. doi: 10.1093/brain/awv052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mignot C, et al. EuroEPINOMICS-RES MAE working group Genetic and neurodevelopmental spectrum of SYNGAP1-associated intellectual disability and epilepsy. J Med Genet. 2016;53(8):511–522. doi: 10.1136/jmedgenet-2015-103451. [DOI] [PubMed] [Google Scholar]
- 30.Waltz S, Christen HJ, Doose H. The different patterns of the photoparoxysmal response—A genetic study. Electroencephalogr Clin Neurophysiol. 1992;83(2):138–145. doi: 10.1016/0013-4694(92)90027-f. [DOI] [PubMed] [Google Scholar]
- 31.Szabó CÁ, Knape KD, Leland MM, Williams JT. Electroclinical phenotypes in a pedigreed baboon colony. Epilepsy Res. 2013;105(1-2):77–85. doi: 10.1016/j.eplepsyres.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ákos Szabó C, et al. Modeling the effective connectivity of the visual network in healthy and photosensitive, epileptic baboons. Brain Struct Funct. 2016;221(4):2023–2033. doi: 10.1007/s00429-015-1022-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Serpell JA, Duffy DL. Aspects of juvenile and adolescent environment predict aggression and fear in 12-month-old guide dogs. Front Vet Sci. 2016;3(June):49. doi: 10.3389/fvets.2016.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scott JP, Fuller JL. Genetics and the Social Behavior of the Dog. 1st Ed Univ of Chicago Press; Chicago: 1965. [Google Scholar]
- 35.Miklósi Á. Dog Behaviour, Evolution, and Cognition. 1st Ed Oxford Univ Press; Oxford: 2015. [Google Scholar]
- 36.Joslyn S, et al. Effect of delayed acquisition times on gadolinium-enhanced magnetic resonance imaging of the presumably normal canine brain. Vet Radiol Ultrasound. 2011;52(6):611–618. doi: 10.1111/j.1740-8261.2011.01847.x. [DOI] [PubMed] [Google Scholar]
- 37.Kasteleijn-Nolst Trenité DG, et al. Consensus on diagnosis and management of JME: From founder’s observations to current trends. Epilepsy Behav. 2013;28(1) Suppl 1:S87–S90. doi: 10.1016/j.yebeh.2012.11.051. [DOI] [PubMed] [Google Scholar]
- 38.Beniczky S, et al. Modulation of epileptiform EEG discharges in juvenile myoclonic epilepsy: an investigation of reflex epileptic traits. Epilepsia. 2012;53(5):832–839. doi: 10.1111/j.1528-1167.2012.03454.x. [DOI] [PubMed] [Google Scholar]
- 39.Caraballo RH, et al. Myoclonic epilepsy in infancy: an electroclinical study and long-term follow-up of 38 patients. Epilepsia. 2013;54(9):1605–1612. doi: 10.1111/epi.12321. [DOI] [PubMed] [Google Scholar]
- 40.Steinlein OK, Hoda J-C, Bertrand S, Bertrand D. Mutations in familial nocturnal frontal lobe epilepsy might be associated with distinct neurological phenotypes. Seizure. 2012;21(2):118–123. doi: 10.1016/j.seizure.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 41.Shen M, et al. Farnesyltransferase and geranylgeranyltransferase I: Structures, mechanism, inhibitors and molecular modeling. Drug Discov Today. 2015;20(2):267–276. doi: 10.1016/j.drudis.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 42.Ye X, Carew TJ. Small G protein signaling in neuronal plasticity and memory formation: The specific role of ras family proteins. Neuron. 2010;68(3):340–361. doi: 10.1016/j.neuron.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gyurkó MD, Csermely P, Sőti C, Steták A. Distinct roles of the RasGAP family proteins in C. elegans associative learning and memory. Sci Rep. 2015;5:15084. doi: 10.1038/srep15084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bergom C, et al. The tumor-suppressive small GTPase DiRas1 binds the noncanonical guanine nucleotide exchange factor SmgGDS and antagonizes SmgGDS interactions with oncogenic small GTPases. J Biol Chem. 2016;291(12):6534–6545. doi: 10.1074/jbc.M115.696831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Z, Neff RA, Berg DK. Sequential interplay of nicotinic and GABAergic signaling guides neuronal development. Science. 2006;314(5805):1610–1613. doi: 10.1126/science.1134246. [DOI] [PubMed] [Google Scholar]
- 46.Becchetti A, Aracri P, Meneghini S, Brusco S, Amadeo A. The role of nicotinic acetylcholine receptors in autosomal dominant nocturnal frontal lobe epilepsy. Front Physiol. 2015;6(22):22. doi: 10.3389/fphys.2015.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Conti V, et al. Nocturnal frontal lobe epilepsy with paroxysmal arousals due to CHRNA2 loss of function. Neurology. 2015;84(15):1520–1528. doi: 10.1212/WNL.0000000000001471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Helbig I, et al. 15q13.3 microdeletions increase risk of idiopathic generalized epilepsy. Nat Genet. 2009;41(2):160–162. doi: 10.1038/ng.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matiasek K, et al. International veterinary epilepsy task force recommendations for systematic sampling and processing of brains from epileptic dogs and cats. BMC Vet Res. 2015;11(1):216. doi: 10.1186/s12917-015-0467-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lange K, et al. Mendel: The Swiss army knife of genetic analysis programs. Bioinformatics. 2013;29(12):1568–1570. doi: 10.1093/bioinformatics/btt187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pellegrino FC, Sica RE. Canine electroencephalographic recording technique: Findings in normal and epileptic dogs. Clin Neurophysiol. 2004;115(2):477–487. doi: 10.1016/s1388-2457(03)00347-x. [DOI] [PubMed] [Google Scholar]
- 53.James FMK, et al. Investigation of the use of three electroencephalographic electrodes for long-term electroencephalographic recording in awake and sedated dogs. Am J Vet Res. 2011;72(3):384–90. doi: 10.2460/ajvr.72.3.384. [DOI] [PubMed] [Google Scholar]
- 54.Elvers I, et al. Exome sequencing of lymphomas from three dog breeds reveals somatic mutation patterns reflecting genetic background. Genome Res. 2015;25(11):1634–1645. doi: 10.1101/gr.194449.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ahonen SJ, Arumilli M, Lohi H. A CNGB1 frameshift mutation in Papillon and Phalène dogs with progressive retinal atrophy. PLoS One. 2013;8(8):e72122. doi: 10.1371/journal.pone.0072122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6(2):80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kyöstilä K, et al. A missense change in the ATG4D gene links aberrant autophagy to a neurodegenerative vacuolar storage disease. PLoS Genet. 2015;11(4):e1005169. doi: 10.1371/journal.pgen.1005169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.