Significance
We anticipate that our proposed single-neuron mass spectrometry technology may provide not only the possibility to study single-cell metabolism, but also the chance to investigate chemical changes in the content and metabolism of intracellular small molecules at the single-cell level during physiological processes, such as growth and development, or pathological processes, such as depression, dementia, inflammation, cancer, and other diseases.
Keywords: single neuron, mass spectrometry, metabolism, patch clamp
Abstract
The use of single-cell assays has emerged as a cutting-edge technique during the past decade. Although single-cell mass spectrometry (MS) has recently achieved remarkable results, deep biological insights have not yet been obtained, probably because of various technical issues, including the unavoidable use of matrices, the inability to maintain cell viability, low throughput because of sample pretreatment, and the lack of recordings of cell physiological activities from the same cell. In this study, we describe a patch clamp/MS-based platform that enables the sensitive, rapid, and in situ chemical profiling of single living neurons. This approach integrates modified patch clamp technique and modified MS measurements to directly collect and detect nanoliter-scale samples from the cytoplasm of single neurons in mice brain slices. Abundant possible cytoplasmic constituents were detected in a single neuron at a relatively fast rate, and over 50 metabolites were identified in this study. The advantages of direct, rapid, and in situ sampling and analysis enabled us to measure the biological activities of the cytoplasmic constituents in a single neuron, including comparing neuron types by cytoplasmic chemical constituents; observing changes in constituent concentrations as the physiological conditions, such as age, vary; and identifying the metabolic pathways of small molecules.
Single-cell genomic, proteomic, and metabolic studies have attracted increasing attention with the development of single-cell technologies, such as flow cytometry (1–3), single-cell PCR (4, 5), single-cell RNA sequencing (6–8), immunofluorescence (9, 10), and electrochemistry (11, 12). Single-cell analysis has become increasingly important in biology, especially in neuroscience research, because neurons are specialized cells with a huge diversity of shapes, connections, and anatomical, electrophysiological, and biochemical properties (13–17). Single-cell chemical profiling could further the current understanding of biological variability and differential susceptibility to disease and treatment, as well as heterogeneity among similar cells (18, 19). Most of currently available single-cell techniques require the chemical of interest (usually large molecules, such as DNA, RNA, and proteins) to be labeled, amplified, or electrochemically detectable (4, 6, 10, 12). In addition to these large molecules, small molecules, such as lipids, saccharides, transmitters, and metabolites, play critical roles in cells, including neurons (20). However, detecting small molecules in a single cell remains challenging because the lack of proper labeling methods is still beset with seemingly insurmountable difficulties.
Advances in mass spectrometry (MS), especially in matrix-assisted laser desorption ionization MS (MALDI-MS) (21) and electrospray MS (ESI-MS) (22), have enabled the direct and accurate measurement of a massive number of chemicals simultaneously in biological samples, without the need for specific labeling or amplification techniques. Because the size of cells is quite limited, most MS approaches for single cells are based on MALDI-MS (23–25). MALDI-MS helped to discover hundreds of novel brain peptides, tens of which have been reported to be biologically active and related to animal behavior and functioning in models from arthropods and nematodes to mollusks (26). Indeed, whereas a laser/ion beam can be easily focused on a tiny spot, the utilization of organic chemical matrices could lead to an inability to maintain cell viability before MS measurement (24, 25). Much effort has been devoted to the collection of raw cytoplasm from living cells, such as by directly drawing the cytoplasm from a cell using a micropipette (27–29). The application of patch clamp technique combined with capillary electrophoresis (CE) separation would provide enhanced dynamic range and high sample coverage (30). Although MS analysis of single cells has been reported, greater impact on the biological sciences, especially in neuron research, would be anticipated if vast scale assays of single live cells could be accomplished with a much higher throughput (23, 31). Thus, we aimed to develop a rapid and convenient MS assay for living neurons for in situ chemical interpretation that does not require a chemical matrix, facilitates maintaining cell viability, improves the analysis speed, and monitors physiological activity from the same cell.
In this study, by integrating modified single-cell techniques, such as enhanced nanoESI/MS and patch clamp electrophysiological recording of brain slices, we detected abundant cytoplasmic metabolites. We identified more than 70 of the detected molecules in onion epidermal cells and mouse brain neurons. The key features of the modified nanoESI/MS, termed as induced nanoESI (InESI) (32), are the ability to provide a stable MS signal for concentrated salt solutions and to overcome severe ion suppression effects as well as matrix effects (33); these characteristics are indispensable for in situ ESI/MS of the raw cytoplasm. Moreover, by modifying the composition of the micropipette solution, we established an MS protocol compatible with the patch clamp electrophysiological technique for single-cell chemical profiling and the coinstantaneous recording of neuronal activity. Furthermore, our single-neuron chemical MS technology allowed us to reveal and analyze the metabolic processes of intracellular constituents in single neurons. Finally, this technique may also allow us to investigate chemical changes in the content and metabolism of intracellular small molecules at the single-cell level during physiological processes, such as growth and development, or pathological processes, such as depression, dementia, inflammation, cancer, and other diseases.
Results
Continuous Analysis of Concentrated Salt Solution at Subpicoliter Level.
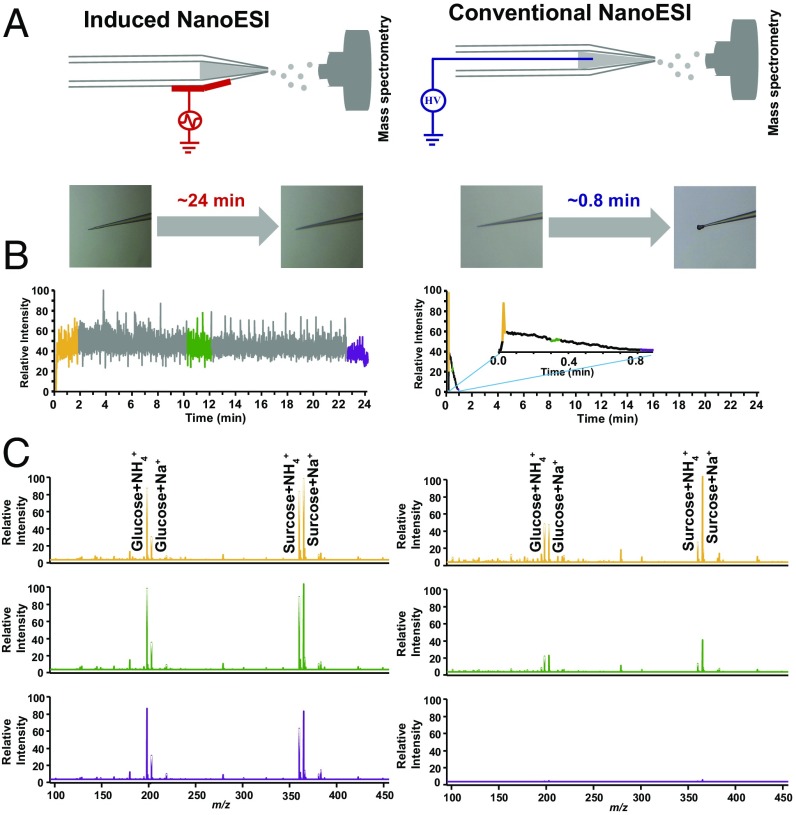
Fig. 1A and SI Appendix, Fig. S1 show the overall concept and workflow of the direct analysis of the cytoplasm of single cells with InESI. A core feature of the InESI technique is to avoid clogging, using concentrated electrolyte solutions, such as cerebrospinal fluid (CSF) solution. The alleviation of the matrix effect (i.e., improved signal-to-noise ratio) is also critical for in situ single-cell analysis. Its lower flow rate, which is more suitable for small-volume samples, would be beneficial for the direct analysis of single-cell cytoplasmic chemical constituents via InESI. We applied an alternating current (AC) voltage with an amplitude of 3 kV at ∼200 Hz at an electrode outside spray emitter to drive the pulsed ESI process (a detailed description of the power supply and protocol can be found in Materials and Methods). An artificial CSF (aCSF) solution diluted with NH4HCO3-NH4Cl (1:1) dramatically decreased the MS signal obtained by conventional nanoESI because total clogging of the spray emitter occurred within ∼60 s. However, using the same aCSF solution, the MS signal produced by InESI remained stable for over ∼24 min (Fig. 1 B and C and SI Appendix, Fig. S2), which is sufficient to allow for the direct MS measurement of cell cytoplasm without sample pretreatment. Using InESI, we measured the chemical constituents of a single epidermal cell from Allium cepa bulbs (SI Appendix, Figs. S3 and S4). The major metabolites we detected were consistent with previous studies (34, 35), indicating that our InESI technology could be applied for single-cell analysis.
Fig. 1.
Overall concept of the direct analysis of the cytoplasm from a single cell with induced nanoESI (InESI, Left) and conventional nanoESI (Right). (A) Schematic (Top) and clogging phenomenon (Down) of InESI and conventional nanoESI. (B) For an aCSF solution diluted with NH4HCO3-NH4Cl (1:1), the MS signal remained stable over ∼24 min for InESI but only ∼60 s for conventional nanoESI. (C) MS spectra of an aCSF solution diluted with NH4HCO3-NH4Cl (1:1) at different times.
Cytoplasmic Chemical Identification and Monitoring of Physiological Activity in the Same Single Neuron.
For single-neuron studies, obtaining both chemical profiles and other biological information in a single assay is desirable. One of the most widely used electrophysiological assays for single neurons—the patch clamp technique—uses a glass micropipette as a recording electrode; such a micropipette is also used in InESI as the spray emitter. Thus, in this study, we combined the patch clamp electrophysiological assay and InESI/MS to achieve both chemical profiling and electrophysiological recording (Fig. 2A).
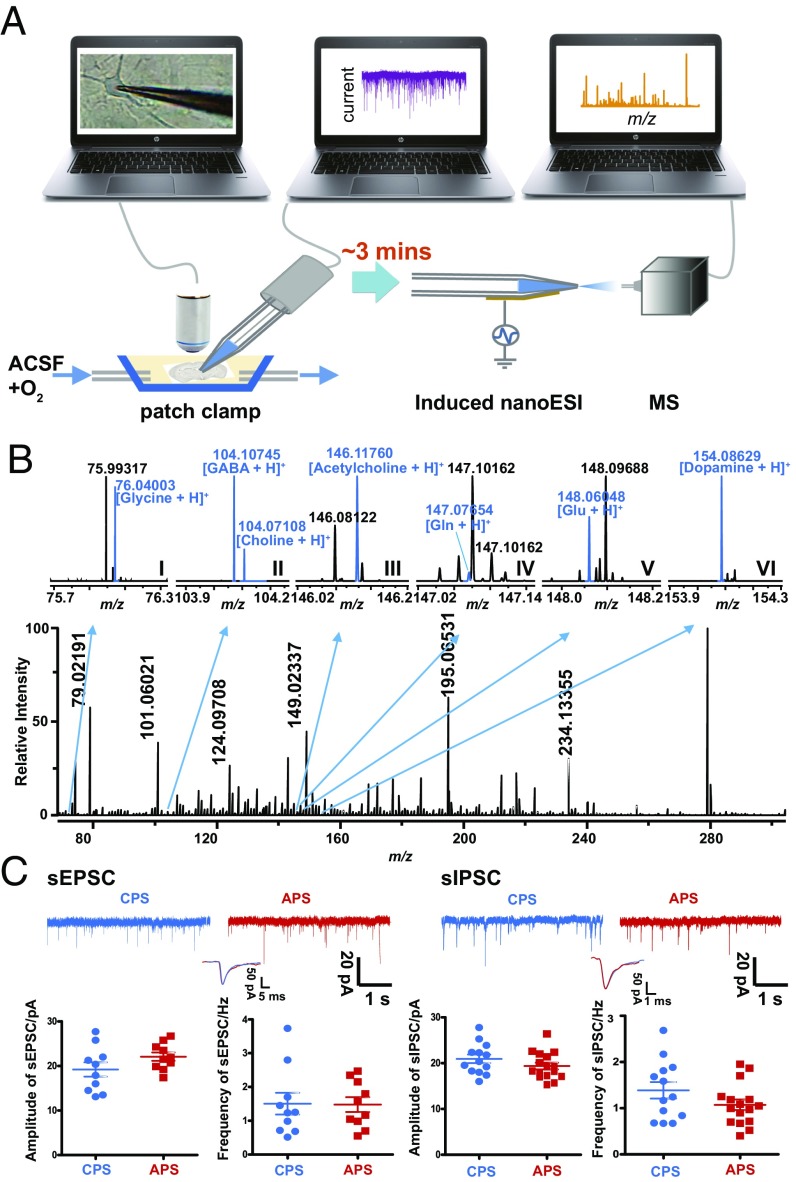
Fig. 2.
Chemical profiling of a single neuron and physiological function recording. (A) Illustration of the workflow for single-neuron sample collection and detection. An upright microscope was used for the electrophysiology experiments with video observation. Following the establishment of the whole-cell configuration, as described in the text, negative pressure was applied to the patch pipette to withdraw the intracellular solution. Then, the patch pipette was removed and directly assayed by InESI/MS. (B) Mass spectra of a single neuron obtained using Orbitrap MS. The expanded views are the mass spectra of glycine (I), gamma-aminobutyric acid (GABA) and choline (II), acetylcholine (III), glutamine (Gln) (IV), glutamate (Glu) (V), and dopamine (VI). (C) (Top) Representative sEPSC and sIPSC traces were recorded in prefrontal cortex (PFC) using conventional pipette solution (CPS, blue) or alternative pipette solution (APS, red). (Bottom) (Left) Summary graphs of sEPSC amplitudes (P = 0.1409 by two-tailed Student’s t test) and frequency (P = 0.9428 by two-tailed Student’s t test) in CPS (blue dots, n = 10 cells) or APS (red dots, n = 10 cells) from three C57 mice. (Right) Summary graphs of the sIPSC amplitude (P = 0.1956 by two-tailed Student’s t test) and frequency (P = 0.1333 by two-tailed Student’s t test) in CPS (blue dots, n = 13 cells) or APS (red dots, n = 16 cells) from three C57 mice.
In brief, this protocol included patching the cell, recording the electrophysiological signal, and performing the MS measurement (a detailed procedure can be found in Materials and Methods). Next, we applied this protocol to single neurons from mouse brain slices. To avoid possible chemical decomposition, total time from cell patch to MS measurement was restricted to ∼3 min. (Fig. 2A). Typical neurotransmitters in a single hippocampal neuron, such as Glu, GABA, dopamine, and acetylcholine, could be confirmed in a single assay. Some other known constituents could also be identified with high-resolution MS, such as glycine, choline, and Gln (Fig. 2B). A total of 54 chemical compounds were confirmed in the cell cytoplasm (SI Appendix, Fig. S6, and SI Appendix, Table S2). The extracellular fluid was drawn into the patch micropipette by applying negative pressure when the pipette tip was placed ∼2 µm from the neurons and used as a control sample; the metabolites noted above were not observed in this sample (SI Appendix, Fig. S7).
Electrophysiological signals from the same single neuron could be obtained before MS measurement (Fig. 2C). However, in this study, the conventional pipette solution was replaced with a NH4HCO3-NH4Cl solution because the presence of high concentrations of inorganic and organic salts, including NaCl, KCl, CsCl, and 4-(2-hydroxyethyl)–1-piperazineethanesulfonic acid (Hepes), in the conventional pipette solution (CPS) induced intensive ion suppression and salt effects on MS detection, which severely decreased the signal intensity of the analytes (SI Appendix, Figs. S8 and S9). To test whether the alternative pipette solution (APS) affected neuronal function during recording, the electrophysiological signals [such as the spontaneous excitatory postsynaptic current (sEPSC) and spontaneous inhibitory postsynaptic current (sIPSC) signals, which reflect neuronal physiological activity] were recorded and compared between the CPS and APS. The results revealed no significant differences in the frequency or amplitude of the sEPSC and sIPSC signals between the two pipette solutions (Fig. 2C and SI Appendix, Fig. S10).
Concentrations of Cytoplasmic Chemical Constituents in Single Neurons from Different Brain Areas and Subtypes.
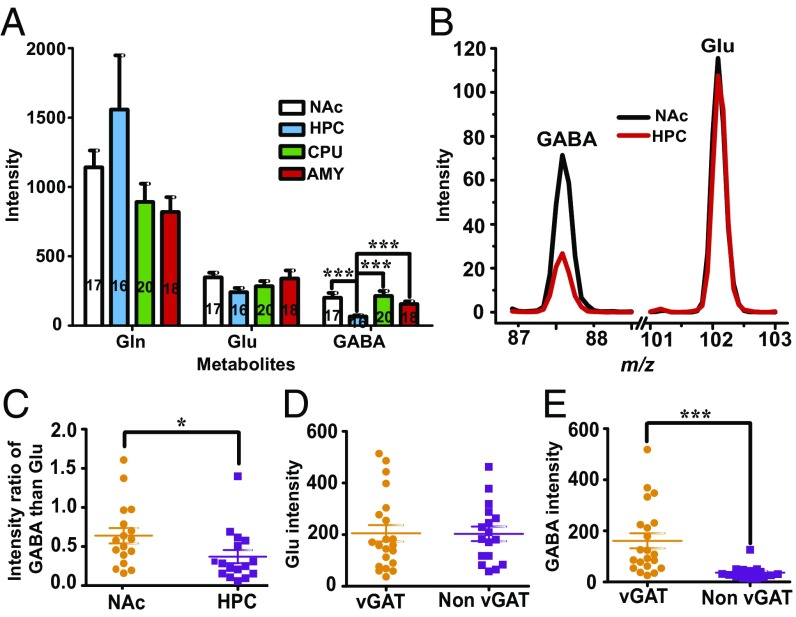
The amino acid Gln and its two metabolites, Glu and GABA, were measured in four brain areas: the nucleus accumbens (NAc), hippocampus (HPC), dorsal striatum (CPU), and the amygdala (AMY) (Fig. 3A and SI Appendix, Fig. S11). No differences in the Gln and Glu concentration in a single neuron were observed in these brain areas. However, the intensity of the GABA signal and the GABA/Glu ratio were significantly lower in the HPC than in the NAc or CPU (Fig. 3 A–C), consistent with the traditional view that most neurons are glutamatergic in the HPC but GABAergic in the NAc and CPU. Surprisingly, the Glu intensities were similar in these brain areas. The single-cell neuronal level of Glu in the HPC was even slightly lower than that in the NAc (Fig. 3A), suggesting that the Glu concentration may be the same in excitatory and inhibitory neurons. Consistent with this idea, in the HPC of transgenic vesicular GABA transporter-green fluorescent protein (vGAT-GFP) mice, both the vGAT-expressing (GABAergic) and non–vGAT-expressing (non-GABAergic) neurons have the same single-neuronal level of Glu after false discovery rate correction. (Fig. 3 D and E and SI Appendix, Fig. S12).
Fig. 3.
Intracellular levels of Gln, Glu, and GABA measured by single-cell MS in various brain areas and neuron subtypes. (A) The intracellular intensities of Gln, Glu, and GABA in the NAc, HPC, CPU, and AMY. (B and C, three mice) The data show a significantly lower level of GABA (B) and ratio of GABA/Glu (C) in the HPC (16 neurons from three mice) than in the NAc (17 neurons from three mice). (D and E) Comparison of the single-cell levels of Glu (D) and GABA (E) in vGAT-expressing (21 neurons from three mice) and non–vGAT-expressing neurons (21 neurons from three mice) in the HPC (P < 0.05 correlated by false discovery rate). *** denotes P < 0.001, * denotes P < 0.05.
Changes in the Levels of Cytoplasmic Chemical Constituents at Different Stages of Brain Development.
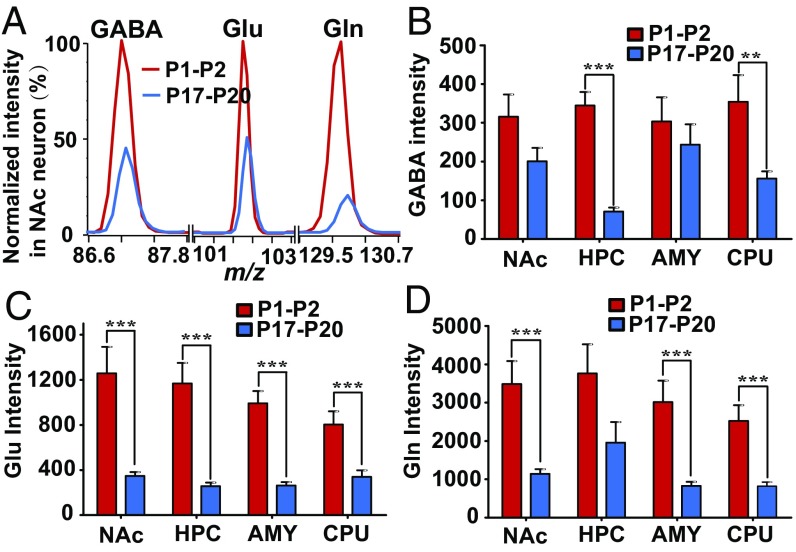
Previous reports suggest that the levels of amino acid neurotransmitters tend to decrease with age (36, 37). Using single-cell MS, we investigated this idea at the single-neuron level. We found that the concentrations of both Gln and Glu significantly decreased from P1-P2 to P17-P20 in almost all brain areas (Fig. 4 A, C, and D and SI Appendix, Fig. S13). However, the single-neuronal levels of GABA decreased significantly in the HPC and CPU but only slightly in the NAc and AMY from P1-P2 to P17-P20 at a false discovery rate (FDR) of q < 0.05. (Fig. 4B and SI Appendix, Fig. S13).
Fig. 4.
Changes in the levels of intracellular chemical with age. (A) Intensities of characteristic fragment ions for GABA, Glu, and Gln in NAc neuron at P1-P2 (red line) and P17-P20 (blue line); all data were normalized to P1-P2 values. Single-cell levels of GABA (B), Glu (C), and Gln (D) at P1-P2 (red bars) and P17-P20 (blue bars) in various brain areas: the NAc (17 neurons at P1-P2 and 17 neurons at P17-P20 from three mice), HPC (17 neurons at P1-P2 and 16 neurons at P17-P20 from three mice), AMY (17 neurons at P1-P2 and 20 neurons at P17-P20 from three mice) and CPU (15 neurons at P1-P2 and 16 neurons at P17-P20 from three mice) (P < 0.05 correlated by false discovery rate). *** denotes P < 0.001, ** denotes P < 0.005.
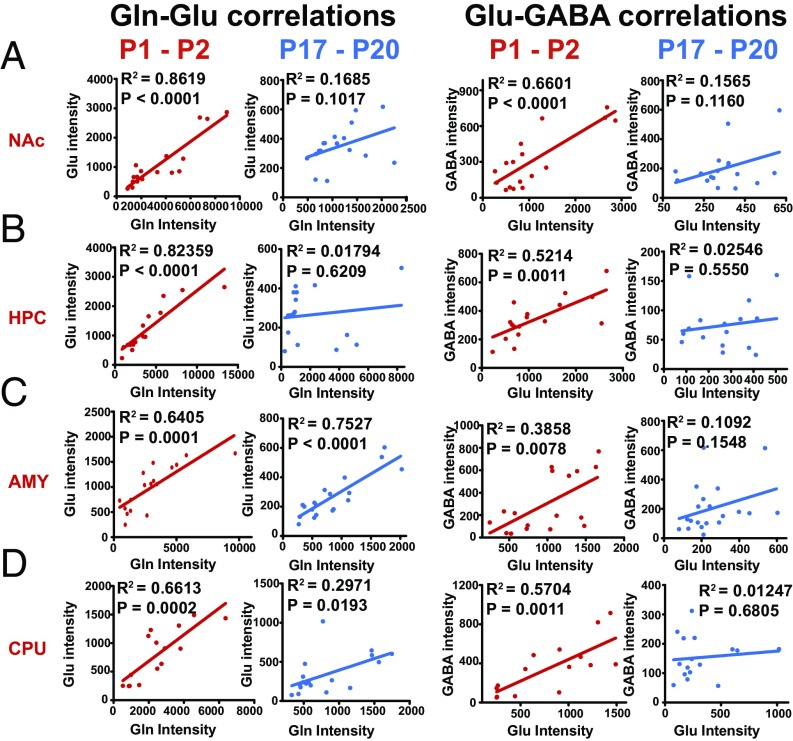
The Gln-Glu-GABA cycling pathway has been thoroughly studied in brain tissues (38, 39). Using single-cell mass spectrometry, we determined the relationship between the single-neuronal levels of Gln and Glu or Glu and GABA in various brain areas (the NAc, HPC, AMY, and CPU) in mice of different ages. Our results indicated that the single-neuronal levels of Gln and Glu or Glu and GABA were well correlated in all four brain areas in newborn mice (P1-P2) but not in child/adolescent mice (P17-P20) (Fig. 5 and SI Appendix, Figs. S16 and S17). Our results suggest that the Gln-Glu-GABA pathway may be the predominant metabolic pathway in Glu and GABA biosynthesis during the early stage but not the late stage of brain development.
Fig. 5.
The relationship between the single-neuronal levels of Gln and Glu (Left eight panels) or Glu and GABA (Right eight panels) at P1-P2 (red dots) and P17-P20 (blue dots) in various brain areas—the NAc (A, 17 neurons at P1-P2 and 17 neurons at P17-P20 from three mice), HPC (B, 17 neurons at P1-P2 and 16 neurons at P17-P20 from three mice), AMY (C, 17 neurons at P1-P2 and 20 neurons at P17-P20 from three mice), and CPU (D, 15 neurons at P1-P2 and 16 neurons at P17-P20 from three mice)—in mice at different ages.
Analysis of Chemical Metabolism in Single Neurons via 13C-Labeling.
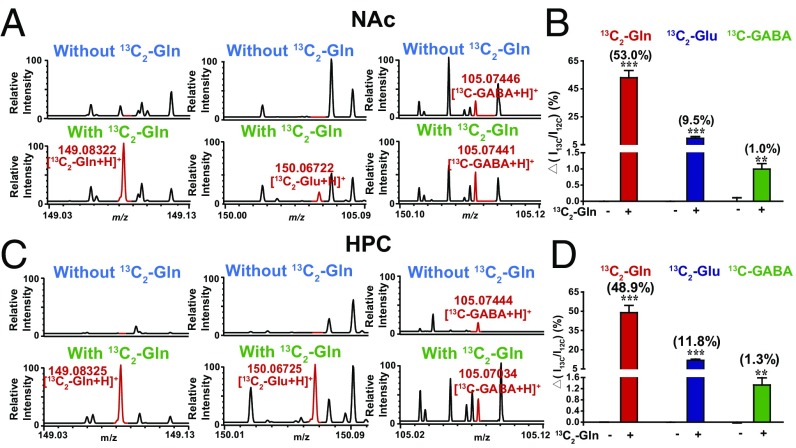
Labeling with 13C-isotope is a powerful tool to study in vivo chemical metabolism. For instance, it could be used to identify the Gln-Glu-GABA metabolic pathway at the single-neuron level. Brain slices were incubated with 1, 2-13C2–labeled Gln (0.4 mM) in aCSF for 2 h. Then, the neurons were patched, and the intracellular solution was extracted and analyzed by InESI/MS. The results revealed a significant increase in the 1, 2-13C2-Glu/Glu ratio and the 13C-GABA/GABA ratio in neurons from NAc and HPC (false discovery rate, q < 0.05) (Fig. 6 and SI Appendix, Figs. S18 and S19), indicating the conversion of Gln to Glu and GABA in these neurons.
Fig. 6.
Identification of the Gln metabolic pathway by 13C-labeling in single neurons. MS spectra of 13C2-Gln, 13C2-Glu, and 13C-GABA in single neurons from the NAc (A) and HPC (C) with or without preincubation of 13C2-Gln. The data shown reflect the percent increases in the 13C2-Gln/Gln, 13C2-Glu/Glu, and 13C-GABA/GABA ratios in single neurons from the NAc (B, 24 neurons without preincubation of 13C2-Gln from three mice and 20 neurons with preincubation of 13C2-Gln from three mice) and HPC (D, 22 neurons without preincubation of 13C2-Gln from three mice and 20 neurons with preincubation of 13C2-Gln from three mice) with or without 2-h incubation of 13C2-Gln (0.4 mM) (P < 0.05 correlated by false discovery rate). *** denotes P < 0.001, ** denotes P < 0.005.
Discussion
Unless some key problems associated with ESI MS are solved, rapid and in situ MS interpretation directly from the cytoplasm would remain difficult, and its impact in the biological sciences, especially in neuroscience, remains limited. Specifically, three major obstacles affect in situ chemical interpretation using ESI MS: concentrated electrolytes (raw cell cytoplasm), the flow rate (small volume samples), and the salt tolerance (complexity of the cell cytoplasm). MS response of cytoplasm obtained by conventional nanoESI technique decreases within ∼1 min (Fig. 1), with unpredictable run-to-run variations. In contrast, the response obtained using InESI MS remained stable for over 20 mins, which was essential for distinguishing differences between cells. Furthermore, this relatively long period of stability would facilitate conducting massive tandem MS identifications in single cells. Thus, in situ MS measurements in the cell cytoplasm could be achieved without any sample pretreatment for either pure solvent (SI Appendix, Fig. S20A) or cytoplasm of a single onion cell (SI Appendix, Fig. S20B).
“Real-time” chemical profiling of single neurons requires rapid sampling and identification. In addition to the MS assay (the identification technique), the sampling technique should also be rapid and accurate. Thus, we applied patch clamp technology, which is typically used to determine physiological activity, at a relatively high sampling rate in single neurons. In addition, information about the cellular status is also critical to avoid artifacts. For the same cell, we accomplished the successful collection of both electrophysiological signals and chemical profiling in “real time” (within ∼300 s). We also found that neurons with abnormal electrophysiological signals could result in atypical chemical profiling, most likely because an unknown biological pathway is active in unhealthy cells.
Recent advances in single-cell technologies, such as transcriptomic sequencing, have produced new insights for the discovery and classification of cell types in the brain (1, 40, 41). However, these neuron phenotypes were classified according to their RNA sequences and related animal behaviors. The present study suggests that the integration of MS with single-cell fluidics may extend the number of parameters that can be measured and used to classify cell types. An example is the diverse intracellular chemical composition found in different brain areas (Fig. 3). Thus, the technology described here may represent a means of classifying different cell types by identifying the chemical compositions and concentrations in single neurons. In this study, we found abundant molecules in individual neurons, but only a few of them (>50) have been identified to date. However, as MS technology continues to develop, increasing numbers of chemicals in cells will be identified, and precise neuron classification will be achievable using our single-cell MS technology.
Of all of the chemicals identified in neurons, Glu and GABA are usually two of the most important excitatory and inhibitory neurotransmitters. Recently, a growing number of studies using a combination of electrophysiological, immunohistochemical, and optogenetic techniques have shown that Glu and GABA may coexist and be coreleased from a single-neuron terminal (42–44). Our findings provided supporting evidence of the coexistence of Glu and GABA in the soma of all neuron types (both excitatory and inhibitory). Certainly, the most direct proof should be the coexistence of Glu and GABA in a single vesicle of the neuron terminals, where the neurotransmitters actually release. However, it needs innovation of patch clamp methods or introduction of other new technologies to directly sample from single vesicles in the neuron terminals.
In addition to the coexistence of Glu and GABA, consistent with previous studies reporting age-dependent changes in multiple neurotransmitter systems in brain tissue (36, 37), we also found that the concentrations of amino acids neurotransmitters, including Glu and GABA, in single cells decrease with age. Interestingly, we observed that the correlation among single-cellular Gln, Glu, and GABA is much more significant in infants than that in children or adolescents. These results may suggest a possibility that the Gln-Glu-GABA pathway may be the predominant biosynthetic pathway for Glu and GABA in the brains of newborn mice, but this situation may change during brain development. However, it should be noted that the concentrations of Glu and GABA, as neurotransmitters, are determined not only by their biosynthesis but also by their terminal release and uptake (45). The glial cells and the process of myelination may also change with age and influence the intracellular levels of transmitters (46, 47). Therefore, further study of relationship between age and the Gln-Glu-GABA biosynthetic pathway is needed.
We then investigated the Glu/GABA conversion pathway in neurons in greater depth. The present method allows us to study the metabolic pathway at the single-cell level using 13C-labeling, which is a conventional method in metabolomics (39, 48, 49). However, the 13C-labeling approach has generally been used for tissue analysis (for a large number of cells). In this study, we introduced 13C-labeling for studying single-neuron metabolism. To this end, we added 13C-labeled Gln to the extracellular solution and eventually observed 13C-labeled Glu and GABA in a single neuron. Our experiments revealed that 13C-labeling can be a powerful tool for validating a known metabolic pathway or identifying metabolites at the single-cell level. Further developing this technology and identifying more intracellular chemicals will eventually allow for the study of single-cell metabolomics.
In summary, a modified patch clamp setup was combined with InESI/MS. The modified patch clamp setup allowed monitoring of the neuronal physiological activity, which indicated the viability of the target cell for subsequent MS analysis. InESI facilitates direct MS analysis without sample pretreatment, thereby greatly helping the throughput. The initial results obtained in this study indicate that this technique is suitable for rapid and accurate chemical profiling of the cytoplasmic constituents of a single neuron and its biological activity. Thus, this method can be used to study neurochemistry, such as localization, alteration, and metabolism, in single living cells rather than in cell lysates. Further developing the patch clamp assay and MS measurement will eventually allow our single-cell technology to be established as a useful tool in neuroscience research, for example, for neuron classification, single-cell metabolome identification, and biomarker screening for brain disorders. Furthermore, it might potentially be beneficial for life science studies as a whole, including cell biology, pharmacology, pathology, and toxicology, at the single-cell level.
Materials and Methods
Animals.
All animal experiments were approved by the Institutional Animal Care and Use Committee of the University of Science and Technology of China (USTC). All mice were group-housed in groups of 2 to 5 with a 12-h light/dark cycle (lights off at 7 PM) and food and water available ad libitum. Male C57BL6/J mice were used for both the sampling and recording experiments. In contrast, male vGAT-ChR2-EYFP line 8 mice (P17 to P20) were used for the sampling experiments only. The C57Bl6/J mice were purchased from the Slack Laboratory Animal LLC. vGAT-ChR2-EYFP line 8 (vGAT-ChR2) mice with the C57Bl6/J genetic background were obtained from Zhi Zhang of University of Science and Technology of China. Genotyping of the VGAT-ChR2-EYFP line 8 mice was performed using the following primers: forward: 5′ACCCTTCTGTCCTTTTCTCC-3′; reverse: 5′-GCAAGGTAGAGCATAGAGGG-3′. All experimental mice were produced from breeding pairs of C57Bl6/J or vGAT-ChR2 mice. Experimenters were blinded to all experimental conditions until all data were collected. Unless otherwise specified, the mice were randomly assigned to experimental groups.
Chemicals.
All pharmacological agents were purchased from Sigma-Aldrich. A paclitaxel (PTX) stock solution (100 mM) was prepared in dimethyl sulfoxide (DMSO) and diluted in aCSF to a final concentration of 100 µM before use. The PTX stock was kept at ‒20 °C. Kynurenic acid [a solid at room temperature (RT)] was directly added to the aCSF and diluted by ultrasonic treatment. A solution of 1,2-13C2-Gln was diluted in aCSF before use. Slices were incubated with drugs in either the recording chambers or the maintenance chambers, as necessary.
Brain Slice Preparation.
The HPC, AMY, CPU, NAc, and PFC slices were prepared from C57BL6/J mice aged P1-P2 and P17-P20. HPC slices were prepared from VGAT-ChR2 mice aged P17-P20. The mice were killed by decapitation, and the brains were quickly removed and immersed in ice-cold preoxygenated cutting solution containing 194 mM sucrose, 30 mM NaCl, 26 mM NaHCO3, 1 mM MgCl2, 10 mM glucose, 4.5 mM KCl, and 1.2 mM NaH2PO4, pH 7.4. Coronal sections (300 μm) were cut using a Vibratome (VT 1200S). The tissue slices were incubated in a holding chamber containing oxygenated (95% O2 and 5% CO2) aCSF, which consisted of 124 mM NaCl, 26 mM NaHCO3, 4.5 mM KCl, 1.2 M NaH2PO4, 1 mM MgCl2, 2 mM CaCl2, and 10 mM glucose (final pH, 7.4; osmolality, 315 mOsm kg−1). The slices were incubated at 32 °C for ∼30 min and then cooled to RT (21–23 °C) at least 30 min before the experiment.
Electrophysiological Recording.
Neurons were visualized using a fixed-stage microscope (BX50WI, OLYMPUS) with differential interference contrast and infrared illumination. Neurons that were clearly visualized and had a smooth surface and good refractivity were chosen for the assay. We obtained all whole-cell patch clamp recordings at 28 °C using borosilicate glass pipettes (long taper: 7–9 mm; opening: 1–2 µm) filled with internal solutions. The micropipette tips were custom-made using filamented borosilicate glass capillaries (BF150-86–10, Sutter Instruments). sEPSCs and sIPSCs were recorded as described previously (50, 51), with detailed settings described in SI Appendix, Fig. S1.
Single-Neuron Sampling Procedure.
After the slices recovered from mechanical injury or drug incubation, they were transferred to the recording chambers. We chose neurons as previously described for electrophysiology recording. When sampling HPC slices of vGAT-ChR2 mice, fluorescent cells were visualized with blue light excited by a mercury lamp (U-RFL-T, OLYMPUS). We designated the HPC fluorescent cells as “vGAT” and other HPC neurons with no fluorescence as “non-vGAT.” We approached the neuron with a micromanipulator (MP-285, Sutter) and patched the neuronal cell body with borosilicate glass pipettes filled with APS by applying negative pressure. After the patched membrane was broken by rapidly applied negative pressure, the neurons were clamped at ‒70 mV as above using glass pipettes filled with APS. After the electrophysiological data were recorded, the cytoplasmic chemical constituents were obtained from the assayed neuron by applying negative pressure. Only neurons with tightly held seals (>1 GΩ) and nonruptured membranes were collected for analysis, ensuring that the intracellular fluid was not diluted by aCSF. Once a sufficient amount of material was withdrawn from the cell, the patch pipette was quickly removed from the slice and then assayed via MS.
Experiments Using 13C2-Gln.
To trace Gln metabolism, 1,2-13C2-Gln was added to another maintenance chamber to a final concentration of 0.4 mM. After the slices recovered from mechanical injury, they were transferred to 1,2-13C2-Gln in aCSF and incubated for 2 h. Drug-treated slices were also recorded without 1,2-13C2-Gln in subsequent sampling experiments. Other experimental procedures were the same as for single-neuron sampling.
Instrument for InESI and MS.
After extraction of the cell cytoplasmic chemical constituents, the capillary was coupled to the InESI device, which had been fabricated as previously described (32) (SI Appendix, Fig. S1). An AC voltage with an amplitude of 3 kV at ∼200 Hz was applied outside of the spray capillary micropipette. The tip of the micropipette was placed ∼5 mm away from the orifice of the MS instrument. Mass measurements were collected using an Orbitrap MS and a LTQ Velos Pro. MS instruments (Q-Exactive, Thermo Scientific). All measurements (InESI and conventional nanoESI) used borosilicate glass pipettes (long taper: 7–9 mm; opening: 1–2 µm).
Electrophysiology Data Analysis.
The electrophysiology data were acquired using an Axon MultiClamp 700B amplifier (Molecular Devices, AXON), filtered at 3 kHz and digitized at 10 kHz using an Axon Digidata 1500A digitizer combined with pCLAMP 10.4 software. Data detection was accomplished using MiniAnalysis software.
MS Data Analysis.
Metabolite identification was based on accurate mass measurements, isotopic profiles, metabolic databases (Plant Metabolic Network database: plantcyc.org/, Human Metabolome Database: www.hmdb.ca/), and matching tandem mass spectral data from endogenous substances with those of chemical standards when available. To exclude data with faulty assignments, the masses of the signals (SI Appendix, Tables S1 and S2) must be present in the spectrum with a mass deviation lower than 15 ppm. In addition to these two methods, chemical assignments were also verified by comparing the MS/MS spectra to those obtained from commercially available standards (MS/MS results of some typical metabolites in neurons are shown in SI Appendix, Fig. S6).
Calibration with External Standards and Reproducibility of the Method.
Estimations of Glu, Gln, and GABA levels in neurons were calculated from calibration curves with external standards as shown in SI Appendix, Fig. S14. All above calibrations were obtained with artificial intracellular solution: potassium gluconate 130 mM, NaCl 6 mM, EGTA 11 mM, Hepes 10 mM, CaCl2 1 mM, NaOH 4 mM, MgCl2 1 mM, Mg-ATP 2 mM, and Na-GTP 0.2 mM (pH was adjusted to 7.3; osmolality was adjusted to 296 mOsm kg−1 with sucrose). The mean levels in NAc neurons of P17-P20 mice were found to be 28.9 μM for GABA, 1.96 mM for Glu, and 92.7 μM for Gln. Nine repeated measurement in continuous 3 days were conducted to illustrate the reproducibility of the present method for GABA, Glu, and Gln, with interday RSDs ranged from 1.10 to 10.92% and intraday RSDs from 2.68 to 4.82%. Discussion about quantitation using internal standards was also included in SI Appendix, Fig. S15.
Statistics.
In all experiments, the cells were evenly suspended and then randomly distributed in each tested well. Average values are expressed as the mean ± SEM. The data distribution was assumed to be normal, but this assumption was not formally tested. Data were statistically compared using an unpaired t test in GraphPad Prism 5.0, as indicated in the specific figure legends. P < 0.05 was considered significant. To account for multiple comparisons, P values were adjusted using the Hochberg and Benjamini false discovery rate (FDR) at q = 0.05 to distinguish statistically significant metabolites (52). The data were collected and processed randomly.
Supplementary Material
Acknowledgments
We thank Z. Zhang (School of Life Sciences, University of Science and Technology of China) for providing VGAT-ChR2-EYFP mice and Y. Pan (National Synchrotron Radiation Laboratory, University of Science and Technology of China) for instrumentation supports. We acknowledge support from the National Key Research and Development Program of China 2016YFC1300500-2 (to W.X.) and 2016YFA0201300 (to G.H.), National Natural Science Foundation of China Grants 21475121(to G.H.) and 91432103 and 91649121 (to W.X.), the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant XDB02010000 to W.X.), the National High-Tech R&D Program of China (863 Program; Grant 2014AA020535 to W.X.), the Fundamental Research Funds for the Central Universities (to G.H. and W.X.), the Major/Innovative Program of Development Foundation of Hefei Center for Physical Science and Technology (W.X.), and Recruitment Program of Global Experts (G.H. and W.X.)
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. S.B.N. is a Guest Editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1615557114/-/DCSupplemental.
References
- 1.Frei AP, et al. Highly multiplexed simultaneous detection of RNAs and proteins in single cells. Nat Methods. 2016;13(3):269–275. doi: 10.1038/nmeth.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blasi T, et al. Label-free cell cycle analysis for high-throughput imaging flow cytometry. Nat Commun. 2016;7(1):10256–10264. doi: 10.1038/ncomms10256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forment JV, Jackson SP. A flow cytometry-based method to simplify the analysis and quantification of protein association to chromatin in mammalian cells. Nat Protoc. 2015;10(9):1297–1307. doi: 10.1038/nprot.2015.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanchez-Freire V, Ebert AD, Kalisky T, Quake SR, Wu JC. Microfluidic single-cell real-time PCR for comparative analysis of gene expression patterns. Nat Protoc. 2012;7(5):829–838. doi: 10.1038/nprot.2012.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dalerba P, et al. Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat Biotechnol. 2011;29(12):1120–1127. doi: 10.1038/nbt.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu AR, et al. Quantitative assessment of single-cell RNA-sequencing methods. Nat Methods. 2014;11(1):41–46. doi: 10.1038/nmeth.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tasic B, et al. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat Neurosci. 2016;19(2):335–346. doi: 10.1038/nn.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gawad C, Koh W, Quake SR. Single-cell genome sequencing: Current state of the science. Nat Rev Genet. 2016;17(3):175–188. doi: 10.1038/nrg.2015.16. [DOI] [PubMed] [Google Scholar]
- 9.Bignami A, Eng LF, Dahl D, Uyeda CT. Localization of the glial fibrillary acidic protein in astrocytes by immunofluorescence. Brain Res. 1972;43(2):429–435. doi: 10.1016/0006-8993(72)90398-8. [DOI] [PubMed] [Google Scholar]
- 10.Lin JR, Fallahi-Sichani M, Sorger PK. Highly multiplexed imaging of single cells using a high-throughput cyclic immunofluorescence method. Nat Commun. 2015;6(5):8390–8396. doi: 10.1038/ncomms9390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chow RH, von Rüden L, Neher E. Delay in vesicle fusion revealed by electrochemical monitoring of single secretory events in adrenal chromaffin cells. Nature. 1992;356(6364):60–63. doi: 10.1038/356060a0. [DOI] [PubMed] [Google Scholar]
- 12.Morris R, Fagan-Murphy A, MacEachern SJ, Covill D, Patel BA. Electrochemical fecal pellet sensor for simultaneous real-time ex vivo detection of colonic serotonin signalling and motility. Sci Rep. 2016;6:23442–23450. doi: 10.1038/srep23442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu Z. Physical connections between different SSVEP neural networks. Sci Rep. 2016;6:22801–22809. doi: 10.1038/srep22801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li L, Gervasi N, Girault JA. Dendritic geometry shapes neuronal cAMP signalling to the nucleus. Nat Commun. 2015;6(1):6319–6333. doi: 10.1038/ncomms7319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ko H, et al. Functional specificity of local synaptic connections in neocortical networks. Nature. 2011;473(7345):87–91. doi: 10.1038/nature09880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cadwell CR, et al. Electrophysiological, transcriptomic and morphologic profiling of single neurons using Patch-seq. Nat Biotechnol. 2016;34(2):199–203. doi: 10.1038/nbt.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bean BP. The action potential in mammalian central neurons. Nat Rev Neurosci. 2007;8(6):451–465. doi: 10.1038/nrn2148. [DOI] [PubMed] [Google Scholar]
- 18.Ma C, et al. A clinical microchip for evaluation of single immune cells reveals high functional heterogeneity in phenotypically similar T cells. Nat Med. 2011;17(6):738–743. doi: 10.1038/nm.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altschuler SJ, Wu LF. Cellular heterogeneity: Do differences make a difference? Cell. 2010;141(4):559–563. doi: 10.1016/j.cell.2010.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Romanova EV, Aerts JT, Croushore CA, Sweedler JV. Small-volume analysis of cell-cell signaling molecules in the brain. Neuropsychopharmacology. 2014;39(1):50–64. doi: 10.1038/npp.2013.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaurand P, et al. Profiling and imaging proteins in the mouse epididymis by imaging mass spectrometry. Proteomics. 2003;3(11):2221–2239. doi: 10.1002/pmic.200300474. [DOI] [PubMed] [Google Scholar]
- 22.Wilm M, et al. Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature. 1996;379(6564):466–469. doi: 10.1038/379466a0. [DOI] [PubMed] [Google Scholar]
- 23.Rubakhin SS, Romanova EV, Nemes P, Sweedler JV. Profiling metabolites and peptides in single cells. Nat Methods. 2011;8(4) Suppl:S20–S29. doi: 10.1038/nmeth.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rubakhin SS, Garden RW, Fuller RR, Sweedler JV. Measuring the peptides in individual organelles with mass spectrometry. Nat Biotechnol. 2000;18(2):172–175. doi: 10.1038/72622. [DOI] [PubMed] [Google Scholar]
- 25.Rubakhin SS, Sweedler JV. Characterizing peptides in individual mammalian cells using mass spectrometry. Nat Protoc. 2007;2(8):1987–1997. doi: 10.1038/nprot.2007.277. [DOI] [PubMed] [Google Scholar]
- 26.Van Veelen PA, et al. Direct peptide profiling of single neurons by matrix-assisted laser spectrometry desorption-ionization mass. Org Mass Spectrom. 1993;28(12):1542–1546. [Google Scholar]
- 27.Lorenzo Tejedor M, Mizuno H, Tsuyama N, Harada T, Masujima T. In situ molecular analysis of plant tissues by live single-cell mass spectrometry. Anal Chem. 2012;84(12):5221–5228. doi: 10.1021/ac202447t. [DOI] [PubMed] [Google Scholar]
- 28.Date S, Mizuno H, Tsuyama N, Harada T, Masujima T. Direct drug metabolism monitoring in a live single hepatic cell by video mass spectrometry. Anal Sci. 2012;28(3):201–203. doi: 10.2116/analsci.28.201. [DOI] [PubMed] [Google Scholar]
- 29.Masujima T. Live single-cell mass spectrometry. Anal Sci. 2009;25(8):953–960. doi: 10.2116/analsci.25.953. [DOI] [PubMed] [Google Scholar]
- 30.Aerts JT, et al. Patch clamp electrophysiology and capillary electrophoresis-mass spectrometry metabolomics for single cell characterization. Anal Chem. 2014;86(6):3203–3208. doi: 10.1021/ac500168d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zenobi R. Single-cell metabolomics: Analytical and biological perspectives. Science. 2013;342(6163):1243259. doi: 10.1126/science.1243259. [DOI] [PubMed] [Google Scholar]
- 32.Huang G, Li G, Cooks RG. Induced nanoelectrospray ionization for matrix-tolerant and high-throughput mass spectrometry. Angew Chem Int Ed Engl. 2011;50(42):9907–9910. doi: 10.1002/anie.201103687. [DOI] [PubMed] [Google Scholar]
- 33.Li G, Yuan S, Pan Y, Liu Y, Huang G. Binding states of protein-metal complexes in cells. Anal Chem. 2016;88(22):10860–10866. doi: 10.1021/acs.analchem.6b00032. [DOI] [PubMed] [Google Scholar]
- 34.Stolee JA, Shrestha B, Mengistu G, Vertes A. Observation of subcellular metabolite gradients in single cells by laser ablation electrospray ionization mass spectrometry. Angew Chem Int Ed Engl. 2012;51(41):10386–10389. doi: 10.1002/anie.201205436. [DOI] [PubMed] [Google Scholar]
- 35.Shrestha B, Vertes A. In situ metabolic profiling of single cells by laser ablation electrospray ionization mass spectrometry. Anal Chem. 2009;81(20):8265–8271. doi: 10.1021/ac901525g. [DOI] [PubMed] [Google Scholar]
- 36.Miranda-Contreras L, Benítez-Diaz PR, Mendoza-Briceño RV, Delgado-Saez MC, Palacios-Prü EL. Levels of amino acid neurotransmitters during mouse cerebellar neurogenesis and in histotypic cerebellar cultures. Dev Neurosci. 1999;21(2):147–158. doi: 10.1159/000017377. [DOI] [PubMed] [Google Scholar]
- 37.Benítez-Diaz P, Miranda-Contreras L, Mendoza-Briceño RV, Peña-Contreras Z, Palacios-Prü E. Prenatal and postnatal contents of amino acid neurotransmitters in mouse parietal cortex. Dev Neurosci. 2003;25(5):366–374. doi: 10.1159/000073514. [DOI] [PubMed] [Google Scholar]
- 38.Patel AB, et al. The contribution of GABA to glutamate/glutamine cycling and energy metabolism in the rat cortex in vivo. Proc Natl Acad Sci USA. 2005;102(15):5588–5593. doi: 10.1073/pnas.0501703102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Albrecht J, Sidoryk-Węgrzynowicz M, Zielińska M, Aschner M. Roles of glutamine in neurotransmission. Neuron Glia Biol. 2010;6(4):263–276. doi: 10.1017/S1740925X11000093. [DOI] [PubMed] [Google Scholar]
- 40.Xu C, Su Z. Identification of cell types from single-cell transcriptomes using a novel clustering method. Bioinformatics. 2015;31(12):1974–1980. doi: 10.1093/bioinformatics/btv088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Usoskin D, et al. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat Neurosci. 2015;18(1):145–153. doi: 10.1038/nn.3881. [DOI] [PubMed] [Google Scholar]
- 42.Shabel SJ, Proulx CD, Piriz J, Malinow R. Mood regulation. GABA/glutamate co-release controls habenula output and is modified by antidepressant treatment. Science. 2014;345(6203):1494–1498. doi: 10.1126/science.1250469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beltrán JQ, Gutiérrez R. Co-release of glutamate and GABA from single, identified mossy fibre giant boutons. J Physiol. 2012;590(19):4789–4800. doi: 10.1113/jphysiol.2012.236372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Noh J, Seal RP, Garver JA, Edwards RH, Kandler K. Glutamate co-release at GABA/glycinergic synapses is crucial for the refinement of an inhibitory map. Nat Neurosci. 2010;13(2):232–238. doi: 10.1038/nn.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Niciu MJ, Kelmendi B, Sanacora G. Overview of glutamatergic neurotransmission in the nervous system. Pharmacol Biochem Behav. 2012;100(4):656–664. doi: 10.1016/j.pbb.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Araque A, Navarrete M. Glial cells in neuronal network function. Philos Trans R Soc Lond B Biol Sci. 2010;365(1551):2375–2381. doi: 10.1098/rstb.2009.0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hamilton NB, Attwell D. Do astrocytes really exocytose neurotransmitters? Nat Rev Neurosci. 2010;11(4):227–238. doi: 10.1038/nrn2803. [DOI] [PubMed] [Google Scholar]
- 48.Metallo CM, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2011;481(7381):380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Son J, et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496(7443):101–105. doi: 10.1038/nature12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cho CH, St-Gelais F, Zhang W, Tomita S, Howe JR. Two families of TARP isoforms that have distinct effects on the kinetic properties of AMPA receptors and synaptic currents. Neuron. 2007;55(6):890–904. doi: 10.1016/j.neuron.2007.08.024. [DOI] [PubMed] [Google Scholar]
- 51.Xiong W, et al. Presynaptic glycine receptors as a potential therapeutic target for hyperekplexia disease. Nat Neurosci. 2014;17(2):232–239. doi: 10.1038/nn.3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J Roy Statist Soc Ser B (Methodological) 1995;57(1):289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.