Significance
The western honey bee, Apis mellifera, provides essential crop pollination services, but for 10 years, US beekeepers have experienced substantial colony losses. Although insecticides have been implicated in these losses, triazole fungicides affect bees by inhibiting cytochrome P450 monooxygenases that detoxify insecticides. These enzymes also detoxify phytochemicals, including the flavonol quercetin, in their nectar- and pollen-based diet. RNA-Seq analysis of bee larvae consuming quercetin revealed that it down-regulates multiple mitochondrion-related nuclear genes involved in energy production. Bees consuming quercetin together with the triazole myclobutanil produce less thoracic ATP and thus less energy for flight muscles. Therefore, agricultural use of triazole fungicides in combination with insecticides can potentially harm bees by compromising their capacity to extract sufficient energy from their natural diet.
Keywords: Apis mellifera, cytochrome P450, flavonol, mitochondria, myclobutanil
Abstract
Cytochrome P450 monooxygenases (P450) in the honey bee, Apis mellifera, detoxify phytochemicals in honey and pollen. The flavonol quercetin is found ubiquitously and abundantly in pollen and frequently at lower concentrations in honey. Worker jelly consumed during the first 3 d of larval development typically contains flavonols at very low levels, however. RNA-Seq analysis of gene expression in neonates reared for three days on diets with and without quercetin revealed that, in addition to up-regulating multiple detoxifying P450 genes, quercetin is a negative transcriptional regulator of mitochondrion-related nuclear genes and genes encoding subunits of complexes I, III, IV, and V in the oxidative phosphorylation pathway. Thus, a consequence of inefficient metabolism of this phytochemical may be compromised energy production. Several P450s metabolize quercetin in adult workers. Docking in silico of 121 pesticide contaminants of American hives into the active pocket of CYP9Q1, a broadly substrate-specific P450 with high quercetin-metabolizing activity, identified six triazole fungicides, all fungal P450 inhibitors, that dock in the catalytic site. In adults fed combinations of quercetin and the triazole myclobutanil, the expression of five of six mitochondrion-related nuclear genes was down-regulated. Midgut metabolism assays verified that adult bees consuming quercetin with myclobutanil metabolized less quercetin and produced less thoracic ATP, the energy source for flight muscles. Although fungicides lack acute toxicity, they may influence bee health by interfering with quercetin detoxification, thereby compromising mitochondrial regeneration and ATP production. Thus, agricultural use of triazole fungicides may put bees at risk of being unable to extract sufficient energy from their natural food.
With 46 cytochrome P450 genes (P450), the honey bee, Apis mellifera, has only one-third to one-half the P450 inventory in most other insect genomes (1) and this detoxification deficit may underlie its susceptibility to interactions among xenobiotics. Honey bees use P450s for detoxification of phytochemicals in honey and pollen (2–4) and mycotoxins (5) in the hive environment, as well as pyrethroid pesticides and in-hive acaricides (4). Owing to their generalized diet, honey bees encounter a broad diversity of pollen and nectar phytochemicals. Moreover, as the world’s premier managed pollinator, honey bees are at risk for encountering a diversity of agrochemicals. Their reduced CYPome suggests that bees may share with humans a dependence on a subset of relatively generalized P450s for most xenobiotic detoxification reactions (6, 7). Just as humans are vulnerable to food–drug interactions, honey bees also may be vulnerable to interactions among ingested dietary and synthetic chemicals.
Potential food–pesticide interactions in bees are significant in the context of massive contamination of hives in North America by agricultural pesticides. Residues of >120 pesticides and metabolites have been detected in pollen and honey, as well as in wax, brood, and adults (8, 9). Additive or synergistic interactions among many of these contaminants have been documented (10), and the adverse effects of such interactions are evidenced by reduced longevity of bees reared in brood comb contaminated with multiple pesticide residues (11). Along with interacting synergistically among themselves, pesticides also may interfere with the ability of bees to metabolize substances in their natural diet, much as certain medications can interfere with xenobiotic metabolism in the human diet (7).
Among the most abundant phytochemicals in the honey bee diet are flavonol aglycones. Quercetin and kaempferol are flavonols that promote pollen germination and tube growth (12, 13) and thus play central roles in regulating plant fertility (14). These ubiquitous pollen constituents are “main flavonols of bee pollen” (15) and together can constitute 2–4% of pollen dry weight (16). In bee-collected pollen, levels have been variously reported to range from 16.22 to 31.76 mg/100 g (0.016–0.032%) and 24.0–529.8 μg/g (0.0024–0.05%) (17, 18). Because quercetin occurs not only in pollen (and hence in beebread), but also in nectar and propolis (2), bees are regularly exposed to high concentrations of quercetin relative to other more idiosyncratically distributed phytochemicals throughout most of their lifespan. At least six of the 28 honey bee P450s in subfamilies CYP6 and CYP9 associated with xenobiotic metabolism (2, 4) metabolize quercetin. This seemingly redundant detoxification capacity suggests that quercetin presents a potential physiological challenge to honey bees.
In many folivorous insects, quercetin has physiological and behavioral effects that include outright toxicity, developmental disruption, and feeding inhibition (19). Direct tests of the effects of quercetin on honey bees are rare, but some studies have suggested that this ubiquitous dietary constituent can interfere with growth and development under certain circumstances. Adult workers, which are normally sterile, initiate ovary development when fed concentrations of quercetin corresponding to high levels in honey (10 mg/100 g of syrup); this response can interfere with caste determination and destabilize the colony (20). Guseman et al. (21) showed that quercetin synergizes the toxicity of the fungicide Pristine (boscalid + pyraclostrobin) and the insecticide acetamiprid. Although the authors attributed the synergism to inhibition of multidrug resistance transporters, an alternative explanation consistent with their findings is that quercetin acts as competitive inhibitor for P450-mediated detoxification of these pesticides (10). Such competitive inhibition by quercetin is well documented in humans (22, 23).
There is one life stage during which honey bees consume only low levels of quercetin and phytochemicals in general: the first 3 d of larval life, during which nurse bees feed all larvae glandular secretions called royal or worker jelly. Nabas et al. (24) measured the total flavonoid content of royal jelly at 1.28 μg/mg (0.128%); flavonols alone occur in pollen at 15-fold higher concentrations (16). During this early stage of development, befitting the low levels of exposure to flavonols, P450-mediated detoxification activity is apparently low, inasmuch as it is not quantifiable by standard assays (personal observation). Accordingly, to determine the potential impacts of quercetin on honey bees, we exposed neonates (the life stage likely most vulnerable to flavonols) to quercetin at ecologically appropriate low levels and conducted RNA-Seq analysis to compare patterns of gene expression and thereby identify the physiological processes most sensitive to disruption by quercetin.
As in older larvae and adults, quercetin consumption in neonates led to the up-regulation of P450 genes associated with detoxification (2, 4, 10). Unexpectedly, however, RNA-Seq analysis revealed that quercetin ingestion affected gene expression in pathways associated with energy production. This finding suggests a potential consequence of interference with quercetin detoxification. Consuming honey and pollen contaminated by pesticides (8, 9, 24), many of which are known P450 inhibitors (e.g., sterol biosynthesis inhibitor fungicides), could conceivably compromise energy production by inhibiting quercetin detoxification. To determine the potential inhibitory capacity of pesticide contaminants of hives on quercetin metabolism, we used in silico docking of 121 pesticides and metabolites into the active pocket of CYP9Q1, a broadly substrate-specific honey bee P450 known to metabolize both quercetin and pesticides individually (4) and thus may be vulnerable to competitive interactions among substrates.
These docking studies identified triazole fungicides as potential competitive inhibitors of P450-mediated detoxification of quercetin. If a fungicide can inhibit P450-mediated metabolism of quercetin, then adults ingesting this flavonol together with the fungicide should display a pattern of gene expression similar to that displayed by 3-d-old larvae exposed to quercetin. To determine whether inhibition of quercetin detoxification by the triazole fungicide myclobutanil results in reduced energy production in adult workers, we examined the effects of ingesting quercetin-myclobutanil combinations on the expression of six mitochondrion-related nuclear genes in adult workers using quantitative RT-PCR (qRT-PCR) (3, 25).
Because qRT-PCR results were consistent with interference with mitochondrial function, we conducted two additional experiments to determine whether ingesting quercetin together with a fungicide interferes with energy production. High rates of electron transfer from mitochondrial respiratory enzyme molecules produced from oxidation of sugars in thoracic muscles (26–28) are essential not only for powering forager flight, but also for enabling colony-level heat production and thus thermoregulation during winter. Therefore, inhibition of energy production in wing muscles by quercetin has the potential to affect both individual and colony performance. Accordingly, we first measured the efficiency of quercetin metabolism by worker bees in the presence and absence of dietary myclobutanil. We then evaluated the potential consequences of accumulated unmetabolized quercetin on energy production by quantifying ATP generation in the thorax of adult workers consuming quercetin along with myclobutanil.
Results
RNA-Seq Analysis of Larvae Consuming Diets With and Without Quercetin.
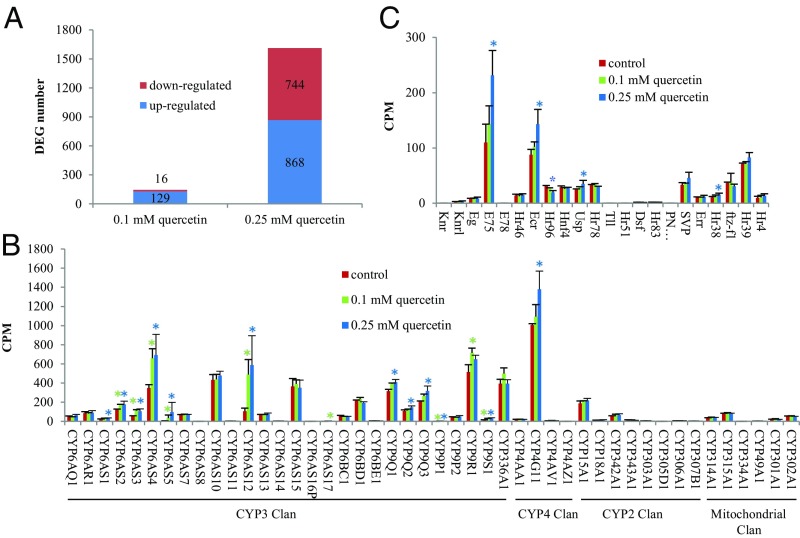
We conducted an RNA-Seq analysis of gene expression in neonate larvae exposed for 3 d to control “bee candy” diets (comprising sucrose and sugar syrup) and diets to which 0.1 mM or 0.25 mM quercetin had been added. Differentially expressed genes (DEGs) between each pair of treatments were identified with edgeR (29). Among the 145 DEGs between control and 0.1 mM quercetin treatment (Dataset S1), 16 were down-regulated (11%) and 129 were up-regulated (89%) (Fig. 1A). However, among the 1,612 DEGs between control and 0.25 mM quercetin treatment (Dataset S2), the numbers of down-regulated and up-regulated genes were nearly equal: 744 (46%) and 868 (54%), respectively (Fig. 1A). Among the 28 P450 genes in the CYP3 clan (to which most detoxifying P450s belong), seven were up-regulated by both quercetin treatments, two were up-regulated by the low-quercetin treatment (CYP6AS17 and CYP9R1), and five (CYP6AS1, CYP9Q1, CYP9Q2, CYP9Q3, and CYP4G11) were up-regulated by the high-quercetin treatment (Fig. 1B). Expression of 17 P450s (including CYP6AS8, CYP6AS16P, CYP4AZ1, CYP305D1, and CYP334A1) was unaffected by quercetin consumption; on diets containing 0.25 mM quercetin, four nuclear receptor genes (Usp, Ecr, E75, and Hr38) were up-regulated and one nuclear receptor gene (Hr96) was down-regulated (Fig. 1C).
Fig. 1.
Ingestion of two concentrations of quercetin (0.1 mM and 0.25 mM) differentially affects gene expression in early instar honey bee larvae. (A) Number of significantly differentially expressed genes in early instar larvae consuming bee candy containing quercetin relative to early instar larvae consuming bee candy alone. (B) Expression levels of cytochrome P450 genes extracted from RNA-Seq data. (C) Expression levels of nuclear receptor genes in RNA-Seq data. Asterisks indicate genes that are differentially expressed between control and 0.1 mM quercetin treatment (green) or 0.25 mM quercetin treatment (blue).
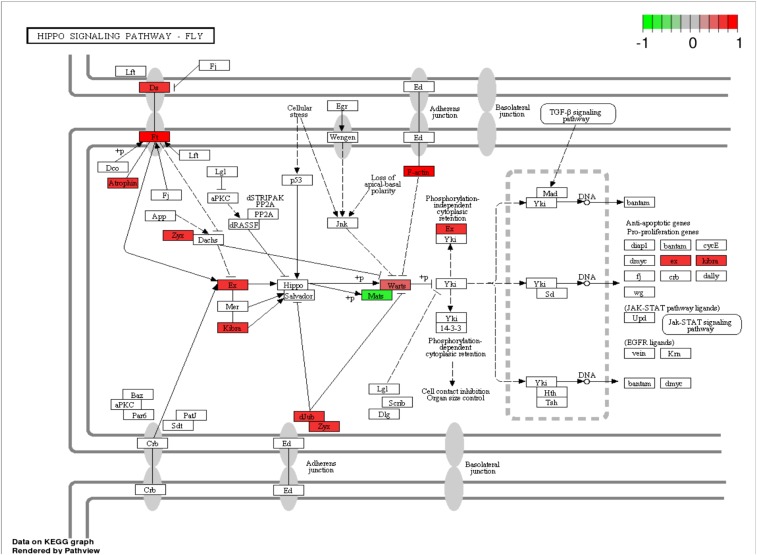
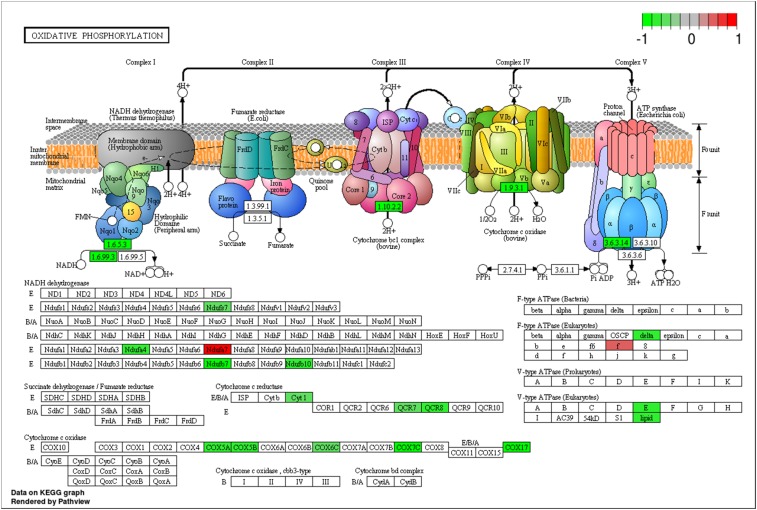
Beyond its effects on xenobiotic-metabolizing P450s, quercetin was a negative transcription regulator of mitochondrion-related nuclear genes. Pathway analyses of the 1,612 DEGs in the 0.25 mM quercetin treatment were carried out against signaling transduction and metabolism pathways extracted from the KEGG pathway database (genome.jp/kegg/pathway.html) with two R packages, Gage (30) and Pathview (31). Two pathways, the Hippo signaling pathway (Fig. S1) and the oxidative phosphorylation pathway (Fig. 2), were overrepresented in the 1,612 DEGs. Among the 11 DEGs in the Hippo signaling pathway, Mats, an activating subunit of Warts kinase, was down-regulated by 1.55-fold, whereas all others were up-regulated by 1.4∼2.0-fold. Although Warts was up-regulated by 1.45-fold, both the down-regulation of Mats and the up-regulation of F-actin and dJub genes likely interfere with Yki phosphorylation by Warts. Moreover, in the oxidative phosphorylation pathway, 15 genes encoding subunits of complex I, complex III, complex IV, and complex V were down-regulated by 1.37∼2.58-fold by quercetin.
Fig. S1.
The Hippo signaling pathway is the only pathway enriched with up-regulated DEGs. The 1,612 DEGs between control and 0.25 mM quercetin treatments were used to identify DEG-enriched pathways from the KEGG pathway database using the two R packages Gage (30) and Pathview (31).
Fig. 2.
The oxidative phosphorylation pathway is the only pathway enriched with down-regulated DEGs following consumption of quercetin. The 1,612 DEGs between control and 0.25 mM quercetin treatment were used to identify DEG-enriched pathways from the KEGG pathway database using the two R packages, Gage (30) and Pathview (31). Differential expression is shown by a color scale ranging from −1 (green, down-regulation) to 1 (red, up-regulation).
In addition to functionally annotating the DEGS of the two quercetin treatments, we performed DAVID functional annotation clustering analysis of the DEGs for each treatment using the FlyBase IDs of their Drosophila melanogaster orthologs. This analysis revealed four enriched clusters among the 208 clusters (Dataset S3). DEGs in cluster 1 are related to larval development, whereas DEGs in cluster 2 and in clusters 3 and 4 are associated with membrane-enclosed lumens, especially mitochondrial and nuclear envelope lumens, and transcription and translation of nuclear and mitochondrial genes, respectively. Among the DEGs in clusters 2–4 are 33 nuclear genes related to mitochondria (Table S1), all of which were down-regulated by 0.25 mM quercetin, including nine genes related to the transport of preproteins and metabolites, 23 genes related to the transcription/translation of mitochondrial genes, and a gene related to mitochondrial ATP synthase biogenesis.
Table S1.
Differentially expressed nuclear-encoded mitochondrial genes in honey bees consuming bee candy with and without 0.25 mM quercetin (identified from the genes in clusters 2–4 of DAVID functional annotation clustering analysis of the 1,612 DEGs between control and 0.25 mM quercetin treatment)
| Group/category | logFC | logCPM | P value | False discovery rate | Gene name | |
| I | GB40810 | −0.69909 | 5.402091 | 0.000115 | 0.001503 | Mitochondrial transmembrane protein 70 (TMM70) |
| II | GB46026 | −0.61376 | 7.521537 | 7.76E-06 | 0.000213 | Translocase of outer membrane 70 (TOM70) |
| GB41686 | −0.83581 | 5.086601 | 1.24E-05 | 0.000304 | Translocase of outer membrane 7 (TOM7) | |
| GB52009 | −0.46277 | 4.993231 | 0.001215 | 0.007815 | Mitochondrial import inner membrane translocase subunit Tim22 | |
| GB51224 | −0.89028 | 5.725054 | 7.53E-07 | 3.72E-05 | Mitochondrial import inner membrane translocase subunit Tim8 | |
| GB41303 | −1.55119 | 4.453304 | 6.86E-08 | 5.60E-06 | Mitochondrial import inner membrane translocase subunit Tim13 | |
| GB48338 | −0.8348 | 4.716947 | 1.15E-06 | 5.19E-05 | Mitochondrial import inner membrane translocase subunit Tim9 | |
| GB41210 | −0.92735 | 3.023617 | 1.41E-05 | 0.000329 | Mitochondrial import inner membrane translocase subunit Tim9B | |
| GB45523 | −0.46461 | 6.193456 | 0.000237 | 0.002509 | Mitochondrial import inner membrane translocase subunit Tim10 | |
| GB51425 | −0.5886 | 5.156958 | 0.000215 | 0.002337 | S-adenosylmethionine mitochondrial carrier protein | |
| III | GB51124 | −0.7261 | 5.72987 | 1.26E-06 | 5.66E-05 | Mitochondrial DNA-directed RNA polymerase |
| GB44671 | −0.60222 | 3.600628 | 0.000483 | 0.004189 | Mitochondrial transcription factor B2 | |
| GB48641 | −0.57184 | 7.087967 | 0.000261 | 0.002698 | Mitochondrial ribosomal protein L9 | |
| GB41068 | −0.47664 | 5.810876 | 0.000448 | 0.003979 | Mitochondrial ribosomal protein L12 | |
| GB45419 | −0.42757 | 5.241461 | 0.001351 | 0.008482 | Mitochondrial ribosomal protein L15 | |
| GB54344 | −0.50983 | 5.53275 | 0.000473 | 0.004153 | Mitochondrial ribosomal protein L20 | |
| GB56016 | −0.55226 | 6.323073 | 0.000456 | 0.004036 | Mitochondrial ribosomal protein L30 | |
| GB46453 | −1.24915 | 1.399884 | 4.26E-05 | 0.000734 | Mitochondrial ribosomal protein L33 | |
| GB42615 | −0.66194 | 4.763463 | 9.05E-05 | 0.001275 | Mitochondrial ribosomal protein L36 | |
| GB45886 | −1.39102 | 0.728716 | 0.001157 | 0.007562 | Mitochondrial ribosomal protein L38 | |
| GB40535 | −1.14895 | 0.855782 | 8.71E-05 | 0.001251 | Mitochondrial ribosomal protein L48 | |
| GB44877 | −0.903 | 6.025927 | 1.79E-05 | 0.0004 | Mitochondrial ribosomal protein L49 | |
| GB50746 | −2.12237 | 1.533952 | 5.90E-08 | 4.90E-06 | Mitochondrial ribosomal protein L52 | |
| GB48201 | −0.74085 | 5.835489 | 0.000245 | 0.002577 | Mitochondrial ribosomal protein L53 | |
| GB46748 | −0.74646 | 6.098186 | 0.000163 | 0.0019 | Mitochondrial ribosomal protein L54 | |
| GB54312 | −1.31916 | 4.571064 | 1.10E-05 | 0.000276 | Mitochondrial ribosomal protein L55 | |
| GB53663 | −0.95587 | 5.444343 | 1.16E-05 | 0.000288 | Mitochondrial GTPase Era | |
| GB50804 | −0.68672 | 5.019691 | 0.00079 | 0.005849 | Mitochondrial ribosomal protein S9 | |
| GB51537 | −0.47101 | 5.079988 | 0.000683 | 0.005287 | Mitochondrial ribosomal protein S11 | |
| GB41359 | −0.85848 | 5.771637 | 0.000336 | 0.003233 | Mitochondrial ribosomal protein S14 | |
| GB48064 | −0.82279 | 6.018325 | 4.10E-05 | 0.000711 | Mitochondrial ribosomal protein S15 | |
| GB43537 | −0.66769 | 5.525932 | 0.000471 | 0.004137 | Mitochondrial ribosomal protein S16 | |
| GB54652 | −0.61878 | 6.022327 | 0.000569 | 0.004696 | Mitochondrial ribosomal protein S18C | |
Docking Hive-Contaminating Pesticides into the Catalytic Site of CYP9Q1.
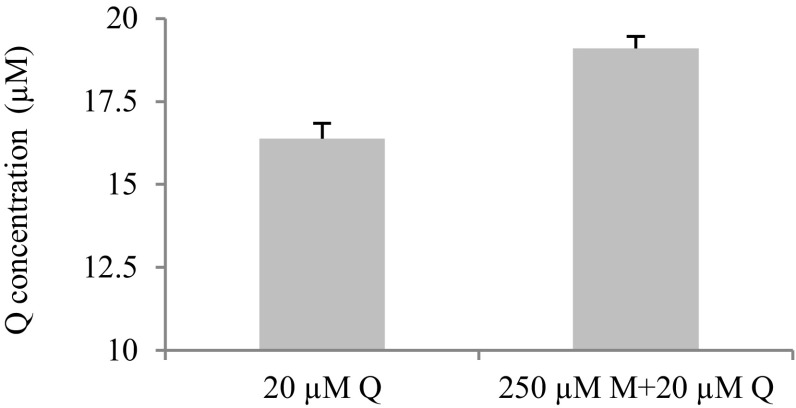
To determine the potential inhibitory capacity of pesticide contaminants of hives, we used in silico docking of 121 pesticides and metabolites (8) into the CYP9Q1 active pocket. Of these 121 pesticides, 68 compounds could be docked into the CYP9Q1 active pocket with a LibDock score >86 (Dataset S4). These included 42 insecticides, of which 16 were pyrethroids and 10 were organophosphates (including the in-hive acaricide coumaphos). Both coumaphos and the pyrethroid in-hive acaricide tau-fluvalinate are metabolized by CYP9Q1 (4). Also docking into the catalytic site were seven herbicides and 18 fungicides, including six conazole fungicides (primarily triazoles) and seven known inhibitors of mitochondrial respiratory chain complex II/III. Previous work has shown that separate pretreatment of adult workers with pyraclostrobin, a mitochondrial inhibitor, and three triazoles (myclobutanil, propiconazole, and fenbuconazole) increases tau-fluvalinate toxicity to adult workers (32), an effect attributed to inhibition of P450-mediated detoxification of this acaricide. In our in vitro P450 assays with intact midguts of adult workers, myclobutanil reduced the amount of quercetin metabolized from 95% to 82%, indicative of a P450-inhibiting effect (Fig. S2).
Fig. S2.
Myclobutanil (M) inhibits quercetin (Q) metabolism in midguts of adult worker bees. Quercetin concentrations in quercetin-myclobutanil treatments were significantly higher than those in treatments containing quercetin alone. n = 3 replicates of 15 individuals, mean ± SE. P < 0.001, two-tailed Student’s t test.
qRT-PCR Analysis to Determine the Effects of Fungicide/Quercetin Ingestion on Mitochondrion-Related Gene Expression.
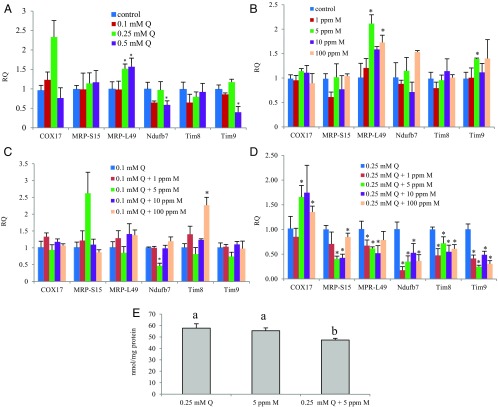
To determine whether the inhibition of quercetin detoxification by myclobutanil (resulting in 13% more unmetabolized quercetin) results in reduced energy production in adult workers, we first examined the effects of ingesting quercetin-myclobutanil combinations on the expression of six mitochondrion-related nuclear genes in adult workers using qRT-PCR (Fig. 3 A–D). Compared with controls, 0.25 and 0.5 mM quercetin up-regulated MRP-L49, whereas 0.5 mM quercetin down-regulated Ndufb7 and Tim9 (Fig. 3A). MRP-L49 was up-regulated by 5 ppm and 100 ppm myclobutanil (Fig. 3B). Tim9 also was more abundant in the 5 ppm myclobutanil treatment. Combinations of 0.1 mM quercetin and myclobutanil in different concentrations had a less dramatic effect on the expression of these genes (Fig. 3C). Whereas Nudfb7 expression was decreased by 0.1 mM quercetin/5 ppm myclobutanil, Tim8 expression was induced by 0.1 mM quercetin/100 ppm myclobutanil. In contrast, expression of all but one of these genes (COX17) was down-regulated by most combinations of 0.25 mM quercetin and myclobutanil (Fig. 3D). COX17 expression was induced by 0.25 mM quercetin/5 ppm myclobutanil and 0.25 mM quercetin/100 ppm myclobutanil, but not by any other combination.
Fig. 3.
Quercetin (Q)-myclobutanil (M) combinations suppress ATP production in adult worker bees. (A–D) Transcript levels of six mitochondrion-related nuclear genes in workers measured by qRT-PCR. Asterisks indicate significant differences compared with controls. n = 3, average ± SD. P < 0.05, two-tailed Student’s t test. (E) ATP concentrations in the thorax of adult workers. Bars with different letters are significantly different from one another. n = 3, average ± SD. P < 0.05, ANOVA with Tukey’s HSD post hoc test.
Quantification of Rates of Metabolism of Quercetin and of ATP Production in the Thorax of Adult Workers Consuming Quercetin in the Presence of Myclobutanil.
The amounts of quercetin remaining unmetabolized in midgut assays were significantly higher for quercetin-myclobutanil treatments than those in treatments containing quercetin alone (three replicates of 15 from the same colony; mean ± SE, 19.10 ± 0.36 vs. 16.37 ± 0.48 μM, respectively; P < 0.001, two-tailed Student’s t test) (Fig. 2). This finding suggests that more unmetabolized quercetin remains in the midgut in the presence of ingested myclobutanil than in its absence. If so, then, consistent with our qRT-PCR findings, bees consuming quercetin with myclobutanil should produce less ATP in their thorax. In quantifying ATP generation in the thorax of adult workers consuming quercetin in the presence of myclobutanil (Fig. 3E), we found that in fact, ATP levels in the thorax of bees consuming 0.25 mM quercetin/5 ppm myclobutanil were depressed relative to levels in bees consuming quercetin alone. This finding is consistent with quercetin-induced interference with mitochondrial regeneration and ATP production via down-regulation of mitochondrion-related nuclear genes.
Discussion
Given its long evolutionary association with flavonol-rich foods, it is not altogether surprising that the honey bee is well endowed with detoxifying enzymes that metabolize quercetin. Absent a highly efficient system, such as in life stages that consume only low quantities, quercetin can act as a negative regulator of mitochondrion-related nuclear gene expression. Taken together, the results of our docking, qRT-PCR, and metabolism experiments suggest that ingesting pollen contaminated by myclobutanil may be a hitherto underappreciated source of stress for honey bees; pesticide inhibition of P450-mediated detoxification may increase the exposure of bees to the biological activities of dietary phytochemicals. The phytochemical quercetin has the capacity to transcriptionally down-regulate not only nuclear genes associated with oxidative phosphorylation and mitochondrial membrane channel construction, but also mitochondrial genes associated with tRNA and oxidative phosphorylation. Given the predictable presence of quercetin in the diet of pollen-eating bees, interference with its P450-mediated detoxification is thus likely to lead to interference with ATP production and depletion of energy available for flight and other activities.
Pollen contamination by pesticides is of particular concern for nurse bees, the chief pollen consumers in the colony. The production of worker and royal jelly to feed brood depends on protein, and thus pollen, intake. Effects of pollen contamination by fungicides on energy production via inhibition of quercetin detoxification may be exacerbated by the toxic properties of the fungicides themselves. Aside from triazoles, other classes of fungicides, including the quinone outside inhibitors (Qo1), owe their toxicity to the inhibition of mitochondrial respiratory chain complex II/III and/or uncoupling of oxidative phosphorylation (33, 34). Such fungicides, including the strobilurins, are likely to suppress ATP production to an even greater extent via simultaneous inhibition of both quercetin-metabolizing P450s and mitochondrial respiratory chain complex II/III.
Collectively, our findings in this study suggest that the dependence of honey bees on P450-mediated detoxification of quercetin may render them particularly vulnerable to pesticide-related inhibition, Such inhibition, particularly of broadly substrate-specific P450s, may reduce the efficiency with which honey bees process not only their plant-based food, but also the flavonol-rich resins on which they rely for producing propolis in many parts of the world (35). The use of P450-inhibiting fungicides in combination with insecticides [e.g., during flowering in almond orchards (20)] thus exposes bees to pesticides that can reduce their ability to detoxify quercetin and to extract energy from their natural food. Whether bees exposed to the combination of myclobutanil and quercetin exhibit altered behavior or performance remains to be determined via in vivo experiments. Moreover, the extent to which other P450-inhibiting fungicides found as contaminants in beebread and pollen alter quercetin metabolism has not yet been characterized. Nonetheless, the multiple lines of evidence presented here suggest that methods for assessing the risks of fungicide exposure to bees may need to be reevaluated in the interest of sustaining the health of A. mellifera and the apicultural enterprise.
Materials and Methods
Chemical Sources.
Fresh-frozen royal jelly (organic) and myclobutanil were purchased from GloryBee Foods and LKT Laboratories, respectively. Quercetin, d-glucose, and d-fructose were obtained from Sigma-Aldrich. Bacto yeast extract was obtained from BD Biosciences.
Honey Bees and Treatments.
All of the honey bees used in these experiments were obtained from the University of Illinois Bee Research Facility in Urbana. Larvae were reared as described previously (21). We chose to examine the effects of quercetin ingestion initially in newly hatched larvae because during the first 3 d of life larvae consume glandular secretions of nurse bees exclusively and in general have very low levels of P450 activity. Thus, these larvae should be sufficiently sensitive to low levels of quercetin to reveal the physiological pathways most affected by its ingestion. In other life stages, efficient quercetin detoxification would preclude identifying its full physiological impacts on bees. We used a range of quercetin concentrations in the various experiments consistent with the concentrations found in pollen (2, 6). For RNA-Seq, we used 0.1 and 0.25 mM (3.02 mg/100 g, or 0.003%, and 7.6 mg/100 g, or 0.0076%); for qRT-PCR, we used 0.1, 0.25, and 0.5 mM (15.1 mg/100 g); and for evaluation of ATP production, we used 25 mM (7.6 mg/100 g) and 0.5 mM (15.1 mg/100 g). These values comport with concentrations of quercetin reported in “bee pollen” (2, 3). Although in solutions, quercetin can potentially transition from a soluble molecule to a colloidal aggregate and at higher concentrations can precipitate out of solution, by incorporating it into bee candy, we could simulate the conditions under which bees encounter this compound in pollen and avoid molecular transition issues. For experiments with myclobutanil, the concentrations used were selected based on the residues reported by Mullen et al. (8), with a maximum of 981 ppb, and by Stoner and Eitzer (36), with a maximum of 4,190 ppb. Accordingly, we chose concentrations of 1 ppm and 5 ppm (roughly corresponding to 981 ppb and 4,190 ppb, respectively).
For experiments on myclobutanil inhibition of quercetin midgut metabolism, 100 newly emerged worker bees in a standard Plexiglas cage were fed water and bee candy, a mixture of powdered sugar and sucrose syrup (2–4), for 3 d. To evaluate ATP production, ∼15 newly emerged worker bees were placed in 9-oz (266-mL) Dixie plastic cups covered with cotton cheesecloth and fed water and bee candy as a control or bee candy containing test compounds as a treatment. Each treatment was independently replicated three times with the same colony. On the fifth day, the workers for each sample were collected and kept in a −80 °C freezer for total RNA and ATP extractions.
RNA Extraction, RNA-Seq Analysis, and qRT-PCR Analyses.
RNA extraction, RNA-Seq analysis, and qRT-PCR analyses were conducted as described previously (3, 21). In brief, first instars were used for RNA-Seq and qRT-PCR. For obtaining eggs, an empty frame that had been occupied previously was equipped with an excluder cage and placed into a hive with a healthy colony at the University of Illinois Bee Research Facility for cleaning by workers. A queen was placed into the excluder cage in the evening of the next day, confined overnight, and then removed. Four mornings later, the frame with its neonate larvae was removed and brought to the laboratory for larval grafting. To evaluate the effects of quercetin acid on larval gene expression, 24 newly hatched larvae for each treatment (0, 0.1, and 0.25 mM) were grafted into a 24-well cell culture plate with 30 μL of the corresponding diet (21) in each well. For RNA-Seq experiments, 12 neonates for each sample for each treatment were transferred, and each treatment was independently replicated three times. The rearing plates were kept in a 34 ± 1 °C dark incubator at 90% relative humidity in a desiccator of 96% (saturated) K2SO4. Excess diet in the wells was removed daily with a vacuum and replaced with fresh diet prewarmed to 34 °C. On the third day, the larvae for the RNA-Seq experiments were collected, frozen in liquid nitrogen, and stored at −80 °C for total RNA extraction.
The following primer sequences were used for qRT-PCR analyses: Tim8, 5′-GCATTAATTCAAGCTCAGAT-3′ (sense), 5′-AGTTAAGCATGTTTCTGTTC-3′ (antisense); Tim9, 5′- TCTGTTCGAGATTTCGTG-3′ (sense), 5′-TTTGCTTTGACATCTCGT-3′ (antisense); MRP_L49, 5′-GGTGTCTGTACGATGTC-3′ (sense), 5′-GATGAATTAGTGCGAACG-3′ (antisense); MRP_L15, 5′- AATTTCAACGAGGCAAAG-3′ (sense), 5′-TTAACTTCTGGAGAACCTC-3′ (antisense); Ndufb7, 5′-TAGAGAACGCAATAGGC-3′ (sense), 5′-TGCTCTTTCACAATCTAATC-3′ (antisense); COX17, 5′-AACCTTGTTGTGCTTGT-3′ (sense), 5′-ATGTGCTTCTATTAAATCCC-3′ (antisense).
Molecular Model and Docking.
Among the honey bee P450s known to detoxify quercetin, CYP9Q1 was selected as an exemplar owing to its high activity relative to the CYP6AS P450s. Assays of activity of heterologously expressed honey bee P450s documented 12.19 pmol substrate.pmol P450.min for CYP9Q1 vs. 0.50 pmol substrate.pmolP450.min for four CYP6AS P450s. In addition, CYP9Q1 is broadly substrate-specific relative to the CYP6AS enzymes, metabolizing both flavonoids and pesticides (4), and, accordingly, is likely vulnerable to competitive interactions among substrates. The homology model previously developed for CYP9Q1 (4) was used in the docking. The database used for docking was constructed with the 121 pesticides and metabolites detected in North American bee hives (9). The homology model was exported as a PDB file to Discovery Studio 3.5 Client (Accelrys); the CYP9Q1-binding site was created using Define and Edit Binding Site tools. Compounds in the database were then docked into the binding site using LibDock, with the conformation method set to high quality and other parameters kept at their default values. The compounds with a LibDock score >86 were considered likely substrates or inhibitors of CYP9Q1.
Assays of Myclobutanil Inhibition of Quercetin Midgut Metabolism.
We conducted in vitro midgut assays to determine whether the fungicide myclobutanil can inhibit quercetin metabolism. For each reaction, midguts of 15 3-d-old worker bees were removed intact and placed in 1 mL of 0.1 M phosphate buffer (pH 7.4). To each of the three reactions for quercetin + myclobutanil treatments, 1 μL of 250 mM piperonyl butoxide dissolved in dimethyl sulfoxide was added; to each of the three reactions for the quercetin treatment, 1 μL of dimethyl sulfoxide was added. All reactions for the two treatments were incubated in a shaking water bath at 35 °C for 10 min. After 1 μL of 20 mM quercetin dissolved in dimethyl sulfoxide was added to each reaction, the reactions were incubated continuously for 60 min, quenched by adding 100 μL of 2N hydrochloric acid, homogenized, and extracted with 1 mL of ethyl acetate. Then 10 µL of the ethyl acetate extract from each reaction was analyzed on a Phenomenex Synergi 4u Fusion-RP 80A column (150 mm × 2.0 mm, 3 μ) using the following gradient run (1 mL/min) with water containing 0.1% formic acid (A) and acetonitrile containing 0.1% formic acid (B): 24% B from 0 to 10 min, followed by 24–48% B from 10 to 18 min. The quercetin content was quantified at 373 nm by comparison with a standard curve, and means were compared using Student’s two-tailed t test.
Thoracic ATP Production Assay.
The thorax and head were removed from each of 10 frozen 4-d-old workers and ground in liquid nitrogen with a mortar and pestle. The homogenized material was transferred to 300 μL (volume/head weight, 2) or 1,200 μL (volume/thorax weight, 3) of 6 M guanidine-HCl in extraction buffer (100 mM Tris-acetate and 2 mM EDTA, pH 7.75) (37). The samples were then centrifuged at 130,000 × g for 10 min at 4 °C, after which the supernatant was diluted 500 times with the extraction buffer. The ATP level in each sample was measured by an ATPlite Luminescence Assay System (PerkinElmer). Means were compared between two samples using Student’s t test and one-way ANOVA with Tukey’s honestly significant difference (HSD) post hoc test, performed in R.
Supplementary Material
Acknowledgments
We thank Gene Robinson and the University of Illinois Bee Research Laboratory for advice and access to bees, Bernarda Calla for help with data deposition, Alvaro Hernandez for assistance with RNA-Seq analysis, and Zachary Huang and Blair Siegfried for their critical evaluation of the manuscript. This project was funded by the US Department of Agriculture (Grant AFRI 2010-03760), a Tyler Prize for Environmental Achievement (to M.R.B.), and the University of Illinois Center for Advanced Study.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE83437).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1614864114/-/DCSupplemental.
References
- 1.Claudianos C, et al. A deficit of detoxification enzymes: Pesticide sensitivity and environmental response in the honeybee. Insect Mol Biol. 2006;15(5):615–636. doi: 10.1111/j.1365-2583.2006.00672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mao W, et al. Quercetin-metabolizing CYP6AS enzymes of the pollinator Apis mellifera (Hymenoptera: Apidae) Comp Biochem Physiol B Biochem Mol Biol. 2009;154(4):427–434. doi: 10.1016/j.cbpb.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 3.Mao W, Schuler MA, Berenbaum MR. Honey constituents up-regulate detoxification and immunity genes in the western honey bee Apis mellifera. Proc Natl Acad Sci USA. 2013;110(22):8842–8846. doi: 10.1073/pnas.1303884110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mao W, Schuler MA, Berenbaum MR. CYP9Q-mediated detoxification of acaricides in the honey bee (Apis mellifera) Proc Natl Acad Sci USA. 2011;108(31):12657–12662. doi: 10.1073/pnas.1109535108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Niu G, Johnson RM, Berenbaum MR. Toxicity of mycotoxins to honey bees and its amelioration by propolis. Apidologie (Celle) 2011;42(1):79–87. [Google Scholar]
- 6.Guengerich FP. Human cytochrome P450 enzymes. In: Ortiz de Montellano PR, editor. Cytochrome P450: Structure, Mechanism, and Biochemistry. 4th Ed. Springer; New York: 2015. pp. 523–785. [Google Scholar]
- 7.Fujita K. Food-drug interactions via human cytochrome P450 3A (CYP3A) Drug Metabol Drug Interact. 2004;20(4):195–217. doi: 10.1515/dmdi.2004.20.4.195. [DOI] [PubMed] [Google Scholar]
- 8.Mullin CA, et al. High levels of miticides and agrochemicals in North American apiaries: Implications for honey bee health. PLoS One. 2010;5(3):e9754. doi: 10.1371/journal.pone.0009754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pettis JS, et al. Crop pollination exposes honey bees to pesticides, which alters their susceptibility to the gut pathogen Nosema ceranae. PLoS One. 2013;8(7):e70182. doi: 10.1371/journal.pone.0070182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berenbaum MR, Johnson RM. Xenobiotic detoxification in the western honey bee Apis mellifera. Curr Opin Insect Sci. 2015;10:51–58. doi: 10.1016/j.cois.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 11.Wu JY, Anelli CM, Sheppard WS. Sub-lethal effects of pesticide residues in brood comb on worker honey bee (Apis mellifera) development and longevity. PLoS One. 2011;6(2):e14720. doi: 10.1371/journal.pone.0014720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mo Y, Nagel C, Taylor LP. Biochemical complementation of chalcone synthase mutants defines a role for flavonols in functional pollen. Proc Natl Acad Sci USA. 1992;89(15):7213–7217. doi: 10.1073/pnas.89.15.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ylstra B, et al. Flavonols stimulate development, germination, and tube growth of tobacco pollen. Plant Physiol. 1992;100(2):902–907. doi: 10.1104/pp.100.2.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahajan M, Ahuja PS, Yadav SK. Post-transcriptional silencing of flavonol synthase mRNA in tobacco leads to fruits with arrested seed set. PLoS One. 2011;6(12):e28315. doi: 10.1371/journal.pone.0028315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rzepecka-Stojko A, et al. Polyphenols from bee pollen: Structure, absorption, metabolism and biological activity. Molecules. 2015;20(12):21732–21749. doi: 10.3390/molecules201219800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wiermann R, Vieth K. Outer pollen wall, an important accumulation site for flavonoids. Protoplasma. 1983;118:230. [Google Scholar]
- 17.Serra Bonvehí J, Soliva Torrentó M, Centelles Lorente E. Evaluation of polyphenolic and flavonoid compounds in honeybee-collected pollen produced in Spain. J Agric Food Chem. 2001;49(4):1848–1853. doi: 10.1021/jf0012300. [DOI] [PubMed] [Google Scholar]
- 18.Kaškonienė V, Ruočkuvienė G, Kaškonas P, Akuneca I, Maruška A. Chemometric analysis of bee pollen based on volatile and phenolic chemical compositions and antioxidant properties. Food Anal Meth. 2015;8:1150–1163. [Google Scholar]
- 19.Liu D, Yuan Y, Li M, Qiu X. Effects of dietary quercetin on performance and cytochrome P450 expression of the cotton bollworm, Helicoverpa armigera. Bull Entomol Res. 2015;105(6):771–777. doi: 10.1017/S0007485315000760. [DOI] [PubMed] [Google Scholar]
- 20.Gao J, Zhao G, Yu Y, Liu F. High concentration of nectar quercetin enhances worker resistance to queen’s signals in bees. J Chem Ecol. 2010;36(11):1241–1243. doi: 10.1007/s10886-010-9866-3. [DOI] [PubMed] [Google Scholar]
- 21.Guseman AJ, et al. Multidrug-resistance transporters and a mechanism-based strategy for assessing risks of pesticide combinations to honey bees. PLoS One. 2016;11(2):e0148242. doi: 10.1371/journal.pone.0148242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sørensen JM. Herb-drug, food-drug, nutrient-drug, and drug-drug interactions: Mechanisms involved and their medical implications. J Altern Complement Med. 2002;8(3):293–308. doi: 10.1089/10755530260127989. [DOI] [PubMed] [Google Scholar]
- 23.Muthiah YD, Ong CE, Sulaiman SA, Tan SC, Ismail R. In-vitro inhibitory effect of Tualang honey on cytochrome P450 2C8 activity. J Pharm Pharmacol. 2012;64(12):1761–1769. doi: 10.1111/j.2042-7158.2012.01551.x. [DOI] [PubMed] [Google Scholar]
- 24.Nabas Z, Haddadin MSY, Haddadin J, Nazer IK. Chemical composition of royal jelly and effects of synbiotic with two different locally isolated probiotic strains on antioxidant activities. Pol J Food Nutr Sci. 2014;64:171–180. [Google Scholar]
- 25.Shendy AH, Al-Ghobashy MA, Mohammed MN, Gad Alla SA, Lotfy HM. Simultaneous determination of 200 pesticide residues in honey using gas chromatography-tandem mass spectrometry in conjunction with a streamlined quantification approach. J Chromatogr A. 2016;1427:142–160. doi: 10.1016/j.chroma.2015.11.068. [DOI] [PubMed] [Google Scholar]
- 26.Mao W, Schuler MA, Berenbaum MR. A dietary phytochemical alters caste determination gene expression in honey bees. Sci Adv. 2015;1:e150079. doi: 10.1126/sciadv.1500795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crozier RH, Crozier YC. The mitochondrial genome of the honeybee Apis mellifera: Complete sequence and genome organization. Genetics. 1993;133(1):97–117. doi: 10.1093/genetics/133.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campbell JB, et al. The fungicide Pristine inhibits mitochondrial function in vitro but not flight metabolic rates in honey bees. J Insect Physiol. 2016;86:11–16. doi: 10.1016/j.jinsphys.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 29.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo W, Friedman MS, Shedden K, Hankenson KD, Woolf PJ. GAGE: Generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics. 2009;10:161. doi: 10.1186/1471-2105-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo W, Brouwer C. Pathview: An R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics. 2013;29(14):1830–1831. doi: 10.1093/bioinformatics/btt285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson RM, Dahlgren L, Siegfried BD, Ellis MD. Acaricide, fungicide and drug interactions in honey bees (Apis mellifera) PLoS One. 2013;8(1):e54092. doi: 10.1371/journal.pone.0054092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang C, Hamel C, Vujanovic V, Gan Y. Fungicide: Mode of action and possible impact on nontarget microorganisms. ISRN Ecol. 2011;2011:130289. [Google Scholar]
- 34.Kanungo JJ. Impact of pyraclostrobin (F-500) on crop plants. Plant Sci Today. 2014;1:174–178. [Google Scholar]
- 35.Bankova V. Recent trends and important developments in propolis research. Evid Based Complement Alternat Med. 2005;2(1):29–32. doi: 10.1093/ecam/neh059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stoner KA, Eitzer BD. Using a hazard quotient to evaluate pesticide residues detected in pollen trapped from honey bees (Apis mellifera) in Connecticut. PLoS One. 2013;8(10):e77550. doi: 10.1371/journal.pone.0077550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwarze SR, Weindruch R, Aiken JM. Oxidative stress and aging reduce COX I RNA and cytochrome oxidase activity in Drosophila. Free Radic Biol Med. 1998;25(6):740–747. doi: 10.1016/s0891-5849(98)00153-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.