Significance
Marine microbes exhibit enormous genetic diversity and drive global biogeochemical processes. Diatoms are some of the most diverse and ecologically influential marine microbes, generating about 40% of global marine primary production. The physical and ecological processes that maintain diversity in marine microbes are widely debated. Here, using empirical evidence from a diatom species, we show that geographic distance between sites does not correlate with genetic divergence. Instead, environmental and ecological selection likely exert a stronger influence than dispersal on the divergence and connectivity of diatom populations throughout the globe. Understanding mechanisms driving divergence and selection of marine microbial populations has enormous potential for improving predictions of global productivity in response to a rapidly changing marine environment.
Keywords: diatom, dispersal, environmental selection, population structure, microsatellites
Abstract
The ability for organisms to disperse throughout their environment is thought to strongly influence population structure and thus evolution of diversity within species. A decades-long debate surrounds processes that generate and support high microbial diversity, particularly in the ocean. The debate concerns whether diversification occurs primarily through geographic partitioning (where distance limits gene flow) or through environmental selection, and remains unresolved due to lack of empirical data. Here we show that gene flow in a diatom, an ecologically important eukaryotic microbe, is not limited by global-scale geographic distance. Instead, environmental and ecological selection likely play a more significant role than dispersal in generating and maintaining diversity. We detected significantly diverged populations (FST > 0.130) and discovered temporal genetic variability at a single site that was on par with spatial genetic variability observed over distances of 15,000 km. Relatedness among populations was decoupled from geographic distance across the global ocean and instead, correlated significantly with water temperature and whole-community chlorophyll a. Correlations with temperature point to the importance of environmental selection in structuring populations. Correlations with whole-community chlorophyll a, a proxy for autotrophic biomass, suggest that ecological selection via interactions with other plankton may generate and maintain population genetic structure in marine microbes despite global-scale dispersal. Here, we provide empirical evidence for global gene flow in a marine eukaryotic microbe, suggesting that everything holds the potential to be everywhere, with environmental and ecological selection rather than geography or dispersal dictating the structure and evolution of diversity over space and time.
Since the 1930s, the tenet of “everything is everywhere but the environment selects” (1) has formed a foundation for theories of microbial biogeography and of questions regarding the potential for endemicity in microbes (2). The ways in which the rapid generation times and high dispersal potential of microbes influence their ecological diversification remain unclear (3, 4). Despite a lengthy debate, conflicting evidence abounds on the role of environmental selection in maintaining microbial biogeographies and whether microbial lineages are geographically ubiquitous, especially in marine environments (5–7). Terrestrial species generally exhibit low dispersal capacity, resulting in strong relationships between geographic distance and genetic relatedness, populations with high levels of genetic structure, and the potential for genetic divergence among populations over small spatial scales (8). In contrast, marine planktonic microbes inhabit a global-scale fluid environment and have enormous dispersal potential with an expected pattern of high gene flow, large and homogenous populations, and little divergence over even large spatial scales. In the ocean, environmental and physical factors vary over time at high frequencies (9), creating a patchy habitat of resource availability, complex hydrodynamic processes (10), dynamic interspecies interactions (11), and biologically mediated fluctuations of pH, light, and nutrients (12, 13). In response to this variability, competitive exclusion, niche differentiation, and environmental selection are thought to act as strong evolutionary forces generating and maintaining diversity within and among species (14). Emerging genomic and transcriptomic evidence points to the functional and metabolic diversity of marine microbes and the potential for niche differentiation in the marine environment (15, 16). However, despite the evolutionary significance of environmental selection, its role in structuring populations within species occupying the high dispersal environment of the global ocean remains unclear. The additional role of dispersal and geography in structuring populations throughout the global ocean has added complexity to the debate (5, 17, 18).
This debate is particularly relevant to phytoplankton, free-floating single-celled primary producers responsible for generating half of the oxygen on Earth and exhibiting enormous intraspecific diversity despite their high potential to disperse throughout the global ocean (4, 19). In theory, enormous dispersal potential should reduce genetic partitioning and homogenize gene pools. Instead, phytoplankton species harbor vast diversity and show evidence for highly structured populations and communities (17, 20–23). Within the phytoplankton, diatoms are among the most productive and speciose eukaryotic microbes in the ocean (4, 24), and are found across the globe, making them useful organisms through which to explore the important links between dispersal, environmental selection, and the evolution of diversity.
It remains a practical challenge to empirically test the influence of dispersal and environmental selection on microbial diversification, especially on global spatial scales. Our view of microbial population structure in the ocean is largely derived from ad hoc samples (often cultures) collected over multiple time points and across locations (25). In analyses, samples collected over time from the same location are often pooled under the assumption that population structure will vary more in relation to geographic distance than it will over time or with changes in the environment. However, the assumption that geographic distance largely controls microbial population structure conflicts with our understanding of high microbial dispersal. Environmental selection may equally influence the evolution of microbial diversity in the ocean and support the divergence of lineages over time.
To tease apart the mechanisms structuring microbial diversity in the ocean, samples must be collected with respect to time as much as space. However, these samples are largely unavailable. Global population structure has only been investigated in the diatom Pseudonitzschia pungens. In this pennate diatom, geographic distance appeared to be a strong barrier to gene flow, suggesting allopatric isolation despite high dispersal potential (17). Individuals of P. pungens collected from the same region but at different times were pooled to represent geographically defined populations, assuming that no genetic changes occurred over time in a given location in this pennate diatom. In contrast, studies of centric diatoms have found extensive shifts in population structure in a single location over time, suggesting that population structure in other diatoms may not necessarily be dominated by allopatric processes (26, 27). Whether allopatric speciation and geographic barriers to gene flow are common processes structuring diatom and marine microbial diversity in general remains unknown.
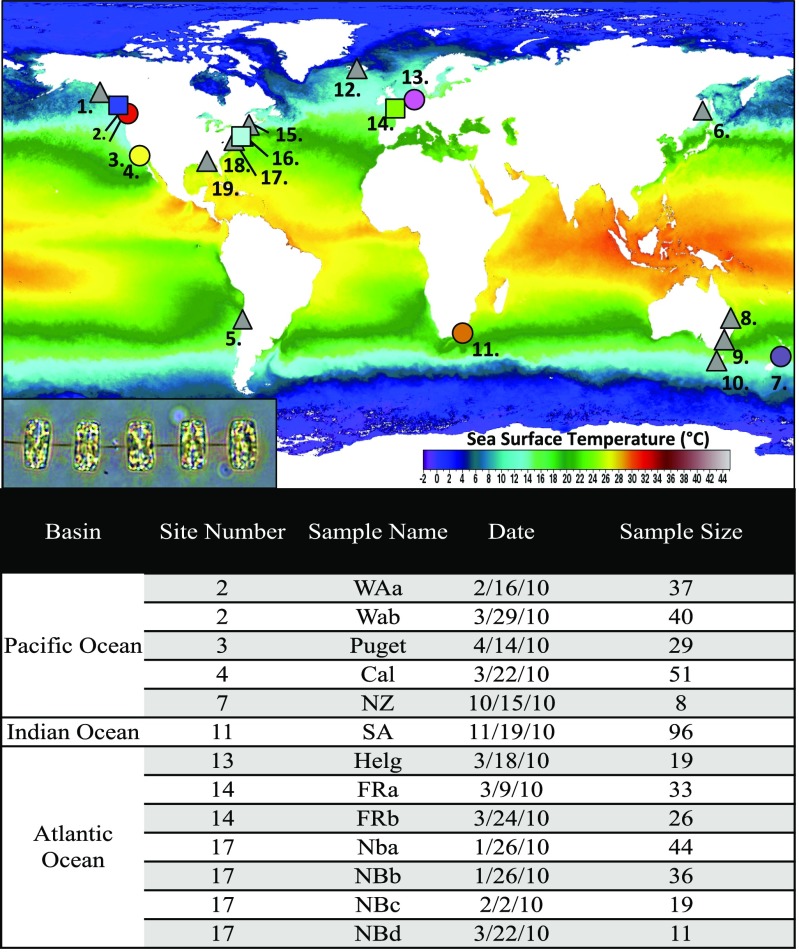
In this study, our sampling approach allowed for a global-scale investigation of population structure in a marine microbial species while controlling for time. We developed six microsatellite markers to identify genetically distinct populations within the globally distributed, bloom-forming diatom species, Thalassiosira rotula. Microsatellites are rapidly evolving and hypervariable population genetic markers widely used in the field of molecular ecology. We isolated individuals of T. rotula wherever present during a global sampling of surface seawater collected within 1- to 2-wk intervals, and sometimes on the same day, during the spring of 2010 in both the northern and southern hemispheres, allowing for a time-controlled analysis of global population structure in this species (Fig. 1 and SI Appendix, Tables 1 and 2). Each water sample was associated with environmental metadata (SI Appendix, Table 3). This time-controlled approach to sampling allowed us to tease apart the relative influence of geographic distance versus environmental variability in structuring global populations of T. rotula. The goal of this work was to examine genetic diversity in the marine diatom T. rotula by capturing variations in genetic composition over time and across the globe and determining its environmental and ecological correlates.
Fig. 1.
Global sampling locations. (A) Sites where whole seawater was sampled and T. rotula cells were isolated one time (circles) and multiple times (squares) in 2010. Northern hemisphere sampling took place during spring, at five time points between January and March, whereas southern hemisphere sampling took place during austral spring, at five time points between October and December. At some sites, water was collected, but T. rotula was not detected (triangles). Base map of composite annual SST from 2010, source: level 3 Terra moderate resolution imaging spectroradiometer (daytime) oceancolor.gsfc.nasa.gov/cgi/l3. (Inset) Light micrograph of T. rotula cells in girdle view. (B) Table indicating sample location, name abbreviation, collection date, and number of cells isolated (sample size). Site numbers refer to map in A. Additional information can be found in SI Appendix, Tables 1 and 2.
Results and Discussion
Isolates of T. rotula were genotyped at six microsatellite loci, revealing enormous diversity. Clonal diversity within water samples was as high as 100%. Among the 449 individuals genotyped, only two genotypes, identical at all six loci, were observed more than once. Both were sampled from site 11. Other diatom species show similarly high levels of clonal diversity (e.g., refs. 21, 28–30) such that single blooms can consist of thousands of clonal lineages (31). Expected heterozygosity (HE) varied between 0.55–0.83 (SI Appendix, Fig. 1 and Tables 4 and 5). Observed heterozygosity (HO) was significantly lower than HE, which appears to be a common genetic characteristic of phytoplankton that undergo both asexual and sexual reproduction and may be attributed to the presence of null alleles or nonrandom mating; however, the mechanism is not fully understood (e.g., refs. 22, 31, 32).
For a subset of isolates from each sample, the internal transcribed spacer 1 ribosomal DNA (ITS1 rDNA) gene was sequenced for lineage-level identification following Whittaker et al. (33) (SI Appendix, Fig. 2 and SI Methods). Isolates belonged to two of the three previously identified lineages (lineages 1 and 3) (33). Both lineages were identified in all three ocean basins investigated (SI Appendix, Fig. 2). Lineages co-occurred in seven water samples [FRb, NBd, SA, WAa, Puget, Cal, and WAb (sample names refer to Fig. 1)], and isolates from different lineages did not exhibit significantly different membership coefficients in STRUCTURE or significant FST values within samples (SI Appendix, Fig. 3 and Table 6), suggesting ongoing gene flow between lineages. Populations did not cluster according to lineage identity, suggesting incomplete lineage sorting at the ITS1 or introgression (e.g., refs. 34–36) and supporting previous research that failed to find conclusive evidence for reproductive isolation among lineages (33). Due to lack of evidence for reproductive isolation and new evidence questioning the strict geographic isolation of these lineages, we examined population structure and the potential for global connectivity in the species as a whole.
Genetic divergence among global samples of T. rotula was calculated using traditional measures of FST. If allopatric or sympatric processes produce restricted gene flow between T. rotula populations, each population will experience independent genetic drift, resulting in a progressive divergence of allele frequencies over time; this divergence is reflected in measures of FST (37, 38). Water samples exhibited a maximum FST divergence of 0.139, indicating a moderate level of differentiation (0.05–0.15), as defined for biallelic loci by Wright (38) (SI Appendix, Table 7). However, it can be conservatively estimated that these water samples contained populations that were strongly differentiated from each other because FST values for highly variable microsatellite loci tend to be smaller than for biallelic loci (39). Divergence in T. rotula was similar to other phytoplankton species sampled over large distances, such as across the Baltic Sea and the NE Atlantic (maximum FST = 0.099) (21, 22) but below the maximum FST (0.76) observed among allopatrically separated populations of the pennate diatom P. pungens (17). Divergence in T. rotula was comparable to population divergence observed in globally distributed, highly migratory marine macrofauna (maximum FST = 0.175) (40–42). Thus, FST observed here for T. rotula is within the range expected for cosmopolitan high-dispersal marine organisms, but lower than those exhibiting strong patterns of allopatry.
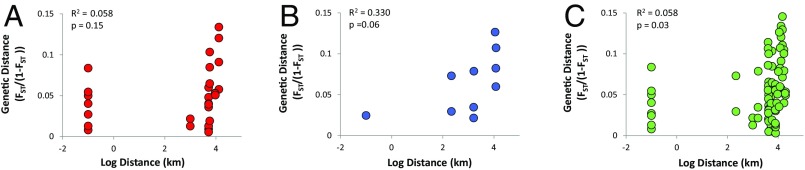
We investigated the potential for isolation by distance (IBD) via statistical (Mantel) tests of correlation between genetic and geographic distances (43). Genetic and geographic distances were not significantly correlated in the Atlantic or Pacific basins (Atlantic: R2 = 0.058 P = 0.15; Pacific: R2 = 0.330 P = 0.06). On a global scale, genetic and geographic distances were significantly correlated, but with an R2 close to 0, indicating weak isolation by distance (global: R2 = 0.058 P = 0.03) (Fig. 2). Weak isolation by distance suggests that gene flow in T. rotula is not limited by geographic distance, even on a global scale. For example, samples collected from the same site could be nearly four times more genetically divergent than samples collected over 7,000 km apart and from different ocean basins. These data suggest that global ocean circulation is sufficient to allow for the global dispersal of individuals, despite the vast distances involved. This study provides empirical evidence for global gene flow in a marine diatom species.
Fig. 2.
Linearized pairwise genetic distance (FST) vs. the log of geographic distance for samples collected in the (A) Atlantic (red circles), (B) Pacific (blue circles), and (C) across the globe (green circles), including the coefficient of determination (R2) and P value for each fitted linear regression.
The observation of global dispersal in T. rotula contrasts with observations of the pennate diatom P. pungens, which exhibits strong geographic limits to gene flow (IBD R2 = 0.70 P < 0.01) (17) (see SI Appendix, Fig. 4 for direct comparison in T. rotula). This contrast may relate to either differences in the potential for survival during dispersal by ocean currents and/or anthropogenic transport, or distinct life history differences between the species. For one, P. pungens, and many other pennate diatoms, are heterothallic, meaning that male and female mating types must be present for successful sexual reproduction (44, 45). Centric diatoms are more typically homothallic, not requiring differentiated mating types, although a diversity of mating systems have evolved across centric diatom species (46). Overall, the timing and triggers of sexual reproduction in diatoms are little understood (46). However, differences in environmental or temporal triggers of sex between the two species may greatly impact patterns of population structure across the wide ecological gradients sampled.
Another hypothesis to explain differences between the species’ population structure may be differences in their ability to form resting spores, or dormant cells, that provide a stepping stone for dispersal in those species that can form them. Life stages of cell quiescence and resting spore formation may determine the ability for cells to survive transport across large distances in the ocean. Resting spores remain viable in environmentally unfavorable conditions, and thus the ability to form resting spores could be essential in facilitating dispersal in these organisms (47, 48). P. pungens does not form resting spores, although the potential for dormant cell stages in pennate diatoms is still debated (49). T. rotula has been documented to form resting spores (50), which may explain a higher dispersal potential in this species and why geographic distance is a greater barrier to gene flow in P. pungens than T. rotula.
A third hypothesis to explain differences in population structure between the species may be thermal tolerance. In diatoms, there is both intra- and interspecific variation of growth rates in response to temperature (28, 33, 51). This variation has been proposed as a mechanism influencing dispersal potential in microbes and thus impacting their evolution and population structure (52). Restricted thermal tolerance of different P. pungens populations would limit their potential for global dispersal and result in a population structure more strongly correlated to geographic distance (53). There is evidence in diatoms that genetically distinct populations have distinct physiological capabilities (28, 33). Overall, dramatically different patterns of population structure in the two species point to the need to expand our understanding of diatom population diversity over larger spatial scales and across more species.
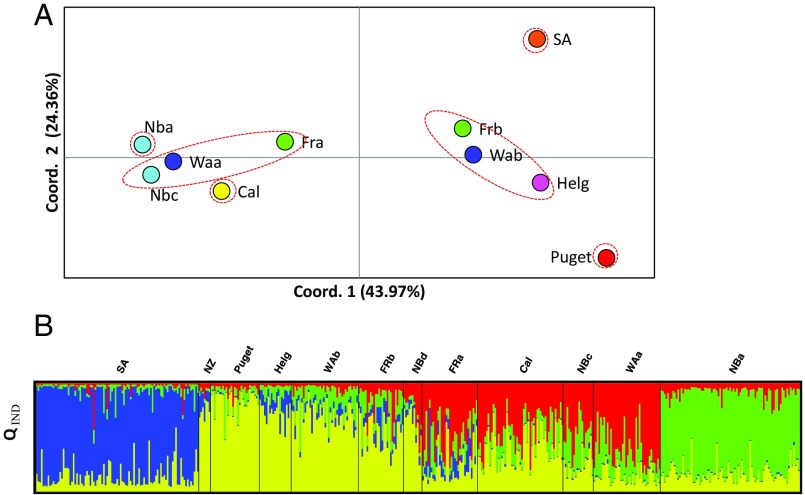
In T. rotula, the magnitude of genetic variation over time in one location was on par with genetic variation over global distances. T. rotula was subdivided into six populations identified using FST, and four populations using Bayesian clustering (STRUCTURE) (Fig. 3 A and B). Assumptions of each method produce slightly different results, but both indicate that multiple populations comprise the species. We define a “population” as consisting of water samples between which no significant genetic divergence was detected. Some populations were identified in samples distributed across ocean basins. For example, one population was sampled from the NE and NW Atlantic and the NE Pacific (NBc, WAa, and FRa). Other populations were identified just once (e.g., Puget, SA, NBa, and Cal). Samples collected over time from one location were significantly diverged (e.g., NBa and NBc, FST = 0.044, P < 0.05), whereas samples collected from different ocean basins were, in some cases, not significantly diverged (e.g., WAa and FRa, FST = 0.002, P > 0.05). In fact, the magnitude of genetic divergence (FST) over time in a single location reached 81% of the genetic divergence observed over the largest distances sampled (>15,000 km). For example, FST between samples collected at different times in Narragansett Bay, RI (site 17) was as great as 0.064, whereas FST was 0.079 between samples collected from the SW Indian Ocean (site 11) and from the NE Pacific (site 3).
Fig. 3.
Genetic subdivision among global samples. (A) Principal coordinates analysis of genetic divergence (FST) among samples for which the number of isolates was larger than 15. Sample names refer to those denoted in Fig. 1B, and colors refer to site markers in Fig. 1A. Relative proximity among samples in the graph represents genetic proximity (FST). Coordinates 1 and 2 explain 43.96% and 24.36%, respectively, of variation among samples. Samples encircled in dashed red lines represent statistically significant populations identified using genotypic tests. (B) Structure diagram displaying membership probability of each individual into K = 4 populations (denoted by different colors). Each individual is depicted by a vertical line partitioned into K colored sections, with the length of each section proportional to the estimated membership coefficient (QIND) of the isolate to each cluster. Sample names refer to those denoted in Fig. 1B.
A population sampled at our single fjord site, site 3, Puget Sound, was not detected elsewhere, suggesting that this population may experience barriers to genetic connectivity. In the Danish Mariager fjord, population stability over 100 y has been observed in the centric diatom species Skeletonema marinoi (54). The Mariager fjord is associated with a remarkably low flushing rate of 8 mo (54). Similarly, the flushing rate of Puget Sound is generally in excess of 1 y (55). These observations of diatom population stability and isolation in slow-flushing fjord systems suggest that populations associated with high hydrographic retention may experience genetic stability over time.
In contrast, significant temporal genetic variation was observed at all three sites where T. rotula was isolated multiple times in the NE and NW Atlantic and NE Pacific, sites 14, 17, and 2, respectively. The detection of significantly diverged populations in samples collected over short periods of time (1–6 wk) contrasts with observations of genetic stability in fjords (54). In contrast to the long hydrographic residence times of fjord systems, Narragansett Bay, site 17, experiences a very short residence time of 10–40 d (56). Our sampling location off the coast of France, site 14, was located within the Western English Channel, which experiences high connectivity with both the North Sea and Atlantic water masses and is dominated by persistent tidal flushing (57). The Washington coast (site 2) experiences dynamic upwelling, and is within a region of high connectivity and convergence among the North Pacific, Alaskan, and California current systems and experiences seasonal eddies. Thus, sites 17, 14, and 2 experience high levels of physical connectivity to surrounding waters. Different populations may be introduced from connected waters and be adapted to specific environmental conditions that can vary widely over short periods of time, leading to the observation of significantly diverged populations at one location over as little as 7 d. Overall, the presence of genetically distinct populations among global samples demonstrates that significant divergence can occur despite the high potential for dispersal in these planktonic organisms. This finding also suggests that sites associated with high hydrographic connectivity may experience dynamic genetic connectivity as well, where population structure may shift rapidly over time. High levels of connectivity in the surface ocean may also explain how individual populations are dispersed between ocean basins over relatively short periods of time (19).
Measures of FST do not differentiate between divergence due to allopatric or sympatric restriction of gene flow or reflect the time scales of divergence. However, the interaction between large population sizes and high dispersal potential suggests that selection maintains divergence between populations of T. rotula, which may persist over time scales longer than global surface seawater connectivity. Planktonic marine microbial census sizes are enormous, with hundreds to thousands of individuals per milliliter of seawater and thousands of clones per population (31). Population genetics theory suggests that neutral mutation, founder effects, and the impact of migration are muted in large populations like those observed in diatoms (58). Thus, under a high-dispersal low-selection scenario, we would expect one panmictic population, a result not observed in T. rotula. The physical connectivity of global surface seawater is estimated to be on the order of decades (19); this time frame aligns with the potential for gene flow in planktonic microbes. If microbes evolve more quickly than global circulation patterns can disperse them, we would expect biogeographic patterns to emerge; the potential for biogeographic patterns deriving from neutral evolution has been demonstrated in recent modeling efforts (18). Because we observed no correlation between geographic distance and population divergence on global scales, but still observed distinct populations in this species, it is likely that divergence observed in T. rotula populations is a product of ongoing selective forces that reduce gene flow among populations and allow them to persist over time frames greater than decadal-scale global surface seawater connectivity and perhaps even longer, given empirical evidence in the diatom S. marinoi, which exhibits stable FST divergence on time scales of 100 y (54).
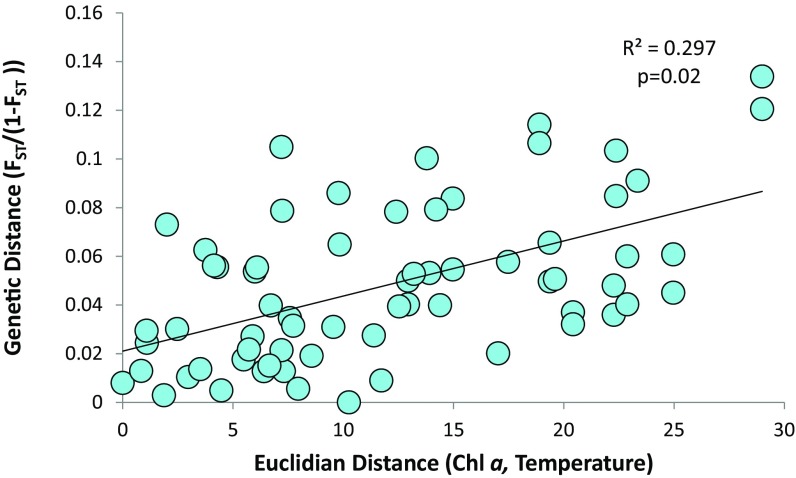
The presence of distinct populations among global samples of T. rotula demonstrates that selective mechanisms exist to support divergence within the species. Here, we uncovered evidence for both environmental and ecological selection in T. rotula populations. We tested correlations between genetic distance (FST) and pairwise differences among samples for variables including temperature, salinity, cell abundance, and whole-community chlorophyll a. These environmental conditions varied widely among samples (SI Appendix, Table 3). A series of Mantel tests revealed that the combination of temperature and chlorophyll a concentration explained 53% of the variance in global population structure (P = 0.02) (SI Appendix, Table 8). Chlorophyll a alone explained 41% of the variance in global population structure. A regression of pairwise FST and the Euclidian distance of chlorophyll a and temperature, calculated using multidimensional scaling, showed a significant (P = 0.02) positive relationship revealing that the most diverged populations were sampled from the most divergent environments (Fig. 4). Correlation between population structure and temperature is not entirely surprising. Temperature is an important factor regulating phytoplankton growth rates both between and within species, and likely plays a role in regulating dispersal capacity (51, 52).
Fig. 4.
Isolation by distance plot displaying the regression of pairwise linearized genetic distance (FST)/(1 − (FST)) and Euclidian distance of environmental factors. Euclidian distance was calculated through multidimensional scaling of environmental variables between sites.
The role of chlorophyll a is less clear. Chlorophyll a is a widely used proxy for whole-community phytoplankton biomass (59). Periods of high and low plankton biomass are associated with chemical and environmental differences that may impart selective pressure on T. rotula populations. Ecological differences include biologically mediated shifts in light regimes and nutrient concentrations, viral activity, interspecific interactions, and transparent exopolymer production; these variables also fluctuate with annual phytoplankton bloom cycles typically initiated in the spring, and progressing over time. These data suggest that the biotic factors associated with phytoplankton abundance (of which chlorophyll a is a proxy) may play an important role in structuring and maintaining T. rotula populations, potentially through the processes of competitive exclusion and/or niche partitioning. The roles of niche partitioning and competitive exclusion are thought to maintain genetic diversity in the field (14), with growing molecular evidence providing support in application to microbial populations (15, 16). This finding suggests that both environmental and ecological selection may shape the population genetic structure over space and time in a microbe with few barriers to dispersal.
Here, we show that, in a marine eukaryotic microbe, genetic differentiation can vary as much temporally as it does spatially. Geographic distance does not limit global gene flow, leading to global scale genetic connectivity. The presence of genetically distinct populations among global samples demonstrates that significant divergence can occur despite the high potential for dispersal in these planktonic organisms. Furthermore, environmental selection may significantly influence the structure and evolution of diatom populations globally. Ecological dynamics associated with whole-community plankton abundance may play a role in structuring and maintaining T. rotula diversity through processes including competitive exclusion and niche partitioning (15, 16). Clear coupling between ecological variability and T. rotula population structure suggests that within-species dynamics of diversity may be key to understanding the ecological and evolutionary response of these important primary producers to a rapidly changing ocean. Our work suggests that “everything has the potential to be everywhere,” with environmental and ecological selection playing important roles in the spatial and temporal distribution of marine microbial species.
Materials and Methods
Field Samples and Isolates.
In 2010, 19 locations across both hemispheres were targeted for T. rotula isolation (Fig. 1 and SI Appendix, Tables 1 and 2). Most samples were collected at established time series sites and thus contained metadata (SI Appendix, Table 3). For other sites, ancillary data (e.g., temperature and salinity) were collected via in situ sensors (e.g., YSI, Xylem Analytics). Whole seawater was rush shipped to the University of Rhode Island up to five times per site, targeting the spring phytoplankton bloom period in both hemispheres. Single cells and chains were isolated from whole seawater wherever present (at 8 sites) and after 1–3 wk of growth, were filtered for DNA extraction (SI Appendix, SI Methods).
Amplification of Field DNA.
Species identity of isolates was confirmed using a T-RFLP screening method (SI Appendix, SI Methods). Isolates were genotyped at six microsatellite loci that were amplified from the gDNA of 449 T. rotula isolates (SI Appendix, Table 9 and SI Methods). Alleles were scored using an ABI 3130xl [T. rotula (TR)1, TR3, TR7] or an ABI 3730xl (TR8, TR10, TR27), and analyzed using the software Gene Mapper 5 (Life Technologies).
Statistical Analysis of Population Structure.
Using microsatellite genotypes and allele frequencies (SI Appendix, SI Methods), hierarchical spatial and temporal population structure was tested. Genetic differentiation among sites was determined using the exact G test and Fisher’s exact probability test (60) in GENEPOP v4.2 (61). Pairwise differentiation between sites, FST, was calculated in GenAlEx 6.5 (62), both separately with each locus, and with all loci. All P values were Bonferroni corrected. Analysis of molecular variance was calculated using GenAlEx 6.5, testing for the presence of genetic structure at global and basin scales. Principal coordinates analysis was used to ordinate pairwise FST among sites with sample size greater than 15 individuals. The Bayesian clustering program STRUCTURE v2.2 was used to examine hierarchical resolution of population structure (SI Appendix, SI Methods).
Correlations were determined between pairwise genetic distance and pairwise Euclidean distance of environmental variables (location, SST, salinity, chlorophyll a, and T. rotula abundance). T. rotula abundance was calculated from triplicate counts of Lugol’s fixed samples using an E800 microscope at 20× (Nikon) and 1-mL Sedgewick-Rafter microscope slides. A series of Mantel tests was performed. Isolation by distance (63) was examined using the software IBDWS v3.16 (64) (SI Appendix, SI Methods). To identify the factors of the ocean environment most correlated with a pairwise FST resemblance matrix, BIOENV analysis was performed in Primer-E (65) using the variables temperature, salinity, chlorophyll a, and cell abundance (SI Appendix, SI Methods).
Supplementary Material
Acknowledgments
We thank the generous researchers who provided whole seawater and environmental data, the NASA Ocean Biology Processing Group for providing sea surface temperature data, and two anonymous reviewers for their thoughtful and constructive feedback. This research was supported by National Science Foundation (NSF) Grants 0727227 (to T.A.R.) and SBE0245039 (to the University of Rhode Island and T.A.R.). Part of the research was conducted using instrumentation supported by NSF’s Experimental Program to Stimulate Competitive Research Grants 0554548 and 1004057.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KY285083–KY285184 and KX538794–KX538799).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1612346114/-/DCSupplemental.
References
- 1.Becking B. Geobiologie of inleiding tot de milieukunde. W.P. Van Stockum & Zoon; The Hague, The Netherlands: 1934. [Google Scholar]
- 2.Foissner W. Biogeography and dispersal of micro-organisms: A review emphasizing protists. Acta Protozool. 2006;45(2):111. [Google Scholar]
- 3.Cohan FM. What are bacterial species? Annu Rev Microbiol. 2002;56(1):457–487. doi: 10.1146/annurev.micro.56.012302.160634. [DOI] [PubMed] [Google Scholar]
- 4.Mann DG, Vanormelingen P. An inordinate fondness? The number, distributions, and origins of diatom species. J Eukaryot Microbiol. 2013;60(4):414–420. doi: 10.1111/jeu.12047. [DOI] [PubMed] [Google Scholar]
- 5.Cermeño P, Falkowski PG. Controls on diatom biogeography in the ocean. Science. 2009;325(5947):1539–1541. doi: 10.1126/science.1174159. [DOI] [PubMed] [Google Scholar]
- 6.Fenchel T, Bland JF. The ubiquity of small species: Patterns of local and global diversity. Bioscience. 2004;54(8):777–784. [Google Scholar]
- 7.Finlay BJ. Global dispersal of free-living microbial eukaryote species. Science. 2002;296(5570):1061–1063. doi: 10.1126/science.1070710. [DOI] [PubMed] [Google Scholar]
- 8.Carr MH. Comparing marine and terrestrial ecosystems: Implications for the design of coastal marine reserves. Ecol Appl. 2003;13(sp1):90–107. [Google Scholar]
- 9.Powell TM. Physical and biological scales of variability in lakes, estuaries, and the coastal ocean. In: Powell TM, Steele JH, editors. Ecological Time Series. Springer; Boston: 1995. pp. 119–138. [Google Scholar]
- 10.Rusconi R, Stocker R. Microbes in flow. Curr Opin Microbiol. 2015;25:1–8. doi: 10.1016/j.mib.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 11.Amin SA, Parker MS, Armbrust EV. Interactions between diatoms and bacteria. Microbiol Mol Biol Rev. 2012;76(3):667–684. doi: 10.1128/MMBR.00007-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stocker R. Marine microbes see a sea of gradients. Science. 2012;338(6107):628–633. doi: 10.1126/science.1208929. [DOI] [PubMed] [Google Scholar]
- 13.Hofmann GE, et al. High-frequency dynamics of ocean pH: A multi-ecosystem comparison. PLoS One. 2011;6(12):e28983. doi: 10.1371/journal.pone.0028983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hardin G. The competitive exclusion principle. Science. 1960;131(3409):1292–1297. doi: 10.1126/science.131.3409.1292. [DOI] [PubMed] [Google Scholar]
- 15.Gifford SM, Sharma S, Booth M, Moran MA. Expression patterns reveal niche diversification in a marine microbial assemblage. ISME J. 2013;7(2):281–298. doi: 10.1038/ismej.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alexander H, Jenkins BD, Rynearson TA, Dyhrman ST. Metatranscriptome analyses indicate resource partitioning between diatoms in the field. Proc Natl Acad Sci USA. 2015;112(17):E2182–E2190. doi: 10.1073/pnas.1421993112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Casteleyn G, et al. Limits to gene flow in a cosmopolitan marine planktonic diatom. Proc Natl Acad Sci USA. 2010;107(29):12952–12957. doi: 10.1073/pnas.1001380107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hellweger FL, van Sebille E, Fredrick ND. Biogeographic patterns in ocean microbes emerge in a neutral agent-based model. Science. 2014;345(6202):1346–1349. doi: 10.1126/science.1254421. [DOI] [PubMed] [Google Scholar]
- 19.Jönsson BF, Watson JR. The timescales of global surface-ocean connectivity. Nat Commun. 2016;7:11239. doi: 10.1038/ncomms11239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rynearson TA, Armbrust EV. DNA fingerprinting reveals extensive genetic diversity in a field population of the centric diatom Ditylum brightwellii. Limnol Oceanogr. 2000;45(6):1329–1340. [Google Scholar]
- 21.Godhe A, et al. Physical barriers and environmental gradients cause spatial and temporal genetic differentiation of an extensive algal bloom. J Biogeogr. 2016;43(6):1130–1142. [Google Scholar]
- 22.Iglesias-Rodriguez MD, Schofield OM, Batley J, Medlin LK, Hayes PK. Intraspecific genetic diversity in the marine coccolithophore Emiliania huxleyi (Prymnesiophyceae): The use of microsatellite analysis in marine phytoplankton population studies. J Phycol. 2006;42:525–536. [Google Scholar]
- 23.Erdner DL, Richlen M, McCauley LAR, Anderson DM. Diversity and dynamics of a widespread bloom of the toxic dinoflagellate Alexandrium fundyense. PLoS One. 2011;6(7):e22965. doi: 10.1371/journal.pone.0022965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Round FE, Crawford RM, Mann DG. The Diatoms: Biology and Morphology of the Genera. Cambridge Univ Press; Cambridge, UK: 1990. [Google Scholar]
- 25.Pedrós-Alió C. Marine microbial diversity: Can it be determined? Trends Microbiol. 2006;14(6):257–263. doi: 10.1016/j.tim.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 26.Godhe A, Härnström K. Linking the planktonic and benthic habitat: Genetic structure of the marine diatom Skeletonema marinoi. Mol Ecol. 2010;19(20):4478–4490. doi: 10.1111/j.1365-294X.2010.04841.x. [DOI] [PubMed] [Google Scholar]
- 27.Rynearson TA, Newton JA, Armbrust EV. Spring bloom development, genetic variation and population succession in the planktonic diatom Ditylum brightwellii. Limnol Oceanogr. 2006;51:1249–1261. [Google Scholar]
- 28.Rynearson TA. Genetic differentiation among populations of the planktonic marine diatom Ditylum brightwellii (Bacillariophyceae) J Phycol. 2004;40(1):34–43. [Google Scholar]
- 29.Evans KM. High levels of genetic diversity and low levels of genetic differentiation in North Sea Pseudo-nitzchia pungens (Bacillariophyceae) populations. J Phycol. 2005;41(3):506–514. [Google Scholar]
- 30.Chen G, Rynearson TA. Genetically distinct populations of a diatom co-exist during the North Atlantic spring bloom. Limnol Oceanogr. 2016;61(6):2165–2179. [Google Scholar]
- 31.Rynearson TA, Armbrust EV. Maintenance of clonal diversity during a spring bloom of the centric diatom Ditylum brightwellii. Mol Ecol. 2005;14(6):1631–1640. doi: 10.1111/j.1365-294X.2005.02526.x. [DOI] [PubMed] [Google Scholar]
- 32.Godhe A, et al. Seascape analysis reveals regional gene flow patterns among populations of a marine planktonic diatom. Proc Biol Sci. 2013;280(1773):20131599. doi: 10.1098/rspb.2013.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whittaker KA, Rignanese DR, Olson RJ, Rynearson TA. Molecular subdivision of the marine diatom Thalassiosira rotula in relation to geographic distribution, genome size, and physiology. BMC Evol Biol. 2012;12(1):209. doi: 10.1186/1471-2148-12-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGuire JA, et al. Mitochondrial introgression and incomplete lineage sorting through space and time: Phylogenetics of crotaphytid lizards. Evolution. 2007;61(12):2879–2897. doi: 10.1111/j.1558-5646.2007.00239.x. [DOI] [PubMed] [Google Scholar]
- 35.Koblmüller S, Egger B, Sturmbauer C, Sefc KM. Rapid radiation, ancient incomplete lineage sorting and ancient hybridization in the endemic Lake Tanganyika cichlid tribe Tropheini. Mol Phylogenet Evol. 2010;55(1):318–334. doi: 10.1016/j.ympev.2009.09.032. [DOI] [PubMed] [Google Scholar]
- 36.Rieseberg LH, Wendel JF. 1993. Introgression and its consequences in plants. Hybrid Zones and the Evolutionary Process, ed Harrison RG (Oxford Univ Press, Oxford), pp 70–109.
- 37.Wright S. The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution. 1965;19(3):395–420. [Google Scholar]
- 38.Wright S. Evolution and the Genetics of Populations. Vol 4. University of Chicago Press; Chicago: 1978. p. 590. [Google Scholar]
- 39.Hedrick PW. Perspective: Highly variable loci and their interpretation in evolution and conservation. Evolution. 1999;53(2):313–318. doi: 10.1111/j.1558-5646.1999.tb03767.x. [DOI] [PubMed] [Google Scholar]
- 40.Bernard AM, et al. Global population genetic dynamics of a highly migratory, apex predator shark. Mol Ecol. 2016;25(21):5312–5329. doi: 10.1111/mec.13845. [DOI] [PubMed] [Google Scholar]
- 41.LeDuc R, et al. Genetic differences between western and eastern gray whales (Eschrichtius robustus) J Cetacean Res Manag. 2002;4(1):1–5. [Google Scholar]
- 42.Laconcha U, et al. New nuclear SNP markers unravel the genetic structure and effective population size of albacore tuna (Thunnus alalunga) PLoS One. 2015;10(6):e0128247. doi: 10.1371/journal.pone.0128247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rousset F. Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics. 1997;145(4):1219–1228. doi: 10.1093/genetics/145.4.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chepurnov VA. Sexual reproduction, mating system, chloroplast dynamics and abrupt cell size reduction in Pseudo-nitzschia pungens from the North Sea (Bacillariophyta) Eur J Phycol. 2005;40(4):379–395. [Google Scholar]
- 45.D’Alelio D, et al. The time for sex: A biennial life cycle in a marine planktonic diatom. Limnol Oceanogr. 2010;55(1):106. [Google Scholar]
- 46.von Dassow P, Montresor M. Unveiling the mysteries of phytoplankton life cycles: Patterns and opportunities behind complexity. J Plankton Res. 2011;33(1):3–12. [Google Scholar]
- 47.McQuoid MR, Hobson LA. Importance of resting stages in diatom seasonal succession. J Phycol. 1995;31(1):44–50. [Google Scholar]
- 48.Rynearson TA, et al. Major contribution of diatom resting spores to vertical flux in the sub-polar North Atlantic. Deep Sea Res Part I Oceanogr Res Pap. 2013;82(0):60–71. [Google Scholar]
- 49.Zhang Y, Lu S, Zhang C, Gao Y. Distribution and germination of viable diatom resting stage cells in sediments of the East China Sea. Acta Oceanol Sin. 2010;29(5):121–128. [Google Scholar]
- 50.Garrison DL. Monterey Bay Phytoplankton. II. Resting spore cycles in coastal diatom populations. J Plankton Res. 1981;3(1):137–156. [Google Scholar]
- 51.Boyd PW, et al. Marine phytoplankton temperature versus growth responses from polar to tropical waters: Outcome of a scientific community-wide study. Plos One. 2013;8(5):e63091. doi: 10.1371/journal.pone.0063091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Doblin MA, van Sebille E. Drift in ocean currents impacts intergenerational microbial exposure to temperature. Proc Natl Acad Sci USA. 2016;113(20):5700–5705. doi: 10.1073/pnas.1521093113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pan Y, Subba Rao DV, Mann K, Li W, Warnock R. Temperature dependence of growth and carbon assimilation in Nitzschia pungens f. multiseries, the causative diatom of domoic acid poisoning. In: Smayda TJ, Shimizu Y, editors. Proceedings of the Fifth International Conference on Toxic Marine Phytoplankton. Elsevier; New York: 1993. pp. 619–624. [Google Scholar]
- 54.Harnstrom K, Ellegaard M, Andersen TJ, Godhe A. Hundred years of genetic structure in a sediment revived diatom population. Proc Natl Acad Sci USA. 2011;108(10):4252–4257. doi: 10.1073/pnas.1013528108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.LeBlond PH. The Strait of Georgia: Functional anatomy of a coastal sea. Can J Fish Aquat Sci. 1983;40(7):1033–1063. [Google Scholar]
- 56.Pilson MQ. On the residence time of water in Narragansett Bay. Estuaries. 1985;8(1):2–14. [Google Scholar]
- 57.Robinson G, Aiken J, Hunt H. Synoptic surveys of the western English Channel. The relationships between plankton and hydrography. J Mar Biol Assoc U K. 1986;66(01):201–218. [Google Scholar]
- 58.Hartl DL, Clark A. Principles of Population Genetics. Sinauer; Sunderland, MA: 2007. [Google Scholar]
- 59.Falkowski P, Kiefer DA. Chlorophyll a fluorescence in phytoplankton: Relationship to photosynthesis and biomass. J Plankton Res. 1985;7(5):715–731. [Google Scholar]
- 60.Raymond M. An exact test for population differentiation. Evolution. 1995;49(6):1280. doi: 10.1111/j.1558-5646.1995.tb04456.x. [DOI] [PubMed] [Google Scholar]
- 61.Raymond M, Rousset F. GENEPOP (Version 1.2): Population genetics software for exact tests and ecumenicism. J Hered. 1995;86(3):248–249. [Google Scholar]
- 62.Peakall R, Smouse PE. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research: An update. Bioinformatics. 2012;28(19):2537–2539. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McRae BH. Isolation by resistance. Evolution. 2006;60(8):1551–1561. [PubMed] [Google Scholar]
- 64.Jensen JL, Bohonak AJ, Kelley ST. Isolation by distance, web service. BMC Genet. 2005;6:13. doi: 10.1186/1471-2156-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Clarke K, Ainsworth M. A method of linking multivariate community structure to environmental variables. Mar Ecol Prog Ser. 1993;92:205–219. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.