Significance
Autophagy contributes to innate immune responses in metazoans by targeted elimination of intracellular pathogens, including viruses, in a process termed “xenophagy.” Whether autophagy has a similar role in plant immunity is unknown. Here we demonstrate that the selective autophagy receptor NEIGHBOR OF BRCA1 (NBR1) binds the viral capsid protein and particles of cauliflower mosaic virus (CaMV) and mediates their autophagic degradation. We further demonstrate that this antiviral xenophagy is counteracted by protective functions of autophagy-resistant CaMV inclusion bodies. Finally, we show that a second, nonselective NBR1-independent autophagy pathway promotes plant viability during infection and serves as a proviral mechanism to extend the timespan for virus production and potential CaMV transmission. Thus autophagy exhibits important pro- and antiviral roles in compatible plant–virus interactions.
Keywords: selective autophagy, plant virus, xenophagy, innate immunity, cauliflower mosaic virus
Abstract
Autophagy plays a paramount role in mammalian antiviral immunity including direct targeting of viruses and their individual components, and many viruses have evolved measures to antagonize or even exploit autophagy mechanisms for the benefit of infection. In plants, however, the functions of autophagy in host immunity and viral pathogenesis are poorly understood. In this study, we have identified both anti- and proviral roles of autophagy in the compatible interaction of cauliflower mosaic virus (CaMV), a double-stranded DNA pararetrovirus, with the model plant Arabidopsis thaliana. We show that the autophagy cargo receptor NEIGHBOR OF BRCA1 (NBR1) targets nonassembled and virus particle-forming capsid proteins to mediate their autophagy-dependent degradation, thereby restricting the establishment of CaMV infection. Intriguingly, the CaMV-induced virus factory inclusions seem to protect against autophagic destruction by sequestering capsid proteins and coordinating particle assembly and storage. In addition, we found that virus-triggered autophagy prevents extensive senescence and tissue death of infected plants in a largely NBR1-independent manner. This survival function significantly extends the timespan of virus production, thereby increasing the chances for virus particle acquisition by aphid vectors and CaMV transmission. Together, our results provide evidence for the integration of selective autophagy into plant immunity against viruses and reveal potential viral strategies to evade and adapt autophagic processes for successful pathogenesis.
Autophagy is a conserved intracellular pathway that engages specialized double-membrane vesicles, called “autophagosomes,” to enclose and transport cytoplasmic content to lytic compartments for degradation and subsequent recycling (1). Autophagosome formation relies on extensive membrane rearrangements and is mediated by the concerted action of a core set of autophagy-related proteins (ATGs) (2, 3). At basal levels, autophagy serves mainly housekeeping functions in cellular homeostasis, whereas stimulated autophagy activity facilitates adaptation to developmental and environmental stress conditions including starvation, aging, and pathogen infection (1, 4). Ample evidence now indicates that autophagy, initially recognized as a mainly bulk catabolic process, is able specifically to target and degrade a multitude of cellular structures ranging from individual and aggregated proteins to entire organelles and invading microbes (5, 6). Selectivity is provided by a growing number of autophagic adaptor or receptor proteins identified in eukaryotic organisms that recruit the cargo to the developing autophagosome through interaction with membrane-associated ATG8/LC3 proteins (7, 8). Several mammalian autophagy receptors have been implicated in the targeting of intracellular bacterial and viral pathogens in a process called “xenophagy” (8–10). For instance, the cargo receptor p62 (SQSTM1) was shown to bind directly to and mediate autophagic clearance of different viral capsid proteins (11–13).
Autophagy is induced by a wide range of viruses and has been ascribed both anti- and proviral roles during animal infections (14, 15). Autophagic mechanisms contribute to various aspects of adaptive and innate immunity, but many viruses have evolved measures to suppress, evade, or even exploit the autophagy pathway for their own benefit (10). Thus, autophagy-dependent processes were found to be adapted for viral replication, particle maturation and release, and the prolongation of cellular lifespan by preventing host cell death (10). In plants, autophagy was shown to control hormone levels and signaling in basal resistance to hemibiotrophic bacteria and necrotrophic fungi and to regulate defense- and disease-associated cell death (16). In addition, the recent identification of an ATG8-binding oomycete effector that antagonizes the autophagy cargo receptor NEIGHBOR OF BRCA1 (NBR1)/JOKA2 suggests an important role of selective autophagy in plant immune responses (17). However, the molecular mechanisms and functions of autophagy in the regulation of immunity and pathogenesis during compatible plant virus infections remain largely unknown.
In this study we investigated how bulk and selective autophagy pathways affect viral accumulation and disease development in response to cauliflower mosaic virus (CaMV; family Caulimoviridae) infection. CaMV is a plant pararetrovirus with a double-stranded DNA genome and is a compatible pathogen of the model plant Arabidopsis thaliana. The cellular infection cycle of CaMV involves nuclear transcription of the circular genome followed by translation into six viral proteins, reverse transcription of pregenomic RNA, and particle assembly that occurs in cytoplasmic inclusions referred to as “viroplasms” or “viral factories” (VFs) (18). Here, we report that NBR1 binds the viral capsid protein and particles to mediate their autophagic degradation, and we show that this antiviral process is antagonized by protective functions of the autophagy-resistant inclusions. Furthermore, we demonstrate that NBR1-independent autophagy facilitates the survival of infected plants and serves CaMV by extending the timespan for particle production and potential viral transmission.
Results
Autophagy Promotes Plant Fitness During CaMV Infection.
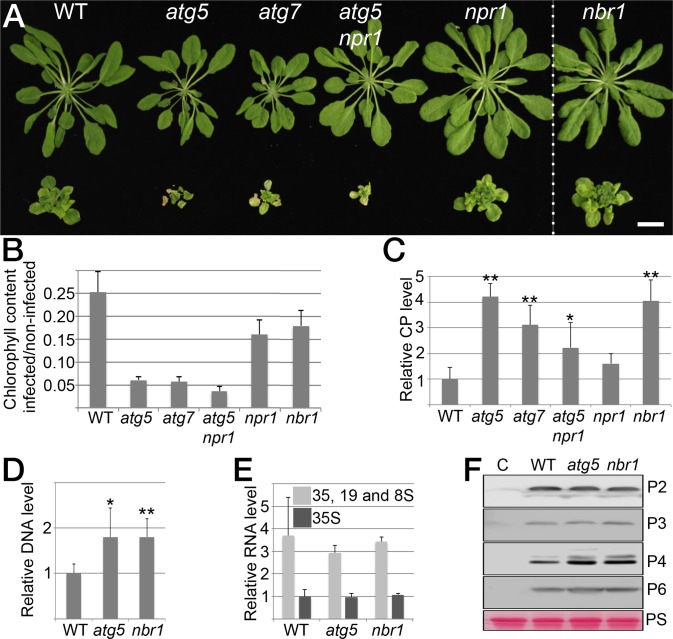
To address whether autophagy affects disease development during CaMV infection, we first evaluated symptom severity in Arabidopsis mutants defective in the core autophagy machinery (atg5, atg7) and the NBR1-mediated selective autophagy pathway (nbr1). Twenty-eight days after inoculation (dai) with the CaMV isolate CM1841, disease symptoms such as stunting, chlorosis, and vein clearing were considerably more pronounced in atg5 and atg7 plants than in nbr1 and Col-0 wild-type (WT) plants (Fig. 1A). In addition, mature leaves of infected atg5 and atg7 plants developed signs of early senescence and tissue necrosis that were absent in nbr1, WT, and noninfected atg mutants. Age- and stress-induced senescence in autophagy-deficient mutants is known to be stimulated by elevated levels of salicylic acid (SA) and to be dampened by knockout of the SA response regulator NONEXPRESSOR OF PR1 (NPR1) in the atg5 npr1 double mutant (19, 20). However, we observed that infected atg5 npr1 plants developed symptoms similar to and occasionally even more severe than the symptoms in atg5 plants (Fig. 1A), suggesting that the increased disease phenotype triggered by autophagy deficiency is not coupled to SA-dependent NPR1 stress responses during CaMV infection. We then analyzed total chlorophyll content as a proxy of biomass accumulation and plant fitness (Fig. 1B). Consistent with the visual phenotype, infection-induced chlorophyll loss was much higher in atg5, atg7, and atg5 npr1 than in WT plants. In nbr1, the effect of CaMV infection on chlorophyll content was less severe than in the atg mutant backgrounds but still was significantly stronger than in WT plants. Together, these results indicate that autophagy has a critical role in maintaining overall plant fitness during CaMV infection and is largely independent of NBR1 and NPR1.
Fig. 1.
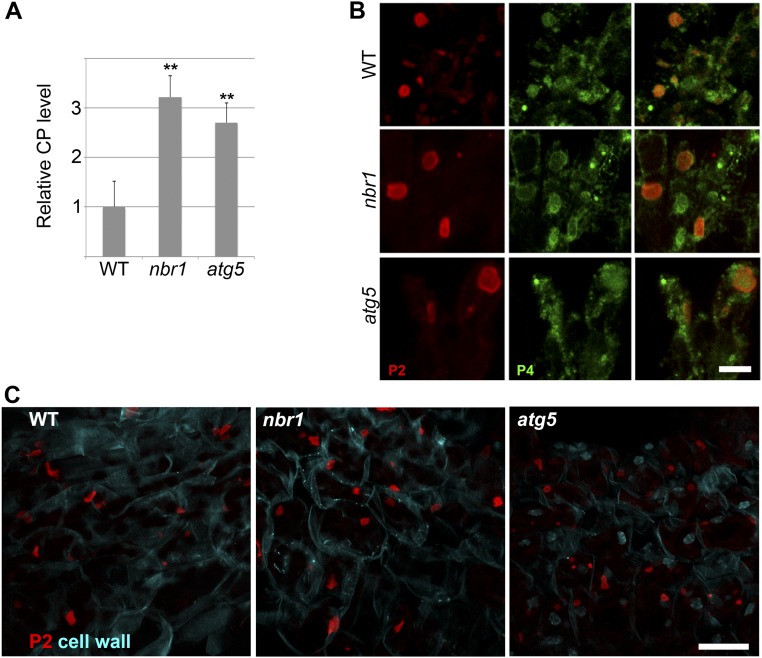
Autophagy promotes plant fitness and suppresses CaMV accumulation. (A) Virus-induced symptoms in WT, atg5, atg7, atg5 npr1, npr1, and nbr1 plants at 28 dai with CaMV strain CM1841 (Lower Row) compared with noninfected controls (Upper Row). (Scale bar, 20 mm.) (B) Ratio of total chlorophyll content in infected and noninfected plants. Error bars represent SD (n = 6). (C) CaMV capsid protein (CP; P4) levels determined by ELISA in systemic leaves of the indicated genotypes at 14 dai. Values are shown as means ± SD (n = 4) and are presented as arbitrary units relative to WT plants. (D) CaMV DNA levels determined by qPCR in systemic leaves of WT, atg5, and nbr1 plants at 14 dai. Values represent means ± SD (n = 4) relative to WT plants and were normalized with 18S ribosomal DNA as the internal reference. (E) CaMV RNA levels determined by RT-qPCR as an individual 35S transcript or via the leader sequence as the sum of 35S, 19S, and 8S transcripts in systemic leaves of WT, atg5, and nbr1 plants at 14 dai. Values are shown as means ± SD (n = 3 biological replicates) relative to the WT 35S transcript level. (F) Immunoblot analysis of CaMV P2, P3, P4, and P6 accumulation in WT, atg5, and nbr1 plants. Total proteins were extracted from whole plants at 14 dai and probed with specific antibodies. Noninfected WT plants served as control (C) for signal background, and Ponceau S (PS) staining verified comparable protein loading. Statistical significance (*P < 0.05; **P < 0.01) was revealed by Student´s t test (compared with WT).
NBR1-Mediated Selective Autophagy Suppresses CaMV P4 and DNA Accumulation.
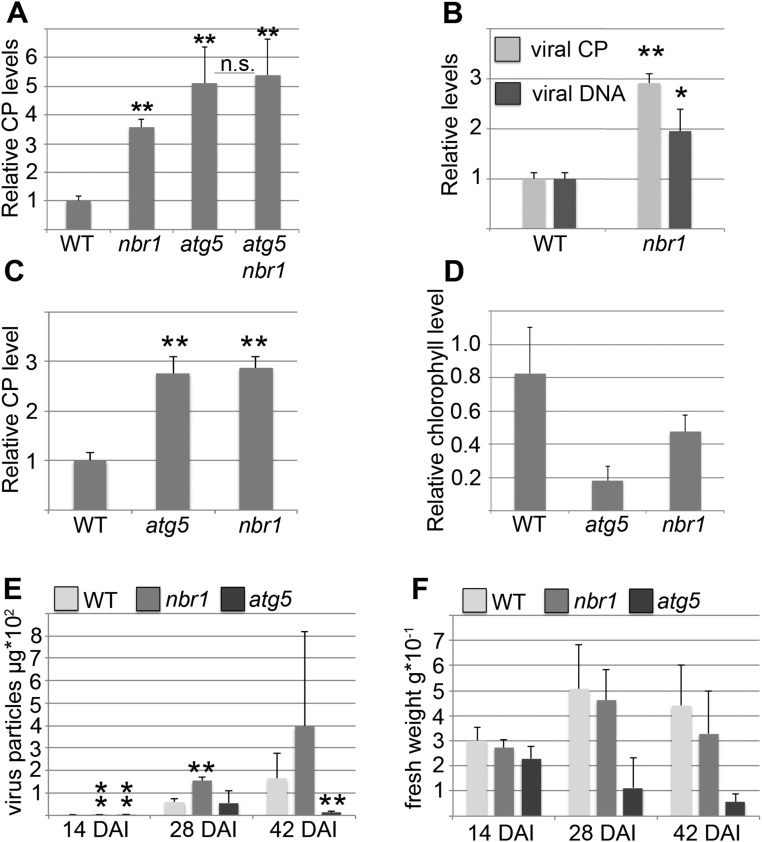
Next, we investigated whether altered symptom severity and plant fitness in autophagy-deficient mutants is accompanied by changes in CaMV accumulation. We used ELISA to determine the abundance of the viral capsid protein P4 in plant lysates at 14 dai and found a two- to fourfold increase in atg5 and atg7 mutants as compared with WT plants (Fig. 1C). Notably, the nbr1 mutant accumulated P4 to similarly high levels as atg5, indicating that the severe infection phenotype of the core autophagy mutants is not directly correlated with virus titer. Reduced capsid protein levels in the strongly symptomatic atg5 npr1 double mutant compared with atg5 further supported this notion (Fig. 1C). Because of the apparent uncoupling of symptom severity and P4 levels in atg5 and nbr1 mutants, we generated an atg5 nbr1 double mutant to assess genetically whether the impact of NBR1 on CaMV accumulation is indeed autophagy dependent. We did not detect an additional increase in P4 levels in atg5 nbr1 as compared with atg5 plants (Fig. S1A), verifying that NBR1-mediated suppression of virus infection acts through the autophagy pathway.
Fig. S1.
NBR1-dependent autophagy restricts CaMV infection in a persistent and isolate-independent manner, whereas bulk autophagy prevents disease-associated senescence and prolongs the timespan for virus production. (A) Capsid protein (CP) levels for CM1841 at 14 dai in nbr1 and atg5 single mutants and in atg5 nbr1 double mutants relative to WT plants. Values represent means ± SD (n = 4 biological replicates). (B) Viral titers of CaMV strain CM1841 in WT and nbr1 plants indicate that CaMV infection is suppressed similarly by NBR1-mediated autophagic processes at 28 dai and at 14 dai (Fig.1 C and D). Capsid protein and genomic DNA levels were determined by ELISA and qPCR, respectively; values represent means ± SD (n = 4) relative to WT plants. (C) Capsid protein levels of CaMV strain Cabb B-JI in WT, atg5, and nbr1 plants at 14 dai reveal that autophagy-mediated effects are not specific to the CM1841 strain (Fig. 1) or caused by the different inoculation methods (mechanical inoculation of purified Cabb B-JI particles vs. agroinfection of CM1841 DNA). Values are means ± SD (n = 4) relative to WT plants. (D) Ratio of total chlorophyll content in infected and noninfected plants. Error bars represent the SD (n = 6). (E) Capsid protein levels per individual CaMV Cabb B-JI–infected WT, atg5, and nbr1 plant at 14, 28, and 42 dai reveal that the total amount of virus accumulation is significantly reduced in atg5 plants at late stages of infection. Values represent means ± SD (n = 6) of virus particles per plant. (F) The fresh weight of plants was determined in parallel with the viral capsid protein levels in E, indicating premature death in atg5 plants. Values represent means of fresh weights for individual plants ± SD (n = 6). Asterisks indicate statistical significance (*P < 0.05; **P < 0.01) determined by Student’s t test (compared with WT); n.s., not significant.
P4 is the major structural protein of the CaMV capsid. The ELISA results thus implicated an increased accumulation of viral particles in autophagy- and NBR1-deficient plants. To analyze these results further, we compared the amounts of virus DNA in atg5 and nbr1 mutants and WT plants using real-time quantitative PCR (qPCR) and found increased amounts of viral DNA in the mutants as compared with WT plants (Fig. 1D). A substantial increase in viral DNA and P4 protein levels was also detectable at a later time point (28 dai) in nbr1 plants (Fig. S1B), demonstrating that NBR1-dependent autophagy persistently restricts CaMV accumulation. We noted that the accumulation of CaMV P4 was higher than the accumulation of CaMV DNA in nbr1 relative to WT plants, suggesting that NBR1 also could mediate the degradation of non–particle-associated P4 (Fig. S1B).
To determine whether the observed phenotypes were isolate specific, we repeated the experiments using a different isolate, Cabb B-JI. As with CM1841, Cabb B-JI accumulated to high levels, and symptom-related chlorophyll loss was increased in both atg5 and nbr1 mutants relative to WT plants (Fig. S1 C and D), indicating that autophagy has a general, isolate-independent role during CaMV infection. However, the analysis of virus production and plant biomass over an extended time course showed that Cabb B-JI–infected atg5 plants have a reduced lifespan and accumulate many fewer particles in total than WT plants and nbr1 mutants (Fig. S1 E and F), revealing the importance of NBR1-independent autophagy in maintaining plant vigor and virus replication.
One of the earliest events in the cellular infection cycle of CaMV is the release of viral genomic DNA from the particle into the nucleus, followed by the transcription of 35S, 19S, and 8S viral RNAs (21). Viral proteins P1–P5 are subsequently translated from the 35S transcript, and P6 is translated from the 19S transcript. To dissect the impact of autophagy on CaMV infection further, we determined the abundance of viral RNAs and proteins in autophagy mutants. qPCR analysis indicated that the levels of viral RNAs were not altered by autophagy deficiency. Similarly, immunoblot analysis revealed no difference in P2, P3, and P6 proteins, but P4 was specifically enhanced in the atg5 and nbr1 mutants as compared with WT plants (Fig. 1F). Unfortunately, P1 and P5 levels could not be determined because of the lack of available antibodies. Nevertheless, unaltered RNA as well as P2, P3 and P6 levels indicated that autophagy does not interfere with CaMV transcription and translation but instead specifically suppresses the accumulation of CaMV P4 and viral DNA at the cellular level.
NBR1-Containing Autophagosomes Are Induced upon CaMV Infection.
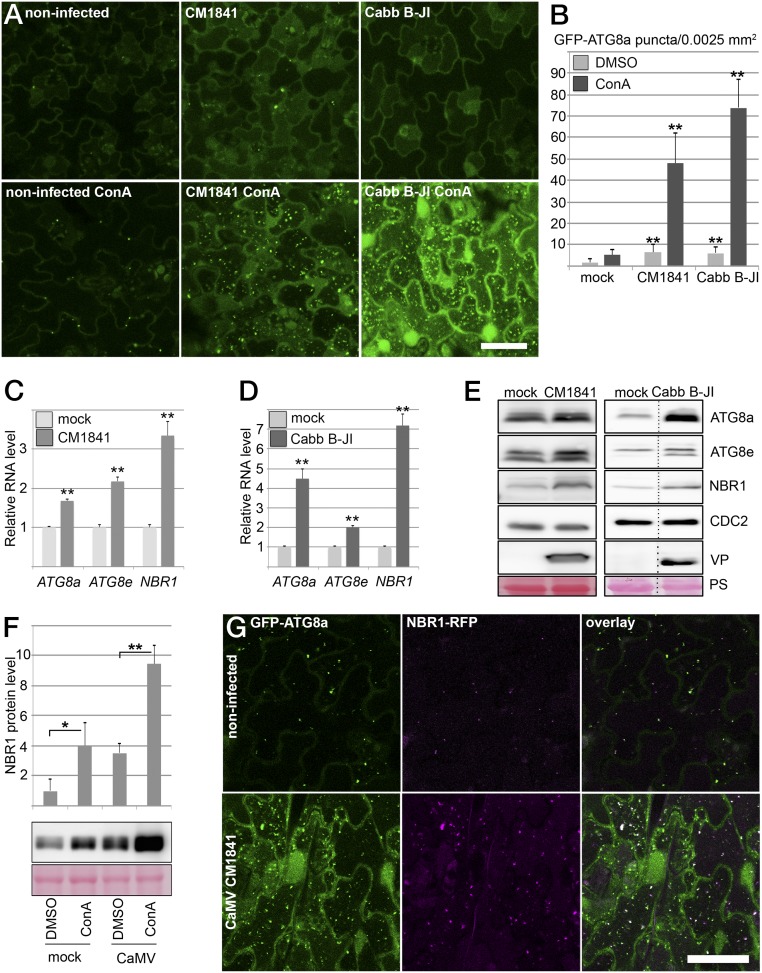
The increased disease development and CaMV accumulation observed in autophagy-deficient mutants prompted us to analyze whether autophagy levels are altered during CaMV infection. Autophagosome formation typically is monitored by GFP fusions with ATG8 isoforms, which bind to autophagosomal membranes and remain associated with the completed vesicles until their lytic destruction in the vacuole. We inoculated a GFP-ATG8a– expressing marker line with CaMV strains Cabb B-JI and CM1841 and found only a modest increase of GFP-labeled autophagosomal structures in systemically infected tissue as compared with noninfected plants (Fig. 2 A and B). However, when we applied concanamycin A (ConA) to inhibit vacuolar acidification and thereby autophagosome turnover, we observed greatly increased numbers of autophagic bodies in CaMV-infected cells. This observation establishes that CaMV infection triggers the formation of autophagosomes, which are efficiently delivered to the vacuole for degradation.
Fig. 2.
NBR1-mediated selective autophagy is induced upon CaMV infection. (A) Transgenic expression of GFP-ATG8a for detection of autophagosome structures in uninfected leaf tissue (control) and in leaf tissue infected with CaMV CM1841 or Cabb B-JI at 14 dai in the absence (Upper Row) or presence of ConA compared with the noninfected control. Images represent single confocal planes from abaxial epidermal cells and were taken with identical microscope settings. (B) Quantification of GFP-ATG8a puncta in mock-infected and infected tissue after DMSO or ConA treatment. The number of puncta was calculated from areas of 0.0025 mm2. Values represent means ± SD (n ≥ 16 independent areas). (C and D) qPCR analysis of ATG8a, ATG8e, and NBR1 transcript levels in WT plants infected with CaMV CM1841 (C) or Cabb B-JI (D) at 14 dai compared with noninfected plants (mock). Values represent the mean ± SD (n = 3 biological replicates) relative to the noninfected control. (E) Immunoblot analysis of the levels of ATG8a, ATG8e, and NBR1 proteins in CaMV-infected and control samples from C and D. The accumulation of the cell-cycle protein CDC2 and Ponceau S staining (PS) were used as loading control, and detection of viral proteins (VP; P6 for CM1841 and P4 for Cabb B-JI) verified successful infection. (F, Upper) Direct ELISA quantitation of NBR1 protein levels in mock-infected and infected tissue (Cabb B-JI; 21 dai) after DMSO and ConA treatment. Values represent means ± SD (n = 3 biological replicates). (Lower) Immunoblot detection of NBR1 in the identical samples; Ponceau S staining served as the loading control. (G) Colocalization of NBR1-RFP and GFP-ATG8a in CaMV CM1841-infected leaves after ConA treatment in comparison with the control. Imaging was done with identical settings, and single confocal planes are shown together with the overlay. (Scale bars in A and G, 20 μm.) Statistical significance (*P < 0.05; **P < 0.01) was revealed by Student´s t test (compared with mock treatment in B, C, and D and with DMSO control in F).
We next monitored the expression of the autophagy core genes ATG8a and ATG8e and the autophagy receptor NBR1, which are all elevated as part of an induced autophagy response during plant heat stress (22). Both CaMV isolates stimulated the accumulation of the autophagic markers’ transcripts and proteins relative to the noninfected control (Fig. 2 C–E). However, the effect seemed to be more pronounced with Cabb B-JI, causing a greater increase of ATG8a and NBR1 transcripts and ATG8a and ATG8e protein levels compared with CM1841 (Fig. 2 C–E). Notably, the strong transcriptional up-regulation of NBR1 in response to Cabb B-JI occurred without concomitant protein accumulation, suggesting enhanced NBR1 turnover and thus autophagic flux (23). To verify this notion, we used direct ELISA to monitor NBR1 levels at steady state and after ConA treatment in Cabb B-JI–infected leaves. We found that ConA-mediated inhibition of vacuolar degradation increased NBR1 protein levels in both infected and control tissue. However, the total amount of NBR1 stabilized by ConA was approximately doubled in response to Cabb B-JI (Fig. 2F), further supporting the observed increase in vacuolar delivery and turnover of autophagosomes upon CaMV infection (Fig. 2B).
During selective autophagy, NBR1 is recruited to the developing autophagosome through interaction with ATG8 proteins (24). Hence, we analyzed the association of NBR1 with infection-induced autophagosomal structures in plants stably expressing NBR1-RFP and GFP-ATG8a after ConA treatment (Fig. 2G). Compared with the noninfected control, NBR1-RFP–labeled punctate structures were more abundant in infected samples and colocalized mainly with GFP-ATG8a. The relatively few NBR1 structures present in noninfected cells were equally colabeled by GFP-ATG8a, in agreement with previous reports (24). Together, these results suggest that the strong induction of NBR1 during CaMV infection is functionally connected to enhanced autophagy levels and flux.
NBR1 Binds to P4 and Viral Particles.
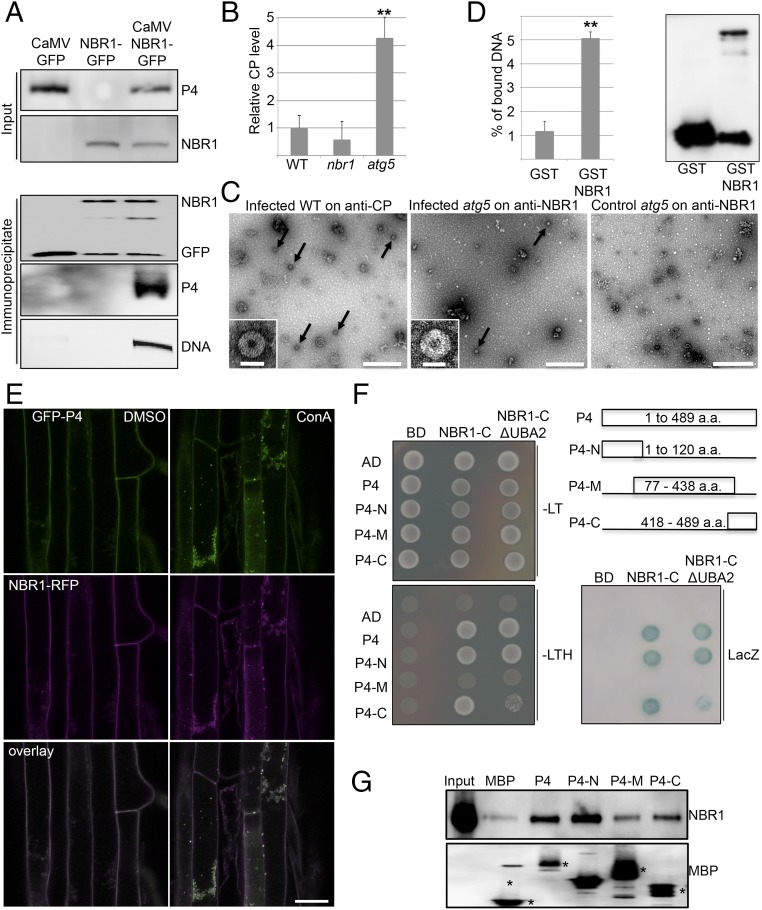
To analyze whether NBR1 directly targets P4 and viral particles in infected plant tissue, we carried out immune pulldown experiments and found that both P4 and viral DNA coimmunoprecipitated with NBR1-GFP in stably transformed transgenic plants (Fig. 3A). We then infected WT, nbr1, and atg5 plants and applied a customized ELISA that used the anti-NBR1 antibody for coating and the anti-P4 antibody for detection. Notably, P4 capture via NBR1 was clearly detectable and was fourfold higher in atg5 mutants than in WT plants (Fig. 3B). We reasoned that NBR1-mediated degradation of viral particles would explain these findings and therefore used immunocapture transmission electron microscopy (IC-TEM) to confirm the presence of NBR1-associated viral particles. TEM grids coated with anti-P4 antibody, which captured the characteristic 50-nm diameter CaMV particles along with other unidentified structures from infected plant lysates (Fig. 3C), were used as a positive control. Intriguingly, anti-NBR1 grids also captured CaMV particles from lysates of infected atg5 mutants (Fig. 3C) but not from lysates of WT plants or nbr1 mutants. This result demonstrated that NBR1 binds to viral particles during infection, and the prevalent accumulation of these complexes in atg5 mutants further supported an efficient clearance by autophagy in WT plants. Finally, we performed an in vitro pulldown experiment to verify that NBR1 is able to bind viral particles directly (Fig. 3D). We used immobilized GST-tagged NBR1 protein that harbored a PB1 domain mutation (K11A) to prevent aggregation (24) and applied CaMV particles that had been purified from infected nbr1 plants to avoid potential interference by particle-associated plant NBR1. We detected a fivefold enrichment of viral DNA in GST-NBR1K11A samples compared with the GST control, demonstrating efficient capture of viral particles by recombinant NBR1.
Fig. 3.
NBR1 binds to CaMV P4 and viral particles. (A) Presence of CaMV P4 in NBR1-GFP immunocomplexes from systemically infected transgenic plants. Immunoprecipitation was performed with anti-GFP monoclonal antibody; mock-treated NBR1-GFP as well as CaMV-infected GFP transgenic served as negative controls. Immunoblot analysis with anti-GFP and anti-P4 antibodies is shown for input and immunoprecipitated fractions, respectively, and viral DNA was detected by PCR using primers specific for the CaMV P1 movement protein. (B) Capture of CaMV P4 from lysates of infected WT, nbr1, and atg5 plants using anti-NBR1 antibody for coating and anti-P4 antibody for detection in a customized ELISA. Error bars represent SD (n = 3). (C) IC-TEM of CaMV particles (arrows) from infected WT and atg5 plants using anti-P4 and anti-NBR1 antibodies, respectively. Noninfected atg5 plants served as a negative control. (Scale bars, 500 nm.) Insets show enlargement of typical 50-nm particle structures. (Scale bars, 50 nm.) (D) In vitro pulldown of viral particles by the nonaggregating mutant GST-NBR1K11A. GST alone and GST-NBR1K11A were coupled to glutathione Sepharose and incubated with viral particles purified from infected nbr1 mutant plants. (Left) The amount of bound viral particles in eluates was quantified by qPCR of genomic DNA and is given as the percent relative to the input. (Right) Immunoblot analysis with anti-GST antibodies indicates the presence of GST proteins in eluates. (E) Colocalization of GFP-P4 and RFP-NBR1 in roots of transgenic Arabidopsis seedlings after treatment with DMSO and 0.5 μM ConA. Confocal images represent single planes of individual channels and the overlays. (Scale bar, 20 μm.) (F) Y2H-based interaction of P4 variants with the NBR1 C terminus that contains (NBR1-C) or lacks (NBR1-C ΔUBA2) the UBA2 and LIR domains. Yeast cells coexpressing P4, P4-N, P4-M, and P4-C as AD fusion and NBR1-C as BD fusion show growth on selective medium [without leucine and tryptophan (−LT) or without leucine, tryptophan, and histidine (−LTH)] and LacZ activity. Combinations with empty BD or AD vectors served as controls for auto-activation. The different P4 fragments and their amino acid coordinates are illustrated. (G) In vitro binding of GST-tagged NBR1K11A to MBP-fused P4 variants. MBP alone and MBP fusions were immobilized on agarose and incubated with recombinant GST-NBR1K11A protein. Immunoblot analysis of the eluates was performed with anti-NBR1 antibodies to indicate specific NBR1 binding (Upper) and with anti-MBP antibodies to demonstrate the amount of matrix-bound MBP proteins (asterisks) (Lower). Statistical significance (**P < 0.01) was revealed by Student’s t test (compared with WT in B and with GST in D).
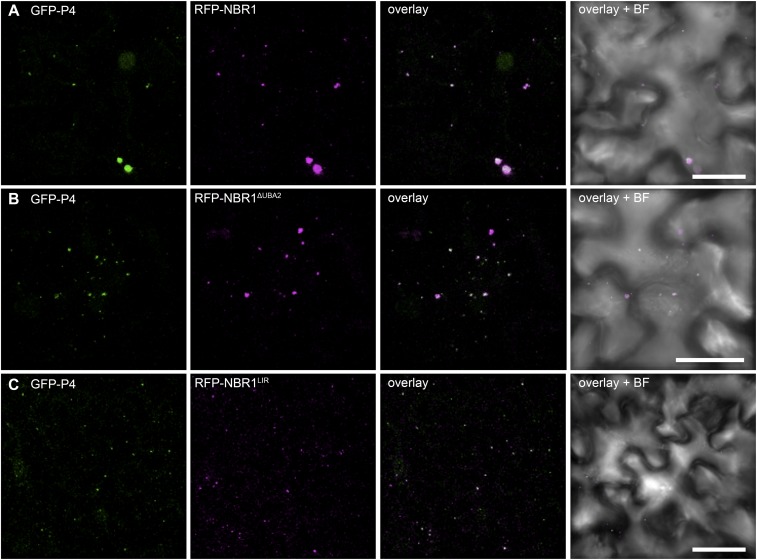
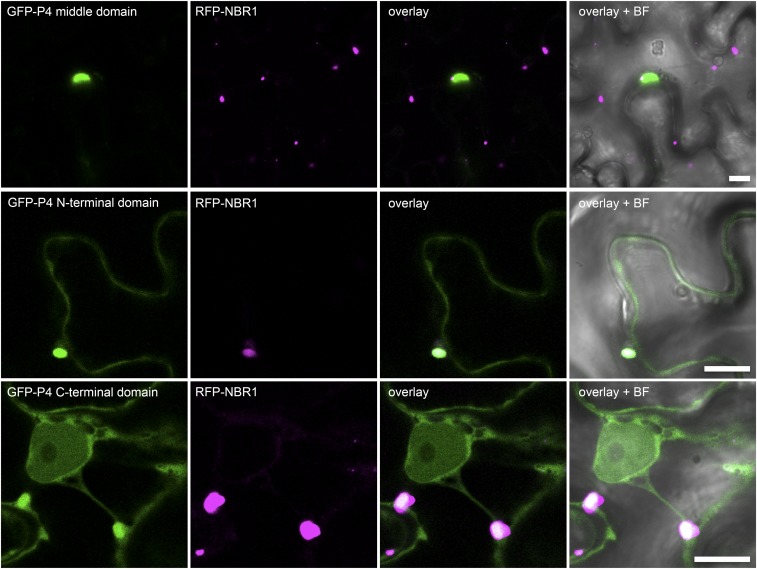
Next, we investigated whether NBR1 could associate with P4 in the absence of virus infection and coordinated particle assembly. NBR1-RFP and GFP-P4 colocalized together in distinct foci upon ConA treatment of transgenic Arabidopsis plants stably coexpressing both proteins, supporting the hypothesis that P4 is targeted by NBR1-mediated autophagy (Fig. 3E). We then addressed the interaction between NBR1 and P4 using the yeast two-hybrid (Y2H) system. Both NBR1 and P4 showed autoactivation of reporter genes when fused to the GAL4 binding domain (BD) and therefore could not be tested. Fortunately, the C-terminal fragment of NBR1 (NBR1-C), which carries the LC3-interacting region (LIR) and two flanking ubiquitin-associated (UBA) domains within amino acids 505–704 (24), did not auto-activate as a BD fusion and showed positive interaction with the activation domain (AD)-fused P4 (Fig. 3F). Of note, P4 has previously been shown to contain instability determinants in the N- and C-terminal domains (25), which potentially could be responsible for NBR1-mediated targeting and subsequent degradation. Indeed, both N- and C-terminal regions, but not the middle (M) part of P4, interacted with NBR1 (Fig. 3F). Deletion of the LIR and main ubiquitin-binding (UBA2) domains from the NBR1-C fragment did not impact binding to full-length P4 and the P4 N-terminal region (P4-N), but the interaction with the P4 C-terminal region (P4-C) was substantially weakened. To assess whether the interaction between P4 and NBR1 was direct, we performed in vitro binding assays with maltose-binding protein (MBP) fusions of the P4 variants and GST-NBR1K11A. Full-length P4 and P4-N displayed significant binding to NBR1K11A compared with the MBP control, whereas P4-C showed reduced interaction, and occasionally there was no interaction with the middle fragment of P4 (P4-M) (Fig. 3G). By using the transient expression system in Nicotiana benthamiana leaves, we verified that colocalization of RFP-NBR1 and GFP-P4 was not affected by mutations in either the LIR or UBA2 domain of NBR1 but was abolished completely by N- and C-terminal deletions of P4 (Figs. S2 and S3). Furthermore, the P4-N and P4-C fragments were sufficient for partial colocalization with NBR1 (Fig. S3).
Fig. S2.
NBR1 targeting of P4 occurs independently of ubiquitin- or ATG8-binding domains. GFP-P4 was coexpressed with RFP-tagged NBR1 (A) or with NBR1 mutants lacking the UBA2 (B) or lacking a functional LIR domain (C). Imaging was carried out 2 d after Agrobacterium-mediated transformation of N. benthamiana leaves. Images are representative maximum projections of z-stacks. (Scale bars, 20 μm.) BF, brightfield.
Fig. S3.
The N- and C-terminal domains of P4 mediate association with NBR1. GFP-tagged N- and C-terminal domains as well as the middle domain of P4 were coexpressed with RFP-tagged NBR1. Imaging was carried out 2 d after Agrobacterium-mediated transformation of N. benthamiana leaves. P4 fragments correspond to those illustrated in Fig. 3F. (Scale bars, 10 μm.) BF, bright field.
Together, these data show that NBR1 can bind nonassembled P4 directly in an ubiquitin-independent manner. This binding may well represent mechanisms underlying particle binding during infection.
Inclusion Bodies Protect Viral Particles from Autophagy.
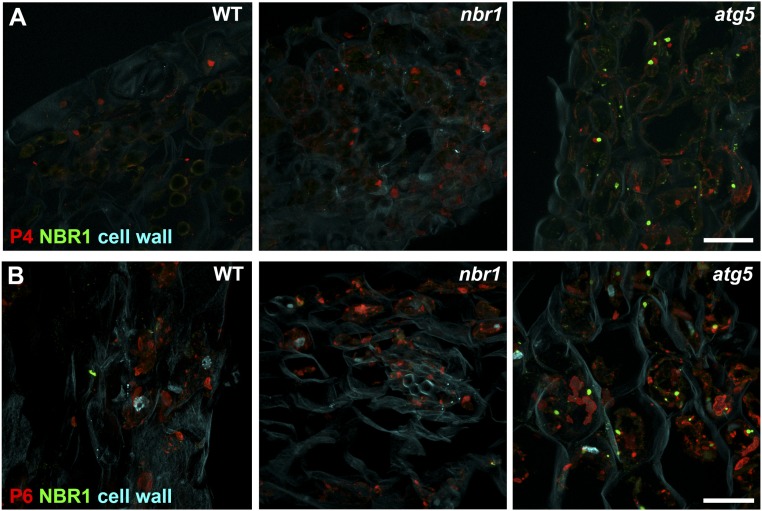
The multifunctional viral P6 protein is the main component of the CaMV-induced VFs, which are sites for viral translation, replication, particle assembly, and storage (18). We used immunofluorescence microscopy to localize NBR1 together with P4 or P6 in infected WT, atg5, and nbr1 plants. NBR1-derived signals remained below the detection limit in WT plants but were clearly visible in the atg5 mutant (Fig. S4). Because atg mutants are known to accumulate NBR1 strongly (20, 24), this observation verified the specificity of the fluorescence signal but also indicated that the ability to visualize NBR1 throughout the cell was limited by the sensitivity of the assay. Nevertheless, we observed that in 14–18% of P4- and P6-labeled structures in atg5, NBR1 was localized in an adjacent but not overlapping manner (Fig. 4 A–C), suggesting that VFs and P4 attract NBR1 in vivo.
Fig. S4.
NBR1 associates with viral particles and VFs. Immunofluorescence analysis of NBR1 together with P4 (A) or P6 (B) in CaMV Cabb B-JI–infected leaf tissue of WT, nbr1, and atg5 plants. Representative maximum projections of z-stacks show NBR1 stained in green, P4 or P6 stained in red, and the cell wall stained in cyan. (Scale bar, 20 μm.)
Fig. 4.
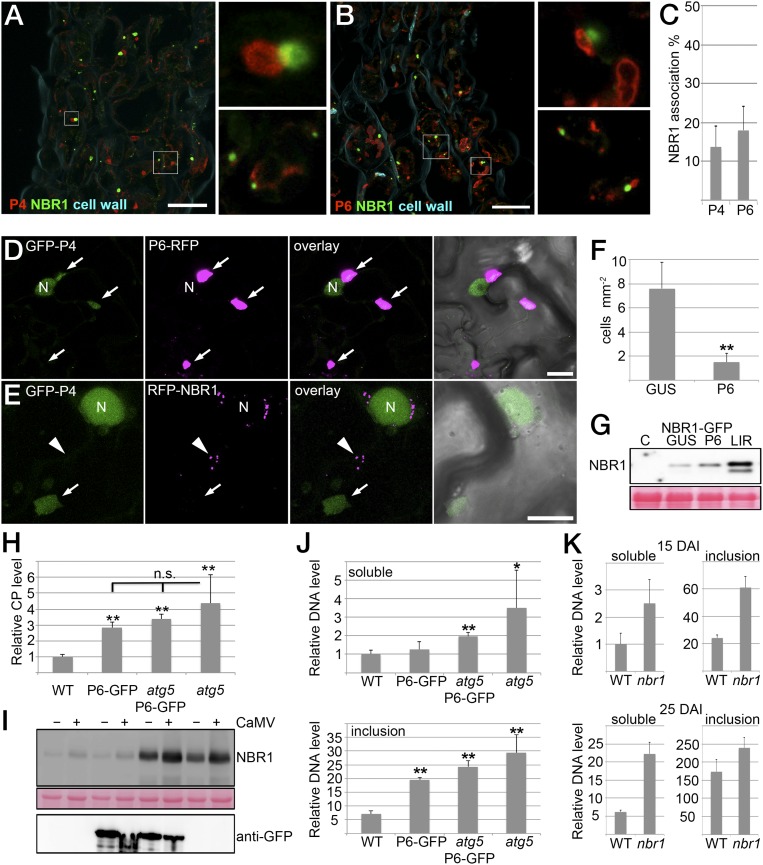
Inclusion bodies protect viral particles from autophagy. (A and B) Immunofluorescence detection of endogenous NBR1 together with viral P4 (A) or P6 (B) proteins in atg5 plants systemically infected by CaMV Cabb B-JI. Main images are projections of z-stacks showing P4 or P6 in red, NBR1 in green, and the cell wall in cyan. Insets are single-plane images of the boxed areas highlighting the association of NBR1 with P4 or P6. (Scale bars, 20 μm.) (C) The percentage of NBR1 structures associated with P4 or P6 structures in atg5 mutants. Values represent means ± SD [n = 14 (P4) or n = 6 (P6) independent sections]. (D) Colocalization analysis of GFP-P4 with P6-RFP upon transient expression in N. benthamiana leaves. Confocal images represent single planes of individual channels and their overlay with brightfield images. Arrows indicate P6 inclusions. N marks the nucleus that also shows a GFP fluorescence signal. (E) Localization of GFP-P4 (arrow) and RFP-NBR1 (arrowhead) upon coexpression of nonlabeled P6 in N. benthamiana leaves. N marks the nucleus also showing a GFP signal. Confocal images are maximum projections of z-stacks except in the brightfield overlay, which represents a single plane. (Scale bars, 20 μm.) (F) The frequency of cells/mm2 showing characteristic GFP-P4 and RFP-NBR1 colabeled foci upon the expression of P6 or control β-glucuronidase (GUS). Values represent means ± SD (n = 12 independent areas). (G, Upper) Immunoblot analysis of GFP-NBR1 transiently expressed with GUS or P6 in N. benthamiana leaves. Transient expression of the GFP-NBR1LIR mutant (LIR) that accumulates NBR1 because of the lack of autophagosomal targeting; noninfiltrated N. benthamiana leaves served as controls (C). (Lower) Ponceau S staining indicates protein loading. (H) CaMV particle levels determined by ELISA at 14 dai in WT and atg5 plants in the absence or presence of constitutively expressed P6-GFP. Values represent means ± SD (n = 4 biological replicates). (I, Upper) Immunoblot analysis of native NBR1 levels in mock-infected (−) and CaMV-infected (+) WT, P6-GFP, P6-GFP/atg5, and atg5 plants (CaMV (+) samples correspond to H) using anti-NBR1 antibodies. (Lower) Ponceau S staining of the anti-NBR1 membrane served as loading control, and immunoreaction with anti-GFP antibodies verified expression of the P6-GFP transgene. (J) Viral DNA levels in soluble and inclusion fractions derived from the CaMV-infected WT, P6-GFP, P6-GFP/atg5, and atg5 plants used for ELISA in H. DNA levels were determined by qPCR, and values are given as means ± SD (n = 4 biological replicates) relative to the amount of DNA in the soluble fraction of WT plants. (K) Viral DNA levels in soluble and inclusion fractions from CaMV-infected WT and nbr1 plants at 15 and 25 dai. Values are given as means ± SD (n = 4) relative to the amount of DNA in the soluble fraction of WT plants at 15 dai. Statistical significance (*P < 0.05; **P < 0.01) was revealed by Student’s t test (compared with GUS in F and with WT in H and J). n.s., not significant.
The localization pattern observed for P4 and NBR1 in the coexpression analysis (Fig. 3E and Fig. S2) was strikingly different from the more adjacent association observed during CaMV infection (Fig. 4 A–C). This difference could be explained by regulatory mechanisms operating during authentic infection, including sequestration and protection of P4 and particles by viral inclusion bodies. Indeed, P4 was recruited to P6 inclusions even in the absence of infection when both proteins were coexpressed in N. benthamiana leaves (Fig. 4D). Localization of P4 in P6 inclusions appeared rather diffuse and clearly differed from the distinct, small puncta of colocalized P4 and NBR1 (Fig. S2). Interestingly, P6 coexpression strongly reduced the number of cells showing characteristic P4/NBR1 colabeled punctate structures (Fig. 4 E and F), suggesting that P6 can counteract the targeting of P4 by NBR1. Expression of P6 had only a minor effect on NBR1 protein levels, which remained low compared with the NBR1LIR mutant that is impaired in autophagosome association and thus accumulated to high levels (Fig. 4G). Hence, P6 does not inhibit NBR1 flux efficiently in the transient system and likely suppresses P4-NBR1 association by other mechanisms.
To address the relevance of this finding in the context of infection, we tested whether constitutive expression of P6 in WT and atg5 plants impacted CaMV accumulation. Notably, P6-GFP transgenic plants and atg5 mutants showed a similarly enhanced level of viral particles as compared with WT plants (Fig. 4H), However, P6-GFP expression did not further increase viral titers in the atg5 background, suggesting that constitutively expressed P6 interfered with the autophagic targeting or degradation of viral particles. Similar to the transient expression of P6, we did not detect significant accumulation of NBR1 in P6-GFP transgenic plants compared with WT plants (Fig. 4I), confirming that P6 counteracted antiviral autophagy by means other than the inhibition of autophagic flux. We reasoned that P6 instead could protect viral particles by sequestering the particles into viral inclusion bodies. To test this notion, we applied a cell fractionation method to separate free and inclusion body-associated viral particles (Fig. 4J). Consistent with the view that CaMV particles are stored mainly in viral inclusions, we found that the majority of viral DNA was present in the inclusion fraction. Although atg5 mutants accumulated significantly more viral DNA in both soluble and inclusion fractions than WT plants, the P6-GFP transgenic line displayed higher levels of viral DNA only in the inclusion fraction, suggesting that P6 antagonizes autophagic degradation of viral particles via protective functions of the viral inclusion bodies. This assumption also implied that soluble free particles are more prone to autophagy degradation. Indeed, the analysis of viral DNA levels in soluble and inclusion fractions of infected WT and nbr1 plants revealed equal suppression of both particle pools by NBR1 early during infection (15 dai) (Fig. 4K). At a later stage of infection (25 dai), however, NBR1 suppression increased for the soluble and decreased for the inclusion pool of viral DNA (Fig. 4K). Together, these findings indicate that antiviral NBR1 flux is functional throughout CaMV infection, and that the formation of inclusion bodies helps protect particles from NBR1-mediated degradation.
NBR1 Suppresses Infection Initiation but Not CaMV Movement Within and from Plants.
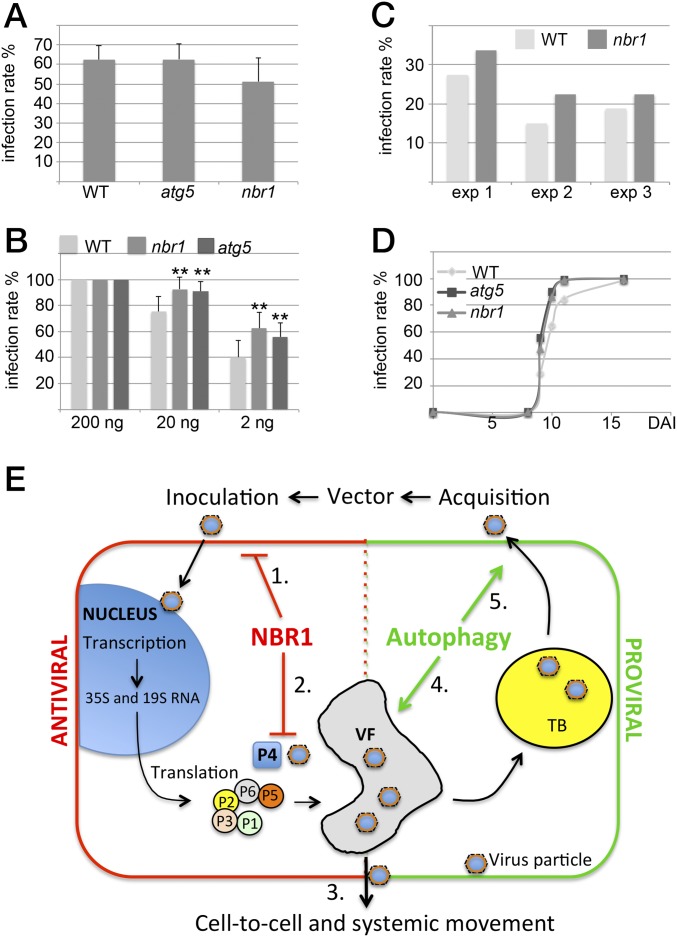
Our final aim was to determine whether NBR1 affects the ability of CaMV particles to initiate infection successfully, to spread within plants, and to engage in transmission. Transmission bodies (TBs) are inclusions that function in aphid transmission and are characterized by the exclusive presence of viral protein P2 (26, 27). To analyze whether autophagy affects TB-related functions, we used a CaMV mutant deleted in P2. CaMV lacking P2 showed similarly elevated virus titers in the atg5 and nbr1 mutants as the Cabb B-JI WT strain (Figs. S1 and S5). In addition, the overall morphology of TBs and the localization of enclosed particles appeared comparable in WT plants and autophagy mutants (Fig. S5). Finally, we investigated the efficiency of virus transmission by aphids feeding on atg5 and nbr1 mutants and found no significant difference from WT plants (Fig. 5A). Together, these results suggest that the properties of TB and their function in aphid transmission are unconnected to the impact of autophagy on CaMV infection.
Fig. S5.
Autophagy does not impact transmission body function. (A) Viral titers of a P2-deleted CaMV Cabb B-JI mutant strain (Cabb B-JI ΔP2) in WT, nbr1, and atg5 plants. Capsid protein levels were determined by ELISA; values represent means ± SD (n = 4) relative to WT. Asterisks indicate statistical significance (**P < 0.01) determined by Student’s t test (compared with WT). (B) Immunofluorescence analysis of P2 together with P4 in CaMV Cabb B-JI–infected leaf tissue of WT, nbr1, and atg5 plants. Representative maximum projections of z-stacks show P4 in green and P2 in red. (Scale bar, 5 μm.) (C) Immunofluorescence analysis of P2 in CaMV Cabb B-JI–infected leaf tissue of WT, nbr1, and atg5 plants. Representative maximum projections of z-stacks show the cell wall in cyan and P2 in red. (Scale bar, 20 μm.)
Fig. 5.
NBR1 contributes to resistance during the establishment of CaMV infection. (A) Aphids were fed on infected WT, atg5, and nbr1 plants and were moved to healthy turnips. The infection rate of 80 turnips for each genotype is given as the mean ± SD from three consecutive experiments (n = 3). (B) Mechanical inoculation of WT, nbr1, and atg5 plants with 200, 20, or 2 ng of isolated CaMV Cabb B-JI particles. The infection rate is given as the mean ± SD (n = 9). Statistical significance (**P < 0.01) was revealed by Student’s t test. (C) Aphids were kept on infected turnips for 2 min to acquire CaMV and then were moved to healthy WT and nbr1 plants. The number of infected plants was scored 3 wk later, and the percentage of infected plants is shown for three consecutive experiments analyzing 80 plants per genotype. (D) The kinetics of symptom appearance in WT, atg5, and nbr1 plants after mechanical inoculation with 200 ng of CaMV Cabb B-JI particles (analyzed in B for infection rates). (E) Model for autophagy roles in CaMV infection. (1) Antiviral NBR1-mediated selective autophagy suppresses the establishment of infection by mechanical or aphid-vectored inoculation. While viral transcription and translation remain unaltered, NBR1 specifically targets P4 and particles for xenophagy (2). However, the build-up of VFs and TBs protects viral particles and thus reduces the potential harm of NBR1 on the spread of virus infection, including cell-to-cell movement (3). Proviral bulk autophagy prevents extensive disease-associated senescence and tissue death, thereby increasing the lifespan of the infected plant, overall virus production (4), and the concomitant probability of aphid acquisition (5).
We finally looked at infection rates in WT and nbr1 plants after aphid inoculation or mechanical inoculation with a dilution series of purified virus particles (Fig. 5 B and C). Initiation of plant virus infections generally depends on the strength of the inoculum; infection rates decrease when the inoculum is diluted (Fig. 5B). The infection rates were 100% with the strongest inoculum, as expected. However, when lower inoculum titers were used, nbr1 and atg5 plants showed significantly higher infection rates, suggesting that they are more susceptible to infection. Similarly, nbr1 mutants showed a trend toward increased susceptibility in our standardized aphid inoculation assay (Fig. 5C). We also recorded the time after inoculation at which symptoms appeared in mechanically inoculated plants and found no difference between WT, atg5, and nbr1 plants (Fig. 5D). The results suggest that NBR1 can suppress some aspect of the early stages (initiation) of infection but does not affect CaMV movement within and aphid transmission from infected plants.
Discussion
Previous studies have established both anti- and proviral functions of autophagy during virus infections in metazoans. Autophagy contributes to host immunity and mediates the selective degradation of viral components, but several animal viruses have evolved mechanisms by which autophagic processes are counteracted or hijacked to promote virulence (10, 14). By analyzing the compatible interaction of the dsDNA virus CaMV with A. thaliana, we provide a seminal description of the key role of autophagy in the establishment of disease and host defense responses during infection by a plant virus. Our findings suggest that (i) NBR1-dependent selective autophagy restricts the establishment of CaMV infection in a process resembling mammalian xenophagy, (ii) the VF protein P6 and CaMV-induced viral inclusions antagonize NBR1 targeting of viral capsid protein and particles, and (iii) NBR1-independent autophagy promotes plant fitness and increases the probability of CaMV transmission by extending the timespan for virus production (Fig. 5E).
Previously, autophagy has been implicated in the degradation of host and viral proteins associated with virus-induced RNA silencing (28, 29), which is regarded as the primary antiviral defense pathway in plants (30). However, because viral RNA levels were not altered in autophagy-deficient mutants, RNA silencing can be largely excluded as the basis for the observed suppression of CaMV infection by autophagy. Instead, we have identified the selective autophagy cargo receptor NBR1 to target specifically both nonassembled and particle-associated P4 but not viral P2, P3, or P6. Because NBR1-linked P4 complexes accumulated in atg5 mutants and were detectable in vacuoles upon chemical inhibition of proteolytic turnover, they likely undergo degradation via xenophagy in WT plants. Thus, autophagy deficiency results in elevated levels of viral particles, detected as the accumulation of P4 and viral DNA, but not of viral RNA or other viral proteins whose levels remain unchanged. In plants, NBR1 is the sole homolog and functional hybrid of the paralogous p62 and NBR1 gene pair in mammals (24). Recent reports suggested that plant NBR1 binds to polyubiquitinated substrates similar to the binding in mammalian counterparts, although the identity of individual plant proteins targeted by NBR1 for autophagic destruction has yet to be determined (22, 24). We found direct ubiquitin-independent interaction of NBR1 with CaMV P4. Notably, both ubiquitin-dependent and -independent targeting of capsids by p62 has been demonstrated for diverse animal viruses, including the dsDNA virus herpes simplex virus (HSV-1) and the positive-stranded RNA viruses foot-and-mouth disease virus (FMDV), chikungunya virus (CHIKV), and sindbis virus (SINV) (11–13, 31, 32). Together, our results advance NBR1/p62-mediated xenophagy of viral capsids to a cross-kingdom function in antiviral immunity.
NBR1-mediated particle xenophagy could impact infection very negatively because the epidemiology of CaMV critically depends on virus particle functions during the establishment of new plant infections, systemic movement within plants, and transmission to new host plants (33). However, nbr1 and atg5 mutants did not show alterations in symptom appearance or aphid transmission rates when used as feeding source, arguing against any significant ability of NBR1 to suppress the spread of infection within and from plants. Both systemic movement and virus particle acquisition by aphids occurs from infected cells and is tightly linked to viral inclusion body functions (VFs and TBs) (27, 34, 35). Indeed, we propose, based on several observations, that viral particles stored in inclusions are protected from autophagy. First, the major inclusion-forming viral constituents, P6 and P2, were not degraded by autophagy. Second, NBR1 showed limited access to P4 in P6 inclusion bodies, as revealed by the absence of colocalization between P4/P6 and NBR1 during infection. Third, free particles were more prone to NBR1-mediated degradation than those associated with inclusions. Fourth, P6 reduced the occurrence of NBR1-associated P4 structures, a likely consequence of P4 interaction with P6 and its subsequent sequestration into P6 inclusions (36). Fifth, viral particles escaped antiviral autophagy in P6 transgenic lines via specific accumulation in inclusion fractions. Interestingly, NBR1 still associated alongside VFs during infection, leading us speculate that NBR1 directly targets and competes for P4 at sites of translation and assembly before P4 localization within the inclusions.
An additional viral countermeasure against NBR1-mediated host defense may come from the recently proposed suppressive effect of P6 on cellular autophagy through activation of the target-of-rapamycin (TOR) kinase (37). However, our observation of strongly enhanced autophagy flux during CaMV infection and the increased accumulation of P4 and viral DNA in autophagy-deficient mutant plants clearly indicate that P6 does not block autophagy efficiently during CaMV infection. Although different viral strategies eventually may contribute to minimize the impact of NBR1 on viral particle functions, NBR1-deficient plants showed increased susceptibility to CaMV infection following mechanical inoculation at lowered inoculum strength and to transmission by aphids. These findings unequivocally reveal the biological significance of NBR1-mediated xenophagy in suppressing the establishment of infection upon primary CaMV inoculation, likely before inclusion body formation.
In summary, we propose a model that places autophagy in the CaMV infection cycle (Fig. 5E). During transmission, viral particles enter the cells of uninfected plants. NBR1-mediated xenophagy suppresses this stage of the infection, likely because of the absence of effective viral counteractions. However, processes that follow the establishment of primary infection, including systemic plant colonization and acquisition by aphids, are no longer sensitive to NBR1 and suggest that CaMV largely escapes the potential harm of xenophagy by functions of autophagy-resistant inclusion bodies. NBR1 provides antiviral roles in CaMV infection, and bulk autophagy significantly extends the lifespan of infected plants and virus particle production. Autophagy has been observed to increase host viability during some metazoan virus infections as well (10, 12, 31). This survival function of autophagy benefits CaMV infection by increasing the likelihood that infected plants will provide particles for transmission.
Materials and Methods
Plant Material and Growth Conditions.
WT plants were A. thaliana ecotype Columbia (Col-0). Mutants atg5-1, atg7-2, nbr1-2, npr1-1, atg5-1 npr1-1, and the GFP-ATG8a transgenic line have been described previously (19, 22, 38, 39). For infection experiments, Arabidopsis plants were grown in soil in a growth cabinet under short-day conditions (8/16-h light/dark cycles), and for transient expression assays N. benthamiana plants were cultivated in a growth room under long-day conditions (16/8-h light/dark cycles) at 150 μE⋅m−2⋅s−1, 21 °C, and 70% relative humidity. Sterile plants were cultivated in vitro on half-strength Murashige and Skoog (MS) medium with a 16-h photoperiod at 150 μE⋅m−2⋅s−1 and 21 °C.
Plasmid Construction, Generation of Transgenic Lines, and Transient Expression.
For stable expression in transgenic plants, a genomic fragment of NBR1 containing 2 kb of the predicted promoter and the coding region without the stop codon was amplified (Table S1 for primer sequences), cloned into pENTR/D-TOPO, and subsequently recombined into pAUL11 (40) to add 3′-end sequences encoding Strep(S)-III and 3xHA tags. The NBR1 fragment containing the 3xHA and SIII extension was reamplified by PCR, cloned into pENTR/D-TOPO, and recombined into pGWB459 or pGWB604 (41) to generate the pNBR1:NBR1-SIII-3xHA-tagRFP and pNBR1:NBR1-SIII-3xHA-GFP constructs, respectively. P6 and P4 of CaMV (Cabb B-JI) were cloned into pENTR/D-TOPO and further recombined into pGWB5 (P6) and pGWB606 (P4), resulting in the fusion constructs 35S:P6-GFP (as described previously in ref. 42) and 35S:GFP-P4, respectively. The binary plasmids were electroporated into Agrobacterium tumefaciens strain GV3101 and were transformed into Col-0 WT or atg5 plants using the floral dip method (43). For colocalization of different markers, the respective transgenic lines were crossed, and the F1 plants were analyzed. For transient expression experiments, P6 of CaMV (Cabb B-JI) was cloned into pENTR/D-TOPO and further recombined into pGWB561 to generate 35S:P6-tagRFP. P4 fragments encoding the N-terminal (amino acids 1–120), middle, or C-terminal (amino acids 418–489) domains were cloned and recombined into pGWB606 as described for full-length P4. The pENTR clones containing the coding sequences of NBR1 WT, PB1 (K11A), UBA2, and LIR domain mutants were described previously (24) and were used for recombination into the binary vector pUBN-DEST-mRFP (44). The plant expression constructs were transformed into A. tumefaciens strain GV3101, and infiltration of N. benthamiana was done at the four- to six-leaf stage.
Table S1.
Primers used in this study
| Method/name | Sequence (5′–3′) | Reference/source |
| qPCR | ||
| CaMV DNA fwd | AGCGGTCAAAATATTGCTTA | (46) |
| CaMV DNA rev | AACTTACCGTATGCTAGATTACCT | |
| Genomic 18S rDNA fwd | CGTGATCGATGAATGCTACC | (46) |
| Genomic 18S rDNA rev | GGGGTTTGTTGCACGTATTA | |
| Chloroplast genomic DNA fwd | TTGCGGTCAATAAGGTAGGG | (56) |
| Chloroplast genomic DNA rev | TAGAGAATTTGTGCGCTTGG | |
| CaMV leader region fwd | TCACCAGTCTCTCTCTACAAATCTATCTC | (57) |
| CaMV leader region rev | GCGAAACCCTATAAGAACCCTAATTCC | |
| CaMV P4 region fwd | GGTGAAGGACCATCTAGATAC | This study |
| CaMV P4 region rev | CAATCCGATTTCTGCTGCCCA | |
| ATG8a fwd | CAAGCTTGGAGCTGAGAAAG | (22) |
| ATG8a rev | GCAACGGTAAGAGATCCAAA | |
| ATG8e fwd | GTACCTTGTGCCATCAGACC | (22) |
| ATG8e rev | AGGAAGCCATCTTCGTCTTT | |
| NBR1 fwd | GAGGACCCAGACCGGAAGG | (22) |
| NBR1 rev | GACAAACACGACGAGGATGC | |
| Cloning | ||
| NBR1 C term for Y2H fwd | CACCATGTTGTCTGGTGG TTCTTCATCT | This study |
| NBR1 C term for Y2H rev | TCAAGCAACGGTAAGAGAT | |
| NBR1 C term ΔUBA2 for Y2H rev | ACAAAGAGCATCAACAGACTGCT | |
| NBR1 SIII-3xHA fwd | CACCTGGAGATCATGTGTGGCATACA | This study |
| NBR1 SIII-3xHA rev | GGCGCGCCTTAGCTGCACTGAGCAGCGTAATCT | |
| genomic NBR1 with promoter fwd | CACCTGGAGATCATGTGTGGCATACA | This study |
| genomic NBR1 with promoter rev | AGCAACGGTAAGAGATCCAAAAGTGT | |
| CaMV P4 for pENTRY fwd | CACCATGGCCGAATCAATTTTAGACAG | This study |
| CaMV P4 for pENTRY rev | ATCTGAGTCTGAGTCTTCAGAAGT | |
| CaMV P4 N terminus fwd | CACCATGGCCGAATCAATTTTAGACAG | This study |
| CaMV P4 N terminus rev | GTATCTAGATGGTCCTTCACCATC | |
| CaMV P4 M domain fwd | CACCATGGCAATAGGAGAAATATCTGAAGAC | This study |
| CaMV P4 M domain rev | GATGTGAGCCTTCTCCGAGCTTTG | |
| CaMV P4 C terminus fwd | CACCATGGAAGGCCATTACGCCAACGA | This study |
| CaMV P4 C terminus rev | ATCTGAGTCTGAGTCTTCAGAAGT | |
| CaMV P6 for pENTRY fwd | CACCATGGAGAACATAGAAAAACTCCTC | This study |
| CaMV P6 for pENTRY rev | ATCCACTTGCTTTGAAGACG | |
C term, C terminus; fwd, forward; rev, reverse.
CaMV Inoculation and Quantification.
The first true leaves of 3-wk-old Arabidopsis plants were inoculated by Agrobacterium-mediated infiltration of the CaMV strain CM1841 or mechanically with particles purified from turnip plants infected with CaMV strains Cabb B-JI and CaMVΔP2 (45). Plants were sampled in biological replicates, each containing three individual plants from which inoculated leaves were removed. For ELISA, four replicate samples per condition were homogenized in 2 mL 100 mM Tris (pH 7.5) and 1 M urea per gram fresh weight, incubated under shaking for 1 h at 4 °C, and diluted 10-fold in 100 mM Tris (pH 7.5), 150 mM NaCl, 2% (wt/vol) polyvinylpyrrolidone (PVP), 0.1% Tween 20, and 2% (wt/vol) skimmed milk powder. Antibodies for CaMV double-antibody sandwich (DAS)-ELISA were purchased from Neogen (product code 1210). Standard curves were generated from serial dilutions of isolated viral particles. For CaMV transcript analysis, total RNA was isolated from three replicates using the RNeasy Plant Mini Kit (Qiagen), and on-column DNA digestion was performed with DNase I (Qiagen). First-strand cDNA was synthesized from 1 μg of total RNA using the Maxima First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). qPCR analysis was done with Maxima SYBR Green/Fluorescein qPCR Master Mix (Thermo Fisher Scientific) using the CFX Connect Real-Time PCR detection system (Bio-Rad) with the gene-specific primers listed in Table S1. Control reactions without reverse transcription were performed to exclude DNA contamination, and normalization was done using PP2A (AT1G69960) and UBQ9 (AT5G37640). For CaMV DNA quantitation, total DNA was isolated from four replicates using the NucleoSpin Plant II Kit (Macherey-Nagel), followed by qPCR with ribosomal DNA as reference essentially as described in ref. 46.
Soluble and inclusion-associated CaMV particles were obtained essentially as described in ref. 47. Briefly, infected plant tissue was homogenized in 100 mM Tris (pH 8.0), 150 mM NaCl, and 1% Triton X-100 and was centrifuged at 17,000 × g for 10 min at 4 °C. The supernatant represented the soluble fraction of free particles, and the pellet contained the inclusion-associated particles. Total DNA was precipitated from both fractions, and CaMV DNA levels were determined by qPCR using primers specific for the chloroplast genome as reference.
Aphid Transmission and Mechanical Inoculation of CaMV for Susceptibility Testing.
Wingless green peach aphids (Myzus persicae Sulzer) were starved for 1 h on Parafilm membranes (Bemis Company, Inc.), and were fed for 2 min on infected Arabidopsis or turnip (Brassica rapa L. Just Right) plants before being placed on turnip seedlings at the cotyledon stage or on Arabidopsis plants at the five-leaf stage for subsequent 1-h inoculation. Then aphids were killed by insecticide treatment, and plants were cultivated as described (48). For assaying the susceptibility to mechanically inoculated CaMV, different amounts of isolated particles were rubbed on a single true leaf of 3-wk-old Arabidopsis plants using a sponge and carborundum as abrasive. Infected plants were identified by visual inspection of typical symptoms.
Confocal Microscopy, Immunofluorescence, and Inhibitor Treatment.
Live cell images were acquired from abaxial leaf epidermal cells using a Zeiss LSM 780 microscope. Excitation/detection parameters for GFP and RFP were 488 nm/490–552 nm and 561 nm/569–652, respectively, and the sequential scanning mode was used for covisualization of both fluorophores. Inhibitor treatment was carried out by syringe-infiltration of mature leaves by or incubation of seedlings in 0.5 μM ConA in half-strength MS medium 14 h before confocal analysis. For immunofluorescence, Arabidopsis leaf segments (approximately 25 mm2) were immersed in fixation buffer (1% glutaraldehyde in 50 mM Hepes, pH 8) for 1 h; then the borders were removed, and the remaining tissue was embedded in 5% (wt/vol) agarose. Vibratome-cut sections (approximately 50 μm) were processed as described (27). Immunolabeling was performed with the primary antibodies rabbit anti-P2 (49), anti-P3 (50), anti-P4 (Neogen), anti-P6 (51), and anti-NBR1 (24) and with mouse anti-P4 and rat anti-P6 (produced by Eurogentec), as well as Alexa 488 and Alexa 594 conjugates (Thermo Fisher), as secondary antibodies. To visualize cell walls, the slides were counterstained with 0.002% Fluostain I (Sigma-Aldrich) before mounting. Immunofluorescence images were acquired with a Zeiss LSM 700 confocal microscope using excitation/detection parameters of 405/415–500 nm for Fluostain I, 488 nm/490–535 nm for Alexa 488 dye, and 555/560–630 nm for Alexa 594 dye. Confocal images were processed with ZEN version 2011 (Zeiss) and Image J version 1.48v software.
NBR1-Dependent IC-TEM and ELISA.
Plants were inoculated with CaMV C1841, and infected tissue was harvested 14 d later along with noninfected controls. Lysates were prepared by homogenizing 1 g of tissue in 2 mL 100 mM Tris (pH 7.5), 150 mM NaCl, 2% (wt/vol) PVP, 0.1% Tween 20, and 2% (wt/vol) skimmed milk powder and were cleared by centrifugation at 1,000 × g for 5 min at 4 °C. The lysates then were applied to carbon-coated TEM grids or ELISA plates coated with anti-NBR1 (1:200) and were incubated overnight at 4 °C. TEM grids were subsequently washed six times for 2 min with PBS and 0.1% Tween 20 and twice for 2 min with double-distilled H2O, followed by staining with 3% (wt/vol) uranyl acetate for 30 s. Ultrastructural analysis and imaging were performed with an FEI Tecnai G2 transmission electron microscope. P4 bound to the anti-NBR1–coated ELISA plates was detected with conjugated anti-P4 (Neogen) and was quantified using the standard curve derived from standard CaMV particle ELISA. NBR1 direct ELISA was performed by binding crude plant extracts directly to ELISA plates after homogenizing tissue in 8 M urea, followed by incubation with primary anti-NBR1 (1:4,000) and secondary anti-rabbit alkaline phospatase (AP) (1:4,000) antibodies. A serial dilution of plant extract functioned as a standard series, and extracts from nbr1 plants served as a negative control.
Immunoblot Analysis.
Proteins were extracted in 100 mM Tris (pH 7.5) with 2% (wt/vol) SDS, boiled for 5 min in Laemmli sample buffer, and cleared by centrifugation. The protein extracts then were separated by SDS/PAGE, transferred to PVDF membranes (Amersham, GE Healthcare), blocked with 5% (wt/vol) skimmed milk in PBS, and incubated with the primary antibodies anti-NBR1 (24), anti-ATG8a (52), anti-ATG8e (53), anti-P2 (49), anti-P3 (50), anti-P4 (Neogen), anti-P6 (54), anti-MBP (New England Biolabs), or anti-GST (MicroMol GmbH) using a 1:1,000 dilution in PBS, 0.1% Tween 20, and secondary horseradish peroxidase (HPR)-conjugated antibodies (1:5,000) in PBS and 0.1% Tween 20 (Amersham, GE Healthcare). The immunoreaction was developed using the ECL Prime kit (Amersham, GE Healthcare) and was detected in a LAS-3000 Luminescent Image Analyzer (Fujifilm).
Immunoprecipitation.
For analysis of NBR1-GFP immunocomplexes, total proteins were extracted from infected pNBR1:NBR1-SIII-3xHA-GFP transgenic plants (CM1841) at 21 dai, and immunoprecipitation was done according to the protocol provided with anti-GFP microbeads (μMACS GFP Isolation Kit; Miltenyi Biotec). Mock-treated NBR1-GFP or infected free-GFP transgenic lines were used as controls. The presence of viral DNA was detected after precipitation of total DNA from the immunoprecipitated samples.
In Vitro Binding Assays.
Several P4 variants including the full-length, N-terminal (amino acids 1–120), middle (amino acids 76–438), and C-terminal (amino acids 418–489) regions were cloned into a gateway-compatible pMAL-c2 (Invitrogen) vector to generate MBP fusions. The nonaggregating NBR1 mutant K11A (NBR1K11A) was recombined into pDEST15 (Invitrogen) to produce a GST fusion construct. Recombinant MBP and GST proteins were expressed using the E. coli strain BL21 DB3.1. MBP proteins were immobilized on amylose resin (New England Biolabs) followed by 2-h incubation under constant rotation at 4 °C with cleared bacterial lysates containing GST-NBR1K11A in buffer solution [20 mM Tris (pH 7.5), 100 mM NaCl, and 1 mM EDTA]. Washing was done four times with two volumes of the same buffer, and matrix-bound protein complexes were eluted with one volume of buffer supplemented with 50 mM maltose. Samples were analyzed by immunoblot analysis in comparison with the MBP control.
For in vitro pull-down of CaMV particles, GST and GST-NBR1K11A were immobilized on glutathione Sepharose (GE Healthcare) and incubated for 2 h under constant rotation at 4 °C with CaMV particles purified from infected nbr1 plants in buffer containing 20 mM Tris (pH 8.0) and 100 mM NaCl. Washing was done four times with two volumes of buffer, and the elution step was performed with one volume of 5 mM Tris (pH 8.0) with 6 mg glutathione/mL. Input and eluate fractions were directly subjected to qPCR analysis to determine the amount of CaMV DNA as a proxy of in vitro-bound particles.
Y2H Analysis.
Y2H techniques were performed according to the Yeast Protocols Handbook (Clontech). The gateway-compatible Y2H vectors described in ref. 55 were used to generate fusions of the C-terminal portions of NBR1 (amino acids 505–704 and amino acids 505–656 lacking the LIR and UBA2 domains) and of the different CaMV P4 fragments (full-length, P4-N, P4-M, and P4-C; see above) with GAL4 BD and AD, respectively. Yeast strain AH109 was cotransformed with the respective plasmid combinations, including empty vector controls, followed by selection on solid SD medium lacking Trp/Leu for 3 d at 28 °C. Interaction was analyzed by growth on Trp/Leu medium lacking His supplemented with 4 mM 3-Aminotriazole (3-AT) (Sigma) and by lacZ assays.
Data Analysis and Presentation.
Data are presented as mean ± SD. Statistical significance was analyzed by Student´s t test; *P < 0.05, **P < 0.01. Significance level with P values <0.001 was not specifically indicated. The number of replicates (n) is given in the figure legends.
Acknowledgments
We thank John Mundy and Morten Petersen (University of Copenhagen) for support and input during the initiation of this project; Steingrim Svenning and Terje Johansson (University of Tromsø) for providing the anti-NBR1 antibody and NBR1-related plasmids; Liwen Jiang (University of Hong Kong) for the kind gift of the anti-ATG8e antibody; James E. Schoelz (University of Missouri) for the anti-P6 antibody; Frederik Börnke (University of Potsdam) for the gateway-compatible MBP and GST expression vectors; and Anders Ahlander at Biological Visualization EM, Science for Life Laboratory, Uppsala University, for assistance with ultrastructural imaging by TEM. This research was funded by grants from the Knut and Alice Wallenberg Foundation (to D.H.), the Swedish University of Agricultural Sciences (to D.H.), and the Swedish Research Council FORMAS (to A.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1610687114/-/DCSupplemental.
References
- 1.Klionsky DJ, Codogno P. The mechanism and physiological function of macroautophagy. J Innate Immun. 2013;5(5):427–433. doi: 10.1159/000351979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 3.Lamb CA, Yoshimori T, Tooze SA. The autophagosome: Origins unknown, biogenesis complex. Nat Rev Mol Cell Biol. 2013;14(12):759–774. doi: 10.1038/nrm3696. [DOI] [PubMed] [Google Scholar]
- 4.Boya P, Reggiori F, Codogno P. Emerging regulation and functions of autophagy. Nat Cell Biol. 2013;15(7):713–720. doi: 10.1038/ncb2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reggiori F, Komatsu M, Finley K, Simonsen A. Autophagy: More than a nonselective pathway. Int J Cell Biol. 2012;2012:219625. doi: 10.1155/2012/219625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Svenning S, Johansen T. Selective autophagy. Essays Biochem. 2013;55:79–92. doi: 10.1042/bse0550079. [DOI] [PubMed] [Google Scholar]
- 7.Stolz A, Ernst A, Dikic I. Cargo recognition and trafficking in selective autophagy. Nat Cell Biol. 2014;16(6):495–501. doi: 10.1038/ncb2979. [DOI] [PubMed] [Google Scholar]
- 8.Zaffagnini G, Martens S. Mechanisms of selective autophagy. J Mol Biol. 2016;428(9 Pt A):1714–1724. doi: 10.1016/j.jmb.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gomes LC, Dikic I. Autophagy in antimicrobial immunity. Mol Cell. 2014;54(2):224–233. doi: 10.1016/j.molcel.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 10.Dong X, Levine B. Autophagy and viruses: Adversaries or allies? J Innate Immun. 2013;5(5):480–493. doi: 10.1159/000346388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Judith D, et al. Species-specific impact of the autophagy machinery on Chikungunya virus infection. EMBO Rep. 2013;14(6):534–544. doi: 10.1038/embor.2013.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shelly S, Lukinova N, Bambina S, Berman A, Cherry S. Autophagy is an essential component of Drosophila immunity against vesicular stomatitis virus. Immunity. 2009;30(4):588–598. doi: 10.1016/j.immuni.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berryman S, et al. Foot-and-mouth disease virus induces autophagosomes during cell entry via a class III phosphatidylinositol 3-kinase-independent pathway. J Virol. 2012;86(23):12940–12953. doi: 10.1128/JVI.00846-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiramel AI, Brady NR, Bartenschlager R. Divergent roles of autophagy in virus infection. Cells. 2013;2(1):83–104. doi: 10.3390/cells2010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackson WT. Viruses and the autophagy pathway. Virology. 2015;479-480:450–456. doi: 10.1016/j.virol.2015.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou J, Yu JQ, Chen Z. The perplexing role of autophagy in plant innate immune responses. Mol Plant Pathol. 2014;15(6):637–645. doi: 10.1111/mpp.12118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dagdas YF, et al. An effector of the Irish potato famine pathogen antagonizes a host autophagy cargo receptor. eLife. 2016;5:e10856. doi: 10.7554/eLife.10856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hohn T, Futterer J. The proteins and functions of plant pararetroviruses: Knowns and unknowns. Crit Rev Plant Sci. 1997;16(1):133–161. [Google Scholar]
- 19.Yoshimoto K, et al. Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell. 2009;21(9):2914–2927. doi: 10.1105/tpc.109.068635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Munch D, et al. Autophagy deficiency leads to accumulation of ubiquitinated proteins, ER stress, and cell death in Arabidopsis. Autophagy. 2014;10(9):1579–1587. doi: 10.4161/auto.29406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haas M, Bureau M, Geldreich A, Yot P, Keller M. Cauliflower mosaic virus: Still in the news. Mol Plant Pathol. 2002;3(6):419–429. doi: 10.1046/j.1364-3703.2002.00136.x. [DOI] [PubMed] [Google Scholar]
- 22.Zhou J, et al. NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet. 2013;9(1):e1003196. doi: 10.1371/journal.pgen.1003196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12(1):1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Svenning S, Lamark T, Krause K, Johansen T. Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy. 2011;7(9):993–1010. doi: 10.4161/auto.7.9.16389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karsies A, Hohn T, Leclerc D. Degradation signals within both terminal domains of the cauliflower mosaic virus capsid protein precursor. Plant J. 2001;27(4):335–343. doi: 10.1046/j.1365-313x.2001.01093.x. [DOI] [PubMed] [Google Scholar]
- 26.Espinoza AM, Medina V, Hull R, Markham PG. Cauliflower mosaic virus gene II product forms distinct inclusion bodies in infected plant cells. Virology. 1991;185(1):337–344. doi: 10.1016/0042-6822(91)90781-6. [DOI] [PubMed] [Google Scholar]
- 27.Martinière A, et al. A virus responds instantly to the presence of the vector on the host and forms transmission morphs. eLife. 2013;2:e00183. doi: 10.7554/eLife.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Derrien B, et al. Degradation of the antiviral component ARGONAUTE1 by the autophagy pathway. Proc Natl Acad Sci USA. 2012;109(39):15942–15946. doi: 10.1073/pnas.1209487109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakahara KS, et al. Tobacco calmodulin-like protein provides secondary defense by binding to and directing degradation of virus RNA silencing suppressors. Proc Natl Acad Sci USA. 2012;109(25):10113–10118. doi: 10.1073/pnas.1201628109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boualem A, Dogimont C, Bendahmane A. The battle for survival between viruses and their host plants. Curr Opin Virol. 2016;17:32–38. doi: 10.1016/j.coviro.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 31.Orvedahl A, et al. Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host Microbe. 2010;7(2):115–127. doi: 10.1016/j.chom.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orvedahl A, et al. Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature. 2011;480(7375):113–117. doi: 10.1038/nature10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hohn T. Plant pararetroviruses: Interactions of cauliflower mosaic virus with plants and insects. Curr Opin Virol. 2013;3(6):629–638. doi: 10.1016/j.coviro.2013.08.014. [DOI] [PubMed] [Google Scholar]
- 34.Bak A, et al. Virus factories of cauliflower mosaic virus are virion reservoirs that engage actively in vector transmission. J Virol. 2013;87(22):12207–12215. doi: 10.1128/JVI.01883-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schoelz JE, Angel CA, Nelson RS, Leisner SM. A model for intracellular movement of Cauliflower mosaic virus: The concept of the mobile virion factory. J Exp Bot. 2016;67(7):2039–2048. doi: 10.1093/jxb/erv520. [DOI] [PubMed] [Google Scholar]
- 36.Himmelbach A, Chapdelaine Y, Hohn T. Interaction between cauliflower mosaic virus inclusion body protein and capsid protein: Implications for viral assembly. Virology. 1996;217(1):147–157. doi: 10.1006/viro.1996.0102. [DOI] [PubMed] [Google Scholar]
- 37.Zvereva AS, et al. Viral protein suppresses oxidative burst and salicylic acid-dependent autophagy and facilitates bacterial growth on virus-infected plants. New Phytol. 2016;211(3):1020–1034. doi: 10.1111/nph.13967. [DOI] [PubMed] [Google Scholar]
- 38.Hofius D, et al. Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell. 2009;137(4):773–783. doi: 10.1016/j.cell.2009.02.036. [DOI] [PubMed] [Google Scholar]
- 39.Munch D, et al. Retromer contributes to immunity-associated cell death in Arabidopsis. Plant Cell. 2015;27(2):463–479. doi: 10.1105/tpc.114.132043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lyska D, Engelmann K, Meierhoff K, Westhoff P. pAUL: A gateway-based vector system for adaptive expression and flexible tagging of proteins in Arabidopsis. PLoS One. 2013;8(1):e53787. doi: 10.1371/journal.pone.0053787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakagawa T, et al. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng. 2007;104(1):34–41. doi: 10.1263/jbb.104.34. [DOI] [PubMed] [Google Scholar]
- 42.Laird J, et al. Identification of the domains of cauliflower mosaic virus protein P6 responsible for suppression of RNA silencing and salicylic acid signalling. J Gen Virol. 2013;94(Pt 12):2777–2789. doi: 10.1099/vir.0.057729-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 44.Grefen C, et al. A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J. 2010;64(2):355–365. doi: 10.1111/j.1365-313X.2010.04322.x. [DOI] [PubMed] [Google Scholar]
- 45.Martinière A, et al. A role for plant microtubules in the formation of transmission-specific inclusion bodies of Cauliflower mosaic virus. Plant J. 2009;58(1):135–146. doi: 10.1111/j.1365-313X.2008.03768.x. [DOI] [PubMed] [Google Scholar]
- 46.Love AJ, Yun BW, Laval V, Loake GJ, Milner JJ. Cauliflower mosaic virus, a compatible pathogen of Arabidopsis, engages three distinct defense-signaling pathways and activates rapid systemic generation of reactive oxygen species. Plant Physiol. 2005;139(2):935–948. doi: 10.1104/pp.105.066803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qiu SG, Wintermantel WM, Sha Y, Schoelz JE. Light-dependent systemic infection of solanaceous species by cauliflower mosaic virus can Be conditioned by a viral gene encoding an aphid transmission factor. Virology. 1997;227(1):180–188. doi: 10.1006/viro.1996.8314. [DOI] [PubMed] [Google Scholar]
- 48.Martinière A, et al. VAPA, an innovative “virus-acquisition phenotyping assay” opens new horizons in research into the vector-transmission of plant viruses. PLoS One. 2011;6(8):e23241. doi: 10.1371/journal.pone.0023241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blanc S, et al. Paracrystalline structure of cauliflower mosaic virus aphid transmission factor produced both in plants and in a heterologous system and relationship with a solubilized active form. Virology. 1993;197(1):283–292. doi: 10.1006/viro.1993.1589. [DOI] [PubMed] [Google Scholar]
- 50.Drucker M, et al. Intracellular distribution of viral gene products regulates a complex mechanism of cauliflower mosaic virus acquisition by its aphid vector. Proc Natl Acad Sci USA. 2002;99(4):2422–2427. doi: 10.1073/pnas.042587799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Khelifa M, et al. Electron-lucent inclusion bodies are structures specialized for aphid transmission of cauliflower mosaic virus. J Gen Virol. 2007;88(Pt 10):2872–2880. doi: 10.1099/vir.0.83009-0. [DOI] [PubMed] [Google Scholar]
- 52.Yoshimoto K, et al. Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell. 2004;16(11):2967–2983. doi: 10.1105/tpc.104.025395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhuang X, et al. A BAR-domain protein SH3P2, which binds to phosphatidylinositol 3-phosphate and ATG8, regulates autophagosome formation in Arabidopsis. Plant Cell. 2013;25(11):4596–4615. doi: 10.1105/tpc.113.118307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schoelz JE, Goldberg KB, Kiernan J. Expression of Cauliflower mosaic-virus (camv) gene-vi in transgenic Nicotiana-Bigelovii complements a strain of camv defective in long-distance movement in nontransformed N-Bigelovii. Mol Plant Microbe Interact. 1991;4(4):350–355. [Google Scholar]
- 55.Maier R, Brandner C, Hintner H, Bauer J, Onder K. Construction of a reading frame-independent yeast two-hybrid vector system for site-specific recombinational cloning and protein interaction screening. Biotechniques. 2008;45(3):235–244. doi: 10.2144/000112897. [DOI] [PubMed] [Google Scholar]
- 56.Rowan BA, Oldenburg DJ, Bendich AJ. A high-throughput method for detection of DNA in chloroplasts using flow cytometry. Plant Methods. 2007;3:5. doi: 10.1186/1746-4811-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blevins T, et al. Massive production of small RNAs from a non-coding region of Cauliflower mosaic virus in plant defense and viral counter-defense. Nucleic Acids Res. 2011;39(12):5003–5014. doi: 10.1093/nar/gkr119. [DOI] [PMC free article] [PubMed] [Google Scholar]