Significance
Physical activities of our body and extremities are achieved by the propagation of electrical signals called action potentials from our brain, through nerves, to skeletal muscles. Voltage-gated sodium channel (Navs) play essential roles in the generation and propagation of action potentials in such excitable cells. Although mammalian Nav function has been studied comprehensively, the precise structural basis for the gating mechanisms has not been fully clarified. In this study, we have used lanthanide-based resonance energy transfer to obtain dynamic structural information in mammalian Nav gating. Our data define a geometrical map of Navs with the bound toxins and reveal voltage-induced structural changes related to channel gating, which lead us further toward an understanding of the gating mechanism in mammalian Navs.
Keywords: μ-conotoxin, voltage gating, relaxed state, slow inactivation, β-scorpion toxin
Abstract
Voltage-gated sodium channels (Navs) play crucial roles in excitable cells. Although vertebrate Nav function has been extensively studied, the detailed structural basis for voltage-dependent gating mechanisms remain obscure. We have assessed the structural changes of the Nav voltage sensor domain using lanthanide-based resonance energy transfer (LRET) between the rat skeletal muscle voltage-gated sodium channel (Nav1.4) and fluorescently labeled Nav1.4-targeting toxins. We generated donor constructs with genetically encoded lanthanide-binding tags (LBTs) inserted at the extracellular end of the S4 segment of each domain (with a single LBT per construct). Three different Bodipy-labeled, Nav1.4-targeting toxins were synthesized as acceptors: β-scorpion toxin (Ts1)-Bodipy, KIIIA-Bodipy, and GIIIA-Bodipy analogs. Functional Nav-LBT channels expressed in Xenopus oocytes were voltage-clamped, and distinct LRET signals were obtained in the resting and slow inactivated states. Intramolecular distances computed from the LRET signals define a geometrical map of Nav1.4 with the bound toxins, and reveal voltage-dependent structural changes related to channel gating.
Voltage-gated sodium channels (Navs) play an essential role in the generation and propagation of action potentials in excitable cells (1). Eukaryotic Navs are composed of a pore-forming α subunit and auxiliary β subunits. The α subunit is a large single-polypeptide chain organized in four different domains (DI–DIV), each of which has a voltage-sensing domain (VSD; S1–S4 segments) and a pore-forming domain (S5–S6 segments). Each domain has a different amino acid composition, pointing to some level of functional specialization. Site-directed fluorimetry shows that the VSDs in DI, DII, and DIII of the rat skeletal muscle voltage-gated sodium channel (Nav1.4) are activated by depolarization faster than in DIV (2). From this observation, it has been hypothesized that DI–III VSDs control the pore opening of the mammalian Nav, whereas the DIV VSD governs its fast inactivation (2–5).
Although mammalian Nav function has been studied comprehensively, the precise structural basis for the gating mechanisms has not been fully elucidated. The crystal structures of several prokaryotic Navs have been solved recently; however, in contrast to the mammalian Navs, they are homotetrameric, and thus structurally more closely related to the organization of voltage-gated potassium channels (Kvs) (6–9). The structure of a human L-type voltage-gated calcium channel type 1.1 (Cav1.1), which is a large single polypeptide composed of four different domains similar to mammalian Navs, has been resolved using cryoelectron microscopy (10, 11). However, functional studies have shown that gating mechanisms of mammalian Cav channels are indeed different from gating mechanisms in mammalian Navs (12). Furthermore, such structural studies only provide static snapshots of the channels in one of many possible conformational states. Therefore, techniques that provide dynamic structural information are needed to elucidate the mechanisms of voltage-sensing operation in mammalian Navs. In earlier functional studies on skeletal muscle Navs, we used derivatives of μ-conotoxin GIIIA to obtain approximate estimates of distances within the pore vestibule (13), and of a “mean” distance moved by gating charges during activation (14), obtained from electrostatic calculations in both cases.
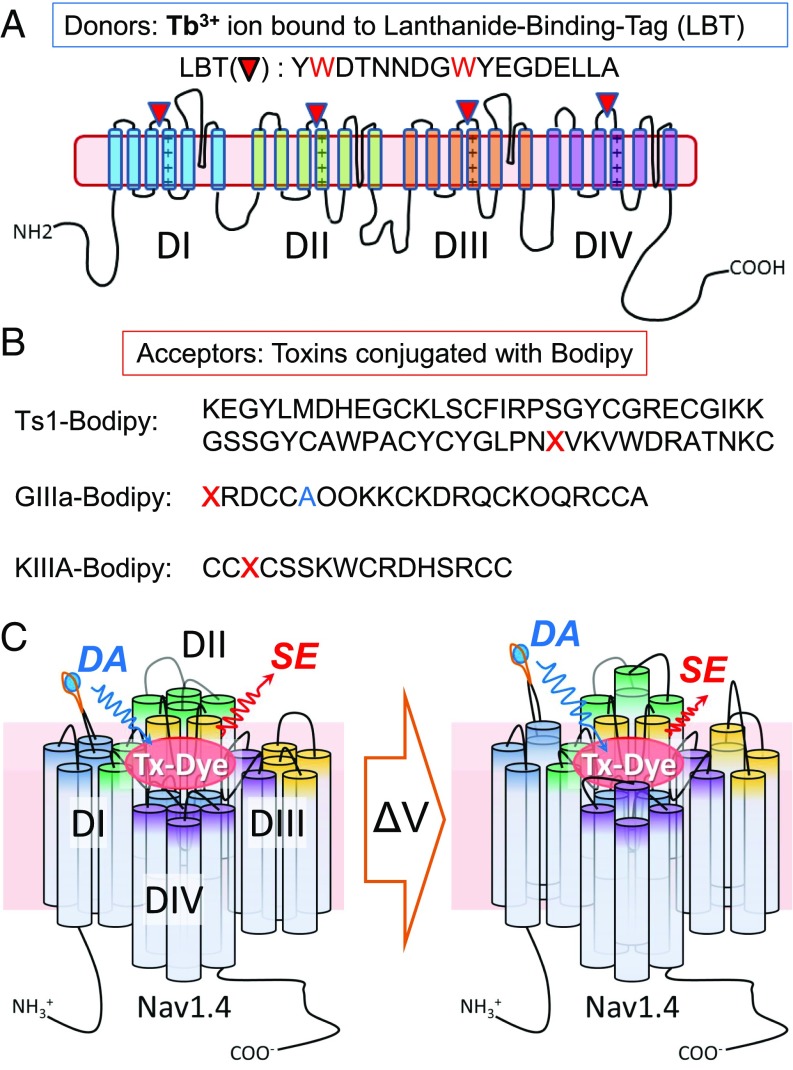
The studies described below provide specific distance measurements of Nav1.4 by lanthanide-based resonance energy transfer (LRET) as an intramolecular ruler, resulting in voltage-driven conformational changes in functioning toxin–channel complexes. LRET measures energy transfer between a luminescent lanthanide [LRET donor; here, a terbium ion (Tb3+)] tightly bound to a lanthanide-binding tag (LBT; Fig. 1A) and a spectroscopic acceptor (LRET acceptor; Fig. 1B), which enables distance measurements between them (15–20). Three different toxins conjugated with the fluorescent Bodipy dye (toxin-dyes) were synthesized to function as LRET acceptors: a β-scorpion toxin (Ts1)-Bodipy analog, as well as two conotoxin analogs, KIIIA-Bodipy and GIIIA-Bodipy (Fig. 1B). The function and binding sites in mammalian Navs for these three toxins have been well characterized by electrophysiological experiments and computational modeling; therefore, they function as trustworthy references for dynamics of the donor movement (21–26). We generated a total of 12 different Nav1.4 clones as LRET donors, which retained functional activity. Each clone has one inserted LBT, which binds a Tb3+ ion with high affinity in one of three different strategic locations on the extracellular side of the S4 segment on DI, DII, DIII, or DIV of Nav1.4 (Nav-LBTs). Multiple distances obtained from the Tb3+ in Nav-LBTs to each toxin-dye constrain the intramolecular geometrical maps of Nav1.4 with bound toxins. In addition, we observed changes in the distance, from either DI or DIV LBTs to the bound KIIIA-Bodipy, between the resting and relaxed states (27), usually associated with a slow inactivated state of the pore under voltage clamp conditions. (Relaxed and slow inactivated states will be used interchangeably). Our results provide direct evidence for voltage-dependent structural changes in mammalian sodium channels (Fig. 1C).
Fig. 1.
Experimental design for LRET in mammalian Nav. (A) Schematic representation of Nav-LBT donor clones. The Nav 1.4 α subunit is composed of four domains: DI (blue), DII (green), DIII (orange), and DIV (purple). The red inverted triangle represents the LBT, whose sequence is also shown. Each Nav-LBT mutant has only a single LBT at the top of S4 in one of the four domains: DI-LBT, DII-LBT, DIII-LBT, and DIV-LBT. Red-highlighted tryptophans (W) in LBT work as antennae to sensitize Tb3+ emission. (B) Chemically synthesized peptide toxins conjugated with the fluorescent Bodipy dye as LRET acceptor. Amino acid sequences of three synthetic toxins are shown: Ts1-Bodipy, KIIIA-Bodipy, and GIIIA-Bodipy. In Ts1-Bodipy, the red X indicates an amino acid substitution from tryptophan to an unnatural amino acid, Pra, for the conjugation with Bodipy through click chemistry. Similarly, the red X’s in the KIIIA-Bodipy and GIIIA-Bodipy sequences indicate an amino acid substitution to the unnatural amino acid beta-azidoalanine from asparagine (N) in KIIIA, or the addition of beta-azidoalanine to the N-terminus of GIIIA for Bodipy conjugation. The blue A in GIIIA-Bodipy is an additional substitution from threonine (T) to alanine (A), and the O’s are trans-4-hydroxyproline residues. (C) Schematic representation of LRET experiments under voltage clamp. Changes in membrane potential induce a conformational change in the VSDs of Nav-LBTs, which can be recorded as distance changes between Tb3+ bound to LBT (the donor) and the toxin-dye acceptor. SE reflects the changes in energy transfer induced by the distance changes. DA indicates the donor signal in the presence of acceptor. Tx-Dye, toxin conjugated to fluorescent dye.
Results
Generating Optimal Nav-LBT Clones as LRET Donors.
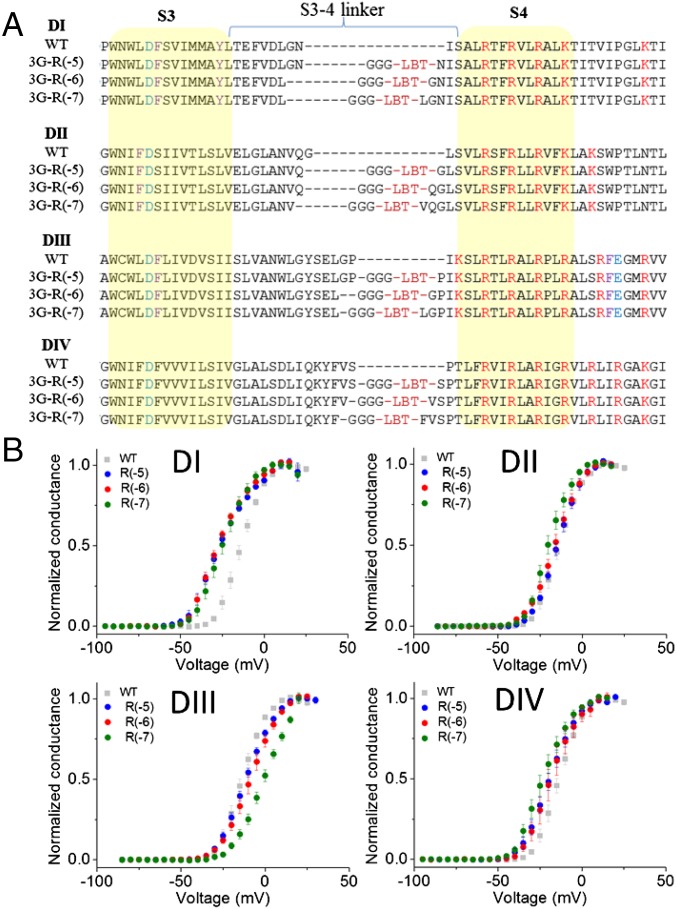
To obtain stable Nav-LBT clones that produce a robust donor (D) signal, we optimized the insertion site of the LBT. First, we generated eight different clones with the LBT in domain I (DI-LBTs), from DI-LBT-R(0) to DI-LBT-R(−7), with the LBT located zero to seven residues from the first S4 gating charge “R1” of DI. The numbers in parentheses indicate the number of residues counting back from the first gating charge, R; thus, DI-LBT-R(0) has the LBT inserted immediately before the first charge and DI-LBT-R(−7) has seven residues between the LBT and the first charge (SI Appendix, Fig. S1A). D-only signals were measured from each clone, and the signal decay time constants were evaluated. As shown in SI Appendix, Fig. S1B, constructs DI-LBT-R(0) to DI-LBT-R(−3) did not show robust D signals. On the other hand, constructs DI-LBT-R(−4), DI-LBT-R(−5), DI-LBT-R(−6), and DI-LBT-R(−7) produced robust D signals with time constants that are longer than 2.2 ms. Among them, the oocytes expressing DI-LBT-R(−4) had a lower survival rate (30–50%) than the others [>65% for DI-LBT-R(−5), DI-LBT-R(−6), and DI-LBT-R(−R7)]. These results are consistent with the idea that the LBT motif must maintain its conformation to exclude water from the chelated terbium, which may be affected depending on where the motif is inserted. It is empirically assumed that introduction of sequential glycine residues increases polypeptide flexibility while conserving the appropriate conformation of the LBT to chelate the Tb3+ ion. Therefore, in an attempt to improve the magnitude of the D signal, we created Nav1.4-LBT constructs containing additional glycine residues attached to the N terminus of the LBT in DI-LBT-R(−5). We found that a triple-glycine insertion optimized the D signal of the DI-LBT clones (SI Appendix, Fig. S1C). This engineered motif, here referred to as 3G-LBT, was implemented in each of the three positions of S4 in all other Nav1.4 domains (DII, DIII, and DIV) for a total of 12 functional donor clones (Fig. 2A). All constructs were functionally characterized under cut-open oocyte voltage clamp (COVC), and confirmed that most of them maintained the voltage-dependent properties of the Na+ conductance, although some [e.g., DIV R(−6), DIII R(−7) in SI Appendix, Fig. S2] showed altered voltage-dependent activation and/or modified fast inactivation (Fig. 2B and SI Appendix, Fig. S2).
Fig. 2.
Overview of Nav-LBT constructs. (A) Sequence alignments of all Nav-LBT constructs. All clones have the LBT inserted at the top of S4 and three consecutive glycines inserted before the LBT. The nomenclature is as follows: Dx-3GLBT-R(−y), where “x” indicates the Nav domain number, in roman numerals, and “y” is the number of amino acids counting back from the first S4 charge of the domain. Where an original glycine exists in the insertion site of 3G before LBT, only two glycines were inserted. Sequences highlighted in yellow indicate putative transmembrane segments. (B) Normalized conductance as a function of voltage (G-V) for all Nav-LBTs. Gray squares correspond to wild-type (WT) Nav1.4. Circles indicate Dx-3GLBT-R(−5) in blue, Dx-3GLBT-R(−6) in red, and Dx-3GLBT-R(−7) in green. Error bars are SEMs.
Pharmacological Properties of Acceptor Toxins.
Ts1 is a β-scorpion toxin isolated from the Brazilian scorpion, Tityus serrulatus (28). Such scorpion toxins bind to Navs through the S1–S2 and the S3–S4 linkers of the VSD in DII and the S5–S6 pore linker in DIII (25, 26). Their mechanism of action has been described as a “voltage sensor trapping effect,” wherein the β-scorpion toxin traps the DII VSD in an activated state, facilitating Nav opening (23, 24). Consequently, β-scorpion toxins shift the threshold of Nav activation in the direction of hyperpolarization.
We prepared Bodipy site-specifically labeled Ts1 toxin as previously reported (29). Briefly, Ts1 bearing the mutation W50 l-propargylglycine (Pra) was chemically synthesized as Ts1-W50Pra, which enabled the bioconjugation with fluorescent dye using “click chemistry” (Fig. 1B). The basic pharmacological and optical properties of Ts1-Bodipy were reported previously (29).
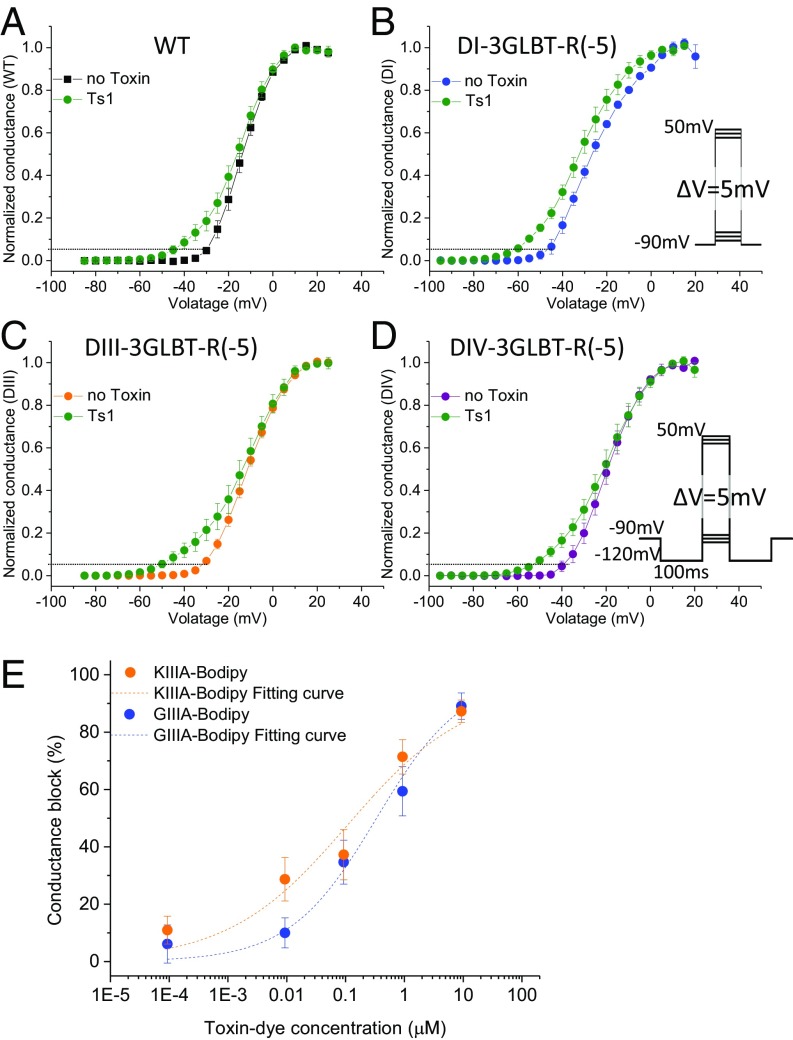
Interestingly, whereas most toxins reported to date that bind to Nav1.4 do so reversibly, Ts1-W50Pra’s binding was essentially irreversible in our hands; therefore, we tested whether this property is preserved for Nav-LBTs. Here, we defined the activation threshold as the voltage where the conductance is 5% of the full conductance. The activation threshold of wild-type Nav 1.4 channels is −30 mV (black symbols in Fig. 3A). After pretreatment with 1 μM Ts1-W50Pra, the activation threshold shifted to approximately −45 mV, even after washing with toxin-free recording solution (green symbols in Fig. 3A). This result indicates that the threshold of Nav1.4 activation was shifted in the hyperpolarizing direction by 15 mV, which is equivalent to the saturating effect caused by wild-type Ts1 (Fig. 3A). Similar results were obtained for all three Nav-LBTs tested: DI-3GLBT-R(−5), DIII-3GLBT-R(−5), and DIV-3GLBT-R(−5), which indicates that the pharmacological properties of the Nav1.4–Ts1 interaction are preserved in the Nav-LBT constructs (Fig. 3 B–D). We tested whether this property is maintained with the labeled Ts1. Although we tested only one cell because of the limited amount of Ts1-Bodipy available, the effect of Ts1-Bodipy remained stable even after washing with toxin-free solution. This property enabled us to conduct LRET measurements in toxin-free solution after pretreatment with Ts1-Bodipy, which has the advantage of decreasing nonspecific transfer in LRET signals.
Fig. 3.
Pharmacological properties of fluorescently labeled peptide toxins. (A–D) Activation plots for WT Nav1.4 and LBT clones preincubated with 1 μM Ts1-W50Pra. Ts1-W50Pra stays bound to Nav1.4, maintaining its activity after washout. Normalized conductance without toxin pretreatment is shown for WT Nav1.4 (A, black), DI-3GLBT-R(−5) (B, blue), DIII-3GLBT-R(−5) (C, orange), and DIV-3GLBT-R(−5) (D, purple). Green circles indicate normalized conductance in toxin-free solution after treatment with 1 μM Ts1-W50Pra. Dotted lines indicate 5% of the maximum conductance, used here to illustrate the change in activation threshold. Error bars indicate SEMs. The assay using DII-3GLBT was not conducted because the preliminary experiments using DII-3GLBT-R(−5) did not show any effect, which indicates that LBT insertion in DII prevents Ts1 binding. (E) WT Nav1.4 ionic current block by KIIIA-Bodipy (orange circles; IC50 = 108.9 ± 50.4 nM, n = 3–6 cells) and GIIIA-Bodipy (blue circles; IC50 = 336.4 ± 75.4 nM, n = 3–5 cells).
The μ-conotoxins are peptide toxins isolated from the venom duct of fish-hunting marine cone snails (30). The μ-conotoxin family is composed of several derivatives with diverse amino acid sequences, including μ-GIIIA and μ-KIIIA (31, 32), here referred to as GIIIA and KIIIA for convenience. GIIIA is a 22-aa polypeptide that binds with high affinity to Nav1.4, causing a complete block of Na+ ion conductance. Conversely, KIIIA is a shorter peptide of only 16 aa that also blocks Na+ ion conductance through Nav1.4. However, the blockage by KIIIA is incomplete, allowing guanidinium toxins like tetrodotoxin (TTX) access to a deeper pocket of site 1 of most Nav isoforms, as indicated by experiments where complete block is achieved when TTX and KIIIA are applied simultaneously (21, 22, 30, 32).
We synthesized two μ-conotoxins analogs conjugated with Bodipy, KIIIA-Bodipy, and GIIIA-Bodipy, with a similar strategy to the one used for Ts1-Bodipy. Briefly, a β-azidoalanine residue was inserted in place of Asn3 in KIIIA or was added to the N terminus of the GIIIA-T5A mutant. After folding and disulfide formation, Bodipy-alkyne was conjugated via click chemistry to the azide-containing polypeptides to generate KIIIA-Bodipy and GIIIA-Bodipy (SI Appendix, Supplemental Experimental Procedures and Figs. S3 and S4). We verified the pharmacological properties of KIIIA-Bodipy and GIIIA-Bodipy by functional recording under COVC on wild-type Nav1.4. We found IC50 values of 108.9 ± 50.4 nM for KIIIA-Bodipy and of 336.4 ± 75.4 nM for GIIIA-Bodipy (Fig. 3E). Although these apparent affinities are somewhat lower than the unlabeled μ-conotoxins, these derivatives remain well-suited for LRET experiments.
Geometrical Parameters of Toxin Bodipy-Bound Nav1.4-LBT.
Intact, unclamped oocytes were used to perform LRET measurements between the Nav-LBT constructs and each Bodipy-labeled toxin. In parallel experiments using identical solutions, resting potentials were measured and found to be in the range −20 to −15 mV. Therefore, a significant population of the Navs described in this type of experiments is assumed to be mostly in the slow inactivated state.
In LRET, the donor/acceptor distance, R, is obtained from the Förster relation:
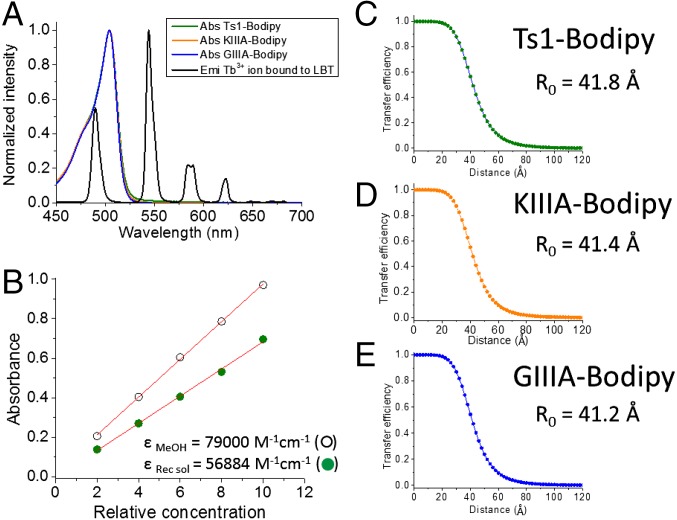
where R0 is the distance for 50% energy transfer efficiency, and τDA and τD are the lifetimes of Tb3+ luminescence decay in the presence and absence of an acceptor, respectively (20). The τSE is the sensitized emission (SE) lifetime or fluorescence decay from the acceptor dye (here, Bodipy). The R0 values for each toxin-dye/LBT Tb3+ pair were: 41.8 Å for Ts1-Bodipy; 41.4 Å for KIIIA-Bodipy, and, 41.2 Å for GIIIA-Bodipy (Fig. 4).
Fig. 4.
Optical properties for Ts1-Bodipy, KIIIA-Bodipy, and GIIIA-Bodipy. (A) Overlap between the emission (Emi) spectrum of Tb3+ bound to LBT (black solid trace) and the absorption (Abs) spectra of Ts1-Bodipy (green trace), KIIIA-Bodipy (orange trace), and GIIIA-Bodipy (blue trace). Note that the green, orange, and blue traces overlay almost exactly, but may be seen between ∼520 and 550 nm. (B) Corrected extinction coefficient of BODIPY FL-γ-azido-Abu in LRET recording solution (Recsol) obtained by the Beer–Lambert law. Energy transfer efficiency is shown as a function of distance in the pair LBT-bound Tb3+ and toxin-dyes, Ts1-Bodipy (C), KIIIA-Bodipy (D), and GIIIA-Bodipy (E). Respective calculated R0 values, a ruler for LRET experiments, are also shown.
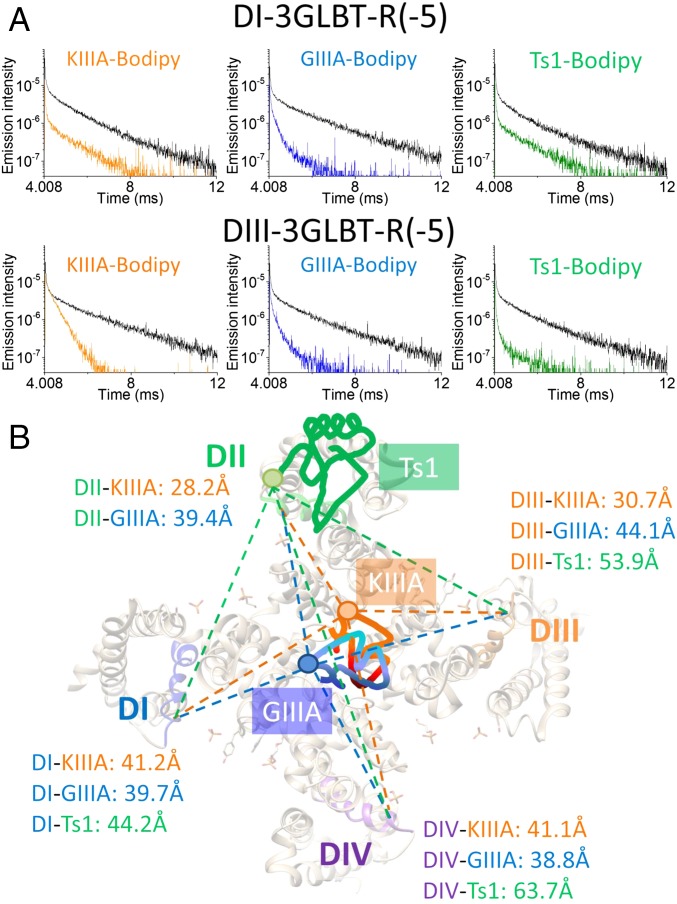
LRET signals from Ts1-Bodipy were obtained as follows. First, we measured all oocytes expressing Nav-LBTs and selected cells showing robust D signals (τD > 2.25 ms). The selected oocytes were pretreated with 1 μM Ts1-Bodipy for 15 min. After rinsing the oocytes twice, using fresh standard oocyte saline (SOS) and then fresh recording solution, SE signals in toxin-free LRET recording solution were measured. Representative LRET decays, including the D (black trace) and SE (green traces) signals, are shown in Fig. 5A (Right). Based on the time constant of the SE signal decay, the distance between DI-3GLBT-R(−5) and Ts1-Bodipy was calculated as 44.2 ± 0.3 Å. Similarly, the distance between DIII-3GLBT-R(−5) and Ts1-Bodipy was 53.9 ± 1.2 Å. The SE signal of DIV-3GLBT-R(−5) was too small to determine reliable time constants (SI Appendix, Fig. S5). Instead, the distance between DIV-3GLBT-R(−5) and Ts1-Bodipy was calculated using the time constant of D in the presence of acceptor decay (DA) signal, resulting in 63.7 ± 1.8 Å. LRET assays using DII-LBT clones and Ts1, on the other hand, were not conducted because preliminary experiments using DII-3GLBT-R(−5) showed no Ts1 effect, suggesting that LBT insertion in DII prevents Ts1 binding. Even though the distance between DIV-3GLBT-R(−5) and Ts1-Bodipy falls within a region where LRET is relatively less sensitive, all three distances serve to provide the general position of the toxin, given that the distances measured were sufficiently different between them. However, because two of the three distances measured with Ts1-Bodipy do not fall within a region of high sensitivity in LRET (i.e., within ∼5 Å from the R0 value in both directions; R0 ∼ 42 Å; Fig. 4C), and because the LRET signals are also smaller, Ts1-Bodipy was less suitable than the other toxin-dyes to serve as an acceptor for measurements of small movements during gating transitions among different conformations under voltage clamp (discussed in the following section).
Fig. 5.
Intramolecular geometrical map of toxin-binding sites in Nav1.4. (A) Representative LRET signal traces using DI-3GLBT-R(−5) (Upper) and DIII-3GLBT-R(−5) (Lower). Black traces represent D signals. SE signals are shown in orange for KIIIA-Bodipy, in blue for GIIIA-Bodipy, and in green for Ts1-Bodipy. (B) Intramolecular geometrical location of KIIIA (orange), GIIIA (blue), and Ts1 (green) in Nav. The depicted Nav structure is modified from the NavAb structure (Protein Data Bank ID code 4ekw). Segment 4 in each domain is highlighted in blue for DI, green for DII, orange for DIII, and purple for DIV. The numbers in orange indicate averaged values of distances from KIIIA-Bodipy to R(−5), R(−6), and R(−7). Similarly, the numbers in blue indicate averages values of distances from GIIIA-Bodipy to R(−5), R(−6), and R(−7). The numbers in green indicate the measured distance from Ts1-Bodipy to R(−5). Given that the oocytes in these experiments were not voltage-clamped, with resting potentials in the range of −20 to −15 mV, the mapping is expected to represent the slow inactivated state (main text).
To obtain SE signals from KIIIA-Bodipy, we first recorded D signals in the same way as described above for Ts1-Bodipy. Then, we measured SE signals in the presence of 0.85 μM KIIIA-Bodipy in the LRET recording solution to avoid toxin dissociation (concentration of approximately eightfold IC50, ∼90% of channels bound). Similarly, we obtained SE signals from GIIIA-Bodipy in the presence of 0.85 μM GIIIA-Bodipy (concentration of approximately threefold IC50, ∼75% of channels bound). Representative LRET signals are shown in Fig. 5A. The SE transfer signals from KIIIA-Bodipy to DII-3GLBT or DIII-3GLBT clones showed faster decay than the SE transfer signals from GIIIA-Bodipy to the same donor constructs (Fig. 5A and SI Appendix, Fig. S5). This result indicates that, comparatively, KIIIA-Bodipy “sits” closer to DII-VSDs and DIII-VSDs than GIIIA-Bodipy. The time constant values measured and the computed distances from all donor/acceptor pairs measured are shown in Tables 1–3. Collectively, these results establish the geometrical locations of toxin-dyes bound to Nav1.4 as shown in Fig. 5B.
Table 1.
LRET time constants and estimated distances from intact, unclamped oocytes with KIIIA-Bodipy
| KIIIA-Bodipy (n) | τD, ms | τSE, ms | Distance, Å |
| D1-3GLBT-R(−5) (6) | 2.44 ± 0.03 | 1.27 ± 0.03 | 42.0 ± 0.2 |
| D1-3GLBT-R(−6) (5) | 2.44 ± 0.02 | 1.20 ± 0.03 | 41.2 ± 0.4 |
| D1-3GLBT-R(−7) (10) | 2.48 ± 0.02 | 1.14 ± 0.03 | 40.3 ± 0.3 |
| D2-3GLBT-R(−5) (5) | 2.48 ± 0.05 | 0.19 ± 0.01 | 27.3 ± 0.3 |
| D2-3GLBT-R(−6) (6) | 2.37 ± 0.04 | 0.21 ± 0.003 | 28.2 ± 0.03 |
| D2-3GLBT-R(−7) (8) | 2.39 ± 0.02 | 0.27 ± 0.02 | 29.2 ± 0.5 |
| D3-3GLBT-R(−5) (9) | 2.46 ± 0.03 | 0.37 ± 0.01 | 30.9 ± 0.1 |
| D3-3GLBT-R(−6) (8) | 2.48 ± 0.03 | 0.34 ± 0.02 | 30.4 ± 0.3 |
| D3-3GLBT-R(−7) (6) | 2.46 ± 0.03 | 0.35 ± 0.01 | 30.7 ± 0.2 |
| D4-3GLBT-R(−5) (7) | 2.39 ± 0.02 | 1.27 ± 0.03 | 42.2 ± 0.4 |
| D4-3GLBT-R(−6) (7) | 2.47 ± 0.03 | 1.11 ± 0.03 | 40.0 ± 0.2 |
| D4-3GLBT-R(−7) (7) | 2.41 ± 0.02 | 1.16 ± 0.05 | 41.0 ± 0.7 |
The number of determinations (n), for each channel construct, is shown in the first column; τD indicates the value of the time constant of donor decay in the absence of acceptor; and τSE is the decay time constant of the sensitized emission.
Table 3.
LRET time constants and estimated distances from intact, unclamped oocytes with Ts1-Bodipy
| Ts1-Bodipy (n) | τD, ms | τSE, ms | Distance, Å |
| D1-3GLBT-R(−5) (5) | 2.42 ± 0.04 | 1.41 ± 0.002 | 44.2 ± 0.3 |
| D3-3GLBT-R(−5) (6) | 2.35 ± 0.02 | 1.91 ± 0.04 | 53.9 ± 1.2 |
| D4-3GLBT-R(−5) (4) | 2.38 ± 0.01 | 2.20 ± 0.02 (τDA) | 63.7 ± 1.8 |
The number of determinations (n), for each channel construct, is shown in the first column; τD is the time constant of the donor only signal; and τDA, in parenthesis, is the time constant of donor decay in the presence of acceptor instead of the time constant obtained from the sensitized emission, τSE.
Table 2.
LRET time constants and estimated distances from intact, unclamped oocytes with GIIIA-Bodipy
| GIIIA-Bodipy (n) | τD, ms | τSE, ms | Distance, Å |
| D1-3GLBT-R(−5) (6) | 2.42 ± 0.03 | 1.08 ± 0.07 | 39.7 ± 0.7 |
| D1-3GLBT-R(−6) (7) | 2.48 ± 0.02 | 1.2 ± 0.06 | 40.8 ± 0.6 |
| D1-3GLBT-R(−7) (6) | 2.44 ± 0.05 | 0.97 ± 0.03 | 38.5 ± 0.3 |
| D2-3GLBT-R(−5) (6) | 2.53 ± 0.01 | 1.25 ± 0.02 | 41.0 ± 0.2 |
| D2-3GLBT-R(−6) (4) | 2.42 ± 0.06 | 0.93 ± 0.04 | 38.0 ± 0.4 |
| D2-3GLBT-R(−7) (6) | 2.40 ± 0.03 | 1.02 ± 0.03 | 39.2 ± 0.4 |
| D3-3GLBT-R(−5) (6) | 2.44 ± 0.02 | 1.45 ± 0.03 | 43.9 ± 0.2 |
| D3-3GLBT-R(−6) (8) | 2.48 ± 0.03 | 1.49 ± 0.03 | 44.2 ± 0.4 |
| D3-3GLBT-R(−7) (8) | 2.41 ± 0.02 | 1.44 ± 0.03 | 44.1 ± 0.5 |
| D4-3GLBT-R(−5) (7) | 2.37 ± 0.02 | 1.01 ± 0.02 | 39.2 ± 0.2 |
| D4-3GLBT-R(−6) (7) | 2.49 ± 0.02 | 0.98 ± 0.02 | 38.4 ± 0.2 |
| D4-3GLBT-R(−7) (9) | 2.43 ± 0.02 | 1.01 ± 0.02 | 38.9 ± 0.2 |
The number of determinations (n), for each channel construct, is shown in the first column; τD indicates the value of the time constant of donor decay in the absence of acceptor; and τSE is the decay time constant of the sensitized emission.
Gating-Dependent Conformational Changes in Nav Channel Detected by LRET Between Functional Nav-LBTs and KIIIA-Bodipy.
DI-LBTs and DIV-LBTs as donors and KIIIA-Bodipy as acceptor were chosen for the detection of voltage-dependent changes in LRET signals in functional gating channels. The choice of donor/acceptor pairs for these experiments was guided by the following: (i) computed distances between KIIIA-Bodipy and DI-LBTs or DIV-LBTs for unclamped oocytes were close to the R0 (41–42 Å) that is the most sensitive range for detection of changes in distance with high precision using LRET, and (ii) SE signals from KIIIA-Bodipy were larger than SE signals from other toxin-dyes, which results in better signal resolution.
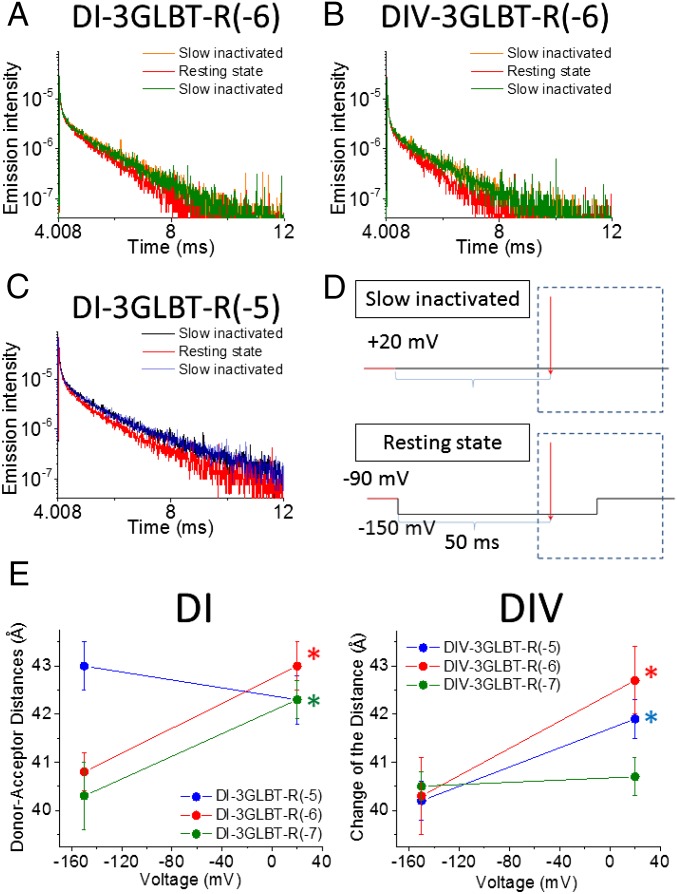
Two functional gating states of the mammalian Nav1.4 were assessed by LRET measurement in voltage-clamped oocytes. The slow inactivated (or relaxed) state and the resting state were induced by changes in the membrane potential under voltage clamp (27, 33–35) (Fig. 6). To ensure that the recorded changes in LRET signals were induced by the membrane potential manipulation and not due to changes in cell morphology by oocyte decay, only oocytes with matching slow inactivated signals at the beginning and end of the run were included in our analysis. A protocol to assess the Nav1.4 resting state was flanked by the two slow inactivation protocols (as shown in Fig. 6D, Right). Briefly, the oocyte was held at +20 mV for over 5 min to induce Nav slow inactivation before LRET signal recordings (orange traces in Fig. 6A). The channels were then relieved of inactivation by holding the membrane potential at −80 to −90 mV for 4 min, and thus driving channels to the resting state. The voltage was held at −90 mV for another minute before recording the LRET signal in the resting state (red traces in Fig. 6A). Finally, the LRET signal corresponding to the slow inactivated state was recorded once again (green traces in Fig. 6A). As shown in Fig. 6A, the SE signals were different depending on the functional state in which the channels were driven by the voltage protocol. However, the D signal from some of the clones also showed voltage-dependent changes, as shown in Fig. 6B, which might be related to direct quenching of the Tb3+ ions (LBT-bound) by voltage-dependent conformational changes. We collected D signal data in the presence of unlabeled KIIIA (KIIIA-N3X) independently and computed all distances using time constants for both D and SE signal decays at each voltage, with error propagation as described previously (18). The voltage-dependent donor/acceptor distance changes in DI-LBTs and DIV-LBTs are summarized in Fig. 6E and in Table 4.
Fig. 6.
LRET under voltage clamp using KIIIA-Bodipy: Voltage-dependent changes in distance for DI and DIV are shown. A representative SE signal in DI-3GLBT-R(−6) (A) and a representative SE signal in DIV-3GLBT-R(−6) (B) in different states are shown. The SE signal in the slow inactivated state recorded first is shown in orange, the SE signal in the resting state is shown in red, and the second recorded SE signal in the slow inactivated state is shown in green. (C) D signal in DI-3GLBT-R(−5) showed a voltage-dependent change. The D signal obtained in the first and second slow inactivated state recordings are shown as black and blue traces, respectively; the resting state D signal is the red trace. (D) Pulse-laser protocols used to induce and record from the slow inactivated and resting states are shown. (E) Distances between KIIIA-Bodipy and DI-LBTs or DIV-LBTs showed significant voltage-dependent changes. Data from Dx-3GLBT-R(−5) clones are shown in blue, data from Dx-3GLBT-R(−6) are shown in red, and data from Dx-3GLBT-R(−7) are shown in green. Error bars represent SEMs. Asterisks indicate significant distance changes (*P < 0.05, unpaired t test).
Table 4.
LRET results using KIIIA-N3X for D signal and KIIIA-Bodipy for SE signal from oocytes under voltage clamp
| Clones | Voltage | τD, ms (n) | τSE, m | Distance, Å |
| D1-3GLBT-R(−5) | −150mV | 2.13 ± 0.03 (6) | 1.19 ± 0.02 (5) | 43.0 ± 0.5 |
| +20mV | 2.42 ± 0.03 (6) | 1.29 ± 0.03 (5) | 42.3 ± 0.5 | |
| D1-3GLBT-R(−6) | −150mV | 2.34 ± 0.01 (5) | 1.11 ± 0.03 (7) | 40.8 ± 0.4 |
| +20mV | 2.37 ± 0.02 (5) | 1.32 ± 0.03 (7) | 43.0 ± 0.5 | |
| D1-3GLBT-R(−7) | −150mV | 2.26 ± 0.06 (5) | 1.04 ± 0.03 (6) | 40.3 ± 0.7 |
| +20mV | 2.35 ± 0.01 (5) | 1.25 ± 0.02 (6) | 42.3 ± 0.4 | |
| D4-3GLBT-R(−5) | −150mV | 2.18 ± 0.04 (5) | 0.99 ± 0.01 (5) | 40.2 ± 0.4 |
| +20mV | 2.30 ± 0.02 (5) | 1.19 ± 0.03 (5) | 41.9 ± 0.4 | |
| D4-3GLBT-R(−6) | −150mV | 2.21 ± 0.08 (6) | 1.01 ± 0.03 (6) | 40.3 ± 0.8 |
| +20mV | 2.38 ± 0.03 (6) | 1.30 ± 0.04 (6) | 42.7 ± 0.7 | |
| D4-3GLBT-R(−7) | −150mV | 2.21 ± 0.04 (7) | 1.03 ± 0.01 (5) | 40.5 ± 0.3 |
| +20mV | 2.39 ± 0.05 (7) | 1.13 ± 0.01 (5) | 40.7 ± 0.4 |
Values shown are averages ± SE of means. n, number of cells measured.
Discussion
We have obtained two major pieces of structural information from functionally active mammalian Nav1.4 in live cells using LRET. First, LRET-based distance estimates, using variants of three different fluorescently labeled peptide toxins as acceptors, are consistent with various other experimental studies and structure-based predictions of the sites of action of these toxins. This structural information regarding sites of action of these toxins has been obtained directly from functioning Nav proteins. Such data have not previously been experimentally accessible. Second, LRET measurements, under voltage clamp, enabled us to directly assess the structural differences between two functional Nav states, resting and slow inactivated. These results will provide important constraints on the functional interpretation of new structural data as they emerge.
Binding sites for Nav-specific toxins, as well as their binding characteristics, have been described functionally by electrophysiological assays, and theoretically by computational molecular modeling based on currently available crystal structures (21–26). Current knowledge for the physical locations of the toxins, however, remains somewhat imprecise. Our results provide details about the locations of toxin-binding sites. For instance, mutations in the voltage sensor of DII, which alter the function of toxin Ts1, imply that this region may be part of the toxin-binding site (25, 26). However, some of the effects of the mutations may be due to long-range allosteric modulations of a distant toxin-binding site. To date, homology modeling has been based on crystal structures of homotetrameric prokaryotic Navs or eukaryotic Kvs because no high-resolution structures of a mammalian Nav have been available. Nonetheless, although the detailed architecture of mammalian Navs may differ somewhat from the current homology models, the currently proposed binding sites and orientations for well-characterized peptide toxins generally show good agreement among different studies.
From mutagenesis-based structure/function studies, the Nav-binding site for the Ts1 toxin has been proposed to be formed mainly by the DII VSD as mentioned above, with a minor contribution from the DIII pore linker (25, 26). The μ-conotoxins are thought to bind to the outer vestibule of the channel and to occlude the Na+ ion-conducting pathway physically (30). For the smallest of these μ-conotoxins, KIIIA, blockade of single-channel conductance is less than complete, and simultaneous occupancy by the classic all-or-none Nav blocker, TTX, is possible. Homology models for Navs indicate that the larger, archetypal GIIIA functionally occludes the ion-conducting pore, whereas KIIIA’s center of mass is slightly shifted toward the VSDs of DII and DIII (21, 22). The positions of the Bodipy acceptor in our toxin-containing dyes are effectively constrained by site-specific conjugation to Ts1-W50X, to the N terminus of KIIIA-N3X, or to GIIIA-T5A-1X (Fig. 1B). Thus, relative to the size of the functional Nav, their positions are defined well. Our experimental LRET data, from the use of multiple toxin-dye/LBT pairs, indicate locations of the bound toxins on Nav1.4 that are in good agreement with those locations in recently proposed toxin docking models (21, 22). Positions of the bound Bodipy-labeled toxins are shown schematically in Fig. 5. In the absence of a high-resolution structure of any mammalian Nav, this study lays a foundation for future studies to elucidate the mechanisms of toxin action in greater detail. When high-resolution structures are available, our data will allow direct comparison of geometric parameters measured in selected functional states with geometric parameters from purely structural studies.
A major goal of this project was to provide distance measurements from functional Navs in different gating states that are determined by voltage. Under voltage clamp, LRET showed significant voltage-dependent changes in the distance between KIIIA-Bodipy and the LBT/Tb3+ in either DI or DIV of Nav1.4. This study reports direct measurements of voltage-dependent distance changes between VSDs and the pore domain (PD), as determined from functional eukaryotic Navs in live cells. Relative to most gating transitions, which occur on the millisecond time scale, the prolonged binding of KIIIA-Bodipy (approximately seconds) provides a stable reference to measure VSD movement.
It is important to reiterate that our method measures distances along a line connecting the donor and acceptor. Thus, the computed distance changes will become smaller if the physical movement occurs perpendicular to the line connecting the donor and acceptor, and will approach zero as the motion becomes symmetrical around the vector connecting the LBT donor and Bodipy acceptor (SI Appendix, Fig. S6). For that reason, the measured distance changes can be considered a lower limit on the distance over which the terbium, chelated to the LBT, and Bodipy, attached to the toxin, approach one another in transitions between resting and relaxed states.
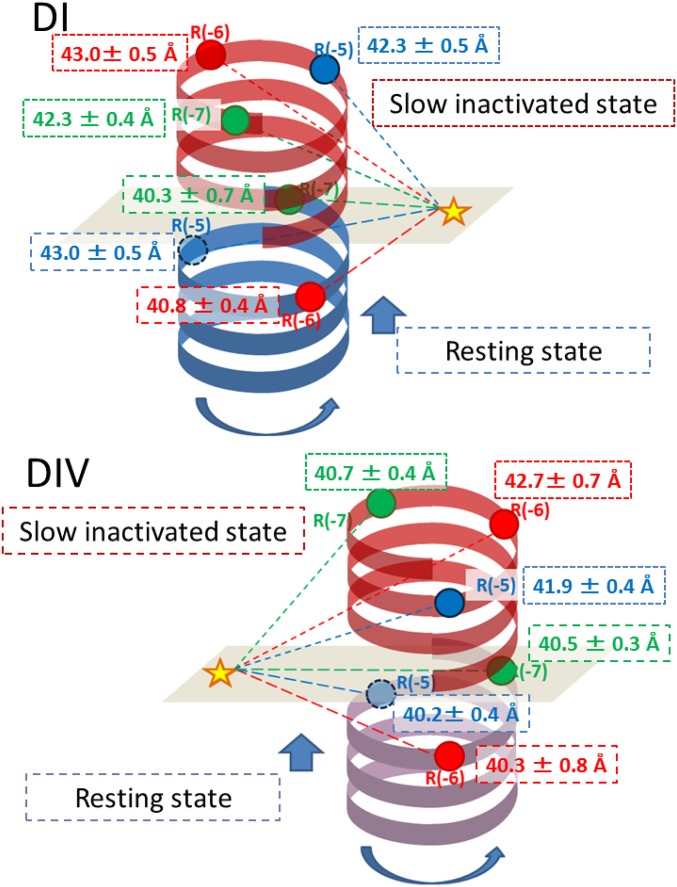
To interpret the LRET results quantitatively in three dimensions, high-resolution structural data from a eukaryotic Nav will indeed be required. At this stage, we can make a semiquantitative assessment of the conformational changes using the following assumptions: (i) The region of S4 preceding the first charge toward the N terminus is helical in structure, as in other voltage-gated ion channels (36), and (ii) the positions of LBTs follow the same helical structure. In SI Appendix, Fig. S1, the detection of D signals showed slight periodicity, which may reflect the helical structure of this region: for DI-LBT-R(−1), the periodicity was more strongly apparent than for DI-LBT-R(0), DI-LBT-R(−2), and DI-LBT-R(−3). Constructs DI-LBT-R(−4) again showed stronger periodicity. Considering all data from three different LBT clones for each domain [R(−5), R(−6) and R(−7)], a simple model containing a rotational and translational movement of the S4 segments is consistent with the LRET results, as represented schematically in Fig. 7. Importantly, our results, most clearly apparent in Fig. 6E, suggest a functional asymmetry of the VSDs in Nav1.4, with respect to KIIIA-Bodipy as follows: (i) For DI, the position of R(−5) in the resting state lies further from KIIIA-Bodipy than the distances of R(−6) and R(−7), whereas there are no significant differences between distances of DI R(−5), DI R(−6), and DI R(−7) with respect to KIIIA-Bodipy in the slow inactivated state, and (ii) for DIV, on the other hand, there is essentially no difference in the distances between KIIIA-Bodipy and R(−5), R(−6), or R(−7) in the resting state, whereas the R(−7) position seems to be closest to KIIIA-Bodipy in the slow inactivated state. This reciprocal change observed between DI and DIV may underlie the different functional roles of DI and DIV VSDs previously proposed based on site-directed fluorimetry (2, 3, 37).
Fig. 7.
Schematic model of Nav1.4 DI and DIV movement that satisfies the LRET data. A simplified model, where the S4 segment does not change its tilt, can be built considering all of our LRET data under voltage clamp from three different LBT clones [R(−5), R(−6), and R(−7)]. The results are consistent with a rotation of the S4 helices in DI and DIV. Significant movements are indicated in the figure, alongside the corresponding distance measurement, for the slow inactivated state. (Upper) Blue helix represents the position of the DI-S4 helix at rest, and red indicates its slow inactivated state. (Lower) Purple helix is the DIV-S4 helix in the resting state, and red indicates its slow inactivated state. Circles indicate the Tb3+ ion position from R(−5) (blue), R(−6) (red), and R(−7) (green), respectively. The star indicates the chromophore of KIIIA-Bodipy. Long dashed lines depict the distance between donor (colored circles) and acceptor (star) in the resting state, and fine dotted lines depict the distance between donor and acceptor in the slow inactivated state.
Another possible implication of our LRET results to be considered is that LBT insertion and/or toxin binding, per se, could modify Nav gating. Most of the 12 Nav-LBTs tested showed gating similar to wild-type Nav1.4, but some showed modified properties, including modest shifts of the voltage dependence of ionic conductance and changes in fast inactivation kinetics, as shown in SI Appendix, Fig. S2.
We conducted LRET analysis, under voltage clamp, in both resting and relaxed states (the latter reflects a slow inactivated state), but we did not obtain clear evidence that Nav-LBTs have exactly the same slow inactivated state as wild-type channels. Possible changes in the relaxed state in Nav-LBTs might generate some underestimation of the distances measured in our LRET analysis. On the other hand, we cannot exclude the possibility that toxin-Bodipy modifies the rate of entry to a slow inactivated state. There is evidence showing that toxins can modify Nav gating currents (34). Therefore, our LRET data might be partly determined by the presence of bound toxin-Bodipy. If so, this effect must be relatively small, and our results should mainly represent the intrinsic Nav gating process.
Other approaches to the estimation of conformational movements, which are subject to different constraints, might offer additional insights. For example, one possibility is to estimate the distance that VSD charges would have to move to account for voltage sensitivity of activation or measured gating currents. Energetic calculations, based on activation shifts resulting from discrete perturbations in the local electric field sensed by the VSDs when a GIIIA derivative binds to and dissociates from the pore, are consistent with a net movement of the center of the gating charge of a few angstroms (14) but require assumptions of the shape of the potential profile across the VSD and the resting position of the center of the gating charge. The present results are consistent with those previous estimates, but further experiments will be needed to construct a complete picture of mammalian Nav VSD charge movement and gating dynamics.
Technically diverse approaches to the measurement of spatial rearrangements in the coupled VSD-PD system will, in general, fall most conveniently on different coordinate systems. For LRET measurements, the estimated distances lie directly along a vector connecting the donor and acceptor, but, a priori, we know neither the spatial locations of the probes in a given functional state nor the direction of movement during the transition between two states. Our calculated changes are ∼2 Å, but amplitudes of movements between different well-defined reference points could be either larger or smaller, and would depend on the direction of movement of the molecular probes used in a particular assay (compare SI Appendix, Fig. S6). Nevertheless, our explicit distance measurements, and their voltage dependence, will provide clear and critical constraints on interpretation (38) of future structural and functional results, and thus will help to lead us closer to a comprehensive understanding of the intricate coupling involved in voltage-dependent gating in eukaryotic Navs.
Conclusions
The voltage sensor segments of Nav1.4 DI and DIV occupy distinguishably different locations in resting and slow inactivated eukaryotic Navs. Voltage clamp-LRET constitutes an invaluable technique to assess structural dynamics of functional ion channels in live cells. The approach presented here provides an important step toward understanding the voltage-driven structural dynamics of mammalian Nav gating; the results will be especially pertinent as structures of a mammalian Nav are solved in additional specific conformations.
Experimental Procedures
Molecular Biology.
The α and β1 subunits of Nav1.4 were cloned into pBSTA vectors. For protein expression, cRNAs for Nav1.4 α and β1 subunits were transcribed in vitro using the T7 mMESSAGE cRNA kit (Ambion) and injected at a 1:1 molar ratio into Xenopus laevis oocytes. For electrophysiological experiments, freshly isolated oocytes were injected with 1 ng of cRNA and kept in SOS solution [96 mM NaCl, 2 mM KCl, 1 mM MgCl2, 1.8 mM CaCl2, 10 mM Hepes, 200 mg/L sodium pyruvate (pH 7.4)] for 1–2 d at 18 °C. For LRET experiments, the oocytes were injected with 75 ng of cRNA and incubated in SOS for 5–6 d at 12 °C, after which they were incubated for 1–2 d at 18 °C.
Chemical Synthesis of Dye-Conjugated Peptide Toxins.
Ts1-Bodipy.
Ts1-Bodipy was prepared as previously described (29). Bodipy was attached at position 50, after replacing Trp50 with Pra yielding the labeled Ts1 W50Pra-Bodipy.
KIIIA, KIIIA-Bodipy, and GIIIA-Bodipy.
Site-specific incorporation of Bodipy was achieved by click-conjugation of an azide-containing polypeptide precursor with Bodipy-alkyne (Lumiprobe). The X building block was incorporated in position 3 of KIIIA or added to the N terminus of GIIIA-T5A. In preliminary tests, the IC50 of GIIIA-T5A (9.8 nM) was found to be similar to the IC50 reported for the wild-type GIIIA (6.3 nM) (SI Appendix, Supplemental Experimental Procedures). NMR spectroscopy was used to confirm the correct fold of the synthetic analogs. NMR experiments on all analogs were performed on a Bruker Avance 600 spectrometer equipped with a cryoprobe. The obtained Hα-chemical shifts provide a sensitive probe of peptide secondary and tertiary structure and were compared with literature values reported previously for wild-type KIIIA and GIIIA (39, 40) (SI Appendix, Figs. S3 and S4). More details of the synthesis are provided in SI Appendix, Supplemental Experimental Procedures.
Preparation of Dye-Labeled Toxins.
All Bodipy-conjugated toxins were diluted in external recording solution for electrophysiology (Experimental Procedures, Electrophysiology) or in the LRET recording medium (Experimental Procedures, LRET Recording). Unlabeled toxin concentrations were obtained by spectrophotometry (Nanodrop; Thermo Scientific). Concentration of toxins conjugated with Bodipy were calculated based on the extinction coefficient values and the peak values of their absorption spectra obtained with a Cary 60 UV-Vis spectrophotometer (Agilent Technologies) (also Experimental Procedures, Extinction Coefficient Measurements in the Recording Solution).
Electrophysiology.
Ionic currents were recorded using a COVC setup (41). The external solution contained 57.5 mM N-methylglucamine (NMG)-methylsulfonate (MS), 57.5 mM Na-MS, 10 mM Hepes, and 2 mM Ca-MS (pH 7.4). The internal solution contained 103.5 mM NMG-MS, 11.5 mM Na-MS, 10 mM Hepes, and 2 mM EGTA (pH 7.4). Ts1 derivatives were dissolved in external solution containing 0.5–1% BSA. Oocytes were pretreated with 1 μM Ts1 derivatives for 15 min in a glass dish followed by two washes in SOS solution and two washes in the recording solution before the measurement. To avoid Ts1 derivative loss by retention to plastic ware, we used glass dishes during the whole process. After thorough washing of oocytes with toxin-free solution, they were mounted on the COVC setup, and Nav1.4 ionic currents in toxin-free recording solution were measured. For pharmacological assays of KIIIA-Bodipy or GIIIA-Bodipy, the toxins were dissolved in external solution without BSA. Control recordings were performed in the absence of toxins before the solution in upper and guard chambers of the COVC setup was replaced with toxin-bearing solution at the appropriate concentrations. The recordings were done after 15–20 min of incubation in the presence of the toxins. For KIIIA-Bodipy or GIIIA-Bodipy experiments, we determined the incubation time by following the appearance of SE signal in LRET. It was estimated that 15–20 min of incubation time was sufficient to achieve saturation.
The ionic currents were recorded with a PC44 board and digitized on the 16-bit A/D converter. Ionic currents were sampled at 10 μs per point. The data acquisition program was developed in-house. Linear leak and membrane capacitive currents were subtracted using a P/6 protocol from a subtracting holding potential of −100 mV. All data, except for pharmacological assay for KIIIA-Bodipy and GIIIA-Bodipy, were obtained at 12–14 °C. Pharmacological assays for KIIIA-Bodipy and GIIIA-Bodipy were performed at 20 °C.
Conductance was calculated as G(v) = Ipeak(v)/(V − Erev), where the reversal potential, Erev, was measured experimentally for each oocyte. Statistical significance was determined using an unpaired t test. Errors indicate SEMs.
LRET Recording.
LRET is a FRET technique that takes advantage of the properties of lanthanides, particularly Tb3+, which has a long lifetime; dark regions between emission peaks in its spectrum; and, most importantly, isotropic emission (20). Briefly, the donor is Tb3+-bound to a genetically encoded LBT motif with the sequence YIDTNNDGWYEGDELLA, which binds Tb3+ with high affinity (Kd = 57 nM) (42). For these experiments, we used a modified LBT that has an additional tryptophan (underlined), YWDTNNDGWYEGDELLA, to improve the Tb3+ D signal of Nav-LBT constructs (Fig. 1A). The optical setup for LRET measurements has been described previously (17–19). The LRET recording solution contained 92 mM NMG-MS, 23 mM Na-MS, 10 mM Hepes, 2 mM Ca-MS, and 5 μM TbCl3 (pH 7.4). During the experimental process, each oocyte was placed in a separate well.
The emission spectrum of Tb3+ ions bound to the LBT-purified peptide was measured in the laboratory. The absorption spectra of Ts1-Bodipy, KIIIA-Bodipy, and GIIIA-Bodipy were obtained by spectrophotometry (Agilent Cary 60; Agilent Technologies). The R0 values for the pair of Tb3+ ions bound to LBT (donor) and toxin-dye (acceptor) were obtained from the extinction coefficient of the acceptor and the spectral overlap between donor emission and acceptor absorbance (Fig. 4A). Because the extinction coefficient value is affected by the solvent in the solution, we obtained the corrected values using the Beer–Lambert law (Fig. 4B).
In LRET recordings without voltage clamp, D signals were obtained by averaging 20 measurements collected from oocytes without any treatment. SE signals were obtained by averaging 40 measurements collected from oocytes with Ts1 pretreatment, in the presence of 0.85 μM KIIIA-Bodipy or 0.85 μM GIIIA-Bodipy. For LRET recordings with voltage clamp, D signals were obtained from oocytes in the presence of 0.85 μM unlabeled KIIIA, KIIIA-N3X. The SE signals were obtained from oocytes in the presence of 0.85 μM KIIIA-Bodipy. Both signals were obtained by averaging 20 measurements for the D signal and 40 measurements for the SE signal; measurements under poor voltage control were excluded from the average.
Data were analyzed with an in-house program, DECAY ANALYSIS (17). For D signals and DA signals, a three-exponential fit was applied. The SE signals were fit with two exponentials. The slowest time constants of this fit represent LRET signals. The two other faster components in D and DA signals and the faster component in SE signals are supposed to be the intrinsic signals from the LRET setup and/or oocytes (17).
Extinction Coefficient Measurements in the Recording Solution.
A serial dilution of free dye, BODIPY FL-γ-azido-Abu, was prepared at linearly spaced concentrations in both MeOH and recording solutions. Absorption spectra were obtained from all samples, and the peak values were plotted as a function of the dye relative concentration. Using the correction equation (Beer–Lambert law), we calculated the εRec sol and used this value for the R0 calculation (Fig. 4B).
Supplementary Material
Acknowledgments
We thank Dr. H. Clark Hyde for invaluable discussion and Ludivine Frezza and Li Tang for technical assistance. This work was supported by American Heart Association Postdoctoral Fellowship 13POST14800031 (to T.K.), Australian Research Council Laureate Fellowship FL150100146 (to D.J.C.), Canadian Institutes of Health Research Grant MOP-10053 (to R.J.F.), NIH Grants GM030376 and U54GM087519 (to F.B.), and NIH Grant GM68044-07 (to A.M.C.).
Footnotes
Conflict of interest statement: Baron Chanda and Francisco Bezanilla were coauthors of a paper published in 2013 [Capes DL, et al. (2013) J Gen Physiol 142(2):101–112], but the collaboration leading to said paper ended in 2006.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1700453114/-/DCSupplemental.
References
- 1.Hodgkin AL, Huxley AF. A quantitative description of membrane current and its application to conduction and excitation in nerve. J Physiol. 1952;117(4):500–544. doi: 10.1113/jphysiol.1952.sp004764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chanda B, Bezanilla F. Tracking voltage-dependent conformational changes in skeletal muscle sodium channel during activation. J Gen Physiol. 2002;120(5):629–645. doi: 10.1085/jgp.20028679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Capes DL, Goldschen-Ohm MP, Arcisio-Miranda M, Bezanilla F, Chanda B. Domain IV voltage-sensor movement is both sufficient and rate limiting for fast inactivation in sodium channels. J Gen Physiol. 2013;142(2):101–112. doi: 10.1085/jgp.201310998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldschen-Ohm MP, Capes DL, Oelstrom KM, Chanda B. Multiple pore conformations driven by asynchronous movements of voltage sensors in a eukaryotic sodium channel. Nat Commun. 2013;4:1350. doi: 10.1038/ncomms2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lacroix JJ, Campos FV, Frezza L, Bezanilla F. Molecular bases for the asynchronous activation of sodium and potassium channels required for nerve impulse generation. Neuron. 2013;79(4):651–657. doi: 10.1016/j.neuron.2013.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCusker EC, et al. Structure of a bacterial voltage-gated sodium channel pore reveals mechanisms of opening and closing. Nat Commun. 2012;3:1102. doi: 10.1038/ncomms2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Payandeh J, Gamal El-Din TM, Scheuer T, Zheng N, Catterall WA. Crystal structure of a voltage-gated sodium channel in two potentially inactivated states. Nature. 2012;486(7401):135–139. doi: 10.1038/nature11077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Payandeh J, Scheuer T, Zheng N, Catterall WA. The crystal structure of a voltage-gated sodium channel. Nature. 2011;475(7356):353–358. doi: 10.1038/nature10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang X, et al. Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel. Nature. 2012;486(7401):130–134. doi: 10.1038/nature11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu J, et al. Structure of the voltage-gated calcium channel Cav1.1 at 3.6 Å resolution. Nature. 2016;537(7619):191–196. doi: 10.1038/nature19321. [DOI] [PubMed] [Google Scholar]
- 11.Wu J, et al. Structure of the voltage-gated calcium channel Cav1.1 complex. Science. 2015;350(6267):aad2395. doi: 10.1126/science.aad2395. [DOI] [PubMed] [Google Scholar]
- 12.Pantazis A, Savalli N, Sigg D, Neely A, Olcese R. Functional heterogeneity of the four voltage sensors of a human L-type calcium channel. Proc Natl Acad Sci USA. 2014;111(51):18381–18386. doi: 10.1073/pnas.1411127112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hui K, Lipkind G, Fozzard HA, French RJ. Electrostatic and steric contributions to block of the skeletal muscle sodium channel by mu-conotoxin. J Gen Physiol. 2002;119(1):45–54. doi: 10.1085/jgp.119.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.French RJ, et al. Interactions between a pore-blocking peptide and the voltage sensor of the sodium channel: An electrostatic approach to channel geometry. Neuron. 1996;16(2):407–413. doi: 10.1016/s0896-6273(00)80058-6. [DOI] [PubMed] [Google Scholar]
- 15.Posson DJ, Ge P, Miller C, Bezanilla F, Selvin PR. Small vertical movement of a K+ channel voltage sensor measured with luminescence energy transfer. Nature. 2005;436(7052):848–851. doi: 10.1038/nature03819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Posson DJ, Selvin PR. Extent of voltage sensor movement during gating of shaker K+ channels. Neuron. 2008;59(1):98–109. doi: 10.1016/j.neuron.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hyde HC, et al. Nano-positioning system for structural analysis of functional homomeric proteins in multiple conformations. Structure. 2012;20(10):1629–1640. doi: 10.1016/j.str.2012.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kubota T, Lacroix JJ, Bezanilla F, Correa AM. Probing α-3(10) transitions in a voltage-sensing S4 helix. Biophys J. 2014;107(5):1117–1128. doi: 10.1016/j.bpj.2014.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sandtner W, Bezanilla F, Correa AM. In vivo measurement of intramolecular distances using genetically encoded reporters. Biophys J. 2007;93(9):L45–L47. doi: 10.1529/biophysj.107.119073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Selvin PR. Principles and biophysical applications of lanthanide-based probes. Annu Rev Biophys Biomol Struct. 2002;31:275–302. doi: 10.1146/annurev.biophys.31.101101.140927. [DOI] [PubMed] [Google Scholar]
- 21.Korkosh VS, Zhorov BS, Tikhonov DB. Folding similarity of the outer pore region in prokaryotic and eukaryotic sodium channels revealed by docking of conotoxins GIIIA, PIIIA, and KIIIA in a NavAb-based model of Nav1.4. J Gen Physiol. 2014;144(3):231–244. doi: 10.1085/jgp.201411226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McArthur JR, et al. Interactions of key charged residues contributing to selective block of neuronal sodium channels by μ-conotoxin KIIIA. Mol Pharmacol. 2011;80(4):573–584. doi: 10.1124/mol.111.073460. [DOI] [PubMed] [Google Scholar]
- 23.Campos FV, Chanda B, Beirão PS, Bezanilla F. beta-Scorpion toxin modifies gating transitions in all four voltage sensors of the sodium channel. J Gen Physiol. 2007;130(3):257–268. doi: 10.1085/jgp.200609719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cestèle S, et al. Voltage sensor-trapping: Enhanced activation of sodium channels by beta-scorpion toxin bound to the S3-S4 loop in domain II. Neuron. 1998;21(4):919–931. doi: 10.1016/s0896-6273(00)80606-6. [DOI] [PubMed] [Google Scholar]
- 25.Zhang JZ, et al. Structure-function map of the receptor site for β-scorpion toxins in domain II of voltage-gated sodium channels. J Biol Chem. 2011;286(38):33641–33651. doi: 10.1074/jbc.M111.282509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang JZ, et al. Mapping the interaction site for a β-scorpion toxin in the pore module of domain III of voltage-gated Na(+) channels. J Biol Chem. 2012;287(36):30719–30728. doi: 10.1074/jbc.M112.370742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Villalba-Galea CA, Sandtner W, Starace DM, Bezanilla F. S4-based voltage sensors have three major conformations. Proc Natl Acad Sci USA. 2008;105(46):17600–17607. doi: 10.1073/pnas.0807387105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Possani LD, Becerril B, Delepierre M, Tytgat J. Scorpion toxins specific for Na+-channels. Eur J Biochem. 1999;264(2):287–300. doi: 10.1046/j.1432-1327.1999.00625.x. [DOI] [PubMed] [Google Scholar]
- 29.Dang B, Kubota T, Correa AM, Bezanilla F, Kent SB. Total chemical synthesis of biologically active fluorescent dye-labeled Ts1 toxin. Angew Chem Int Ed Engl. 2014;53(34):8970–8974. doi: 10.1002/anie.201404438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.French RJ, Yoshikami D, Sheets MF, Olivera BM. The tetrodotoxin receptor of voltage-gated sodium channels--perspectives from interactions with micro-conotoxins. Mar Drugs. 2010;8(7):2153–2161. doi: 10.3390/md8072153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cruz LJ, et al. mu-conotoxin GIIIA, a peptide ligand for muscle sodium channels: Chemical synthesis, radiolabeling, and receptor characterization. Biochemistry. 1989;28(8):3437–3442. doi: 10.1021/bi00434a043. [DOI] [PubMed] [Google Scholar]
- 32.Zhang MM, et al. Structure/function characterization of micro-conotoxin KIIIA, an analgesic, nearly irreversible blocker of mammalian neuronal sodium channels. J Biol Chem. 2007;282(42):30699–30706. doi: 10.1074/jbc.M704616200. [DOI] [PubMed] [Google Scholar]
- 33.Bezanilla F, Taylor RE, Fernández JM. Distribution and kinetics of membrane dielectric polarization. 1. Long-term inactivation of gating currents. J Gen Physiol. 1982;79(1):21–40. doi: 10.1085/jgp.79.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lacroix JJ, Labro AJ, Bezanilla F. Properties of deactivation gating currents in Shaker channels. Biophys J. 2011;100(5):L28–L30. doi: 10.1016/j.bpj.2011.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vargas E, Bezanilla F, Roux B. In search of a consensus model of the resting state of a voltage-sensing domain. Neuron. 2011;72(5):713–720. doi: 10.1016/j.neuron.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450(7168):376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 37.Capes DL, Arcisio-Miranda M, Jarecki BW, French RJ, Chanda B. Gating transitions in the selectivity filter region of a sodium channel are coupled to the domain IV voltage sensor. Proc Natl Acad Sci USA. 2012;109(7):2648–2653. doi: 10.1073/pnas.1115575109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vargas E, et al. An emerging consensus on voltage-dependent gating from computational modeling and molecular dynamics simulations. J Gen Physiol. 2012;140(6):587–594. doi: 10.1085/jgp.201210873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khoo KK, et al. Distinct disulfide isomers of μ-conotoxins KIIIA and KIIIB block voltage-gated sodium channels. Biochemistry. 2012;51(49):9826–9835. doi: 10.1021/bi301256s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lancelin JM, et al. Tertiary structure of conotoxin GIIIA in aqueous solution. Biochemistry. 1991;30(28):6908–6916. doi: 10.1021/bi00242a014. [DOI] [PubMed] [Google Scholar]
- 41.Stefani E, Bezanilla F. Cut-open oocyte voltage-clamp technique. Methods Enzymol. 1998;293:300–318. doi: 10.1016/s0076-6879(98)93020-8. [DOI] [PubMed] [Google Scholar]
- 42.Nitz M, et al. Structural origin of the high affinity of a chemically evolved lanthanide-binding peptide. Angew Chem Int Ed Engl. 2004;43(28):3682–3685. doi: 10.1002/anie.200460028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.