Significance
Multicellular organisms must have complex immune systems to detect and defeat pathogens. Plants rely on nucleotide binding site leucine rich repeat (NLR) intracellular receptors to detect pathogens. For hundreds of years, plant breeders have selected for disease-resistance traits derived from NLR genes. Despite the molecular cloning of the first NLRs more than 20 y ago, we still do not understand how these sensors function at a mechanistic level. Here, we identified a truncated NLR protein that activates cell death in response to a specific pathogen effector. Understanding how truncated NLRs function will provide a better mechanistic understanding of the plant immune system and an expanded toolkit with which to engineer disease resistance rationally in crops.
Keywords: plant immunity, NLR, Toll–interleukin-1 receptor homology domain, oligomerization, type III secretion
Abstract
Detection of pathogens by plants is mediated by intracellular nucleotide-binding site leucine-rich repeat (NLR) receptor proteins. NLR proteins are defined by their stereotypical multidomain structure: an N-terminal Toll–interleukin receptor (TIR) or coiled-coil (CC) domain, a central nucleotide-binding (NB) domain, and a C-terminal leucine-rich repeat (LRR). The plant innate immune system contains a limited NLR repertoire that functions to recognize all potential pathogens. We isolated Response to the bacterial type III effector protein HopBA1 (RBA1), a gene that encodes a TIR-only protein lacking all other canonical NLR domains. RBA1 is sufficient to trigger cell death in response to HopBA1. We generated a crystal structure for HopBA1 and found that it has similarity to a class of proteins that includes esterases, the heme-binding protein ChaN, and an uncharacterized domain of Pasteurella multocida toxin. Self-association, coimmunoprecipitation with HopBA1, and function of RBA1 require two previously identified TIR–TIR dimerization interfaces. Although previously described as distinct in other TIR proteins, in RBA1 neither of these interfaces is sufficient when the other is disrupted. These data suggest that oligomerization of RBA1 is required for function. Our identification of RBA1 demonstrates that “truncated” NLRs can function as pathogen sensors, expanding our understanding of both receptor architecture and the mechanism of activation in the plant immune system.
Plants lack an adaptive immune system and thus must rely on a limited innate immune system to detect and defeat potential pathogens. Exactly how plant innate immune receptors form a functional immune system is not well understood. There are two large classes of plant immune receptors (1). The first contains extracellular domains that detect microbial-associated molecular patterns (MAMPs) and activate MAMP-triggered immunity (MTI). These MTI receptors are sufficient to induce resistance to most microbes. However, evolutionarily adapted pathogens have evolved sophisticated systems to suppress MTI. Gram-negative bacterial pathogens such as Pseudomonas syringae use a molecular needle, the type III secretion system, to inject a set of bacterial virulence proteins (type III effectors; T3Es) directly into the host cytoplasm to suppress MTI and promote pathogen proliferation (2). In response, plants evolved a second class of immune receptors, the intracellular nucleotide-binding site leucine-rich repeat (NBS-LRR) receptors (NLRs) (3). NLRs directly or indirectly detect the presence of T3Es. The reference Arabidopsis genome (from the inbred accession Col-0) contains roughly 160 NLR proteins (4). These receptors are characterized by a multidomain architecture consisting in plants of either an N-terminal coiled-coil (CC) or Toll–interleukin (TIR) domain, a central AAA ATPase nucleotide-binding site domain (NBS), and C-terminal leucine-rich repeats (LRRs). Current models propose that NLR proteins switch from a closed “off” conformation to an open “on” conformation. The off conformation is thought to be maintained by intramolecular folding of C-terminal NBS and LRR domains to regulate signaling via the N-terminal TIR or CC domain negatively (5, 6). Upon pathogen detection, intramolecular negative regulation is released by unknown mechanisms, resulting in conformational changes in the NBS associated with nucleotide exchange. Homodimerization of the N-terminal domain is thought to activate downstream effector-triggered immunity (ETI). The mechanistic activation of NLRs and how they trigger downstream ETI remain obscure, although recent structural studies of animal analogs have begun to provide important details and support NLR oligomerization as a key feature of activation (7–9).
Both animal and plant TIR domains form homo- and heterodimers. In the case of animal transmembrane Toll-like receptors (TLRs), intracellular TIR homodimerization forms a nucleating site that recruits additional TIR domain-containing adaptors to transduce immune signals (10). In the case of plant intracellular NLRs, TIR-domain homodimers are often sufficient to signal for immune responses (11). Plant and animal TIR-domain crystal structures support the existence of a variety of dimer interfaces (12). There are two distinct interfaces from crystal structures of plant TIRs, defined by the homodimeric flax NLR L6 TIR domain and the homo- and heterodimeric Arabidopsis RPS4/RRS1 TIR domains (13–16). Overexpression of either the L6 or RPS4 TIR domain is sufficient to trigger cell death (13, 17). Mutation of the putative dimer interface in either L6 or RPS4 demonstrates that the corresponding dimer orientation is required for their function (13, 14). Although structural and genetic data are consistent with TIR domain dimerization, whether NLR proteins are limited to dimerization or higher-order oligomeric structures exist remains an open question. How these structurally distinct dimers (or any NLRs, for that matter) become activated by effectors and signal downstream to activate defense responses remains unknown.
The function of many bacterial T3Es is still not understood, but several either suppress important parts of the host immune system or alter host metabolism to promote pathogen success (18, 19). Because T3Es are evolved tools, understanding their targets and function should reveal unique insights into the host’s biology. We screened a large collection of P. syringae effectors to uncover effector-dependent phenotypes on a wide range of Arabidopsis genotypes. We discovered that the T3E HopBA1 is recognized by a TIR-only immune receptor that we call “RBA1” (for “response to HopBA1”) and that the ensuing immune response requires canonical TIR-NBS-LRR (TNL) signaling components. Additionally, we solved the crystal structure of HopBA1 and found striking resemblance to a bacterial heme scavenger protein. We characterized the HopBA1–RBA1 interaction and provide genetic data suggesting that a plant TIR domain can simultaneously engage multiple self-association interfaces. Our findings demonstrate immune function for a previously uncharacterized “truncated” NLR, thus expanding the known capabilities of plant intracellular receptors to recognize signals of microbial invasion.
Results
Screening Natural Variations of Arabidopsis Revealed a Polymorphic Response to HopBA1.
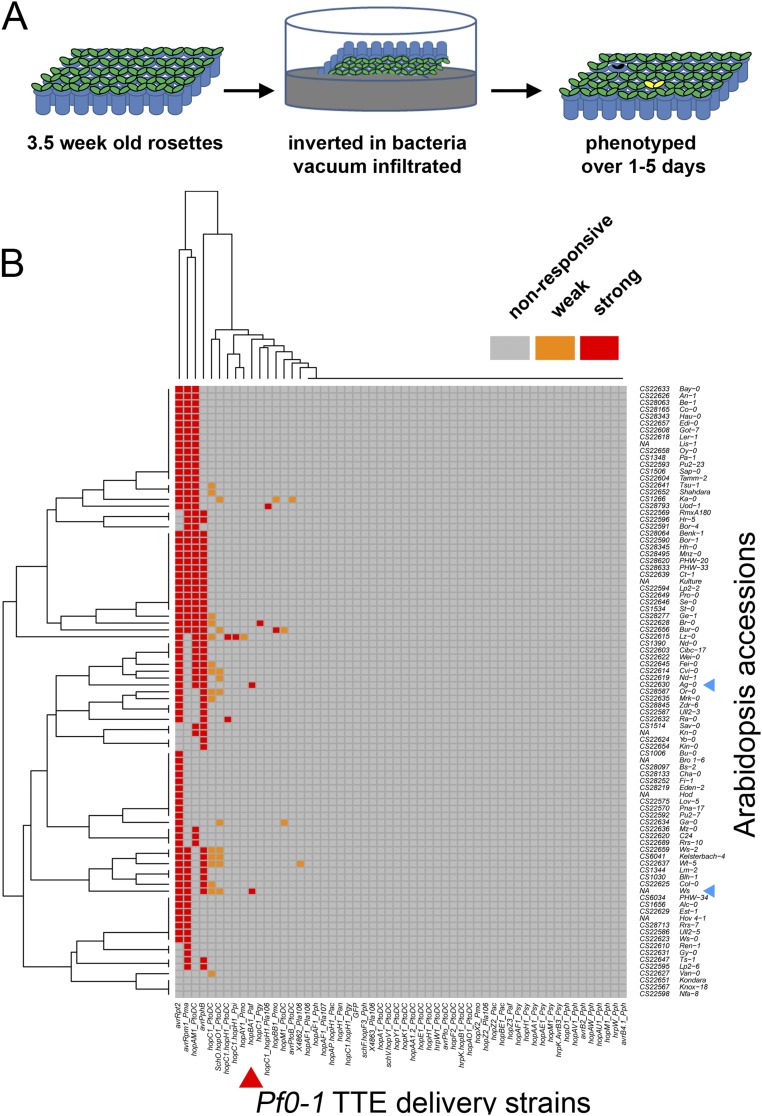
Previously, we identified a large collection of P. syringae T3Es (20, 21). We sought to identify host-polymorphic phenotypes by delivering each T3E individually to leaves from a panel of inbred Arabidopsis accessions. We delivered the T3Es from a modified Pseudomonas fluorescens strain that lacks endogenous effectors but expresses a heterologous P. syringae type III delivery system (Pf0-1 EtHAn, hereafter Pf0-1) (22). We screened 58 effectors across 89 inbred Arabidopsis accessions (Fig. S1). HopBA1, previously undescribed, triggered a cell-death phenotype on two accessions of Arabidopsis. The strongest and most consistent cell-death response to HopBA1 was in the accession Ag-0 (Fig. 1A).
Fig. S1.
Pf0-1-EtHAn screening of Arabidopsis accession collection (data related to Fig. 1). (A) Cartoon representation of screening of accession collection. Three- to four-week-old rosettes were grown in a grid (48 genotypes per grid), which was inverted, submerged, and vacuum-infiltrated with bacterial solution. Plants were phenotyped over the next 5 d for visible symptoms. (B) Heat map of responses of Arabidopsis accessions vacuum-infiltrated with Pf0-1 expressing individual effectors. Strong necrosis and weaker chlorosis symptoms are indicated by red and orange, respectively. HopBA1 is indicated by a red arrowhead. HopBA1-responsive accessions (Ag-0 and Ws) are indicated by blue arrowheads.
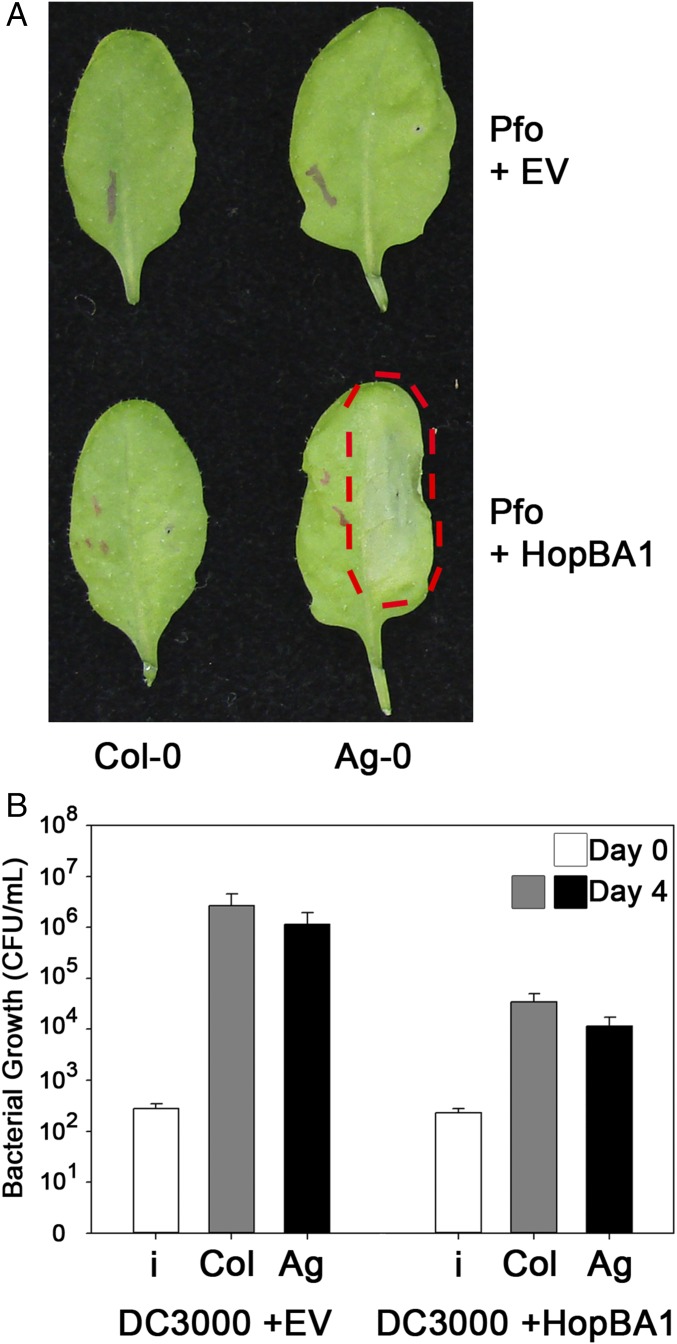
Fig. 1.
HopBA1 triggers host genotype-dependent cell death. (A) Delivery of HopBA1 via Pf0-1 triggers cell death in Arabidopsis accession Ag-0 but not Col-0. Bacteria were injected at OD 0.1 in 10 mM MgCl2 on one half of each leaf (across the midrib from the indicating ink marks). The red dashed line highlights the region of HopBA1-triggered necrosis in Ag-0. Images were taken 24 h postinoculation. (B) HopBA1 restricts the growth of Pto DC3000 on both Ag-0 and Col-0. Day 0 and Day 4, 0 and 4 d after inoculation; i, inoculum. Pto DC3000 was injected at OD 0.0002 (∼105 cfu) in 10 mM MgCl2. See also Fig. S1.
HopBA1 Is Recognized by the Host Immune System.
HopBA1 triggered cell death on Ag-0 leaves when delivered from the Pf0-1 system. To test if HopBA1 had an effect when delivered from a plant pathogen, we performed growth assays using the virulent strain P. syringae pv. tomato (Pto) DC3000 (virulent on Arabidopsis). Pto DC3000 expressing HopBA1 grew 10- to 100-fold less than Pto DC3000 containing an empty vector (EV) (Fig. 1B). Surprisingly, HopBA1 was able to restrict pathogen growth on both the Ag-0 and Col-0 accessions, despite the lack of a visible cell-death phenotype on leaves of the latter (Fig. 1B).
Phylogenetic Distribution of hopBA1.
HopBA1 was originally defined as a T3E from two P. syringae strains isolated from wheat (pv. japonica) or sugar beet (pv. aptata), respectively (20). To expand our collection of alleles, we searched public databases for variants of HopBA1 or HopBA1-similar proteins (Table S1; alignment available on request). In addition to P. syringae, we identified HopBA1-related proteins in multiple species of the plant pathogen genus Erwinia. We found more divergent HopBA1 family members in two strains of enterobacteria: in Ag1, a mosquito gut microbiome strain (23), and in Cedecea davisae, an emerging pathogen from immunocompromised humans (24). We cloned several of the plant pathogen HopBA1 variants and tested their ability to trigger cell death in Ag-0. All P. syringae alleles tested were recognized, but more divergent alleles remain ambiguous because they were not translocatable using a P. syringae type-III secretion system (Table S2).
Table S1.
Joint Genome Institute-Integrated Microbial Genomes accession numbers for HopBA1 and HopBA1-like proteins
| Accession no. | Organism |
| 2541308765 | Cedecea davisae 005, DSM 4568 |
| 2599262943 | Cedecea neteri M006 |
| 2520395437 | Enterobacter sp. Ag1 |
| 2579586825 | Erwinia mallotivora BT-MARDI |
| 646983737 | Erwinia pyrifoliae DSM 12163 |
| 646577976 | Erwinia pyrifoliae Ep1/96 |
| 650415283 | Erwinia sp. Ejp617 |
| 2588704606 | Ewingella americana C.024, ATCC 33852 |
| 2599328671 | Pseudomonas sp. NFACC10-1 |
| 2612253752 | Pseudomonas syringae Alf3 |
| 2543172195 | Pseudomonas syringae BRIP34876 |
| 2543180053 | Pseudomonas syringae BRIP34881 |
| 2543185915 | Pseudomonas syringae BRIP39023 |
| 2543187255 | Pseudomonas syringae BRIP39023 |
| 2506892820 | Pseudomonas syringae CC1458 |
| 2506958508 | Pseudomonas syringae CC1543 |
| 2525063307 | Pseudomonas syringae CC1629 |
| 2506880073 | Pseudomonas syringae CC440 |
| 2506887414 | Pseudomonas syringae CC457 |
| 2547779750 | Pseudomonas syringae FF5 |
| 2547780841 | Pseudomonas syringae FF5 |
| 2505603619 | Pseudomonas syringae PavISPaVe013 (Psy117) |
| 2508774915 | Pseudomonas syringae pv. aptata DSM 50252 |
| 2508774925 | Pseudomonas syringae pv. aptata DSM 50252 |
| 2508776813 | Pseudomonas syringae pv. aptata DSM 50252 |
| 2574722659 | Pseudomonas syringae pv. atrofaciens DSM 50255 |
| 2586892371 | Pseudomonas syringae pv. atrofaciens LMG 5095 |
| 2536469993 | Pseudomonas syringae pv. avellanae ISPaVe013 |
| 2507339069 | Pseudomonas syringae pv. japonica M301072PT |
| 2564019397 | Pseudomonas syringae pv. panici LMG 2367 |
| 2523800455 | Pseudomonas syringae pv. syringae B64 |
| 2617427584 | Pseudomonas syringae pv. syringae HS191 |
| 2525310816 | Pseudomonas syringae pv. syringae SM |
Table S2.
HopBA1 mutant and HopBA1 allele phenotypes
| Genotype | Cell death | Translocation |
| HopBA1 | Y | Y |
| H56A* | N | Y |
| H56F* | N | Y |
| E83A | N | N |
| F84A | N | N |
| W112A* | N | Y |
| R114A | Y | Not tested |
| H115A | Y | Not tested |
| H115F | Y | Not tested |
| K143A | Y | Not tested |
| M114A | Y | Not tested |
| I146A | Y | Not tested |
| D147A | Y | Not tested |
| Q148A | Y | Not tested |
| K150A | Y | Not tested |
| K150M | Y | Not tested |
| K155A | Y | Not tested |
| K155M | Y | Not tested |
| Y158A* | N | Y |
| Y158F* | N | Y |
| M159A | Y | Not tested |
| R162A* | N | Y |
| W169A | N | N |
| K184R | Y | Not tested |
| H193A | N | N |
| S195A | Y | Not tested |
| N196A | Y | Not tested |
| H229A | Y | Not tested |
| HopBA1_Japonica | Y | Y (20) |
| HopBA1_FF5 | Y | Not tested |
| HopBA1_B76 | Y | Not tested |
| HopB1_DC3000 | N | Y (21) |
Loss-of-function mutants resulting in cell death but retaining translocation into the plant.
Genetic Analysis in Arabidopsis Defines a TIR-Containing, Truncated NLR Protein That Encodes RBA1.
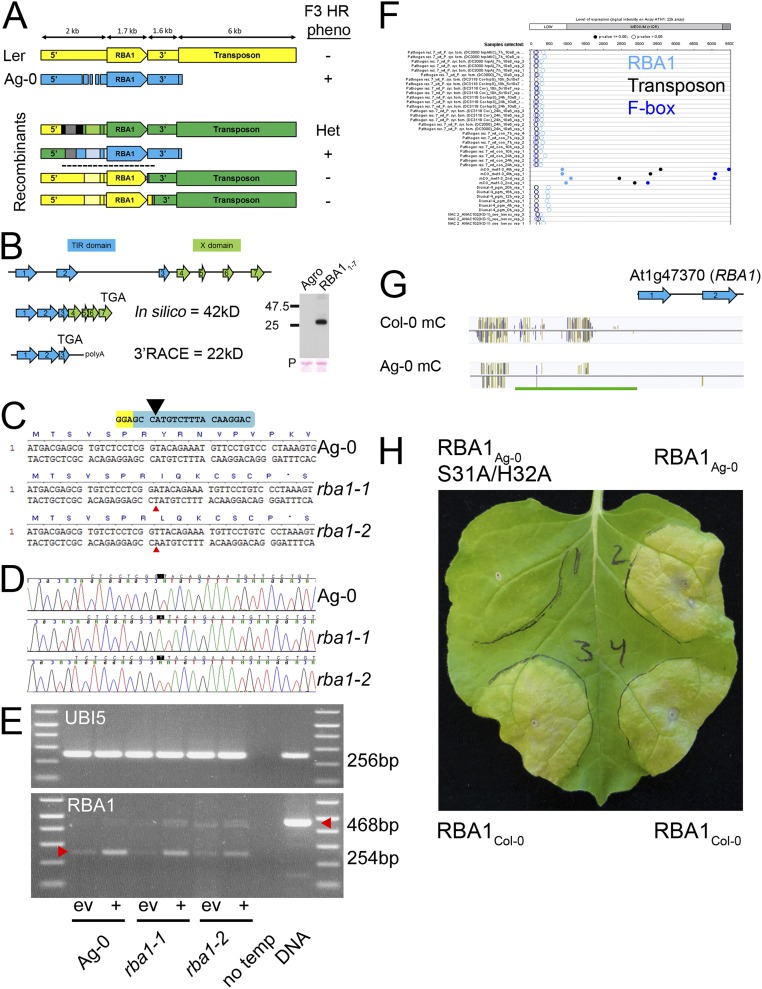
To determine what genes are required for Arabidopsis to recognize HopBA1, we positionally cloned RBA1 using interaccession variation in the cell-death response as the mapping phenotype. In an F2 population of ∼3,100 Ag-0 × Ler-0 individuals, cell death segregated as a single, dominant Mendelian trait to an interval containing a single gene: At1g47370 (Fig. S2A). Sequencing of Ag-0 revealed some structural variations relative to the reference genome Col-0 (Fig. S2A) but no novel genes, and it was consistent with subsequent public resequencing data (25). At1g47370 was predicted to encode a novel TIR-X protein in the reference Col-0 genome containing a NLR-like TIR domain followed by an unknown “X domain” lacking homology to known proteins (Fig. S2B). Our molecular reannotation of At1g47370 in Ag-0 and Col-0 indicated that it encodes a TIR-only protein, lacking any “X domain” or NB-LRR domains (Fig. 2A and Fig. S2B). Homology modeling indicates that RBA1 is structurally most similar to the TIR domain of the immune receptor RPS4, a full-length TIR-NBS-LRR protein (Phyre2 100% confidence, 41% sequence identity) (Fig. 2B).
Fig. S2.
Mapping and reannotation of RBA1; CAS9 mutagenesis; and public methylation data (data related to Fig. 2). (A) Summary of synteny and fine mapping of the RBA1 locus (At1g47370). The RBA1 locus is structurally identical in Ler-0 (yellow) and the reference genome Col-0. The RBA1 locus in Ag-0 (blue) is differentiated by structural variation in the promoter (deletion relative to Ler-0/Col-0) and the absence of a downstream 6-kb transposon. The four Ag-0/Ler-0 F2 recombinants used to define the RBA1 mapping interval (black dashed line) are color-coded according to genotype: Ler-0, yellow; Ag-0, blue; heterozygous, green. Recombination points after SSLP fine mapping were determined by Sanger sequencing limited by SNP. Lighter shades indicate that the genotype was assumed based on the flanking sequence. Gray regions are identical in Ag-0 and Ler-0; black regions were not determined. Phenotypes for F3 progeny of the F2 recombinants are listed on the right: +, all progeny displayed Ag-0–like cell death; −, a Ler-0–like null response; Het, a 3:1 ratio of cell death to null response. The upper two recombinants place the left flank slightly 5′ of the polymorphic promoter region. The lower two recombinants place the right flank near the stop codon of the TIR-only form of RBA1 (exon 3, see B below) The RBA1 mapping interval is indicated by a dashed line. (B) In silico annotation of RBA1 in Col-0 predicts a seven-exon 42-kDa TIR-X protein; 3′ RACE analysis predicts a shorter, three-exon 22-kDa TIR-only protein (“TGA” indicates location of the stop codon). Consistent with this prediction, transient expression of the seven-exon Ag-0 genomic construct in N. benthamiana produces a TIR-only sized protein (22 kDa + 7 kDa HA epitope tag). (C) DNA and corresponding predicted truncated mutant protein sequences of rba1-1 and rba1-2 mutants. Sequences begin at the initiating methionine of RBA1. Guide RNA is shown in blue; protospacer adjacent motif is shown in yellow. The black caret shows the expected CAS9 cleavage site. Red carets indicate the position of single base-pair insertions. (D) Electropherograms of CAS9-generated Ag-0 rba1 mutants. The positions of single base-pair insertion alleles are highlighted. (E) RT-PCR indicates that the RBA1 transcript is present in rba1 mutants following induction with Pf0-1 expressing hopBA1 (+) relative to empty vector (ev). Spliced RBA1 transcripts should produce a 254-bp product, whereas genomic DNA should produce a 468-bp product. Equal amplification of UBQ5 indicates roughly equal template concentrations. (F) As is consistent with CG methylation contributing to silencing in this region, RBA1, the 3′ transposon, and a further 3′ F-box protein are expressed only in the met1-3 mutant (public data from Genevestigator browser). (G) Browser shot of C-methylation at the RBA1 locus in Col-0 and Ag-0 accessions. The green bar indicates an 830-bp region immediately upstream of RBA1 that has reduced cytosine methylation in Ag-0 relative to Col-0. Data are from the 1001 Project; experiment numbers are Col 6909_pub, and Ag-0 6897_pub. (H) The RBA1 Col-0 allele is autoactive when transiently expressed in N. benthamiana. Injections were RBA1Ag-0 (Upper Right), an inactive S31A/H32A mutant of RBA1Ag-0 (Upper Left), and RBA1Col-0 (Bottom Left and Bottom Right). Constructs were 35S YFP:RBA1 TIR-only genomic clones (exons 1–3) delivered by A. tumefaciens strain GV3101 at OD 0.8.
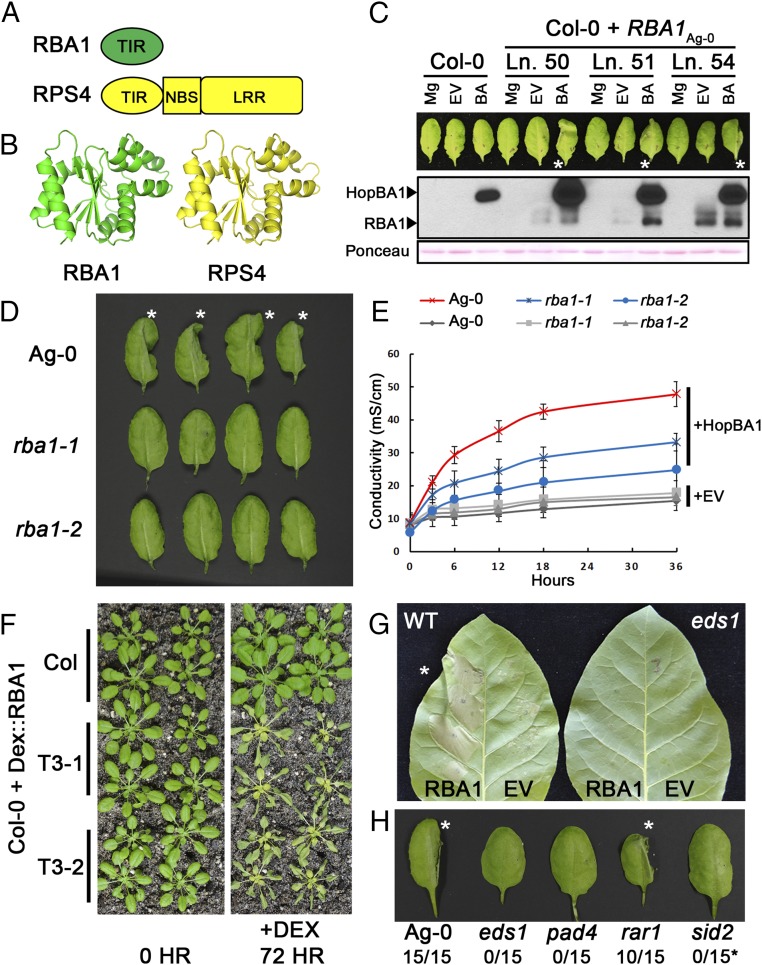
Fig. 2.
RBA1 encodes a TIR-only truncated NLR-like protein. (A) Full-length RBA1 (At1g47370) is equivalent to the TIR domain of TIR-NBS-LRR immune receptors. (B) Homology modeling of RBA1 indicates that it is most similar to the RPS4 TIR domain crystal structure (PDB ID code 4C6R). (C) A native promoter-driven, N-terminal 1×HA-tagged genomic fragment of RBA1 from Ag-0 complements Col-0’s lack of cell-death response to Pf0-1 expressing HopBA1. Pf0-1 was injected at OD 0.1 in 10 mM MgCl2, and leaves were imaged after 24 h. Asterisks indicate leaves with cell death in the injected half. Native promoter HA-RBA1 Col-0 transgenics accumulate RBA1 in response to Pf0-1 expressing HopBA1. BA, Pf0-1 expressing HopBA1; EV, Pf0-1 empty vector control; Ln. #, genetically independent transgenic lines; Mg, MgCl2 control. Total protein loading was assessed by Ponceau S staining of the membrane. (D) Two independent CAS9-generated Ag-0 rba1 mutants showed reduced cell death 24 h after inoculation with Pf0-1 expressing HopBA1. Asterisks indicate WT leaves with cell death in the injected half. (E) Quantitative measurement of decreased conductivity in response to HopBA1 in rba1 mutants. (F) Dexamethasone-inducible RBA1 triggers cell death in Col-0. (G) Agrobacterium-delivered RBA1 ectopic cell death is suppressed in EDS1-silenced N. tabacum plants. The asterisk indicates a WT leaf with cell death in the RBA1-expressing half. (H) RBA1-HopBA1 cell death in Arabidopsis is dependent on known immune-related genes. Plants are representative genotyped double-mutant F3 progeny of F2 plants previously fixed for RBA1Ag-0 and heterozygous for an immune mutant in the Col-0 background. Asterisks indicate leaves with cell death in the injected half. Numbers indicate the number of collapsed leaves relative to the total number of leaves. The sid2 mutant displayed suppressed cell death but had a weak chlorosis phenotype in response to HopBA1. See also Fig. S2.
The TIR-Only Ag-0 RBA1 Allele Complements Col-0 for Cell Death in Response to HopBA1.
We generated a complementation construct that contains 725 bp of upstream promoter region, a single HA epitope tag, and 1,733 bp of genomic Ag-0 DNA corresponding to the TIR-only form of RBA1. When transformed into Col-0, this construct conferred a cell-death response in response to HopBA1 delivered from Pf0-1, validating our reannotation of the locus (Fig. 2C and SI Materials and Methods). In addition, we generated two rba1 mutants with nonsense alleles in the first exon via CAS9 mutagenesis in the Ag-0 genome (Fig. S2 C–E). Both rba1 mutants displayed a decreased cell-death response to HopBA1 at 24 h postinfection, as assessed qualitatively (by leaf collapse) and quantitatively (measured via ion leakage) (Fig. 2 D and E). Collectively, these results indicated that RBA1 is a TIR-only NLR-like protein, which, despite the lack of canonical NBS and LRR domains, functions to activate cell death in response to a pathogen virulence effector. Public expression data (Genevestigator) of the Col-0 accession indicated that RBA1Col-0 is likely a pseudogene, potentially controlled via cytosine methylation, because it is expressed in the met1-3 mutant that is deficient in transcription-repressing CG-methylation (Fig. S2F). Consistent with this interpretation, public methylation data indicate that the ∼830 bp section immediately upstream of RBA1 is methylated to a lower extent in Ag-0 than in Col-0 (Fig. S2G) (26). This region contains the 725-bp promoter used to drive our native promoter RBA1 construct and thus is sufficient for function. We speculate that the distinct Col-0/Ag-0 RBA1 phenotypic response to HopBA1 is an expression polymorphism. Consistent with this interpretation, we were able to amplify RBA1 cDNA from Ag-0 but not from Col-0. Additionally, as is consistent with expression being the causal difference between the alleles, the protein sequence of RBA1Col-0 is highly conserved relative to RBA1Ag-0 (98.4% identical) and was functional when expressed ectopically with the 35S promoter in Nicotiana benthamiana (Fig. S2H).
RBA1 Is Autoactive and Enhanced Disease Susceptibility 1-Dependent When Overexpressed.
The TIR domains of several typical TIR-NLR immune receptors (L6, RPS4, RPP1) are autoactive, triggering cell death in the absence of pathogens when ectopically overexpressed as truncated TIRs (13, 17, 27). Ectopic dexamethasone- or 35S promoter-driven overexpression of RBA1 in Arabidopsis, N. benthamiana, and Nicotiana tabacum resulted in host cell death in the absence of HopBA1, suggesting that RBA1 functions similarly to TIR domain truncations (Figs. 2 F and G and 3 C and D). The ectopic cell-death phenotypes driven by overexpression of TIR-NLR and/or derived single-domain TIRs typically are dependent on the Enhanced disease susceptibility1 (EDS1) signaling module (17, 28). We found that RBA1-mediated cell death was also EDS1 dependent in the context of either transient overexpression (in eds1-silenced N. tabacum) or when expressed from its endogenous locus in an Arabidopsis eds1 mutant background (Fig. 2 G and H). RBA1-mediated cell death also was dependent on PAD4, which is part of the EDS1 signaling complex (29), and was partially dependent on SID2, but was independent of RAR1, a cochaperone protein required for some, but not all, NLR-based disease-resistance responses (Fig. 2H) (30).
Fig. 3.
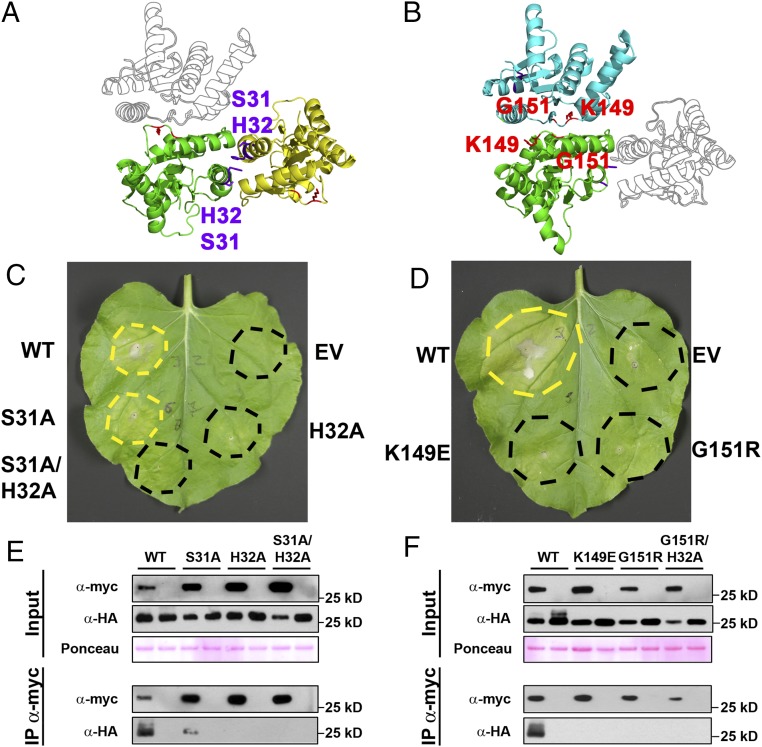
RBA1 requires two distinct self-association interfaces for function. (A) RBA1 homology model in the RPS4 (PDB ID code 4C6R) homodimer interface orientation (L6-orientation monomer shown in outline). Purple and red residues are required for the proposed RPS4-type and L6-type RBA1 dimers, respectively (see below). (B) RBA1 homology model in the L6 (PDB ID code 3OZI) homodimer interface orientation (RPS4-orientation monomer shown in outline). (C and D) Both RPS4-type and L6-type dimer mutants in RBA1 show loss of function for ectopic 35S promoter-driven cell death in N. benthamiana. (E and F) Both RPS4-type and L6-type dimer interfaces in RBA1 are required for RBA1–RBA1 coimmunoprecipitation in N. benthamiana. RBA1 constructs are 35S promoter-driven 3xHA-RBA1 and 4xmyc-RBA1. Lysates were immunoprecipitated with anti-myc beads and then were immunoblotted for both anti-myc and anti-HA to assess input, immunoprecipitation, and coimmunoprecipitation. Total protein loading was assessed by Ponceau S staining of the membrane. See also Figs. S3–S5.
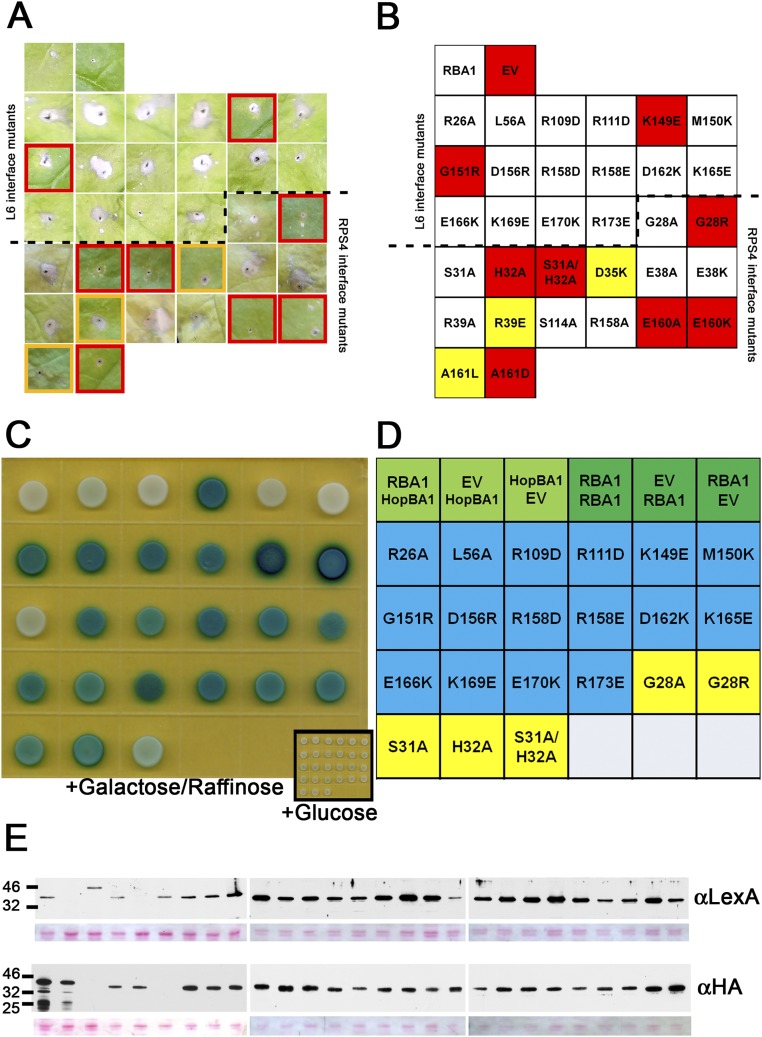
Predicted RBA1 Dimer Interface Mutants Affect Both Function and Homotypic Interaction.
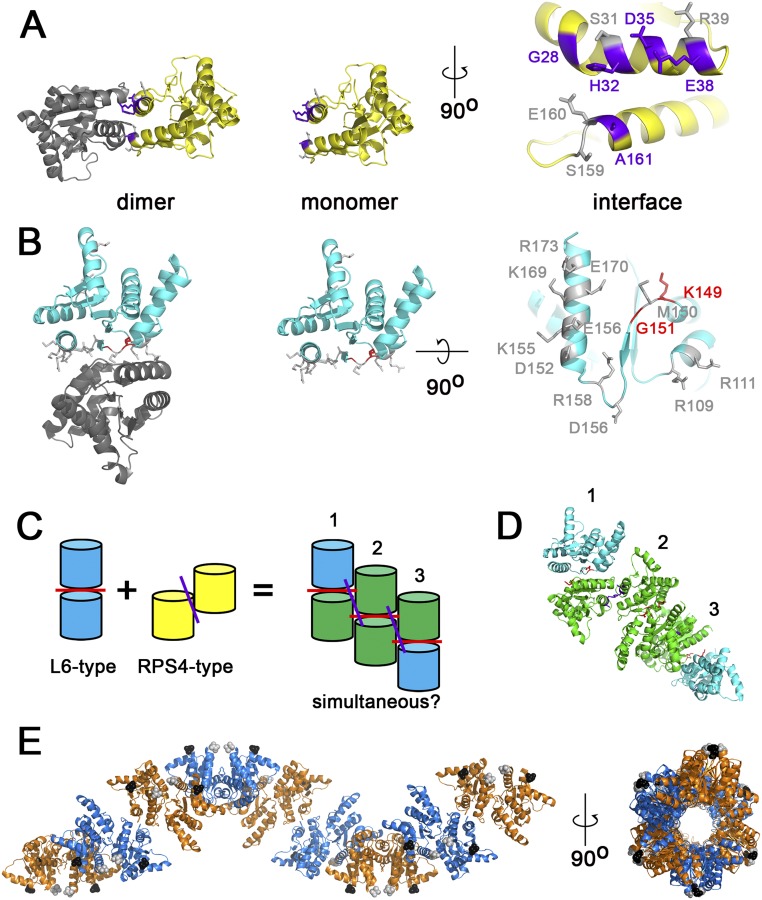
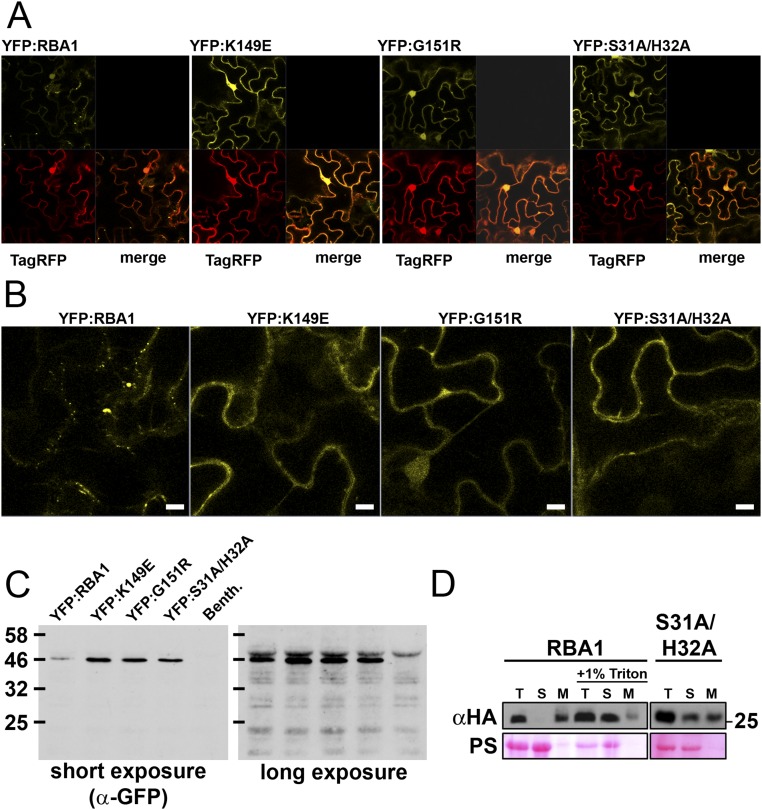
To date, we have been unable to purify RBA1 protein and determine its structure experimentally. To put RBA1 into the context of existing TIR structures, we generated homology models with published RPS4 and L6 TIR domain structures. Modeling of RBA1 generated high-confidence tertiary structure predictions consistent with both RPS4 and L6 TIR dimer interface types (Fig. S3). Similar to the RPS4 and L6 TIR domains, RBA1 can self-associate, as measured by both yeast two-hybrid (Y2H) and coimmunoprecipitation assays of differentially epitope-tagged RBA1 proteins (Fig. 3 and Fig. S4). To determine which interface was used for RBA1 self-association, we generated RBA1 mutations predicted to be at or near either the RPS4-type or the L6-type dimer interface (Table 1 and Fig. S3). Interestingly, mutation of particular RBA1 residues at either interface blocked RBA1-dependent cell death in transient overexpression assays (Fig. 3, Table 1, and Fig. S3). Mutation of conserved residues required for the function of RPS4/RRS1 (H34/H26) and L6 (G201) also resulted in loss of function for RBA1 (H32A and G151R). RBA1 S31A had a weaker effect than H32A, as is consistent with equivalent mutants’ effects on RRS1 function (14). The nonconserved RBA1 L6-type mutant K149E (L199 in L6) was also required for autoactivity. As is consistent with the loss of autoactivity, RBA1 dimer interface mutants also lost self-association as measured by coimmunoprecipitation (Fig. 3). Some, but not all, RBA1 mutants that lost the ability to be coimmunoprecipitated also lost Y2H interactions (Table 1 and Fig. S4). We also observed changes in localization in the RBA1 mutants. Functional YFP-RBA1 fusion proteins formed aggregate-like cyto-nucleoplasmic puncta (Fig. S5). However, nonfunctional YFP-RBA1 dimer interface mutants became delocalized and indistinguishable from coexpressed TagRFP (Fig. S5). YFP-RBA1 loss-of-function mutants accumulated higher levels of protein than WT YFP-RBA1 (Fig. S5C). Microsomal fractionation of transiently overexpressed RBA1 in N. benthamiana indicated that RBA1 was associated with a detergent-sensitive membrane fraction, whereas much of the RBA1 interface S31A H32A double mutant accumulated in the soluble fraction (Fig. S5D). This result was consistent with the differential localization of the fluorescently tagged RBA dimer interface mutants.
Fig. S3.
PyMOL modeling of RBA1 dimer mutants; hypothetical higher-order TIR complexes are sterically feasible (data related to Fig. 3). (A) L6-type RBA1 mutations. The RBA1 residues shown in purple lose autoactivity function when mutated. The residues shown in gray were dispensable for autoactivity. (B) RPS4-type RBA1 mutations. The RBA1 residues shown in red lose autoactivity function when mutated. The residues shown in gray were dispensable for autoactivity. (C) The TIR dimers for L6 and RPS4 interact on distinct surfaces. (D and E) Hypothetical models of RBA1 interacting simultaneously using both L6 and RPS4 interfaces. D shows a three-dimer chain color-coded as in C. In E, nine dimers form a helical structure around a central channel. Monomers from L6-type dimers share colors (either orange or marine); L6-type dimers are connected across colors by RPS4-type interfaces. The N and C termini of each monomer are indicated by black and gray spheres, respectively.
Fig. S4.
Phenotypes of RBA1 mutants (data related to Figs. 3 and 5). Mutations were made in residues predicted to affect either L6- or RPS4-type dimer interfaces. (A) Summary of transient N. benthamiana-Agrobacterium assays. Red outlines indicate nonresponsive mutants with phenotypes similar to the EV control. Yellow outlines indicate weak, partial loss-of-function phenotypes. (B) Identification of mutants producing the leaf phenotypes in A. Red boxes indicate strong loss-of-function phenotypes; orange boxes have partial loss-of-function phenotypes. Images were taken 5–7 d postinoculation. (C) Y2H assay. (D) Identification of mutants in the Y2H assay in C. (E) Western blot of Y2H.
Table 1.
Summary ofRBA1 mutant phenotypes
| Genotype | Putative interface | Cell death | Y2H self | Co-IP self | Co-IP HopBA1 |
| WT | Na | + | + | + | + |
| R26A | Na | + | + | Nt | Nt |
| G28A | RPS4 | + | + | Nt | Nt |
| G28R | RPS4 | − | + | Nt | Nt |
| S31A | RPS4 | + | + | + | + |
| H32A | RPS4 | − | + | − | − |
| S31A/H32A | RPS4 | − | − | − | Nt |
| D35K | RPS4 | ± | Nt | Nt | Nt |
| E38A | RPS4 | + | Nt | Nt | Nt |
| E38K | RPS4 | + | Nt | Nt | Nt |
| R39A | RPS4 | + | Nt | Nt | Nt |
| R39E | RPS4 | ± | Nt | Nt | Nt |
| L56A | Na | + | + | Nt | Nt |
| R109D | L6 | + | + | Nt | Nt |
| R111D | L6 | + | + | Nt | Nt |
| S114A | L6 | + | Nt | Nt | Nt |
| K149E | L6 | − | + | − | − |
| M150K | L6 | + | + | Nt | Nt |
| G151R | L6 | − | − | − | − |
| D156R | L6 | + | + | Nt | Nt |
| R158A | L6 | + | Nt | Nt | Nt |
| R158D | L6 | + | + | Nt | Nt |
| R158E | L6 | + | + | Nt | Nt |
| E160A | RPS4 | − | Nt | Nt | Nt |
| E160K | RPS4 | − | Nt | Nt | Nt |
| A161D | RPS4 | − | Nt | Nt | Nt |
| A161L | RPS4 | +/− | Nt | Nt | Nt |
| D162K | L6 | + | + | Nt | Nt |
| K165E | L6 | + | + | Nt | Nt |
| E166K | L6 | + | + | Nt | Nt |
| K169E | L6 | + | + | Nt | Nt |
| E170K | L6 | + | + | Nt | Nt |
| R173E | L6 | + | + | Nt | Nt |
“Putative interface” indicates that the residue lies in a hypothetical RPS4- or L6-like dimer interface. Data for cell death and Y2H self-association are from Fig. S4. Data for co-IP self are from Fig. 3. Data for co-IP HopBA1 are from Fig. 5. Co-ip, coimmunoprecipitation; Na, mutations not in predicted RPS4- or L6-type interfaces; Nt, not tested.
Fig. S5.
Localization of RBA1 (data related to Figs. 3 and 5). (A) Confocal imaging (magnification, 40×) of N. benthamiana-Agrobacteria transient expression of 35S YFP:RBA variants (yellow) and 35S TagRFP (red) in epidermal cells 24 h postinoculation (before visible symptoms). (B) Enlarged YFP images from the lower right quadrant of YFP images from A. (Scale bars, 40 uM.) (C) Western blot (anti-GFP) demonstrating that localization images reflect full-length fusion proteins. The short exposure demonstrates that mutants accumulate at higher levels. The long exposure shows no detectable free GFP, indicating that nuclear localization is reflective of full-length protein. Nonspecific plant cross-reacting bands indicate roughly equal loading among samples. (D) Microsomal fractionation of transiently overexpressed HA-RBA1 in N. benthamiana. M, microsomal pellet; S, soluble lysate; T, total lysate after centrifugation. Lanes are equally loaded with cell equivalents. PS, Ponceau stain to indicate protein loading of the membrane.
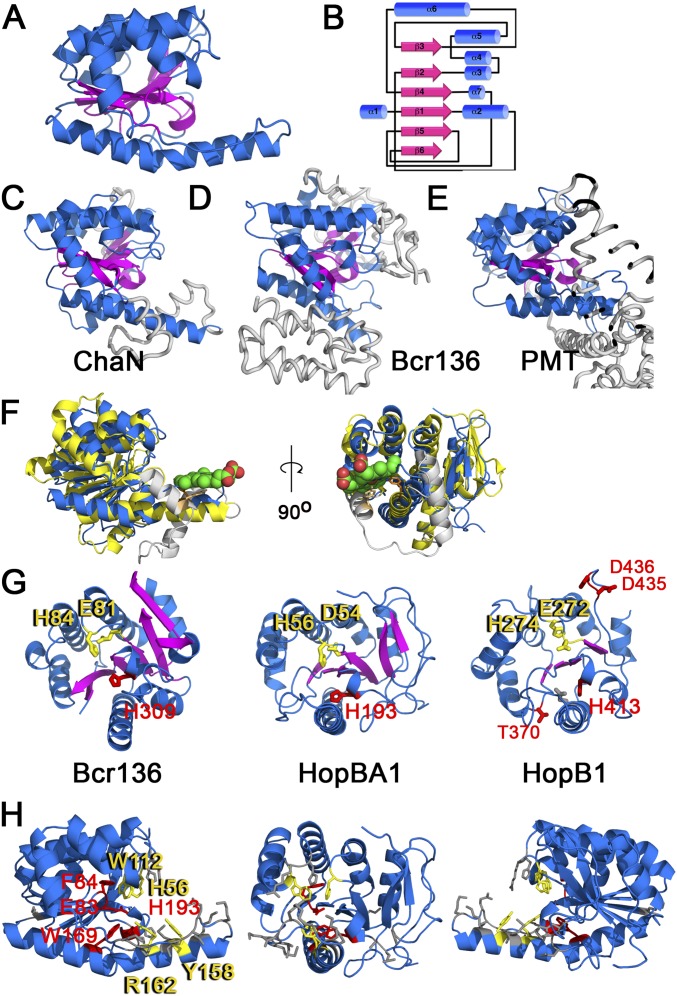
HopBA1 Is Structurally Similar to ChaN, a Heme-Binding Protein, and an Uncharacterized Domain of PMT, a Type III-Delivered Pasteurella multocida Toxin.
Because the primary sequence of HopBA1 was uninformative, and T3Es are prokaryotic proteins that often evolve to mimic eukaryotic folds, we determined the 3D structure of HopBA1 to a resolution of 2.0 Å (SI Materials and Methods, Fig. S6 and Table S3). We used the HopBA1 structure to search the Protein Data Bank for structural homologs using the DALI server. This search indicated that HopBA1 is structurally related to the erythromycin esterase (EreA)-like/ChaN superfamily of proteins, sharing a similar α/β fold with an all–β-strand core as defined by SCOP (31). This superfamily (Fig. S6) consisted of ChaN, a heme-binding protein from Campylobacter jejuni (32), the EreA-like esterase Bcr136 from Bacillus cereus (33), and a portion of the C2 domain of Pasteurella multocida dermonecrotic toxin (PMT) (34). Modeling of HopBA1-like P. syringae effector proteins revealed that the T3E HopB1 (Fig. S6G) is also similar to ChaN/Bcr136/PMT-C2. Interestingly, excluding its disordered N-terminal type three secretion signal, HopBA1 represents a minimal fold present in all family members (Fig. S6 C–E).
Fig. S6.
HopBA1 structure and similarity to known proteins (data related to Fig. 4). (A) The structure of HopBA1 (PDB ID code 5T09). (B) Schematic of HopBA1 structure. (C–E) Structures of proteins similar to HopBA1: ChaN (C), Bcr136 (D), and PMT (E). Sequences not found in HopBA1 are shown in gray. (F) HopBA1 (blue) is structurally similar to ChaN, a heme-binding protein (yellow). Heme is shown in green. The position of the ChaN heme-binding residue Y148 (light orange) is shown relative to HopBA1 Y158 (dark orange). ChaN helices not present in HopBA1 are shown in gray. (G) Comparison of Bcr136, HopBA1, and the homology model of HopB1 (on PMT). Bcr136 H84 and E81 comprise the proposed esterase catalytic dyad. HopBA1 (H56/D54) and HopB1 (H274/E272) contain similar residues (in yellow). Proposed HopB1 catalytic residues are shown in red. The position of HopB1 H413 is similar to that of Bcr136 H309 and HopBA1 H193. (H) Residues mutated in HopBA1, color-coded according to functional data (Table S3): gray, no effect; red, loss of translocation; yellow, loss of function in planta. Both HopBA1 H193 and H56 described above in G lose the ability to trigger cell death, although H193 is not translocated into the plant.
Table S3.
HopBA1 crystallography statistics
| Crystal | HopBA1 | HopBA1 SeMet |
| Data collection | ||
| Space group | P3121 | P3121 |
| Cell dimensions | ||
| a, b, c, Å | 64.9, 64.9, 96.4 | 64.78, 64.78, 95.01 |
| α, β, γ, degrees | 90.00, 90.00, 120.00 | 90.0, 90.0, 120.0 |
| Wavelength, Å | 0.9794 | 0.97227 |
| Resolution, Å | 32.13–2.01 (2.03–2.01) | 28.05–2.34 (2.36–2.34) |
| Rmerge | 5.4 (100.0) | 6.0 (79.9) |
| I/σI | 49.9 (1.9) | 24.5 (1.6) |
| Unique reflections | 16,048 (423) | 18,759 (464) |
| Completeness, % | 99.9 (100.00) | 99.8 (99.1) |
| Redundancy | 9.0 (9.1) | 4.8 (4.3) |
| Refinement | ||
| Resolution, Å | 32.13–2.01 (2.14–2.01) | |
| No. reflections | 16,014/800 (2,474/122) | |
| Rwork/Rfree | 19.94/24.03 (26.71/30.56) | |
| No. of atoms (nonhydrogen) | ||
| All | 1676 | |
| Protein | 1635 | |
| Ions | 3Cl−, 1K+ | |
| Water | 37 | |
| B factors, Å2 | ||
| All | 62.18 | |
| Protein | 62.38 | |
| Ions | 73.6 | |
| Water | 52.32 | |
| Rmsd | ||
| Bond lengths, Å | 0.011 | |
| Bond angles, degrees | 1.19 | |
| Ramachandran | ||
| Favored, % | 99.1 | |
| Generally allowed, % | 0.9 | |
| Disallowed, % | 0 | |
| Missing residues | 1–22 | |
Numbers in parentheses are statistics for the highest-resolution shell.
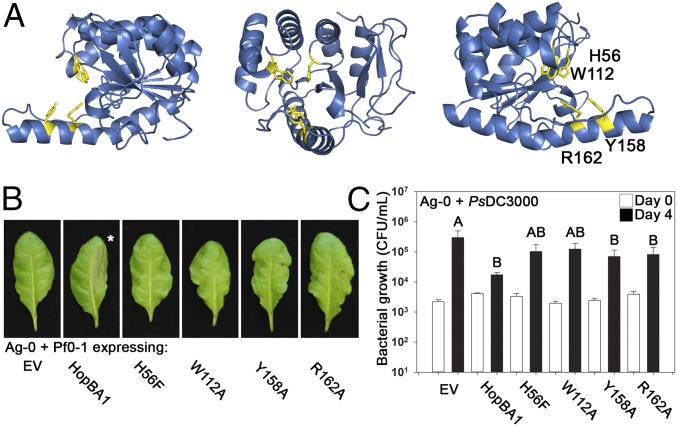
Structurally Informed Mutagenesis of HopBA1 Identifies Residues Required for RBA1-Mediated Host Cell Death.
To test the relevance of the HopBA1 structural resemblance to Bcr136 and ChaN, we identified residues in HopBA1 that were in positions similar to those of the putative catalytic residues in Bcr136 and the heme-binding residues from ChaN (Fig. S6 F–H). We mutagenized these residues to alanine or to more conservative alternate residues and assayed the mutants for the ability to trigger cell death in Ag-0 after delivery from Pf0-1. Mutation of putative Bcr136 catalytic residues and several sites near the ChaN heme-binding–related surface resulted in the loss of the ability to trigger host cell death (Fig. 4 A and B and Table S2). The four HopBA1 loss-of-function mutants that translocated into the host plant cell (H56F, W112A, Y158A, and R162A) (Table S2) were of particular interest, because their defect occurs in planta. As compared with the strong loss of cell-death phenotype, these four mutants had a weaker effect on pathogen growth restriction (Fig. 4C).
Fig. 4.
Mutagenesis of HopBA1 reveals residues required for the Ag-0 RBA1 cell-death response. (A) Mutagenesis of HopBA1 revealed four residues, shown in yellow, required for function in planta. (B) HopBA1 mutants lose the ability to trigger cell death in accession Ag-0. The asterisk indicates a leaf with WT HopBA1-triggered cell death in the injected half. (C) HopBA1 mutants (genotype indicated below) partially lose the ability to restrict the growth of Pto DC3000 on Ag-0. White bars show growth on the day of inoculation; black bars show growth 4 d after inoculation. See also Fig. S6.
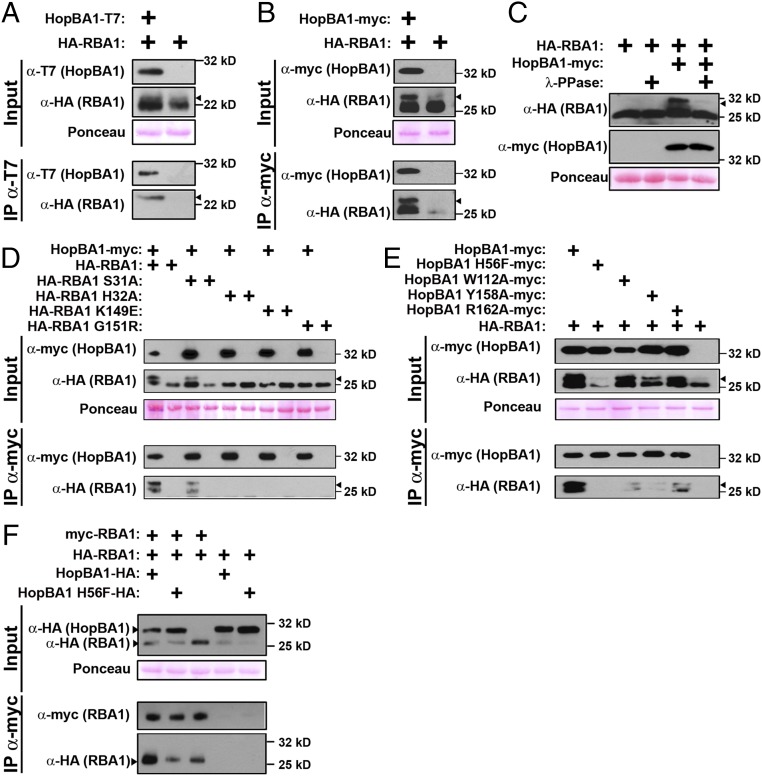
HopBA1 Coimmunoprecipitates RBA1 and Enhances RBA1 Self-Association in Planta.
NLRs can detect pathogen effectors either directly, as nominal ligands, or indirectly, via the activity of an effector on a host target or the decoy of a target (35). To test whether RBA1 directly or indirectly sensed HopBA1, we assayed for physical interactions between the two proteins. HopBA1 and RBA1 did not interact directly when assayed in a Y2H system (Fig. S4C). However, HopBA1 was able to coimmunoprecipitate RBA1 either in transient coexpression experiments in N. benthamiana or from Arabidopsis tissue infected with Pf0-1 (HopBA1) (Fig. 5 A and B). Although these results suggest a close physical proximity, they do not rule out the possibility of bridging molecules in a larger complex that contains both RBA1 and HopBA1. We noted that RBA1 displays a molecular weight shift on SDS/PAGE gels that is enhanced by (but not dependent upon) the presence of HopBA1 (Fig. 5B). In coimmunoprecipitations from Arabidopsis, HopBA1 preferentially associates with the higher molecular weight form of RBA1 (Fig. 5A). The RBA1 size shift is correlated with function and is absent in RBA1 dimer mutants (it is still present in the functional, autoactive S31A single mutant) (Fig. 5D). To test if this shift could be caused by phosphorylation, cell lysates from N. benthamiana expressing RBA1 and HopBA1 were incubated with or without lambda phosphatase (λ-PPase), which has activity for phosphorylated serine, threonine, and tyrosine. The addition of λ-PPase to samples containing both RBA1 and HopBA1 effectively eliminated the observed larger molecular weight band, strongly suggesting that RBA1 is phosphorylated (Fig. 5C). Coimmunoprecipitation of RBA1 and HopBA1 was abrogated by the loss-of-function RBA1 dimer interface mutations (Fig. 5D). Conversely, the ability to coimmunoprecipitate WT RBA1 was reduced by loss-of-function HopBA1 mutations (Fig. 5E). This partial loss of interaction is consistent with the partial loss of disease resistance we observed for the HopBA1 mutants (Fig. 4). Based on the requirement of RBA1 self-association for its interaction with HopBA1, we measured RBA1 self-association in the presence of HopBA1. Transient coexpression and subsequent coimmunoprecipitation of RBA1 with HopBA1 promoted enhanced RBA1 self-association relative to controls that either lacked the effector or carried the inactive HopBA1 H56F mutant (Fig. 5F).
Fig. 5.
HopBA1 interacts with RBA1 and enhances RBA1 self-association. (A and B) Stable transgenic, native promoter-driven 1×HA-RBA1 coimmunoprecipitates with Pf0-1–delivered T7-HopBA1 in Arabidopsis (A) and, when transiently coexpressed as 35S-driven 3xHA-RBA1, in N. benthamiana (B). Arrowheads indicate the RBA1 size shift associated with activity. (C) The RBA1 mobility shift is removed following treatment with protein phosphatase. (D) RBA1 self-association mutants are unable to coimmunoprecipitate HopBA1 when transiently coexpressed in N. benthamiana. (E) HopBA1 loss-of-response mutants partially lose association with RBA1 when transiently expressed in N. benthamiana. (F) RBA1 self-association is enhanced in the presence of WT Myc-HopBA1 but not HopBA1 H56F. See also Figs. S3–S5.
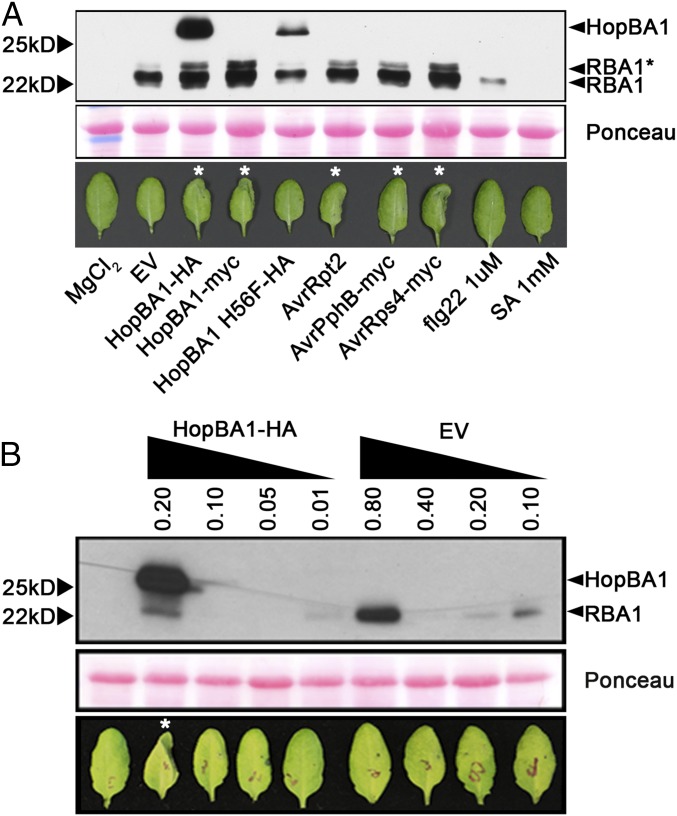
RBA1 Accumulates During Immune Responses Activated by a Variety of NLR Proteins.
We noted that native promoter-driven RBA1 protein is undetectable in transgenic plants (Fig. 2C). However, RBA1 protein accumulated to high levels after Pf0-1 delivery of HopBA1 (Figs. 2C and 6A). Accumulation of RBA1 after Pf0-1 delivery of the loss-of-function allele HopBA1 H56F was equivalent to that triggered by Pf0-1 (EV) control. To address whether RBA1 accumulation is HopBA1 specific, we used Pf0-1 to deliver other ETI-inducing T3Es (AvrRpt2 to activate RPS2, AvrRps4 to activate RPS4, and AvrPphB to activate RPS5). All ETI-inducing treatments induced RBA1 accumulation (Fig. 6). Pf0-1 weakly induced RBA1 accumulation, but flg22 elicitors were able to induce RBA1 accumulation only slightly, and SA elicitors were unable to do so (Fig. 6A). These results show that RBA1 accumulation is correlated with cell death triggered broadly during ETI, with MTI triggers having a much weaker effect.
Fig. 6.
Accumulation of RBA1 protein is induced during effector-triggered immunity; HopBA1 contributes to RBA1 cell death posttranslationally. (A, Upper) Western blot (anti-HA) showing accumulation of HA-RBA1 in Col-0 RBA1Ag-0 native promoter transgenic (lower band, ∼22 kDa) and HopBA1-HA (upper band, ∼25 kDa) in response to various treatments. (Lower) Images taken 24 h after injection of bacteria, flg22, or SA. Asterisks indicate leaves with cell death in the injected half. AvrRpt2 triggered strong cell death earlier than hopBA1, whereas AvrPphB-injected leaves had collapsed at 24 h postinoculation. (B) High levels of HA-RBA1 induced by high-dose Pf0-1 EV are not sufficient to trigger cell death in Arabidopsis in the absence of HopBA1. The asterisk indicates a leaf with cell death in the injected half.
Because RBA1 was autoactive at high expression levels in both N. benthamiana and Arabidopsis, we asked whether HopBA1 triggers RBA1 activity at lower expression levels or if cell death was merely correlated with higher levels of RBA1 protein accumulation. Therefore we compared RBA1 expression after high-titer inoculation with Pf0-1 (EV) with that induced by lower-level treatments of Pf0-1 (HopBA1) (Fig. 6B). Although Pf0-1 (HopBA1) triggered leaf collapse at an Agrobacterium inoculation density of OD 0.2, Pf0-1 (EV) was unable to trigger collapse even at OD 0.8. It is notable that the lack of response after high-dose Pf0-1 (EV) inoculation was accompanied by an induced accumulation of RBA1 protein well in excess of that induced by the cell-death–triggering lower-dose inoculation of Pf0-1 (HopBA1) (Fig. 6B, Lower Band). Thus, increased expression of RBA1 driven by its native promoter is insufficient in this context to activate cell death in the absence of HopBA1.
SI Materials and Methods
High-Throughput Plant Phenotyping.
Plants were grown in plastic cones (Ray-Leach Super Stubby Cells; Hummert Co.). The 48-well trays contained 48 unique ecotypes. Seeds of each ecotype were sprinkled into meshed cones to allow inversion. After cold treatment for 2–3 d, trays were transferred to growth chambers. Seedlings were thinned to three to four plants per ecotype after 2 wk in growth chambers. Vacuum infiltration was applied to 3.5-wk-old plants using a large vacuum container (Techni-Dome 360 Vacuum Desiccator; Bel-Art Products) Vacuum was maintained at 29 psi for 1 min and then was released slowly to ambient air pressure. Plants were returned to the growth chamber and phenotyped over the course of 5 d.
Positional Cloning of RBA1.
Crosses between responsive (Ag-0 and Ws-2) and nonresponsive (Col-0 and Ler-0) accessions indicated that an Ag-0 × Ler-0 F2 population gave the most robust and unambiguous phenotypes in response to Pf0-1–delivered HopBA1. Preliminary bulk segregant analysis was done using Col/La-er simple sequence-length polymorphism (SSLP) markers found to be polymorphic between Ag-0 and La-er. After rough mapping of RBA1 to the top of the bottom arm of chromosome 1, we screened 3,100 F2 plants for recombinants between At1g46192 and At1g47520, an ∼230-Mb interval. Phenotypes of recombinants were tested in the F2 population and then were verified in an F3 population. Fine mapping proceeded using polymorphisms identified from resequencing of Ag-0. To verify synteny with the reference genome, Ag-0 genomic DNA corresponding to 17360774–17372859 of the Col-0 reference genome (spanning the entire fine-mapping interval) was PCR amplified and Sanger sequenced.
Fine Structure of the RBA1 Locus.
We compared the functional RBA1 allele present in Ag-0 with the reference and nonfunctional Col-0 allele. RBA1 was not supported by Col-0 cDNA data and is not expressed in public Affymetrix databases, as is consistent with pseudogenization. A survey of public sequencing data found that RBA1 is present in only 5 of 17 libraries at a level of 3–19 transcripts per million (28). The most striking structural difference between the genomes of Col-0 and Ag-0 in this region is the absence in Ag-0 of a large transposon that is found immediately downstream of RBA1 in Col-0. In Ag-0, this transposon is absent, bringing the 5′ region of an F-box protein close to the 3′ end of RBA1. This structural difference led us to explore how RBA1 is spliced in Ag-0 and Col-0. As consistent with the hypothesis of pseudogenization, we were unable to isolate RT-PCR fragments across putative intron–exon junctions from Col-0–derived cDNA. We were able to isolate Ag-0–derived cDNA fragments consistent with a TIR-only protein. We were unable to validate either the annotated TIR-X isoform from Ag-0 or Col-0 or the presence of a TIR-F-box splice product in Ag-0. As is consistent with this interpretation, an N-terminally tagged TIR-X genomic construct from Ag-0 expressed in N. benthamiana produced a 25-kDa TIR-only protein rather than the predicted 40-kDa TIR-X protein (Fig. S2B).
Generation of CAS9 Mutants.
Arabidopsis mutants were generated via CAS9 mutagenesis. AtU6 promoter-driven guide RNAs were generated to target the first exon of RBA1 and were recombined with LR Clonase II (Invitrogen) into the pCUT6 Ubi10promoter:CAS9 binary vector (62). This vector was stably transformed into Arabidopsis via floral dip. T1 plants were selected for BASTA resistance and then were screened for somatic CAS9 cutting at the expected site. Plants that displayed frameshifts via Sanger sequencing were selfed and taken to the T2 generation. To identify germline mutations in RBA1, T2 plants that had segregated away the transgene (BASTA-sensitive) were reanalyzed at the cut site. We identified two independent, transgene-minus, heterozygous rba1 mutants. These were self-fertilized, and homozygotes were selected in the T3 generation.
Determination of HopBA1 Crystal Structure.
Expression and purification of recombinant HopBA1.
GST-tagged HopBA1 was expressed in Novagen Rosetta E. coli BL21(DE3) cells. Cells were grown in 1 L Terrific Broth (ampicillin 100 μg/mL, chloramphenicol 33 μg/mL) at 37 °C to an OD600 of 0.5, incubated in an ice-water bath for 1 min, and then induced with 0.1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) at 19 °C overnight. Purification was achieved by tandem GST affinity chromatography in buffer A [20 mM Tris (pH 7.5), 150 mM NaCl, 2 mM DTT, and 5% (vol/vol) glycerol]. Elution was effected with 40 mM GST in buffer A. Subsequently, the GST tag was removed by tobacco etch virus (TEV) in buffer A. Purified HopBA1 was then obtained by size-exclusion chromatography in buffer A. Elution of HopBA1 during size-exclusion chromatography was compatible with a particle that has the hydrodynamic radius of an ∼25-kDa globular protein, thus indicating that HopBA1 exists as a monomer in solution under the conditions used. HopBA1 then was concentrated to 13 mg/mL and sent to crystallization (the excess was stored at −80C).
Crystallization of HopBA1.
Initial crystallization hits were obtained using commercially available crystallization screens (63) and then were optimized. Crystals suitable for structure determination were ultimately obtained by vapor diffusion (hanging-drop mode) using 6 μL protein solution and 6 μL crystallization solution [either 0.5 M potassium sulfate, 0.1 M Hepes (pH 7.0), and 30% (vol/vol) PEG 400 or 0.5 M lithium sulfate, 0.1 M Tris (pH 7), and 30% (vol/vol) PEG 400] equilibrated over 0.5 mL of crystallization solution. Crystals were harvested, dipped in glycerol, cooled by plunging into liquid nitrogen, and stored for further X-ray diffraction experiments. Final HopBA1 crystals were ∼100 μm in length/width/height and diffracted X-rays to about 2.8 Å using an in-house X-ray source.
Expression of the HopBA1 selenomethionine variant.
The GST-HopBA1 construct was transformed into the methionine auxotroph E. coli BL21 834 (DE3) and expressed in the presence of selenomethionine. The purification protocol was unaltered from that of recombinant HopBA1 described above. Crystals were obtained in the same manner as for HopBA1, with a crystallization solution of 0.5 M potassium sulfate, 0.1 M Hepes (pH 7.0), and 30% (vol/vol) PEG 400, and had a final size of ∼120 μm in length/width/height. Optimal crystals were produced at a concentration of 10.7 mg/mL.
Crystal structure determination.
The crystal structure of HopBA1 was determined by single-wavelength anomalous diffraction (SAD) using data to a resolution of 3.3 Å collected at the Advanced Photon Source Southeast Regional Collaborative Access Team (SER-CAT) beamline 22-ID (Argonne, IL). An initial model was obtained using the program CRANK from the CCP4 suite (64) and was completed using iterative model building with COOT (65) interspersed by refinement with the program REFMAC (66, 67). The resulting model was used for molecular replacement using diffraction data to a resolution of 2.01 Å (collected at SER-CAT beamline 22-ID) to obtain a model for native HopBA1 that then was refined with the program suite Phenix (68). The final model has excellent stereochemistry as assessed by the program MolProbity (69, 70) with 99.1% of the residues falling into the preferred and 0.9% of the residues falling into the allowed regions of a Ramachandran plot (Table S1). The model of HopBA1 has been deposited in the PDB database (PDB ID code 5T09).
Y2H Assays.
Protein interactions were assayed using Gateway-modified versions of the traditional pEG202 and pJG4-5 vectors. pDONR207 Gateway Entry clones containing ORFs were recombined into pEG202 and pJG4-5 using LR Clonase II. The resulting vectors were sequence verified and then were transformed into EGY48 (pEG202) and RFY206 containing the LacZ reporter pSH18-34 (pJG4-5). The resulting strains were mated, and diploids were selected on synthetic medium (SC-HUW). Diploids were spotted onto SC-HUW +X-gal (80 μg/mL). Protein accumulation of fusion proteins in diploids was verified with standard Western blotting using anti-LexA (rabbit polyclonal Ab; Abcam) and anti-HA (rat mAb; Roche) antibodies.
Confocal Microscopy.
Leaf discs (7 mm diameter) of 4- to 5-wk-old N. benthamiana leaves were collected 24 h after Agro infiltration (as described above). The abaxial side of the leaves was imaged using a C-Apochromat 40×/NA1.2 water immersion lens on a Zeiss LSM710 confocal laser-scanning microscope. Images were taken with standardized excitation intensities and photomultiplier gains. XFP fluorescence was imaged using an Argon/2 laser and the photomultiplier tube (PMT) detector to collect emissions. Excitation wavelength/emission bandwidth were set at 514/519–560 nm for YFP and 561/594–651 nm for tRFP.
Microsomal Fractionation.
Leaf discs (as described above; 100–200 mg) were ground in 500 μL cold sucrose buffer [20 mM Tris (pH 8), 0.33 M sucrose, 1 mM EDTA, 5 mM DTT, and 1× Sigma plant protease inhibitors]. Samples were spun at 4 °C in a microcentrifuge at 2 × g for 5 min to remove debris. The supernatant here is the total lysate fraction (T). A portion of the total lysate was then spun at 4 °C at 20 × g for 30 min. The resulting supernatant is the soluble fraction (S), and the pellet was resuspended in an equal volume of sucrose buffer to yield the microsomal fraction (M).
Discussion
The ability to monitor signals of microbial invasion is universally the first step in mounting an immune response. In plants, the intracellular “sensor” proteins of the NLR family are critical components for activating plant immunity. These sensors monitor a reliable indicator of pathogenesis: the presence of pathogen effector proteins. These pathogen effector proteins can be thought of as evolved tools that manipulate critical, and often conserved, components required for host immunity (36).
Here we define and characterize a type of NLR-like protein that consists of only a functional TIR domain, RBA1. We provide a crystal structure for the T3E protein, HopBA1, that triggers an RBA1-dependent cell-death response and show that it resembles a bacterial heme-scavenger protein. Mutations in and around the putative heme-binding site of HopBA1 result in loss of coimmunoprecipitation with RBA1 and loss of RBA1-dependent cell death. HopBA1 enhances RBA1 self-association, which is necessary for ectopic autoactivation of host cell death. Surprisingly, mutations in either of two previously characterized TIR dimerization interfaces abolished RBA1 autoactivity, its self-association, and its interaction with HopBA1. Our data demonstrate that neither of the previously characterized plant TIR dimer interfaces alone is sufficient for effective RBA1 signaling.
To identify components of the host immune system, we used a genetic approach exploiting natural variations in both the host and the pathogen. We identified HopBA1 as a trigger of RBA1 activity. Like many pathogen virulence effectors, HopBA1’s biochemical function is not revealed by its primary protein sequence. In combination with a genetic dissection of HopBA1-triggered responses in the host, we took a structural approach and determined the crystal structure of HopBA1. Bacterial virulence proteins are often structural mimics of eukaryotic proteins. However, the structure of HopBA1 that we solved was most similar to the bacterial heme-binding protein ChaN and the bacterial esterase Bcr136. We were not able to demonstrate any heme-binding or esterase activity using in vitro-purified HopBA1 protein. However, using ChaN and Bcr136 as a guide, we generated HopBA1 mutants that lose the ability to trigger RBA1 cell death in planta.
Recently, the Pto DC3000 effector HopB1 was proposed to be a novel type of serine protease (37). Secondary structure analysis in Li et al. (37) showed no homology to known proteases. However, our structure-based homology searches indicated that HopB1 is actually a HopBA1-like protein most similar to the PMT-C2 domain (Fig. S6G). Li et al. proposed that HopB1 H413, T370, D435, and D436 form a catalytic core. Based on the HopB1 homology model, the proposed HopB1 catalytic histidine (H413) is conserved in HopBA1 (H193), in the heme scavenger ChaN (H220), and in the EreA-like esterase Bcr136 (H309), but not in PMT (L1045). The HopB1 catalytic threonine and aspartates (T370/D435/D436) proposed by Li et al. are not conserved in HopBA1/ChaN/Bcr136 and do not appear to be in a position likely to promote catalysis involving HopB1 H413, based on modeling to the structure of the PMT-C2 domain (Fig. S6G). In contrast to HopB1, structural and biochemical analyses of EreA/Bcr136 by Morar et al. (33) indicate that the Bcr136 proposed catalytic histidine is likely to be H84, rather than H309, and that its catalysis is not dependent on serine or threonine. This proposed Bcr136 catalytic histidine is also conserved across the family of HopBA1-like proteins. The residue in HopB1 (H274) corresponding to the proposed Bcr136 catalytic histidine was not tested for HopB1 catalytic activity in Li et al. (37). Both HopBA1 H56 (analogous to the proposed Bcr136 esterase catalytic residue) and H193 (proposed HopB1 serine protease catalytic residue) are required for HopBA1-triggered cell death. Despite considerable conservation at both of these histidines across the family, HopBA1-like proteins appear to have quite diverse biochemical functions. These proteins also have been proposed to be ancient relatives of the human TIKI metalloprotease (38).
HopBA1-like proteins may be important virulence factors for both plant and animal pathogens. Intriguingly, the structure of HopBA1 is similar to a domain of the Pasteurella multocida toxin, PMT. PMT has acetyltransferase activity that modifies the animal cytoskeleton. However, PMT is a large, multidomain protein, and its HopBA1/Bcr136/ChaN-like domain has no described function. The presence of a HopBA1-like protein in a mosquito microbiome strain also hints at a conserved role for HopBA1-like proteins as effectors in both plants and animals. The precise role of HopBA1 in virulence remains obscure. To date, we have been unable to demonstrate a virulence function for hopBA1, by either the addition of hopBA1 into the Arabidopsis/Pto DC3000 system or the deletion of hopBA1 from the sugar beet/P. syringae pv aptata pathosystem. Recently, deletion of hopBA1 was shown to reduce P. syringae growth on wheat, especially in the context of a hopBA1/hopA2/hopAZ1 triple mutant (39). A full understanding of the mechanism of HopBA1 function will require genetic and biochemical characterization of additional components beyond HopBA1 and RBA1, including HopBA1 virulence targets and unknown genes required for immune perception and activation (see discussion below).
A critical unsolved question in plant innate immunity is how NLRs actually signal downstream to activate cell death and disease resistance as measured by pathogen growth restriction. Although the direct and indirect pathogen-recognition events activating NLRs are increasingly well understood, the downstream events remain obscure (40). In the case of TIR-NLRs, ectopic cell death triggered by the expression of truncated TIR domains in the absence of pathogens indicates that they are likely sufficient to trigger disease resistance (13, 17). A requirement for dimerization is typically reported for TIR-truncation phenotypes, even though the reported dimers (L6 and RPS4/RRS1) are structurally distinct (13, 14). Homology modeling to structures of TIR domains from the full-length NLRs RPS4 and L6 guided our RBA1 mutagenesis. Mutations in RBA1 that reflect alterations in either the L6 or the RPS4 dimerization interfaces resulted in the loss of RBA1 activity and self-association. This genetic evidence strongly suggests that a WT RPS4 interface is insufficient to support self-association in the former set of mutants and that a WT L6 interface is insufficient to support self-association in the latter. Additionally, RBA1 mutants in either of the two interfaces expressed in trans do not self-associate (Fig. 3F).
Our results are consistent with speculative models in which RBA1 either has a unique TIR structure or forms functional multimers that are dependent on both interfaces (Fig. S3). RBA1 self-association could be stabilized by cooperativity, as reported for TIR domains during activation of the Toll signaling complex (41). The requirement for both TIR interfaces in plants is likely a generalizable observation, because equivalent RPS4 dimer interface mutations made in the L6 TIR domain and vice versa result in loss of function, indicating the general importance of both interfaces (42). Like the RBA1 mutants, these L6 and RPS4 mutants present a result not expected from the published crystal structures. The hypothesis that interfaces are simultaneously engaged could resolve the puzzle presented by the distinct dimer interfaces found in the L6 and RPS4 crystals. Potentially, these crystals are incomplete, and each represents only part of a larger complex that is bigger than simple dimers. Consistent with this model, Zhang et al. (42) have generated two new plant TIR domain crystal structures [of Successor of npr1-1, constitutive 1 (SNC1) and RPP1] that support our speculation that two interaction surfaces are engaged simultaneously. Consistent with these results, Hyun et al. (43) also found both L6 and RPS4-like interfaces within a SNC1 TIR crystal. To test if a TIR multimer is at least sterically feasible, we generated a model of a hybrid RBA1 TIR oligomer structure containing simultaneous L6 and RPS4-type interfaces (Fig. S3 C–E). Although the structure of full-length NLRs remains obscure, this hybrid TIR structure also would be sterically feasible for full-length NLRs because it distributes both the N and C termini around the outer surface of the helix (Fig. S3E). Whether such a structure actually occurs remains to be determined, but in animals TIR domains are known to oligomerize and heterodimerize to transmit signals (44). Analogous to this hypothetical RBA1 oligomer, the animal TIR protein MyD88 is proposed to form a helical homo-oligomer (with multiple, distinct interaction surfaces) as part of a larger complex of TIR-containing proteins (45). Thus, it is possible that plant TIR domain oligomerization and/or heterodimerization regulate the formation of functional signaling macrocomplexes.
Importantly, the loss of RBA1 self-association is correlated with the loss of RBA1 interaction with HopBA1. Coimmunoprecipitation is not sufficient to demonstrate a direct interaction between HopBA1 and RBA1; thus we cannot rule out the possibility that additional host-derived molecules may bridge HopBA1 and RBA1. Indeed, Y2H assays failed to demonstrate a direct HopBA1–RBA1 interaction. Exactly how HopBA1 triggers RBA1 remains unknown. The absence of typical NLR negative regulatory domains in RBA1 presents a puzzle as to how RBA1 is regulated. Treatment of native promoter-driven RBA1 transgenics with high-titer Pf0-1 (EV) induces RBA1 expression. However, this expression does not trigger cell death as efficiently as the weaker RBA1 accumulation triggered by lower titers of Pf0-1 (HopBA1). Thus, a low level of RBA1 in the presence of HopBA1 triggers cell death more strongly than a high level of RBA1 in the absence of the effector. This finding strongly supports a direct mechanistic link between HopBA1 recognition and RBA1 activation. The presence of HopBA1 leads to a molecular weight shift in RBA1 that is sensitive to λ-PPase. This sensitivity suggests that HopBA1 enhances RBA1 phosphorylation and that phosphorylation may be required for full RBA1 activity. We observed that other ETI-inducing treatments (and Pf0-1, to a lesser extent) also lead to a molecular weight shift in RBA1, so the shift is likely caused by the activation of an endogenous phosphorylation pathway downstream of pathogen recognition rather than by a specific response to HopBA1. Consistent with this notion, the lack of phosphorylation in the RBA1 self-association mutants suggests that phosphorylation is either coincident with or downstream of RBA1 self-association. Intriguingly, we found in Arabidopsis that HopBA1 interacted preferentially with the higher molecular weight, presumably phosphorylated, form of RBA1. This observation suggests that the RBA1 size shift is functionally relevant to activation; to test this hypothesis, we are pursuing mass spectrometry to identify the exact modifications. Future identification of which residues of RBA1 are modified, and how, is likely a fertile ground for understanding downstream events at or after NLR activation. We note that the full-length TIR-NLR RPP1 also may be posttranslationally modified (27), indicating that posttranslational modification of TIR domains is not an RBA1-specific event but rather may be a general event during NLR activation and/or signaling. The promotion of RBA1 self-association by HopBA1 suggests a simple model wherein HopBA1 lowers a threshold for RBA1 oligomerization, dependent on both the RPS4- and L6-type interfaces, to activate cell death.
Given the recent example of RPS4/RRS1 TIR–TIR heterodimers in plants (14) and the animal TIR signalosome (46), it is a distinct possibility that another NLR facilitates RBA1 function (or is itself regulated by RBA1) via TIR–TIR interactions. The existence of unknown genes related to HopBA1 recognition and RBA1 function is strongly supported by our genetic data. Col-0 lacks the RBA1-dependent HopBA1-induced cell death response; however, the growth of Pto DC3000 on Col-0 is still restricted by HopBA. Whether RBA1 modifies this RBA1-independent recognition or functions independently remains to be determined. Although there are no tightly linked full-length TNLs near RBA1 in the genome, NLR heteromerization offers an alternative model wherein RBA1 forms a complex with an unknown full-length NLR. Although hypersensitive cell death and disease resistance are often correlated, the mechanistic contribution and importance of hypersensitive cell death to disease resistance remains obscure. Pto DC3000 containing HopBA1 is still growth restricted on Col-0, indicating a decoupling of cell death and disease resistance. Decoupling of cell death and disease resistance has been previously reported (47–49). HopBA1 is not present naturally in Pto DC3000, so better understanding its contribution to disease resistance might require future studies in its endogenous strains (P. syringae pv. Aptata and P. syringae pv. Japonica) in their hosts of isolation (sugar beet and wheat, respectively).
In this work, we demonstrate that a TIR-only protein can determine the response to a pathogen effector. In addition to RBA1, there are ∼50 TIR, TIR-X, and TIR-NB genes present in the Arabidopsis Col-0 reference genome (50). Although RBA1 appears to be a pseudogene in the Col-0 genome, most of the TIR-X and TIR-NB genes are expressed (50). Interestingly, several of these are transcriptionally induced by the defense hormone salicylic acid, suggesting a role in disease resistance (28). Like RBA1 or TIR domains truncated from full-length TNLs, overexpression of TIR-X and TIR-NB proteins can trigger EDS1-dependent cell death (28). Loss-of-function mutations in TN2, a TIR-NB protein lacking LRRs, were isolated as genetic suppressors of cell death triggered by loss-of-function alleles of EXO70B1, a component of the exocyst vesicle trafficking complex (51). These phenotypes are consistent with a model wherein EXO70B1 is a guardee, monitored by the immune receptor TN2 and targeted by as yet unknown pathogen effectors. This model is analogous to the classical RPS2/RIN4 NLR/guardee model, except that the NLR is a truncation lacking the canonical LRR domain (52, 53). There is also precedence for TIR-NB proteins regulating full-length TNLs. The chs1-2 (a TIR-NB) mutant phenotype of temperature-dependent cell death requires the neighboring full-length TNL gene SOC1 (54). Truncated NLR proteins also have been reported to arise from a single full-length TNL gene via alternative splicing (55, 56). In the case of the tobacco N gene, analysis of alternative cDNA products suggests that a truncated TIR-NB splice variant is required for full function (56). Our studies of RBA1 provide a conclusive answer that TIR-containing truncated NLRs can regulate immune responses in response to pathogens and furthermore suggest a more complex model of TIR oligomerization using distinct structural interfaces. These observations expand our view of the diversity of the plant immune system in terms of both receptor architecture and mechanisms of activation.
Materials and Methods
Plant Materials and Growth Conditions.
Arabidopsis was grown in walk-in rooms maintained at 21 °C/18 °C (day/night) with a 9-h/15-h day/night cycle. Tobacco was grown in walk-in growth rooms maintained at 26 °C/22 °C with a 12-h/12-h day/night cycle. Transgenic Arabidopsis strains were generated using standard floral dip techniques (57). Ag-0 defense mutant plant lines were generated by introgressing mutant alleles from Col-0 into Ag-0. The Ag-0 mutant plants used were progeny of plants fixed for RBA1 Ag-0 and heterozygous for the defense gene, thus allowing cosegregation analysis. Correlation of genotypes and phenotypes was verified using standard PCR markers (oligonucleotides are available upon request).
Generation of Expression Plasmids.
Gateway-compatible Entry clones and Destination clones were generated by BP and LR cloning (Invitrogen) or by direct synthesis (GenScript). Site-directed mutants were generated by overlap extension PCR or site-directed, ligase-independent mutagenesis (SLIM) (58). Oligonucleotides used for cloning are available upon request. RBA1 is N-terminally epitope tagged throughout the paper, because C-terminal tags have reduced function. Agrobacteria 35S-promoter expression plasmids included pGWB615 (3×HA), pGWB618 (4×MYC), pGWB642 (eYFP), and pGWB661 (TagRFP). Pseudomonas expression plasmids used were pJC531 (native promoter:1×HA) and pJC532 (native promoter: ΔavrRpt2). The RBA1 native promoter HA:RBA1 genomic complementation construct was generated in pGWB616. Dexamethasone-inducible RBA1 was expressed from the pBUD vector. pGWB vectors are from the Nakagawa laboratory (59).
Bacterial Strains and Growth Conditions.
Escherichia coli Top10 and Agrobacterium tumefaciens strain GV3101/pMP90 were grown in LB medium at 37 °C and 28 °C, respectively. Pseudomonas strains were grown at 28 °C in King’s B medium. The E. coli antibiotic concentrations used (in micrograms per milliliter) were ampicillin 100, kanamycin 30, gentamycin 25, and spectinomycin 50. Agrobacterium antibiotic concentrations used (in micrograms per milliliter) were gentamycin 50, kanamycin 100, rifampicin 100, and spectinomycin 100. Pseudomonas antibiotic concentrations used (in micrograms per milliliter) were gentamycin 25, kanamycin 30, and rifampicin 50.
Bacterial Assays and Conductivity Measurements.
P. fluorescens (Pf0-1) effector delivery assays were performed as described (22). Typically, Pf0-1 was grown overnight, washed, and diluted in 10 mM MgCl2 to an OD600 of 0.1. These cultures were hand-injected with needleless syringes into 4- to 6-wk-old Arabidopsis rosette leaves between 10 AM and noon and were phenotyped 24, 36, and/or 48 h after infiltration. P. syringae bacterial growth assays were performed as described (60). Briefly, Pto DC3000 was grown overnight and washed in 10 mM MgCl2, resuspended to OD = 0.0002, and then injected as described above. Leaves were cored (no. 4 cork borer), ground, and dilution plated to assess recovered colony-forming units. Each experiment contained six biological replicates per genotype, and statistical significance was assessed using a one-way ANOVA and post hoc Tukey’s honestly significant difference (HSD) test (P ≤ 0.05) (SigmaPlot). To measure conductivity, four leaf discs were collected with a no. 4 corer from four independent plants infiltrated 18 h earlier. Leaf disks were added to clear tubes with 6 mL of distilled water at room temperature under continuous light (three replicates per sample). Changes in water conductivity were measured at the indicated time points with an Orion Model 130. Agrobacterium (GV3101/pMP90) transient assays were performed similarly but were resuspended in 10 mM MgCl2 amended with 10 mM Mes (pH 5.0) and 150 μM acetosyringone. Agrobacteria were injected at a total OD of 0.8 (including OD 0.1 of 35S:P19) into 5- to 6-wk-old N. benthamiana leaves. Ectopic RBA1 chlorosis and tissue collapse typically appeared ∼36–48 h postinoculation.
Structural Modeling of RBA1.
Homology modeling of RBA1 was done using the Phyre2 suite (61). When using the intensive modeling setting, the RPS4 TIR domain [Protein Data Bank (PDB) ID code 4C6R] is the highest hit returned (100% confidence, 41% sequence identity), and the L6 TIR domain (PDB ID code 3OZI) is the second (100% confidence, 30% sequence identity). Hypothetical RBA1 dimers and oligomer were generated by using the ALIGN function of PyMOL (version 1.4.1; Schrodinger, LLC).
Coimmunoprecipitation and Western Blots.
A combination of Agrobacterium strains harboring the correct vectors were infiltrated into two separate halves of N. benthamiana leaves that were subsequently flash frozen 36 h postinoculation. Frozen leaf tissue was collected and ground in a mortar and pestle with liquid nitrogen and resuspended in 2 mL of extraction buffer [50 mM Hepes-KOH (pH 7.5), 50 mM NaCl, 10 mM EDTA (pH 8.0), 0.2% Triton X-100, 5 mM DTT with 1× plant protease inhibitor mixture (Sigma-Aldrich)]. Soluble supernatants were cleared twice by centrifugation at 10,000 × g for 10 min at 4 °C and were incubated for 2 h with end-over-end turning at 4 °C with 50 μL of α-myc– or α-HA–conjugated magnetic beads (Miltenyi Biotec) for Myc-HopBA1 or HA-RBA1 precipitation or with 100 μL of α-T7 agarose beads (Novagen) for T7-HopBA1. Samples were captured with MACS separation columns (Miltenyi Biotec) and were washed three times with washing buffer (extraction buffer with 0.1% Triton X-100 and 150 mM NaCl). Bound proteins were eluted in elution buffer [50 mM Tris⋅HCl (pH 6.8), 50 mM DTT, 1% SDS, 1 mM EDTA (pH 8.0), 0.005% bromophenol blue, and 10% glycerol]. T7 agarose beads were collected by centrifugation at 1,000 × g for 5 min at 4 °C and were washed three times with washing buffer. The bound proteins were eluted in 100 μL of elution buffer. Samples were resolved by electrophoresis on 12% SDS/PAGE, transferred to PVDF, and blotted with primary antibodies overnight at 4 °C in TBS with 1% Tween (TBST) and 5% nonfat dry milk. The following concentrations were used: α-Myc, 1:1,000 (Santa Cruz Biotechnology); α-HA, 1:1,000 (Sigma); α-T7, 1:10,000 (Novagen).
Acknowledgments
We thank Robin Allen, Victor Baillif, and Joseph Woodruff for technical assistance; Cris Argueso for access to Colorado State University facilities; and Drs. Sarah Grant and Freddy Monteiro for critical reading of the manuscript. Tim Denny provided P. syringae B76. David Baltrus provided P. syringae FF5. Barbara Baker provided eds1-silenced N. tabacum. Detlef Weigel provided Ag-0 prerelease genome resequencing data. This work was supported by National Science Foundation Grant IOS-1257373 (to J.L.D.). J.L.D. is an Investigator of the Howard Hughes Medical Institute (HHMI) and is supported by the HHMI and by Gordon and Betty Moore Foundation Grant GBMF3030. R.G.A. was supported by NIH Ruth L. Kirschstein National Research Service Award Fellowship F32GM108226.
Footnotes
The authors declare no conflict of interest.
Data deposition: Crystallography, atomic coordinates, and structure factors reported in this paper have been deposited in the Protein Data Bank database (PDB ID code 5T09).
See Commentary on page 2445.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1620973114/-/DCSupplemental.
References
- 1.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444(7117):323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 2.Lindeberg M, Cunnac S, Collmer A. Pseudomonas syringae type III effector repertoires: Last words in endless arguments. Trends Microbiol. 2012;20(4):199–208. doi: 10.1016/j.tim.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 3.Jacob F, Vernaldi S, Maekawa T. Evolution and conservation of plant NLR functions. Front Immunol. 2013;4:297. doi: 10.3389/fimmu.2013.00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell. 2003;15(4):809–834. doi: 10.1105/tpc.009308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duxbury Z, et al. Pathogen perception by NLRs in plants and animals: Parallel worlds. BioEssays. 2016;38(8):769–781. doi: 10.1002/bies.201600046. [DOI] [PubMed] [Google Scholar]
- 6.Takken FL, Goverse A. How to build a pathogen detector: Structural basis of NB-LRR function. Curr Opin Plant Biol. 2012;15(4):375–384. doi: 10.1016/j.pbi.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 7.Hu Z, et al. Structural and biochemical basis for induced self-propagation of NLRC4. Science. 2015;350(6259):399–404. doi: 10.1126/science.aac5489. [DOI] [PubMed] [Google Scholar]
- 8.Zhang L, et al. Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization. Science. 2015;350(6259):404–409. doi: 10.1126/science.aac5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bentham A, Burdett H, Anderson PA, Williams SJ, Kobe B. Animal NLRs provide structural insights into plant NLR function. Ann Bot. 2016 doi: 10.1093/aob/mcw171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Botos I, Segal DM, Davies DR. The structural biology of Toll-like receptors. Structure. 2011;19(4):447–459. doi: 10.1016/j.str.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maekawa T, Kufer TA, Schulze-Lefert P. NLR functions in plant and animal immune systems: So far and yet so close. Nat Immunol. 2011;12(9):817–826. doi: 10.1038/ni.2083. [DOI] [PubMed] [Google Scholar]
- 12.Valkov E, et al. Crystal structure of Toll-like receptor adaptor MAL/TIRAP reveals the molecular basis for signal transduction and disease protection. Proc Natl Acad Sci USA. 2011;108(36):14879–14884. doi: 10.1073/pnas.1104780108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bernoux M, et al. Structural and functional analysis of a plant resistance protein TIR domain reveals interfaces for self-association, signaling, and autoregulation. Cell Host Microbe. 2011;9(3):200–211. doi: 10.1016/j.chom.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams SJ, et al. Structural basis for assembly and function of a heterodimeric plant immune receptor. Science. 2014;344(6181):299–303. doi: 10.1126/science.1247357. [DOI] [PubMed] [Google Scholar]
- 15.Chan SL, Mukasa T, Santelli E, Low LY, Pascual J. The crystal structure of a TIR domain from Arabidopsis thaliana reveals a conserved helical region unique to plants. Protein Sci. 2010;19(1):155–161. doi: 10.1002/pro.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ve T, Williams SJ, Kobe B. Structure and function of Toll/interleukin-1 receptor/resistance protein (TIR) domains. Apoptosis. 2015;20(2):250–261. doi: 10.1007/s10495-014-1064-2. [DOI] [PubMed] [Google Scholar]
- 17.Swiderski MR, Birker D, Jones JDG. The TIR domain of TIR-NB-LRR resistance proteins is a signaling domain involved in cell death induction. Mol Plant Microbe Interact. 2009;22(2):157–165. doi: 10.1094/MPMI-22-2-0157. [DOI] [PubMed] [Google Scholar]
- 18.Macho AP, Zipfel C. Targeting of plant pattern recognition receptor-triggered immunity by bacterial type-III secretion system effectors. Curr Opin Microbiol. 2015;23:14–22. doi: 10.1016/j.mib.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 19.Le Fevre R, Evangelisti E, Rey T, Schornack S. Modulation of host cell biology by plant pathogenic microbes. Annu Rev Cell Dev Biol. 2015;31:201–229. doi: 10.1146/annurev-cellbio-102314-112502. [DOI] [PubMed] [Google Scholar]
- 20.Baltrus DA, et al. Dynamic evolution of pathogenicity revealed by sequencing and comparative genomics of 19 Pseudomonas syringae isolates. PLoS Pathog. 2011;7(7):e1002132. doi: 10.1371/journal.ppat.1002132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang JH, et al. A high-throughput, near-saturating screen for type III effector genes from Pseudomonas syringae. Proc Natl Acad Sci USA. 2005;102(7):2549–2554. doi: 10.1073/pnas.0409660102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas WJ, Thireault CA, Kimbrel JA, Chang JH. Recombineering and stable integration of the Pseudomonas syringae pv. syringae 61 hrp/hrc cluster into the genome of the soil bacterium Pseudomonas fluorescens Pf0-1. Plant J. 2009;60(5):919–928. doi: 10.1111/j.1365-313X.2009.03998.x. [DOI] [PubMed] [Google Scholar]
- 23.Jiang J, Alvarez C, Kukutla P, Yu W, Xu J. Draft genome sequences of Enterobacter sp. isolate Ag1 from the midgut of the malaria mosquito Anopheles gambiae. J Bacteriol. 2012;194(19):5481. doi: 10.1128/JB.01275-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peretz A, et al. A rare bacteremia caused by Cedecea davisae in patient with chronic renal disease. Am J Case Rep. 2013;14:216–218. doi: 10.12659/AJCR.889285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao J, et al. Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat Genet. 2011;43(10):956–963. doi: 10.1038/ng.911. [DOI] [PubMed] [Google Scholar]
- 26.Kawakatsu T, et al. 1001 Genomes Consortium Epigenomic diversity in a global collection of Arabidopsis thaliana accessions. Cell. 2016;166(2):492–505. doi: 10.1016/j.cell.2016.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schreiber KJ, Bentham A, Williams SJ, Kobe B, Staskawicz BJ. Multiple domain associations within the Arabidopsis immune receptor RPP1 regulate the activation of programmed cell death. PLoS Pathog. 2016;12(7):e1005769. doi: 10.1371/journal.ppat.1005769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nandety RS, et al. The role of TIR-NBS and TIR-X proteins in plant basal defense responses. Plant Physiol. 2013;162(3):1459–1472. doi: 10.1104/pp.113.219162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rietz S, et al. Different roles of Enhanced Disease Susceptibility1 (EDS1) bound to and dissociated from Phytoalexin Deficient4 (PAD4) in Arabidopsis immunity. New Phytol. 2011;191(1):107–119. doi: 10.1111/j.1469-8137.2011.03675.x. [DOI] [PubMed] [Google Scholar]
- 30.Holt BF, 3rd, Belkhadir Y, Dangl JL. Antagonistic control of disease resistance protein stability in the plant immune system. Science. 2005;309(5736):929–932. doi: 10.1126/science.1109977. [DOI] [PubMed] [Google Scholar]
- 31.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: A structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247(4):536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 32.Chan AC, et al. Cofacial heme binding is linked to dimerization by a bacterial heme transport protein. J Mol Biol. 2006;362(5):1108–1119. doi: 10.1016/j.jmb.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 33.Morar M, Pengelly K, Koteva K, Wright GD. Mechanism and diversity of the erythromycin esterase family of enzymes. Biochemistry. 2012;51(8):1740–1751. doi: 10.1021/bi201790u. [DOI] [PubMed] [Google Scholar]
- 34.Kitadokoro K, et al. Crystal structures reveal a thiol protease-like catalytic triad in the C-terminal region of Pasteurella multocida toxin. Proc Natl Acad Sci USA. 2007;104(12):5139–5144. doi: 10.1073/pnas.0608197104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cui H, Tsuda K, Parker JE. Effector-triggered immunity: From pathogen perception to robust defense. Annu Rev Plant Biol. 2015;66:487–511. doi: 10.1146/annurev-arplant-050213-040012. [DOI] [PubMed] [Google Scholar]
- 36.Mukhtar MS, et al. European Union Effectoromics Consortium Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science. 2011;333(6042):596–601. doi: 10.1126/science.1203659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li L, et al. Activation-dependent destruction of a co-receptor by a Pseudomonas syringae Effector dampens plant immunity. Cell Host Microbe. 2016;20(4):504–514. doi: 10.1016/j.chom.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 38.Bazan JF, Macdonald BT, He X. The TIKI/TraB/PrgY family: A common protease fold for cell signaling from bacteria to metazoa? Dev Cell. 2013;25(3):225–227. doi: 10.1016/j.devcel.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dudnik A, Dudler R. Virulence determinants of Pseudomonas syringae strains isolated from grasses in the context of a small type III effector repertoire. BMC Microbiol. 2014;14:304. doi: 10.1186/s12866-014-0304-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qi D, Innes RW. Recent advances in plant NLR structure, function, localization, and signaling. Front Immunol. 2013;4:348. doi: 10.3389/fimmu.2013.00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bovijn C, et al. Identification of binding sites for myeloid differentiation primary response gene 88 (MyD88) and Toll-like receptor 4 in MyD88 adapter-like (Mal) J Biol Chem. 2013;288(17):12054–12066. doi: 10.1074/jbc.M112.415810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang X. Multiple functional self-association interfaces in plant TIR domains. Proc Natl Acad Sci USA. 2017;114:E2046–E2052. doi: 10.1073/pnas.1621248114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hyun KG, Lee Y, Yoon J, Yi H, Song JJ. Crystal structure of Arabidopsis thaliana SNC1 TIR domain. Biochem Biophys Res Commun. 2016;481(1-2):146–152. doi: 10.1016/j.bbrc.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 44.Gay NJ, Symmons MF, Gangloff M, Bryant CE. Assembly and localization of Toll-like receptor signalling complexes. Nat Rev Immunol. 2014;14(8):546–558. doi: 10.1038/nri3713. [DOI] [PubMed] [Google Scholar]
- 45.Vyncke L, et al. Reconstructing the TIR side of the myddosome: A paradigm for TIR-TIR interactions. Structure. 2016;24(3):437–447. doi: 10.1016/j.str.2015.12.018. [DOI] [PubMed] [Google Scholar]
- 46.Guven-Maiorov E, et al. The Aachitecture of the TIR domain signalosome in the Toll-like receptor-4 signaling pathway. Sci Rep. 2015;5:13128. doi: 10.1038/srep13128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heidrich K, et al. Arabidopsis EDS1 connects pathogen effector recognition to cell compartment-specific immune responses. Science. 2011;334(6061):1401–1404. doi: 10.1126/science.1211641. [DOI] [PubMed] [Google Scholar]
- 48.Iakovidis M, et al. Effector-triggered immune response in Arabidopsis thaliana is a quantitative trait. Genetics. 2016;204(1):337–353. doi: 10.1534/genetics.116.190678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Coll NS, Epple P, Dangl JL. Programmed cell death in the plant immune system. Cell Death Differ. 2011;18(8):1247–1256. doi: 10.1038/cdd.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meyers BC, Morgante M, Michelmore RW. TIR-X and TIR-NBS proteins: Two new families related to disease resistance TIR-NBS-LRR proteins encoded in Arabidopsis and other plant genomes. Plant J. 2002;32(1):77–92. doi: 10.1046/j.1365-313x.2002.01404.x. [DOI] [PubMed] [Google Scholar]
- 51.Zhao T, et al. A truncated NLR protein, TIR-NBS2, is required for activated defense responses in the exo70B1 mutant. PLoS Genet. 2015;11(1):e1004945. doi: 10.1371/journal.pgen.1004945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mackey D, Belkhadir Y, Alonso JM, Ecker JR, Dangl JL. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell. 2003;112(3):379–389. doi: 10.1016/s0092-8674(03)00040-0. [DOI] [PubMed] [Google Scholar]
- 53.Axtell MJ, Staskawicz BJ. Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell. 2003;112(3):369–377. doi: 10.1016/s0092-8674(03)00036-9. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Y, et al. Temperature-dependent autoimmunity mediated by chs1 requires its neighboring TNL gene SOC3. New Phytol. 2016;213(3):1330–1345. doi: 10.1111/nph.14216. [DOI] [PubMed] [Google Scholar]
- 55.Zhang XC, Gassmann W. Alternative splicing and mRNA levels of the disease resistance gene RPS4 are induced during defense responses. Plant Physiol. 2007;145(4):1577–1587. doi: 10.1104/pp.107.108720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dinesh-Kumar SP, Baker BJ. Alternatively spliced N resistance gene transcripts: Their possible role in tobacco mosaic virus resistance. Proc Natl Acad Sci USA. 2000;97(4):1908–1913. doi: 10.1073/pnas.020367497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 58.Chiu J, March PE, Lee R, Tillett D. Site-directed, Ligase-Independent Mutagenesis (SLIM): A single-tube methodology approaching 100% efficiency in 4 h. Nucleic Acids Res. 2004;32(21):e174. doi: 10.1093/nar/gnh172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nakagawa T, et al. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng. 2007;104(1):34–41. doi: 10.1263/jbb.104.34. [DOI] [PubMed] [Google Scholar]
- 60.Morel JB, Dangl JL. Suppressors of the arabidopsis lsd5 cell death mutation identify genes involved in regulating disease resistance responses. Genetics. 1999;151(1):305–319. doi: 10.1093/genetics/151.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10(6):845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peterson BA, et al. Genome-wide assessment of efficiency and specificity in CRISPR/Cas9 mediated multiple site targeting in Arabidopsis. PLoS One. 2016;11(9):e0162169. doi: 10.1371/journal.pone.0162169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McPherson A. Preparation and Analysis of Protein Crystals. Wiley; New York: 1982. p. vii, 371 pp. [Google Scholar]
- 64.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 67.Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003;374:300–321. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- 68.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Davis IW, et al. MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35(Web Server issue):W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]