Significance
Organisms evolve and adapt via changes in their genomes that improve survival and reproduction in the context of their environment. Few experiments have examined how these genomic signatures of adaptation, which may favor mutations in certain genes or molecular pathways, vary across a set of similar environments that have both shared and distinctive characteristics. We sequenced complete genomes from 30 Escherichia coli lineages that evolved for 2,000 generations in one of five environments that differed only in the temperatures they experienced. Particular “signature” genes acquired mutations in these bacteria in response to selection imposed by specific temperature treatments. Thus, it is sometimes possible to predict aspects of the environment recently experienced by microbial populations from changes in their genome sequences.
Keywords: experimental evolution, genome evolution, mutation, natural selection, temperature
Abstract
Isolated populations derived from a common ancestor are expected to diverge genetically and phenotypically as they adapt to different local environments. To examine this process, 30 populations of Escherichia coli were evolved for 2,000 generations, with six in each of five different thermal regimes: constant 20 °C, 32 °C, 37 °C, 42 °C, and daily alternations between 32 °C and 42 °C. Here, we sequenced the genomes of one endpoint clone from each population to test whether the history of adaptation in different thermal regimes was evident at the genomic level. The evolved strains had accumulated ∼5.3 mutations, on average, and exhibited distinct signatures of adaptation to the different environments. On average, two strains that evolved under the same regime exhibited ∼17% overlap in which genes were mutated, whereas pairs that evolved under different conditions shared only ∼4%. For example, all six strains evolved at 32 °C had mutations in nadR, whereas none of the other 24 strains did. However, a population evolved at 37 °C for an additional 18,000 generations eventually accumulated mutations in the signature genes strongly associated with adaptation to the other temperature regimes. Two mutations that arose in one temperature treatment tended to be beneficial when tested in the others, although less so than in the regime in which they evolved. These findings demonstrate that genomic signatures of adaptation can be highly specific, even with respect to subtle environmental differences, but that this imprint may become obscured over longer timescales as populations continue to change and adapt to the shared features of their environments.
The origin and diversification of species often occurs when isolated populations evolve and accumulate genetic and phenotypic differences as they adapt to distinctive aspects of their local environments (1, 2). However, if populations derived from the same ancestral population are isolated but experience the same or similar environments, they might or might not diverge from one another. On the one hand, stochastic processes including mutation and drift tend to promote divergence. On the other hand, natural selection often produces similar adaptations to similar environments. Such parallel phenotypic evolution may sometimes arise from repeated changes in the same genes or genetic networks (3). As a result of these and other opposing forces favoring divergence and convergence, it is unclear to what extent environmental differences and commonalities will be reflected in evolution at the genomic level. It is possible that the history of genetic changes in a lineage might be used to predict retrospectively key features of its past environments. Alternatively, this type of hindcasting (i.e., retrospective prediction) might be precluded by divergent responses of populations that experienced the same environment, parallel adaptations to shared features of different environments, or both.
Temperature is a ubiquitous factor that shapes the geographic distributions of species (4), as organisms can survive and reproduce only in environments within a restricted thermal range. Adaptation for improved thermotolerance has important implications for plant growth (5) and crop productivity (6), especially in light of climate change (7). Directed evolution of microorganisms, such as yeast, to function effectively at higher or lower temperatures is used to make industrial processes, including alcohol fermentation and biofuel production, more efficient (8, 9). Adaptation to new thermal niches has also been implicated in the emergence of some fungal pathogens, which must acquire the ability to replicate at the elevated temperatures within warm-blooded organisms (e.g., humans) to pose a health risk (10).
To examine the processes governing evolutionary convergence and divergence, 30 populations of Escherichia coli were propagated in the laboratory for 2,000 generations, six each in five different temperature regimes (11–13). These environments included optimal (constant 37 °C) and moderate (constant 32 °C) temperatures well within the thermal range of the ancestral strain. They also included temperatures at the low (constant 20 °C) and high (constant 42 °C) ends of its thermal range, defined by the ability of the bacteria to sustain a population in the face of 100-fold daily dilutions, as well as a treatment in which the bacteria alternated between moderate and stressful temperatures (designated “Switch” with alternating days at 32 °C and 42 °C). These populations evolved increased competitive fitness in the thermal regime that they experienced during the experiment (11, 12). In most cases, their fitness also increased in the other environments, although this was not always the case. For example, the populations evolved at 42 °C exhibited increased fitness, on average, at both 37 °C and 32 °C (11), but the populations evolved at constant 20 °C, 32 °C, or 37 °C were less fit than the ancestor at 42 °C (11, 12).
Here, we sequenced and analyzed the complete genomes of one endpoint clone isolated from each temperature-evolved population to examine the genetic specificity of evolution. We found characteristic signatures of divergence driven by adaptation to the thermal regimes. Mutations in specific genes were strongly associated with each of the four constant-temperature treatments, and even some uncommon mutations in other genes were significantly biased with respect to the selective environment. We also constructed pairs of strains that differ only by the presence or absence of mutations in two genes that showed temperature-specific evolution. Competitive fitness assays indicate that these mutations are broadly beneficial under multiple temperature regimes, but more so under the particular conditions in which they evolved. Moreover, during a longer evolution experiment at the optimal temperature (37 °C), mutations in the signature genes that were associated with different temperature treatments were eventually incorporated into the winning lineage after thousands to tens of thousands of generations of further evolution. Thus, it appears that changing the thermal regime did not radically alter the pathways by which these populations adapted, which depended not only on temperature but also on all of the other environmental factors that were kept constant across the treatments. Rather, our results indicate that each thermal regime modulated the benefits of a common set of mutations, such that mutations in different genes dominated the initial stages of adaptation.
Results
Retention of Genetic Markers Characteristic of the Progenitor.
The 30 populations in the 2000-generation temperature-evolution experiment (TEE) were founded from a progenitor strain of E. coli, designated REL1206 (Fig. 1A). REL1206 was isolated from a population that had previously evolved for 2,000 generations at 37 °C in the same glucose-limited medium and other culture conditions as the TEE as part of the long-term evolution experiment (LTEE); the LTEE has now been running for over 50,000 total generations in this environment (14–16). During the 2,000 generations of evolution in the LTEE, REL1206 accumulated seven mutations relative to its progenitor, strain REL606. The complete genome sequence of REL606 has been determined (17), along with its derivation from the E. coli B strain used by Luria and Delbrück (18) and its genomic similarities and differences relative to other B derivatives and to E. coli K-12 (19). The seven LTEE-derived mutations in REL1206 include point mutations in spoT, topA, and yeiB; a 1-bp insertion upstream of glmU; insertion sequence IS150 elements interrupting fimA and pykF; and an IS150-associated deletion of ∼7 kb in the rbs operon.
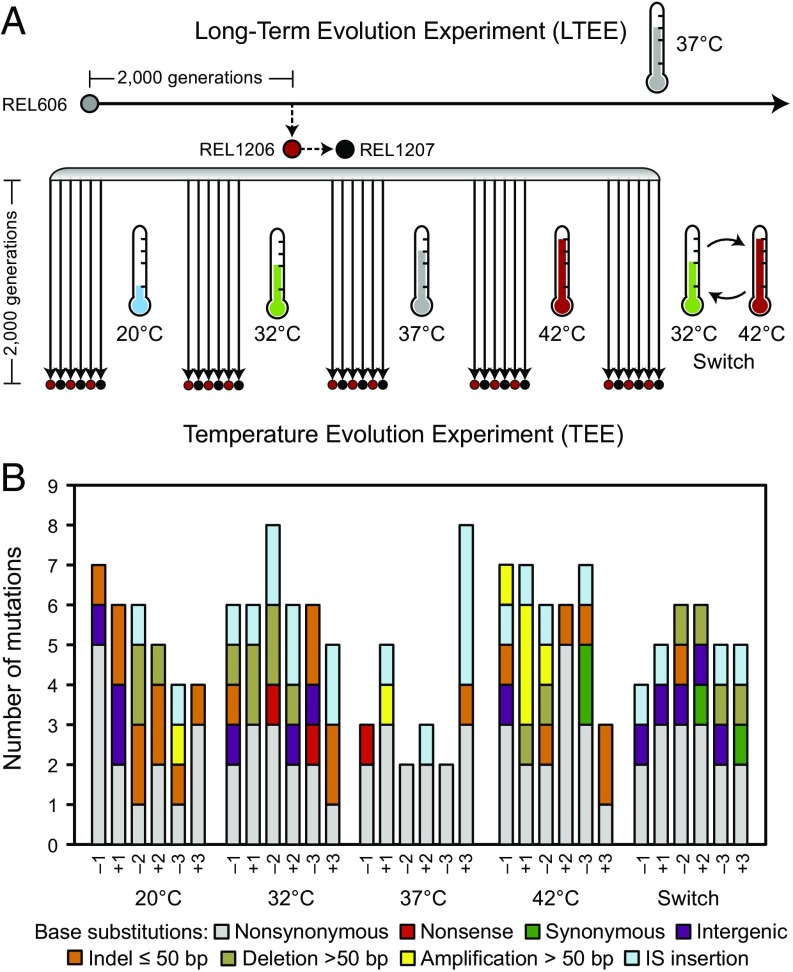
Fig. 1.
Genomic changes in E. coli evolved under different temperature regimes. (A) Overview of the TEE. E. coli clone REL1206 was isolated from the LTEE after 2,000 generations of evolution at 37 °C. In the TEE, six populations evolved for 2,000 generations in each of five different temperature treatments. Three populations in each treatment started from Ara– clone REL1206 (designated −1, −2, −3) and three from Ara+ clone REL1207 (+1, +2, +3), which differs from REL1206 by a single point mutation that is neutral with respect to fitness. (B) Summary of the 159 derived mutations observed in the 30 sequenced TEE genomes by the type of genetic change.
All 30 sequenced TEE clones retained all of these mutational differences relative to the REL606 sequence. Half of the 30 TEE populations (three in each treatment) were founded directly from REL1206, whereas the other half were founded from REL1207. REL1206 cannot grow on arabinose (Ara−), whereas REL1207 is an Ara+ mutant of REL1206 that was obtained by plating numerous cells on minimal-arabinose agar (14). Clones derived from REL1207 share only one additional mutation relative to REL1206, a nonsynonymous base substitution in the araA gene. The arabinose phenotype served as a readily identifiable marker to distinguish ancestral and derived bacteria during competition assays that were performed to measure changes in fitness (20). There was strict alternation between the Ara− and Ara+ populations during the daily serial transfers in the evolution experiment to maximize the likelihood that inadvertent cross-contamination would be detected as a change in the marker. As expected, all 15 TEE clones derived from REL1207 (and none derived from REL1206) have the same point mutation in codon 92 of the araA gene, changing it from encoding aspartic acid (GAC) to alanine (GCC). The retention of all these markers confirms that the 30 TEE clones evolved without outside contamination or cross-contamination that would compromise the genomic analyses.
Rates of Genomic Evolution in Different Thermal Regimes.
The genomes of the 30 TEE clones harbor a grand total of 159 new mutations, with the number ranging from a low of two in two clones to a high of eight in two others (Fig. 1B). Under the hypothesis that many of these mutations underlie adaptations to altered thermal conditions, one would expect fewer changes in the clones that evolved at 37 °C because that group had continued in the same thermal regime that the common progenitor, REL1206, had already evolved in for the preceding 2,000 generations. Indeed, competition assays performed in an earlier study indicate that the 37 °C populations, as a group, showed significantly smaller average fitness gains than did the other four groups (20). We did find that the six 37 °C lines accumulated significantly fewer mutations, an average of 3.8 per sequenced clone, compared with the other groups, which averaged 5.7 mutations per clone (P = 0.0157, one-tailed Mann–Whitney test). The difference between the average number of mutations in the 37 °C treatment and any of other four groups was not significant, however, when they were considered alone (all four P > 0.05, two-tailed Mann–Whitney tests). The number of mutations also did not differ significantly among the four other thermal treatments (P = 0.2259, Kruskal–Wallis test).
Of all mutations in the TEE clones, 57.2% (91/159) are single-base substitutions, of which 86.8% (79/91) occur in protein-coding sequences (Fig. 1B). Only four of the 79 base substitutions in protein-coding regions are synonymous, whereas the others are nonsynonymous or nonsense substitutions that change the corresponding amino acid sequences. This proportion of synonymous mutations is significantly less than the 24.9% expected by chance, given the distribution of different base changes observed, under the null hypothesis that nonsynonymous and synonymous substitutions are equally likely to persist (P = 3.1 × 10−6, one-tailed binomial test). Thus, there is a strong overall signal of adaptive evolution across all temperature regimes.
The other 42.8% (68/159) of the mutations in the TEE lines are insertions, deletions, and rearrangements (Fig. 1B). Of these, 32.4% (22/68) are small insertions or deletions (indels) of ≤50 bp. Of the single-base indels (n = 10), many are at potentially hypermutable sites on the boundaries of insertion sequence (IS) elements (n = 5). Of the indels of 4–50 bp (n = 6), all three of the insertions create tandem duplications of existing single-copy sequences, and one of the three deletions removes one of two ancestral copies of a four-base tandem repeat. Large duplications (n = 7), ranging in size from 129 to 60,769 bp, account for 10.3% of the mutations. Another 35.2% are new insertions of IS elements, including IS150 (n = 15), IS1 (n = 4), IS186 (n = 2), IS3 (n = 2), and ISRso11 (n = 1). The remaining 22.1% of mutations are large deletions (n = 15) of 84–32,853 bp, most of which were mediated by IS elements (n = 10) or by other recently duplicated genes or repeat sequences in the ancestral genome (n = 3).
A previous study used pulsed field gel electrophoresis (PFGE) to detect large-scale chromosomal changes—defined as those altering restriction enzyme fragment sizes by ≥10 kb—in 24 of the 30 TEE clones (all except those from the 20 °C treatment) (21). We recovered all seven of the duplications and deletions found by the PFGE analysis. We also identified a previously unreported 60-kb amplification. However, short-read DNA sequencing data are unable to detect certain types of chromosomal rearrangements, particularly inversions involving long repeat sequences (22). These limitations are unlikely to meaningfully impact our conclusions regarding genome evolution, as only 1 of the 24 TEE genomes analyzed by PFGE, a clone from the 42 °C treatment designated 42+1 (REL2042), was found to have an inversion (21).
Together the large deletions removed a sum total of 198,893 bp from 12 different strains across every regime except the 37 °C treatment. More of these deletions were in the lines that only experienced lower temperatures, with 9 of the 15 deletions clustered into the 20 °C and 32 °C treatments, whereas the remaining six were found in either the 42 °C or Switch treatments. Conversely, the seven large amplifications added a total of 171,144 bp, with five in the 42 °C treatment and only one in the 20 °C and 32 °C treatments. Overall, the average net change in genome size that resulted from all mutations was +25,240 bp for the six clones from the 42 °C treatment and −8,886 bp for the 12 clones from the 20 °C and 32 °C treatments. This difference, biased in the direction of larger evolved genome sizes at the higher temperature compared with the two lower temperatures, was marginally significant (P = 0.0311, two-tailed Mann–Whitney test).
Hypermutability has been shown to commonly arise in laboratory evolution experiments, because it improves the chances of an asexual lineage sampling beneficial mutations (16, 23). As many as 3 of the 30 TEE lines may have evolved hypermutable phenotypes via previously reported means, but none of them shows an excess of total mutations that would substantially affect our analysis. The clone from the 42 °C treatment designated 42–3 (REL2050) has an IS150 element inserted in the dinI gene, which encodes an SOS-induced protein that, when defective, causes moderately increased mutability (24). Interestingly, this same clone also harbors two of the four synonymous mutations observed in this study, although it only has seven total mutations. The rate of accumulation of neutral mutations should be proportional to the underlying mutation rate (25), and previous studies have confirmed the predicted increase in synonymous substitutions (most of which are likely to be neutral) in experimental populations that have evolved mutator phenotypes (16). The clone 32+1 (REL2038) from the 32 °C treatment has a nonsynonymous mutation in mutM, a DNA glycosylase important for base excision repair of oxidized guanine (26); however, it has no synonymous mutations or any apparent excess of other mutations relative to other lineages, as it has only six mutations in total. Finally, the clone 42−1 (REL2047) from the 42 °C treatment has a nonsynonymous mutation in rmuC (yigN). Mutations in this gene have been reported to increase the rates of inversions and other chromosomal rearrangements (27). Although there was a 37,530-bp amplification in 42−1, two of the other five 42 °C genomes each had more large chromosomal rearrangements than did 42−1, suggesting that the rmuC mutation did not increase the rate at which these mutations accumulated.
Specificity of Genomic Evolution with Respect to Thermal Regime.
We next compared the similarity of mutations between replicate lines within the same treatment and across different treatments to evaluate the specificity of genomic evolution with respect to the thermal regime. For these calculations, we included only those mutations that unambiguously impact a single gene; these “qualifying” mutations include nonsynonymous point mutations and also smaller deletions, duplications, and insertions (including IS element insertions). We excluded synonymous mutations as well as deletions and amplifications that overlapped multiple genes for this analysis. Mutations occurring within intergenic regions were grouped with the nearest flanking gene so long as the mutation was upstream and within 150 bp of the gene start. A total of 132 mutations qualify based on these criteria.
We computed Dice’s Coefficient of Similarity, S, for each pair of evolved clones, where S = 2 |X ∩ Y|/(|X| + |Y|). Here, |X| and |Y| are the sets of genes that have qualifying mutations in two clones, and |X ∩ Y| is the set that harbors mutations in both clones. This coefficient thus ranges from 0, when the two clones have no qualifying mutated genes in common, to 1, when both clones have qualifying mutations in exactly the same set of genes.
Across all 30 evolved clones, the grand mean similarity, Sm, is 0.065. The mean within-treatment similarity, Sw, for clone pairs from the same thermal treatment is 0.168, whereas the mean between-treatment similarity, Sb, for clones that evolved under different regimes was only 0.043. Thus, two clones that independently evolved under the same thermal treatment had almost 17% of their mutated genes in common, whereas those that evolved under different temperature regimes shared <5% of their mutated genes.
We performed a randomization test to evaluate whether this difference between Sw and Sb was significant. Each clone was used in 29 pair-wise calculations of similarity. There are a total of 435 (30 × 29/2) values of S for all combinations of 30 clones, but those values are not independent. Instead, each population is the appropriate unit of independent observation. The randomization test therefore scrambled the clones associated with the five treatments but did not alter the number or identity of mutations in any clone. We performed one million randomizations, and for each one we calculated the difference between Sw and Sb. None of these trials produced a value as high as or higher than the observed difference. Therefore, the higher genomic similarity within rather than between evolutionary treatments is highly significant (P < 10−6).
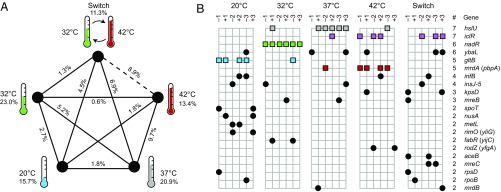
We next sought to determine whether this genomic signature extended to most or all of the treatment pairs or to only a few of them. To that end, we repeated the same calculations and randomization test as above except now on each of the 10 pairs of treatments separately. To account for the fact that each treatment was involved in four significance tests, we applied the sequential Bonferroni correction (28). Almost all treatment pairs show significantly different genomic changes (Fig. 2A). Interestingly, the only exception is the Switch treatment, which evolved under daily alterations between 32 °C and 42 °C. The genes that accumulated mutations in the Switch lines were not significantly different from those of the constant 42 °C lines.
Fig. 2.
Mutations in several signature genes are associated with evolution under different temperature regimes. (A) Temperature treatments and edges connecting them are labeled with Dice similarity scores that represent the average percentage of mutated genes in common among all genome pairs within each treatment (Sw) and the average across all genome pairs between two treatments (Sb), respectively. Only the 132 qualifying mutations that affect a single gene were included in these calculations. The difference between Sb and Sw, indicating a temperature-specific pattern of genomic evolution, is significant (P < 0.05, randomization test with sequential Bonferroni correction) for all treatment pairs except the combination of the Switch (alternating 32 °C and 42 °C) and 42 °C regimes (dashed line). (B) Genes (rows) affected by at least two qualifying mutations in the 30 TEE clones (columns, organized by temperature regime and populations –1, +1, –2, +2, –3, or +3). The five signature mutations, shown as colored squares, are significantly associated with one or two treatments (see Gene Targets That Contribute to Evolved Thermal Specificity). All other mutations are depicted as black circles.
Gene Targets That Contribute to Evolved Thermal Specificity.
The previous section established that genomic evolution was highly specific with respect to the temperature regime under which the 30 populations evolved, but it did not identify the genes that contributed significantly to that specificity. Here, we perform statistical tests to identify those genes that show an excess of mutational changes under any of the five selective regimes. For the 10 genes that had at least three qualifying mutations (Fig. 2B), we calculated the probability that the mutations would be concentrated, by chance alone, in any particular treatment to the same or greater degree than observed. This conservative approach is similar to the Fisher’s Exact Test comparing the mutations among the lines in one treatment against all of the other lines, except that the first within-treatment mutation is excluded from the calculation because it must fall in some treatment and we had no a priori expectation about which treatment that might be. For example, consider nadR, a gene in which 6 of the 30 evolved strains acquired mutations with all of them in the 32 °C treatment. The probability that the second through sixth mutations would all appear in the same treatment as the first is given by (5/29) × (4/28) × (3/27) × (2/26) × (1/25) = 8.4 × 10−6.
Mutations in a total of four genes show significant associations with a particular evolutionary treatment after performing a Benjamini–Hochberg correction for multiple testing at a false discovery rate of 5% (Fig. 2B). The gene gltB had mutations in four of the six 20 °C evolved clones, along with one 32 °C clone (adjusted P = 0.0258, two-tailed Fisher’s Exact Test excluding the first within-treatment mutation, as described above). This gene encodes the large subunit of the glutamate synthase enzyme, which is involved in nitrogen metabolism. As noted above, nadR acquired mutations in all six clones that evolved at 32 °C and none in any of the other treatments (adjusted P = 8.421 × 10−5). The nadR gene encodes a bifunctional protein that has both regulatory and enzymatic activities that affect the import and synthesis of nicotinamide adenine dinucleotide (NAD) (29). The hslU gene harbors mutations in five clones that evolved at 37 °C as well as in one line at 32 °C and another at 42 °C (adjusted P = 0.0148). This gene encodes the ATPase component of the HslVU protease complex involved in the degradation of misfolded proteins. HslU has been shown to have a role in regulating cell size (30), and previous studies have documented that the LTEE populations have consistently evolved larger cells under the same conditions as those experienced by these 37 °C lines (15). Four of the six lines that evolved at 42 °C acquired mutations in mrdA (pbpA), whereas one 37 °C line also did so (adjusted P = 0.0258). The mrdA gene encodes penicillin-binding protein 2, a transpeptidase involved in peptidoglycan synthesis, and mutations in this gene have also been shown to affect cell size and shape (31).
Thus, each of the evolutionary treatments—except the Switch regime that experienced alternating temperatures—has a “signature” gene that harbors a disproportionate number of mutations in the six lines that evolved at that temperature. Other genes might also contribute to the genomic specificity of adaptation, but they are not associated with any one treatment to such a degree that they are individually significant. Most mutations in the six other genes with three or more qualifying mutations are spread across different treatments, with the notable exception of the seven mutations in the gene iclR, which encodes a transcriptional repressor of the bypass enzymes in the glyoxylate cycle (32). Six of these mutations are divided equally between the 42 °C and Switch treatments. Using the same logic as above, this association with these two conditions, which both included growth at 42 °C, is significant (P = 0.0410, two-tailed Fisher’s Exact Test omitting the first mutations within each of these two groups).
Furthermore, it is very suggestive that in 8 of the 11 cases in which exactly two clones have mutations in the same gene, the two clones with the mutations evolved under the same thermal regime (Fig. 2B). One would expect that correspondence in only ∼17% (5/29) of such cases, and this difference between observed and expected frequencies is highly significant (P = 7.3 × 10−5, two-tailed binomial test). Therefore, many or most of the genes in which two mutations arose in the same evolutionary treatment may also be temperature-specific targets of selection, although the confidence associated with any particular case is low.
To examine more generally the contribution of all qualifying mutations besides those in the four genes associated with a single temperature treatment, we removed those four genes and repeated the full randomization test presented in the previous section (in which all 30 clones were scrambled between the five treatments). The observed difference in Dice’s Coefficient of Similarity computed within and between evolutionary treatments was 0.025, which was still marginally significant (P = 0.0257). This test thus supports the inference that mutations in other genes, besides the four that are individually significant, contribute to the genomic specificity of thermal adaptation.
All of these tests to identify genetic targets of thermal selection were focused on specific genes (and they used only those mutations that unambiguously impact a given gene). These tests, although clear and explicit, might overlook other associations at the level of metabolic pathways and cellular functions. The following relationships between thermal treatments and impacted functions, although more difficult to quantify statistically, are worth highlighting as well.
Eight genes involved in aspects of cell-wall structure and peptidoglycan synthesis and turnover—mrcB, mrdAB, mreBCD, rodZ (yfgA), and anmK (ydhH)—accumulated a total of 17 qualifying mutations. There are several striking features of these data. First, only two of these mutations occur in the 12 populations that evolved at either constant 20 °C or 32 °C, whereas 15 are found among the 18 populations that encountered higher temperatures, including the Switch treatment. This difference is significant (P = 0.0229 two-tailed binomial test); although the breakpoint between the low and high temperature groups is ad hoc, there are only a few possibilities, and so the significance is unlikely to be spurious. Second, the 15 mutations at higher temperatures are spread among 14 different clones; only the clone Sw+2 (REL2055) from the Switch treatment has two of these mutations. In comparison with the expectation based on the Poisson distribution for zero, one, and two or more mutations, the observed uniformity is highly significant (P = 0.0060, two-tailed χ2 test). This uniformity suggests that additional mutations in any of these genes provide smaller fitness gains than the initial mutation. Third, as we have shown, mutations in mrdA are a signature of adaptation to constant 42 °C, with four of the six clones that evolved under this regime having mutations in that gene. The two 42 °C evolved lines that do not have mutations in mrdA each carry a new mutation in rodZ (yfgA). This complementary distribution may indicate some functional interaction between their gene products, as has been discovered for other genes in other evolution experiments (33, 34). Consistent with that possibility, bacterial two-hybrid assays using RodZ constructs show evidence of a physical association with MrdA, as well as with MreB, MreC, and MreD (35).
Although the genes involved in cell-wall synthesis and structure are more prevalent at high temperatures, other functions appear to have been the most important targets of selection at low temperatures. It is worth emphasizing that the populations that evolved at 20 °C were near their thermal limit, below which the cells could not grow fast enough to offset the 100-fold dilution that was imposed every day (11, 12, 20). A disproportionate number of the qualifying mutations observed at lower temperatures involve key aspects of transcription and translation including 11 such mutations in the clones that evolved at 20 °C and four more in the 32 °C clones, with only eight such mutations combined in the treatment groups that evolved at 37 °C, 42 °C, or alternating 32 °C and 42 °C. This difference is again significant (P = 0.0180, two-tailed binomial test), using the same breakpoint between low and high temperature groups as used for the cell-wall functional category. These genes include cspC, nusA, rho, rpoB, rpoC, and rpoS in the category of core transcription functions and deaD, infB, relA, rpsA, rpsD, spoT, rlmH (ybeA), rimO (yliG), and rsmI (yraL) involved in translation, including ribosome structure and the stringent response regulon. Most of these genes have mutations in only one or two clones, and none were identified as a signature gene for any particular temperature regime. Most notable, perhaps, are three mutations in genes involved in the stringent response (36), two in spoT and one in relA, in three different lines that evolved at 20 °C.
Mutations in the TEE Recapitulate Changes That Were Subsequently Observed in the Same Lineage from Which the Progenitor Was Sampled.
The progenitor of the TEE lines was sampled at generation 2,000 from a population that was evolving at 37 °C as part of the LTEE. Other clones from that same LTEE population were previously sequenced as part of another study (37), including one taken at generation 5,000 (which was 3,000 generations after the REL1206 progenitor of the TEE was isolated from this population), one from generation 10,000, and one from generation 20,000 (referred to hereafter as 5K, 10K, and 20K). [This population evolved a hypermutable phenotype between 20,000 and 30,000 generations (16), and therefore, we do not include any later clones in the analysis that follows.] The sets of genes with mutations in these LTEE clones provide interesting comparisons for this study. The six TEE populations in the 37 °C treatment continued to evolve independently under the same conditions as the LTEE population, whereas the 24 TEE lines under the other thermal regimes faced different selection pressures. One might therefore expect the genomic changes in the TEE lines that evolved at 37 °C to be more similar to the mutations that subsequently arose in the LTEE population than would be the mutations seen in the TEE lines that experienced the other thermal regimes. On the other hand, all 30 TEE lines continued to experience the same glucose-limited medium and other culture conditions as the LTEE population, so one might also expect all of the evolved TEE and LTEE clones to share certain common signatures of genomic evolution.
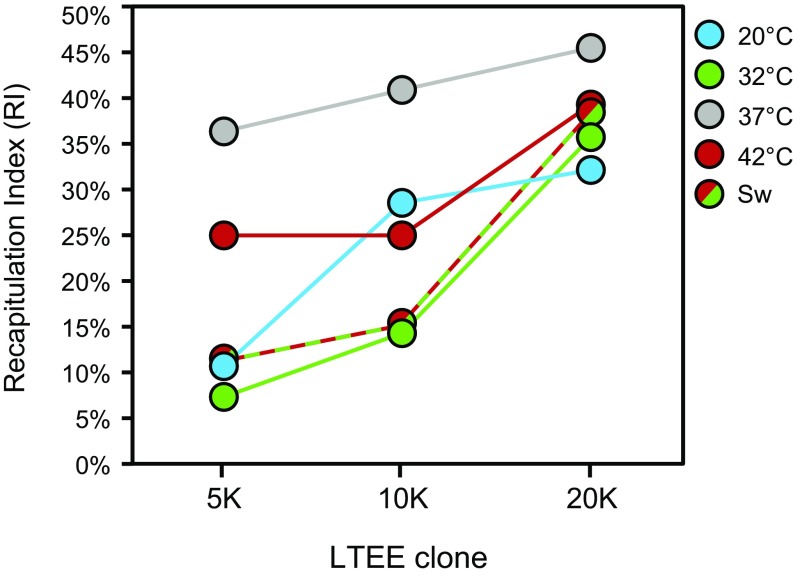
We measured the similarity of genomic evolution in the TEE to the LTEE by calculating a recapitulation index (RI) equal to the percentage of qualifying mutations in a particular thermal group that affected genes that subsequently acquired a qualifying mutation in an LTEE clone from the source population. For example, 10 of the 22 qualifying mutations in the 37 °C TEE group are found in hslU (five lines), ybaL (two lines), iclR, insJ-5, and mrdA, and all five of these genes also harbor qualifying mutations in the 20K clone in the LTEE population from which the progenitor was sampled. The 20K RI is therefore 45% (10/22) for the set of TEE lines that evolved at 37 °C. To count as qualifying mutations for this analysis, we further required genomic changes in each of the LTEE clones to be on the main line of descent within this population (37). Mutations failing this criterion could be considered to have arisen “before their time” because they were outcompeted by lineages with other beneficial mutations. The only LTEE mutation found in the TEE thus excluded was at 5K in the nadR gene. No nadR mutation was present in the 10K clone, and a different nadR mutation that later fixed in the population was present in the 20K clone.
As expected, the TEE clones that evolved at 37 °C have the highest RI at all three time points (Fig. 3). By one-tailed Fisher’s Exact Tests on the proportions of mutations shared with the LTEE, this overrepresentation in the 37 °C TEE treatment compared with all four other treatments is statistically significant at 5K (P = 0.0163) and 10K (P = 0.0463). At 20K, this difference is no longer significant (P = 0.2844), primarily because the RI in all of the other treatments has greatly increased and largely caught up to 37 °C. Strikingly, all of the signature genes with mutations that were significantly associated with specific TEE treatments were eventually mutated in the LTEE: hslU of the 37 °C treatment and mrdA of the 42 °C treatment by 5K, gltB of the 20 °C treatment by 10K, and nadR of the 32 °C treatment and iclR of both the 42 °C and Switch treatments by 20K. Summed over the four groups and 24 lines that evolved under thermal regimes different from the constant 37 °C experienced in the LTEE, 36% (40/110) of the total qualifying mutations were in genes that had also mutated in the source population by 20K. This number is impressively high given that the 20K LTEE clone had accumulated only 30 qualifying mutations in a genome that encodes over 4,000 proteins.
Fig. 3.
Genes that are targets of selection in the TEE often accumulated mutations later in the LTEE at 37 °C. For each TEE treatment, we calculated an RI equal to the percentage of mutations affecting single genes in those six genomes that also experienced mutations in clones from the same LTEE population that produced the TEE progenitor. We calculated RI values for comparisons to LTEE clones sampled at 5K, 10K, and 20K. The TEE began from a 2000-generation clone from this population, so these LTEE clones had evolved for 3,000, 8,000, and 18,000 additional generations at constant 37 °C. As expected, the 37 °C treatment had a significantly higher RI at 5K and 10K than the other four treatments combined (P < 0.05, one-tailed Fisher’s Exact Test). This difference was no longer significant at 20K. This convergence supports the hypothesis that many of the mutations that arose in the different temperature treatments, which all shared the same nutrient and other conditions with the LTEE, also improve fitness at 37 °C, although to a lesser degree, thus accounting for their later emergence at that ancestral temperature.
Furthermore, these RI values likely underrepresent the functional parallelism between these experiments, because they do not take into account cases in which mutations in different genes may affect the same molecular process. In addition to the genomic recapitulation observed for the TEE lines compared with the later evolution of the source LTEE population at 37 °C, it is also striking that four of the five treatment-specific signature genes we identified (Fig. 2B: nadR, hslU, mrdA, and iclR) were among the top nine genes identified as targets of selection based on parallel nonsynonymous substitutions across the 12 LTEE populations as a whole (16). Moreover, the TEE founding strain, REL1206, had already acquired mutations during the LTEE in three of those nine genes (Dataset S1: pykF, spoT, and topA). Therefore, four of the five treatment-specific signature genes we identified in the TEE lines were among the top six remaining targets of selection based on the LTEE.
Our overall interpretation of these results is that mutations in many of the genetic targets hit in the TEE are broadly beneficial, so long as certain aspects of the nutrient environment and culture conditions are preserved. However, each of the different temperature regimes focuses selection on different subsets of these genes, such that mutations in one or a few signature genes are the most beneficial under any particular treatment. As a result, these mutations dominate within the first 2,000 generations in the other temperature regimes, rather than accumulating only on much longer timescales of 10,000 to 20,000 or more generations of evolution at 37 °C.
Some Mutations Evolved in One Temperature Regime Are Also Beneficial in Others.
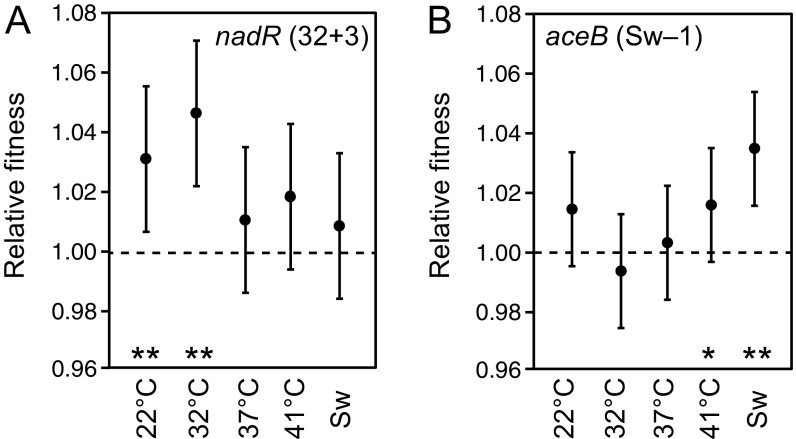
To test more directly the predictions of the recapitulation analysis, we measured the effects of two TEE mutations on the competitive fitness of E. coli in all five temperature treatments. The first was the 32 °C signature mutation in nadR from clone 32+3 (REL2042). We competed a version of the REL1206 strain with its genome edited to include only this new mutation against the REL1207 ancestor strain to measure its effect on fitness (Fig. 4A). This mutation causes a frameshift near the beginning of the nadR gene that presumably results in a nonfunctional protein. Three of the other five mutations observed in nadR (all in the 32 °C treatment) also result in disrupted reading frames via IS insertions or a 25-bp duplication.
Fig. 4.
Temperature specificity of beneficial mutations from two TEE populations. Two mutations that evolved during the TEE were moved into the ancestral genetic background. The resulting constructed strains then competed against a neutrally marked variant of the ancestor in each of five thermal regimes. Temperatures used for these competitions differed slightly from those used in the TEE (Materials and Methods). Error bars show 95% confidence intervals estimated from a one-way ANOVA. For treatments marked with one or two asterisks, the relative fitness of the strain with the evolved mutation was greater than one at P < 0.05 or P < 0.01, respectively (one-tailed t tests). (A) Results for the nadR mutation from the clone designated 32+3 (REL2042), which is a 2-bp deletion that causes a frameshift in the protein-coding sequence at amino acid 105. (B) Results for the aceB mutation from clone Sw−1 (REL2052), which is an A→C base change that causes a glutamate to alanine substitution at amino acid 69. This aceB mutation is expected to have an effect on cellular metabolism similar to that of the iclR mutations that were common in the 42 °C and Switch treatments (see Gene Targets That Contribute to Evolved Thermal Specificity).
The effect of the nadR mutation on fitness hardly varied with thermal regime (one-way ANOVA, F4, 25 = 1.765, P = 0.1676). As expected, the largest benefit was in the 32 °C treatment in which this mutation arose. This 4.6% increase and a smaller increase of 3.1% at 20 °C, the other low-temperature treatment, were both statistically significant (P = 0.0003 and P = 0.0068, respectively, one-tailed t tests). Although the fitness effect of the nadR mutation in the other three temperature treatments was not significant when tested individually, when combined they averaged a 1.3% fitness increase, which is marginally significant (P = 0.0347). Thus, the nadR mutation appears to be broadly beneficial, as suggested by the recapitulation analysis, which found one mutation in this gene in the 37 °C LTEE population that was off the line of descent at 5K and another mutation at 20K that eventually fixed.
The second mutation we tested was in the aceB gene of clone Sw−1 (REL2052). Although not a statistically significant signature gene, aceB mutations occur in two TEE lines that both evolved in the Switch regime where temperatures alternated between 32 °C and 42 °C. Furthermore, aceB mutations likely affect the same metabolic pathway as iclR, which accumulated mutations in three of the four other Switch lines, in three of the six 42 °C lines, and in one 37 °C line. The aceB gene encodes malate synthase, which catalyzes the second bypass step of the glyoxylate shunt, and iclR encodes a transcriptional repressor of the aceBAK operon (32). Mutations in iclR commonly arise in evolution experiments with E. coli that use glucose-limited medium because they improve the utilization of acetate, a transient byproduct of E. coli growth on glucose (38).
We found that the fitness effect of the aceB mutation also varied little across the thermal regimes (one-way ANOVA, F4, 25 = 2.757, P = 0.0501). It conferred the largest benefit of 3.5% in the Switch treatment, where it evolved. This increase was highly significant (P = 0.0005, one-tailed t test). The benefit of 1.6% measured at 41 °C was also marginally significant (P = 0.0484). No significant increase was found when combining all of the measurements obtained in the other three treatments (P = 0.2373). Thus, the aceB mutation showed qualitatively similar treatment-specific improvements in fitness to the nadR mutation, although its benefits were somewhat smaller. This pattern is consistent with the appearance of aceB mutations in only two of the TEE clones and the fact that no aceB mutation was observed in the source population that continued at 37 °C out to 20,000 generations in the LTEE.
Discussion
We found clear genomic signatures of divergent adaptation in 30 E. coli populations that evolved for 2,000 generations in the laboratory under several different thermal regimes. Only temperature was varied in this experiment, whereas the growth medium and other culture conditions were kept constant. Even so, there was a striking degree of genetic specificity in how these populations adapted, such that one could accurately predict which regime a clone came from by looking for mutations in only a few diagnostic signature genes.
However, despite these compelling differences, further evolution of the source population (from which the founding strain used in this study had been isolated) at constant 37 °C for an additional 18,000 generations resulted in that lineage incorporating mutations in all of the signature genes that were associated with the various constant-temperature regimes ranging from 20 °C to 42 °C. This pattern of recapitulation suggested that the mutations in the signature genes were broadly adaptive to the shared environmental conditions other than temperature but that the strength of selection acting on the various targets varied across the thermal regimes. Indeed, we showed that a nadR mutation from the 32 °C treatment and an aceB mutation from the Switch treatment (with daily alternations between 32 °C and 42 °C) were most beneficial under the thermal regimes in which they had evolved, but both were also beneficial in some other regimes and neither was deleterious under any regime. Taken together, these results might suggest that there were no tradeoffs in performance across temperatures (i.e., a net loss of fitness relative to the ancestor, when measured in a different environment) or at least none caused by the mutations that most often contributed to adaptation to each condition.
We also know from previous work that these populations were generally most fit in the thermal environment in which they evolved and that they exhibited a mix of positive and negative correlated responses to the other regimes (11, 12). For example, the populations that evolved at 42 °C were more fit, on average, than their ancestor at 37 °C and 32 °C, whereas populations that evolved at 20 °C, 32 °C, or 37 °C were less fit at 42 °C. Therefore, there are some tradeoffs, and some mutations must cause them. What then could account for the strong genetic signatures of adaptation to these different conditions? There are at least two possibilities, and they are not mutually exclusive. First, some mutations other than those we tested might be responsible for the tradeoffs when they do occur. The genes involved in cell-wall production, including mrdA (a signature gene for the 42 °C treatment), are interesting candidates in this respect. Mutations in these genes occur in 15 of the 18 TEE lines that faced moderate or high temperatures (the 37 °C, 42 °C, and Switch treatments), and they are common in the LTEE populations that evolved at 37 °C (16), but they are present in only 2 of the 12 TEE lines that evolved at constant low temperatures (20 °C and 32 °C). Second, a phenomenon called clonal interference occurs in evolving asexual populations because beneficial mutations that arise in different lineages in the same population cannot be brought together by recombination. As a consequence, lineages with highly beneficial mutations outcompete other lineages with less beneficial mutations, such that only the most beneficial mutations available typically can achieve fixation on the timescale of this experiment (39–41). For example, even though mutations in nadR may be beneficial under all of the temperature regimes, they are able to reach high frequency in competition against other mutations only in the 32 °C environment, whereas mutations in other genes will dominate the initial waves of adaptation at other temperatures.
Therefore, the overall impression is that the different temperature treatments made some adaptive pathways more beneficial and others less so or even maladaptive. On balance, it appears that many potential adaptive pathways are shared across the different temperature regimes. If so, and if the TEE was continued indefinitely, then the populations evolving under the different regimes might eventually show more genetic parallelism than divergence. By contrast, if the different temperature treatments promote radically different adaptive pathways, then we would expect increasing genetic divergence between the treatments over time, instead of the convergence to the 37 °C treatment indicated by the recapitulation analysis.
The convergence scenario requires that antagonistic pleiotropy not be so pervasive that most mutations that are adaptive in one environment are deleterious in other conditions (42, 43). However, as more mutations accumulate in any population, whether by selection, hitchhiking, or drift, there are more chances that a mutation that is deleterious in some other conditions will fix. For example, populations from the LTEE that evolved for 20,000 generations at 37 °C tended to have decreased growth rates at low (20 °C) and high (41–42 °C) temperatures (42). Our genetic results and fitness measurements do not rule out that some of the mutations in the clones evolved in one temperature regime reduced fitness in the other temperature regimes; instead, they imply that many of the most common mutations did not impose these tradeoffs. The loss of performance in alternative environments, where selection has not acted to maintain performance, is especially important in populations that evolve hypermutability because many mutations can accumulate that are neutral or nearly so in the current environment but where each has some chance of reducing fitness in other environments (44, 45). It is also possible that, had the TEE continued for as long as the LTEE, beneficial mutations in the other treatments would generate highly temperature-specific adaptations that preclude genetic convergence with the 37 °C lines.
A study by Tenaillon et al. (46) sequenced the genomes of clones from 115 populations that evolved for 2,000 generations at 42.2 °C, starting from the same REL1206 progenitor strain as used for the TEE that we have analyzed. These 115 populations are therefore similar in their selection history to the six populations in our 42 °C treatment. However, the genes that Tenaillon et al. (46) found to be most commonly mutated in their lines are not temperature-specific in our study. For example, mutations in the ybaL gene occurred in 65 of their lines and in 6 TEE lines spread across four temperature regimes, and mutations in rpoB occurred in 87 of their lines but in only 2 TEE lines from different treatments. The mrdA gene, on the other hand, shows more commonality between the two high-temperature experiments, being present in 23 lines from the study by Tenaillon et al. and in 4 of the 6 TEE lines that evolved at 42 °C (as well as in one that evolved at 37 °C).
That said, 42 °C is very near the thermal limit for sufficient growth of the ancestral strain to maintain itself given the daily dilutions in the minimal medium used in these experiments, and the relative fitness of competitors is extremely sensitive to small deviations in temperature (46). As a consequence, the most important targets of selection may also be highly sensitive to slight variations in experimental conditions at this temperature frontier, which could well explain the substantial differences between the genetic outcomes of the two experiments. Indeed, it was the awareness of this sensitivity, derived from studying the TEE populations, that led Tenaillon et al. (46) to propagate their populations at a precisely controlled 42.2 °C by incubating the cultures in a water bath, as opposed to an airflow incubator as was used for the TEE and this study.
More generally, studies of temperature adaptation using other E. coli strains and culture conditions have shown that mutations in genes affecting a wide array of cellular processes can improve fitness at elevated temperatures. For example, increased chaperonin expression (47), reduced glycerol transport (48), and altered RNA polymerase activity (46, 49) have been found in E. coli strains selected for increased growth at 42 °C or for extending the upper limit on growth to as high as 48.5 °C. Single mutations are often responsible for most of the improvements in any one evolved strain (48, 49), although this pattern would presumably not hold for experiments of longer duration. Some studies have found that E. coli evolved at stressfully high temperature tend to restore their overall gene expression profiles toward the baseline levels corresponding to their ancestor when it is grown the optimal growth temperature of 37 °C (49, 50). This finding fits with the overall prominence of mutations in global regulatory processes in laboratory evolution experiments with microbes (51), including the LTEE (52).
However, none of the signature genes that evolved many times in parallel in the TEE were directly involved in global regulatory processes. Instead, these genes are expected to affect cell size and shape (mrdA, hslU), to alter the regulation of specific pathways related to nutrient utilization (nadR, iclR), and to change the activity of a metabolic enzyme (gltB). We did find a significant concentration of mutations in genes related to transcription and translation but only when they were combined together as a group and then only in the low-temperature treatments. Overall, our results support the evidence from other studies that thermotolerance is a complex trait in E. coli, one that is sensitive to both the genetic background and precise growth conditions.
More generally, we found a clear signal of genomic specificity in how E. coli populations adapted to different temperature regimes. Even though the nutrient environment and all other aspects of the culture conditions were the same, mutations converged on a distinctive set of genes in each treatment. Whether this genomic signal would persist over longer evolutionary timescales is unclear. On the one hand, most of the divergence that we observed was eventually incorporated into the lineage that continued to evolve at 37 °C for another 18,000 generations, so these populations may become more genetically similar over time. On the other hand, it is also possible that if the TEE were continued for another 18,000 generations, mutations in other sets of genes would become diagnostic for each temperature regime as these populations became even better adapted to their particular conditions.
These results may have implications for other fields as well. In the context of microbial forensics, the prevalence of mutations in environment-specific signature genes suggests that even subtle differences in conditions may leave a genomic imprint that could be useful in ascertaining the provenance of a culture (53). Similarly, information about past environments may be found in genome-based phylogenies of microbial pathogens that track transmission and infection across patients. Indeed, evidence of particular selective processes, including the evolution of antibiotic resistance, has been found by examining genetic parallelism in these studies (54, 55), and there may well be additional genomic signals related to host physiology or pathogen persistence in the environment. In short, despite the multitude of diverse mutational pathways by which bacteria might in principle adapt to changing conditions, we observed a striking degree of predictability in how E. coli populations evolved in different thermal environments.
Materials and Methods
Evolution Experiments.
The TEE (11, 12) and LTEE (14, 16) have been fully described elsewhere. Table S1 lists the endpoint E. coli clones isolated from each TEE population at generation 2,000 that were sequenced in this study.
Genome Sequencing.
For each clonal isolate from the TEE, a sample stored frozen at –80 °C in ∼15% (vol/vol) glycerol was used to inoculate 10 mL of LB–Miller broth. After overnight growth at 37 °C, genomic DNA was purified from several milliliters of culture using Qiagen genomic-tip 100/G columns. DNA fragment libraries were prepared from these samples and sequenced on an Illumina Genome Analyzer IIx by the Michigan State University Research Technology Support facility. The resulting FASTQ files of 36 base reads are available from the NCBI Sequence Read Archive (accession no. SRP009429).
Mutation Prediction.
We used breseq (version 0.26.0) (56, 57) to identify mutations in evolved genomes by comparing Illumina reads to a version of the complete genome sequence of REL606 (17) with updated feature annotations (included in Dataset S1). As necessary, we manually curated breseq results to eliminate spurious predictions and to resolve predictions of sequence junctions and changes in read-coverage depth into structural mutations (57). To check mutation predictions, we applied the reported sequence changes to the ancestral genome sequence and reran breseq to verify that there were no further discrepancies between the reads and the mutated reference genome. Table S2 and the Genome Diff (56) files included in Dataset S1 provide full details about all mutations, including those present in the REL1206 and REL1207 ancestors of the TEE.
Statistical Methods.
Calculations of Dice similarity scores and the associated randomization tests to estimate the significance of differences in these scores were implemented in a custom Python script included in Dataset S1. All other statistical tests were performed in R (version 3.1.2) (58).
Fitness Assays.
We used the pKOV allelic replacement method to add the evolved nadR and aceA alleles to the Ara– TEE ancestral strain REL1206 (59). We then performed competition assays between each of these strains and the Ara+ ancestor REL1207 to measure their relative fitness. These assays were performed as previously described (20) with the following modifications. First, the competitions ran for two full 24-h cycles for all temperature treatments. Second, cultures were incubated at 22 °C (room temperature) for the 20 °C treatment and at 41 °C instead of 42 °C. The latter modification was necessary because strains did not reliably grow at the upper limit of their thermal range, as described previously (20, 46). For the Switch treatment, the preconditioning (i.e., acclimation) day was conducted at 32 °C, the first day of competition at 41 °C, and the second day of competition at 32 °C. We performed six replicate fitness assays for each mutant strain in each temperature treatment.
Supplementary Material
Acknowledgments
We thank Neerja Hajela and Rohan Maddamsetti for technical assistance and helpful discussions and acknowledge the Texas Advanced Computing Center (TACC) for providing high-performance computing resources. This research was supported in part by National Institutes of Health Grant R00-GM087550 (to J.E.B.), Cancer Prevention and Research Institute of Texas (CPRIT) Grant RP130124 (to J.E.B.), Army Research Office Grant W911NF-12-1-0390 (to J.E.B.), National Science Foundation (currently) Grants DEB-1451740 and DBI-0939454 (to R.E.L.), and Defense Advanced Research Projects Agency “Fun Bio” Program Grant HR0011-09-1-0055 (to R.E.L.). D.E.D. acknowledges a University of Texas at Austin CPRIT Research Traineeship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the NCBI Sequence Read Archive (accession no. SRP009429).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1616132114/-/DCSupplemental.
References
- 1.Mayr E. Populations, Species, and Evolution. Belknap; Cambridge, MA: 1970. [Google Scholar]
- 2.Grant PR, Grant BR. How and Why Species Multiply: The Radiation of Darwin’s Finches. Princeton Univ Press; Princeton, NJ: 2011. [Google Scholar]
- 3.Stern DL. The genetic causes of convergent evolution. Nat Rev Genet. 2013;14(11):751–764. doi: 10.1038/nrg3483. [DOI] [PubMed] [Google Scholar]
- 4.Merriam CH. Laws of temperature and control of the geographic distribution of terrestrial animals and plants. Natl Geogr Mag. 1894;6:229–238. [Google Scholar]
- 5.Fournier-Level A, et al. A map of local adaptation in Arabidopsis thaliana. Science. 2011;334(6052):86–89. doi: 10.1126/science.1209271. [DOI] [PubMed] [Google Scholar]
- 6.Boyer JS. Plant productivity and environment. Science. 1982;218(4571):443–448. doi: 10.1126/science.218.4571.443. [DOI] [PubMed] [Google Scholar]
- 7.Ciais P, et al. Europe-wide reduction in primary productivity caused by the heat and drought in 2003. Nature. 2005;437(7058):529–533. doi: 10.1038/nature03972. [DOI] [PubMed] [Google Scholar]
- 8.Caspeta L, Nielsen J. Thermotolerant yeast strains adapted by laboratory evolution show trade-off at ancestral temperatures and preadaptation to other stresses. MBio. 2015;6(4):e00431. doi: 10.1128/mBio.00431-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.López-Malo M, et al. Evolutionary engineering of a wine yeast strain revealed a key role of inositol and mannoprotein metabolism during low-temperature fermentation. BMC Genomics. 2015;16:537. doi: 10.1186/s12864-015-1755-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooney NM, Klein BS. Fungal adaptation to the mammalian host: It is a new world, after all. Curr Opin Microbiol. 2008;11(6):511–516. doi: 10.1016/j.mib.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bennett AF, Lenski RE, Mittler JE. Evolutionary adaptation to temperature. I. Fitness responses of Escherichia coli to changes in its thermal environment. Evolution (N Y) 1992;46:16–30. doi: 10.1111/j.1558-5646.1992.tb01981.x. [DOI] [PubMed] [Google Scholar]
- 12.Mongold JA, Bennett AF, Lenski RE. Evolutionary adaptation to temperature. IV. Adaptation of Escherichia coli at a niche boundary. Evolution (N Y) 1996;50:35–43. doi: 10.1111/j.1558-5646.1996.tb04470.x. [DOI] [PubMed] [Google Scholar]
- 13.Bennett AF, Dao KM, Lenski RE. Rapid evolution in response to high-temperature selection. Nature. 1990;346(6279):79–81. doi: 10.1038/346079a0. [DOI] [PubMed] [Google Scholar]
- 14.Lenski RE, Rose MR, Simpson SC, Tadler SC. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am Nat. 1991;138:1315–1341. [Google Scholar]
- 15.Lenski RE, Travisano M. Dynamics of adaptation and diversification: A 10,000-generation experiment with bacterial populations. Proc Natl Acad Sci USA. 1994;91(15):6808–6814. doi: 10.1073/pnas.91.15.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tenaillon O, et al. Tempo and mode of genome evolution in a 50,000-generation experiment. Nature. 2016;536(7615):165–170. doi: 10.1038/nature18959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeong H, et al. Genome sequences of Escherichia coli B strains REL606 and BL21(DE3) J Mol Biol. 2009;394(4):644–652. doi: 10.1016/j.jmb.2009.09.052. [DOI] [PubMed] [Google Scholar]
- 18.Daegelen P, Studier FW, Lenski RE, Cure S, Kim JF. Tracing ancestors and relatives of Escherichia coli B, and the derivation of B strains REL606 and BL21(DE3) J Mol Biol. 2009;394(4):634–643. doi: 10.1016/j.jmb.2009.09.022. [DOI] [PubMed] [Google Scholar]
- 19.Studier FW, Daegelen P, Lenski RE, Maslov S, Kim JF. Understanding the differences between genome sequences of Escherichia coli B strains REL606 and BL21(DE3) and comparison of the E. coli B and K-12 genomes. J Mol Biol. 2009;394(4):653–680. doi: 10.1016/j.jmb.2009.09.021. [DOI] [PubMed] [Google Scholar]
- 20.Bennett AF, Lenski RE. An experimental test of evolutionary trade-offs during temperature adaptation. Proc Natl Acad Sci USA. 2007;104(Suppl 1):8649–8654. doi: 10.1073/pnas.0702117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bergthorsson U, Ochman H. Chromosomal changes during experimental evolution in laboratory populations of Escherichia coli. J Bacteriol. 1999;181(4):1360–1363. doi: 10.1128/jb.181.4.1360-1363.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raeside C, et al. Large chromosomal rearrangements during a long-term evolution experiment with Escherichia coli. MBio. 2014;5(5):e01377–e14. doi: 10.1128/mBio.01377-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sniegowski PD, Gerrish PJ, Lenski RE. Evolution of high mutation rates in experimental populations of E. coli. Nature. 1997;387(6634):703–705. doi: 10.1038/42701. [DOI] [PubMed] [Google Scholar]
- 24.Yasuda T, Morimatsu K, Horii T, Nagata T, Ohmori H. Inhibition of Escherichia coli RecA coprotease activities by DinI. EMBO J. 1998;17(11):3207–3216. doi: 10.1093/emboj/17.11.3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kimura M. The Neutral Theory of Molecular Evolution. Cambridge Univ Press; New York: 1983. [Google Scholar]
- 26.Michaels ML, Pham L, Cruz C, Miller JH. MutM, a protein that prevents G.C----T.A transversions, is formamidopyrimidine-DNA glycosylase. Nucleic Acids Res. 1991;19(13):3629–3632. doi: 10.1093/nar/19.13.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slupska MM, et al. Genes involved in the determination of the rate of inversions at short inverted repeats. Genes Cells. 2000;5(6):425–437. doi: 10.1046/j.1365-2443.2000.00341.x. [DOI] [PubMed] [Google Scholar]
- 28.Holm S. A simple sequentially rejective multiple test procedure. Scand J Stat. 1979;6:65–70. [Google Scholar]
- 29.Raffaelli N, et al. The Escherichia coli NadR regulator is endowed with nicotinamide mononucleotide adenylyltransferase activity. J Bacteriol. 1999;181(17):5509–5511. doi: 10.1128/jb.181.17.5509-5511.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katayama T, Kubota T, Takata M, Akimitsu N, Sekimizu K. Disruption of the hslU gene, which encodes an ATPase subunit of the eukaryotic 26S proteasome homolog in Escherichia coli, suppresses the temperature-sensitive dnaA46 mutation. Biochem Biophys Res Commun. 1996;229(1):219–224. doi: 10.1006/bbrc.1996.1783. [DOI] [PubMed] [Google Scholar]
- 31.Philippe N, Pelosi L, Lenski RE, Schneider D. Evolution of penicillin-binding protein 2 concentration and cell shape during a long-term experiment with Escherichia coli. J Bacteriol. 2009;191(3):909–921. doi: 10.1128/JB.01419-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamamoto K, Ishihama A. Two different modes of transcription repression of the Escherichia coli acetate operon by IclR. Mol Microbiol. 2003;47(1):183–194. doi: 10.1046/j.1365-2958.2003.03287.x. [DOI] [PubMed] [Google Scholar]
- 33.Crozat E, et al. Parallel genetic and phenotypic evolution of DNA superhelicity in experimental populations of Escherichia coli. Mol Biol Evol. 2010;27(9):2113–2128. doi: 10.1093/molbev/msq099. [DOI] [PubMed] [Google Scholar]
- 34.Winkler JD, Garcia C, Olson M, Callaway E, Kao KC. Evolved osmotolerant Escherichia coli mutants frequently exhibit defective N-acetylglucosamine catabolism and point mutations in cell shape-regulating protein MreB. Appl Environ Microbiol. 2014;80(12):3729–3740. doi: 10.1128/AEM.00499-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bendezú FO, Hale CA, Bernhardt TG, de Boer PA. RodZ (YfgA) is required for proper assembly of the MreB actin cytoskeleton and cell shape in E. coli. EMBO J. 2009;28(3):193–204. doi: 10.1038/emboj.2008.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Philippe N, Crozat E, Lenski RE, Schneider D. Evolution of global regulatory networks during a long-term experiment with Escherichia coli. BioEssays. 2007;29(9):846–860. doi: 10.1002/bies.20629. [DOI] [PubMed] [Google Scholar]
- 37.Barrick JE, et al. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature. 2009;461(7268):1243–1247. doi: 10.1038/nature08480. [DOI] [PubMed] [Google Scholar]
- 38.Herron MD, Doebeli M. Parallel evolutionary dynamics of adaptive diversification in Escherichia coli. PLoS Biol. 2013;11(2):e1001490. doi: 10.1371/journal.pbio.1001490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gerrish PJ, Lenski RE. The fate of competing beneficial mutations in an asexual population. Genetica. 1998;102-103(1-6):127–144. [PubMed] [Google Scholar]
- 40.Levy SF, et al. Quantitative evolutionary dynamics using high-resolution lineage tracking. Nature. 2015;519(7542):181–186. doi: 10.1038/nature14279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Good BH, Rouzine IM, Balick DJ, Hallatschek O, Desai MM. Distribution of fixed beneficial mutations and the rate of adaptation in asexual populations. Proc Natl Acad Sci USA. 2012;109(13):4950–4955. doi: 10.1073/pnas.1119910109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cooper VS, Bennett AF, Lenski RE. Evolution of thermal dependence of growth rate of Escherichia coli populations during 20,000 generations in a constant environment. Evolution. 2001;55(5):889–896. doi: 10.1554/0014-3820(2001)055[0889:eotdog]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 43.Bull JJ, Badgett MR, Wichman HA. Big-benefit mutations in a bacteriophage inhibited with heat. Mol Biol Evol. 2000;17(6):942–950. doi: 10.1093/oxfordjournals.molbev.a026375. [DOI] [PubMed] [Google Scholar]
- 44.Cooper VS, Lenski RE. The population genetics of ecological specialization in evolving Escherichia coli populations. Nature. 2000;407(6805):736–739. doi: 10.1038/35037572. [DOI] [PubMed] [Google Scholar]
- 45.Leiby N, Harcombe WR, Marx CJ. Multiple long-term, experimentally-evolved populations of Escherichia coli acquire dependence upon citrate as an iron chelator for optimal growth on glucose. BMC Evol Biol. 2012;12:151. doi: 10.1186/1471-2148-12-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tenaillon O, et al. The molecular diversity of adaptive convergence. Science. 2012;335(6067):457–461. doi: 10.1126/science.1212986. [DOI] [PubMed] [Google Scholar]
- 47.Rudolph B, Gebendorfer KM, Buchner J, Winter J. Evolution of Escherichia coli for growth at high temperatures. J Biol Chem. 2010;285(25):19029–19034. doi: 10.1074/jbc.M110.103374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blaby IK, et al. Experimental evolution of a facultative thermophile from a mesophilic ancestor. Appl Environ Microbiol. 2012;78(1):144–155. doi: 10.1128/AEM.05773-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sandberg TE, et al. Evolution of Escherichia coli to 42 °C and subsequent genetic engineering reveals adaptive mechanisms and novel mutations. Mol Biol Evol. 2014;31(10):2647–2662. doi: 10.1093/molbev/msu209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodríguez-Verdugo A, Tenaillon O, Gaut BS. First-step mutations during adaptation restore the expression of hundreds of genes. Mol Biol Evol. 2016;33(1):25–39. doi: 10.1093/molbev/msv228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Conrad TM, Lewis NE, Palsson BO. Microbial laboratory evolution in the era of genome-scale science. Mol Syst Biol. 2011;7:509. doi: 10.1038/msb.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hindré T, Knibbe C, Beslon G, Schneider D. New insights into bacterial adaptation through in vivo and in silico experimental evolution. Nat Rev Microbiol. 2012;10(5):352–365. doi: 10.1038/nrmicro2750. [DOI] [PubMed] [Google Scholar]
- 53.Lenski RE, Keim P. Population genetics of bacteria in a forensic context. In: Breeze RG, Budowle B, Schutzer SE, editors. Microbial Forensics. Elsevier; San Diego: 2005. pp. 355–369. [Google Scholar]
- 54.Lieberman TD, et al. Genetic variation of a bacterial pathogen within individuals with cystic fibrosis provides a record of selective pressures. Nat Genet. 2014;46(1):82–87. doi: 10.1038/ng.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marvig RL, Sommer LM, Molin S, Johansen HK. Convergent evolution and adaptation of Pseudomonas aeruginosa within patients with cystic fibrosis. Nat Genet. 2015;47(1):57–64. doi: 10.1038/ng.3148. [DOI] [PubMed] [Google Scholar]
- 56.Deatherage DE, Barrick JE. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol Biol. 2014;1151:165–188. doi: 10.1007/978-1-4939-0554-6_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barrick JE, et al. Identifying structural variation in haploid microbial genomes from short-read resequencing data using breseq. BMC Genomics. 2014;15:1039. doi: 10.1186/1471-2164-15-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.R Core Team 2014 R: A Language and Environment for Statistical Computing. Available at: www.r-project.org/
- 59.Link AJ, Phillips D, Church GM. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: Application to open reading frame characterization. J Bacteriol. 1997;179(20):6228–6237. doi: 10.1128/jb.179.20.6228-6237.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.