Abstract
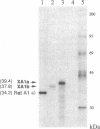
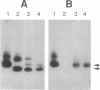
We have identified the Xenopus cognates for the RNA-binding protein A1. This protein has previously been shown to be one of the components of the heterogeneous nuclear ribonucleoprotein (hnRNP) complex in rat and human cells. We have isolated several Xenopus clones from oocyte, tailbud embryo, and leg muscle cDNA libraries and determined their nucleotide sequences. Potentially, two different A1 isoforms are expressed in Xenopus; they have been termed XA1a and XA1b. Besides insertions and deletions, the XA1a protein sequence is 92% identical to the rat protein and suggests very similar secondary structures. There are two segments in the COOH-terminal domain where deletions or insertions are apparent: the rat protein does not have a 48-residue sequence that is present in the frog protein, and the frog protein does not have a 12-residue sequence that is present in the rat protein. We have confirmed that the XA1a protein is larger than rat A1 by in vitro transcription, translation, and gel electrophoresis. The second isoform, XA1b, is very similar to the XA1a isoform, except it has a different COOH terminus due to the absence of a 73-nucleotide region from its cDNA clones. Transcripts representing both isoforms have been detected in various Xenopus RNA preparations by polymerase chain reaction experiments with A1-specific oligonucleotides. Our findings suggest that the isoforms are encoded by one or two genes and are the result of alternative splicing. We discuss the biological implications of having two forms of the A1 component of hnRNP particles.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adam S. A., Nakagawa T., Swanson M. S., Woodruff T. K., Dreyfuss G. mRNA polyadenylate-binding protein: gene isolation and sequencing and identification of a ribonucleoprotein consensus sequence. Mol Cell Biol. 1986 Aug;6(8):2932–2943. doi: 10.1128/mcb.6.8.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreadis A., Gallego M. E., Nadal-Ginard B. Generation of protein isoform diversity by alternative splicing: mechanistic and biological implications. Annu Rev Cell Biol. 1987;3:207–242. doi: 10.1146/annurev.cb.03.110187.001231. [DOI] [PubMed] [Google Scholar]
- Biamonti G., Buvoli M., Bassi M. T., Morandi C., Cobianchi F., Riva S. Isolation of an active gene encoding human hnRNP protein A1. Evidence for alternative splicing. J Mol Biol. 1989 Jun 5;207(3):491–503. doi: 10.1016/0022-2836(89)90459-2. [DOI] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Buvoli M., Biamonti G., Tsoulfas P., Bassi M. T., Ghetti A., Riva S., Morandi C. cDNA cloning of human hnRNP protein A1 reveals the existence of multiple mRNA isoforms. Nucleic Acids Res. 1988 May 11;16(9):3751–3770. doi: 10.1093/nar/16.9.3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Cobianchi F., Karpel R. L., Williams K. R., Notario V., Wilson S. H. Mammalian heterogeneous nuclear ribonucleoprotein complex protein A1. Large-scale overproduction in Escherichia coli and cooperative binding to single-stranded nucleic acids. J Biol Chem. 1988 Jan 15;263(2):1063–1071. [PubMed] [Google Scholar]
- Cobianchi F., SenGupta D. N., Zmudzka B. Z., Wilson S. H. Structure of rodent helix-destabilizing protein revealed by cDNA cloning. J Biol Chem. 1986 Mar 15;261(8):3536–3543. [PubMed] [Google Scholar]
- Crabtree G. R., Kant J. A. Organization of the rat gamma-fibrinogen gene: alternative mRNA splice patterns produce the gamma A and gamma B (gamma ') chains of fibrinogen. Cell. 1982 Nov;31(1):159–166. doi: 10.1016/0092-8674(82)90415-9. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G., Swanson M. S., Piñol-Roma S. Heterogeneous nuclear ribonucleoprotein particles and the pathway of mRNA formation. Trends Biochem Sci. 1988 Mar;13(3):86–91. doi: 10.1016/0968-0004(88)90046-1. [DOI] [PubMed] [Google Scholar]
- Frohman M. A., Dush M. K., Martin G. R. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier J., Osguthorpe D. J., Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978 Mar 25;120(1):97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- Haynes S. R., Rebbert M. L., Mozer B. A., Forquignon F., Dawid I. B. pen repeat sequences are GGN clusters and encode a glycine-rich domain in a Drosophila cDNA homologous to the rat helix destabilizing protein. Proc Natl Acad Sci U S A. 1987 Apr;84(7):1819–1823. doi: 10.1073/pnas.84.7.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Hopp T. P., Woods K. R. Prediction of protein antigenic determinants from amino acid sequences. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3824–3828. doi: 10.1073/pnas.78.6.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg P. A., Melton D. A. In vitro RNA synthesis with SP6 RNA polymerase. Methods Enzymol. 1987;155:397–415. doi: 10.1016/0076-6879(87)55027-3. [DOI] [PubMed] [Google Scholar]
- Lapeyre B., Bourbon H., Amalric F. Nucleolin, the major nucleolar protein of growing eukaryotic cells: an unusual protein structure revealed by the nucleotide sequence. Proc Natl Acad Sci U S A. 1987 Mar;84(6):1472–1476. doi: 10.1073/pnas.84.6.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leser G. P., Escara-Wilke J., Martin T. E. Monoclonal antibodies to heterogeneous nuclear RNA-protein complexes. The core proteins comprise a conserved group of related polypeptides. J Biol Chem. 1984 Feb 10;259(3):1827–1833. [PubMed] [Google Scholar]
- Merrill B. M., Stone K. L., Cobianchi F., Wilson S. H., Williams K. R. Phenylalanines that are conserved among several RNA-binding proteins form part of a nucleic acid-binding pocket in the A1 heterogeneous nuclear ribonucleoprotein. J Biol Chem. 1988 Mar 5;263(7):3307–3313. [PubMed] [Google Scholar]
- Piñol-Roma S., Choi Y. D., Matunis M. J., Dreyfuss G. Immunopurification of heterogeneous nuclear ribonucleoprotein particles reveals an assortment of RNA-binding proteins. Genes Dev. 1988 Feb;2(2):215–227. doi: 10.1101/gad.2.2.215. [DOI] [PubMed] [Google Scholar]
- Preugschat F., Wold B. Isolation and characterization of a Xenopus laevis C protein cDNA: structure and expression of a heterogeneous nuclear ribonucleoprotein core protein. Proc Natl Acad Sci U S A. 1988 Dec;85(24):9669–9673. doi: 10.1073/pnas.85.24.9669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebagliati M. R., Weeks D. L., Harvey R. P., Melton D. A. Identification and cloning of localized maternal RNAs from Xenopus eggs. Cell. 1985 Oct;42(3):769–777. doi: 10.1016/0092-8674(85)90273-9. [DOI] [PubMed] [Google Scholar]
- Riva S., Morandi C., Tsoulfas P., Pandolfo M., Biamonti G., Merrill B., Williams K. R., Multhaup G., Beyreuther K., Werr H. Mammalian single-stranded DNA binding protein UP I is derived from the hnRNP core protein A1. EMBO J. 1986 Sep;5(9):2267–2273. doi: 10.1002/j.1460-2075.1986.tb04494.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs A. B., Bond M. W., Kornberg R. D. A single gene from yeast for both nuclear and cytoplasmic polyadenylate-binding proteins: domain structure and expression. Cell. 1986 Jun 20;45(6):827–835. doi: 10.1016/0092-8674(86)90557-x. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson M. S., Nakagawa T. Y., LeVan K., Dreyfuss G. Primary structure of human nuclear ribonucleoprotein particle C proteins: conservation of sequence and domain structures in heterogeneous nuclear RNA, mRNA, and pre-rRNA-binding proteins. Mol Cell Biol. 1987 May;7(5):1731–1739. doi: 10.1128/mcb.7.5.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk H. E., Werr H., Friedrich D., Kiltz H. H., Schäfer K. P. The core proteins of 35S hnRNP complexes. Characterization of nine different species. Eur J Biochem. 1985 Jan 2;146(1):71–81. doi: 10.1111/j.1432-1033.1985.tb08621.x. [DOI] [PubMed] [Google Scholar]