Significance
Cell content analysis has rapidly become one of the most important new tools for measuring cell phenotype and behavior. However, the central limitation of current sampling technologies is they are destructive and must lyse the cells to measure the contents. This destruction prevents knowledge of prior or future states of the cell, which is particularly important for dynamic cell processes, such as development and differentiation. Here, we show a nondestructive longitudinal sampling and analysis platform that can sample repeatedly and accurately from the same single cell or group of cells over a long time period. We demonstrate sampling of both proteins and mRNA for cell lines as well as human-derived cardiomyocytes and astrocytes.
Keywords: sampling, nanotechnology, molecular biology, cellular biology
Abstract
Here, we report a method for time-resolved, longitudinal extraction and quantitative measurement of intracellular proteins and mRNA from a variety of cell types. Cytosolic contents were repeatedly sampled from the same cell or population of cells for more than 5 d through a cell-culture substrate, incorporating hollow 150-nm-diameter nanostraws (NS) within a defined sampling region. Once extracted, the cellular contents were analyzed with conventional methods, including fluorescence, enzymatic assays (ELISA), and quantitative real-time PCR. This process was nondestructive with >95% cell viability after sampling, enabling long-term analysis. It is important to note that the measured quantities from the cell extract were found to constitute a statistically significant representation of the actual contents within the cells. Of 48 mRNA sequences analyzed from a population of cardiomyocytes derived from human induced pluripotent stem cells (hiPSC-CMs), 41 were accurately quantified. The NS platform samples from a select subpopulation of cells within a larger culture, allowing native cell-to-cell contact and communication even during vigorous activity such as cardiomyocyte beating. This platform was applied both to cell lines and to primary cells, including CHO cells, hiPSC-CMs, and human astrocytes derived in 3D cortical spheroids. By tracking the same cell or group of cells over time, this method offers an avenue to understand dynamic cell behavior, including processes such as induced pluripotency and differentiation.
Quantitative analyses of intracellular proteins and mRNA provide crucial information to decipher cellular behavior related to disease pathogenesis (1), cellular senescence (2), development and differentiation (3). Increasingly sensitive and even single-cell mRNA and protein detection methods have been developed, leading to new insights into cell function, phenotype heterogeneity, and noise in cellular systems (4, 5). Although powerful, these methods are hampered by the need to lyse the cell to extract the intracellular contents, providing only a single snapshot in time without information about prior or future states. This limitation is particularly problematic when studying dynamic transformations, including induced pluripotency (3) and differentiation (6), or stochastic noise in gene expression (7–9) at the single-cell level. Phenotype heterogeneity and fluctuations in single cells imply that cells in parallel cultures often are not representative, highlighting the need for nondestructive sampling from the same set of cells repeatedly over time.
Time-resolved, longitudinal monitoring (sampling the same population periodically) has been possible to some extent with intracellular fluorescence techniques. Genetically encoded fluorescent protein (FP)-based biosensors are a useful tool to follow intracellular enzymatic activity, spatiotemporal localization, and dynamics of FP-fused proteins nondestructively at a subcellular level (10). Although monitoring two to five species in living cells is possible with FRET biosensors (11) and bimolecular fluorescence complementation (BiFC) (12, 13), the number of intracellular targets is still limited because of spectral overlap (14). The presence of the FP label also may interfere with the function of the fused protein, and validating the specificity of the sensor is crucial. Further, the transfection of the FP gene is itself an intrusive process (12, 13). Genetically encoded biosensors, such as quantum dot-labeled antibodies (15–17) and molecular beacons (18), are also used for intracellular detection but are challenging to deliver intracellularly, and perturbation of the cell caused by the labeling methods and the presence of the label is still a significant concern (15). Overall, even with the availability of FP methods, longitudinal studies are relatively rare.
Nanotechnology provides an alternative approach by taking advantage of nanoscale dimensions to introduce sensors into cells or to extract small quantities of cellular contents nondestructively. Several groups have demonstrated the insertion of high aspect-ratio nanostructures for molecular delivery with minimal cell perturbation, suggesting that nanomaterials may pass through the cell membrane without dire consequences (19–24). Extending this approach, Na et al. (25) showed a nanowire “sandwich assay” in which nanowires approximately 100 nm in diameter functionalized with antibodies penetrate the cell with limited toxicity and can bind specific enzymes for extraction. Actis et al. (26) demonstrated a “nanobiopsy” that can extract less than 50 fL from the cytoplasm compartments of a single cell without cell cytotoxicity and with a success rate of approximately 70%.
Recently, the nano-sampling approach was extended using an atomic force microscopy-based sampling platform with controlled picoliter volume extraction, followed by a single-time point mRNA analysis (27). This significant advance was found to be largely nondestructive, with 86% cell viability, demonstrating that cells may lose a fraction of their volume without apoptosis. Intracellular protein sampling was also possible using magnetized carbon nanotubes coated with poly-l-tyrosine to extract GFP from a cell culture, with better than 70% cell viability (28). These promising results indicate insertion and sampling at a single time-point is possible. However, none of these approaches repeatedly sampled from the same set of cells to follow their expression over time or provided quantitative assessment of the measured quantities compared with the actual intracellular contents. Here, we developed the nanostraw extraction (NEX) process to sample a small quantity of intracellular proteins and mRNA nondestructively and periodically from the same single cell or cells over an extended period.
Results
The NEX Process.
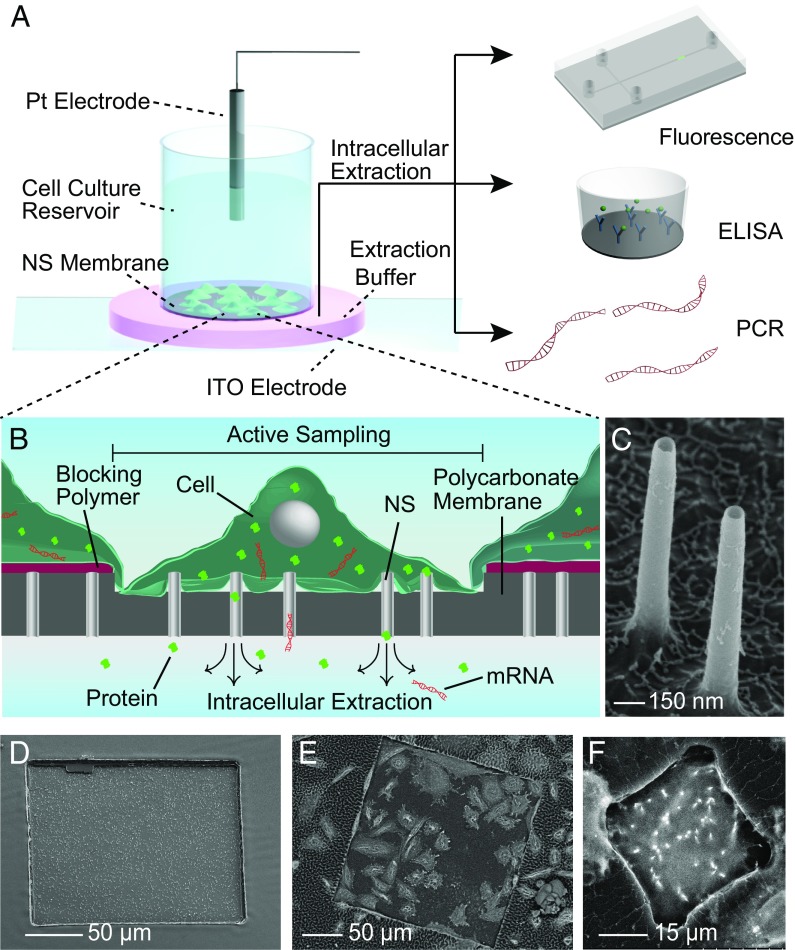
The NEX process is based on diffusively sampling material from inside the cell using a nanostraw (NS)-embedded substrate. Cells of interest are cultured on a polymer membrane with a defined region of hollow NS extending through the polymer and protruding from the surface (Fig. 1) (23, 29, 30) Cells grow normally over the entire polymer membrane, so that cells within the sampling region interact with surrounding cells, avoiding cell isolation. Intracellular samples are collected by applying an electrical voltage through the NS, locally opening small holes in the cell membrane near the NS tip. During the subsequent 2- to 5-min interval when these pores are open (29, 31, 32), approximately 5–10% of the proteins, mRNA, and small molecules diffuse from cells, through the NS, and into an extraction solution below the culture well (Fig. 1 A and B). The identity of the extraction buffer was found not to be important as long as it was reasonably osmolality-matched to the cell; typically a 1× PBS solution was used. After 10 min for diffusion, the extraction buffer is pipetted from beneath the culture well and analyzed conventionally, including fluorescence, mRNA detection, or ELISA. The cell-culture well then is returned to an incubator until a new sample is required.
Fig. 1.
Design and operation of the NEX sampling system. (A) The system consists of a polymer membrane with protruding 150-nm diameter NS attached to the bottom of a cell-culture dish. Sampling is performed by temporarily electroporating the cells cultured on the NS, allowing cellular content to diffuse through the NS and into the underlying extraction buffer (pink). An aliquot of the buffer then is aspirated with a standard pipette and analyzed conventionally, using fluorescence imaging, ELISA, or qPCR. (B) During sampling, intracellular species within the cell diffuse through the NS and into the extraction buffer below the membrane. The size of the sampling region can be defined lithographically so that only the cells that grow in the active regions are sampled. (C and D) Tilted view (45°) SEM images of the 150-nm-diameter NS (C) and the 200 × 200 µm active sampling region (D). Cells outside this window are unaffected by the sampling process. (E and F) SEM images of cells cultured on) a 200 × 200 µm active sampling region containing 42 cells (E) and a 30 × 30 µm sampling region used to isolate and sample from a single cell (F).
Here, we present the NEX process and evaluate its ability to extract and analyze protein and mRNA contents both statically and longitudinally for several cell types. We find that the system is nondestructive and provides quantitatively useful information about intracellular contents for mRNA sequences and proteins. Notably, the method had >95% cell viability that enabled multiple, real-time sampling over extended time periods and was well tolerated over 20 d by astrocytes derived from human induced pluripotent stem cells (hiPSCs). Equally important is that the sampling process extracted species throughout the cell, providing a comprehensive view of expression rather than a single-site extraction location. The system was less accurate for some, but not all, larger nucleic acid molecules (>16,000 nt), likely reflecting these molecules’ slower diffusion and limited cytosolic accessibility. NEX sampling was successful even for single cells, although the small quantities of material extracted at this level restricted applicable analytical methods, such as commercial ELISA and PCR. With continued improvement of single-cell assays, this limitation may soon be overcome. Overall, the NEX process appears to be a straightforward method to follow temporal dynamics of cellular protein and mRNA contents nondestructively over time.
Platform Design and Operation.
The NEX platform is based on a polycarbonate membrane with 150-nm-diameter inorganic NS extending through the polymer and protruding 1–3 microns above the surface (Fig. 1 A and B). This NS membrane is mounted on the bottom of a glass cylinder 2–5 mm in diameter that fits into a 48- or 96-well plate for cell culture. Nanostraw membranes were fabricated as previously described (refs. 23, 29; also Methods), producing flat polycarbonate membranes with the NS extending from the surface (Fig. 1 C and D). The NS heights could be controlled by the amount of polycarbonate etched from the surface. Specific cell-sampling regions were defined by blocking the remainder of NS membrane with photolithography-patterned polymers (Fig. S1). During cell culture, only the cells that grow in the selected regions with exposed NS will be sampled, leaving cells in the blocked area unaffected (Fig. 1 D and E). The size of the sampling window can be adjusted from <1 μm to millimeters on a side, allowing scalable sampling from a single cell to 105 cells while maintaining cell-to-cell connectivity and communication (Fig. 1 E and F).
Fig. S1.
Patterned NS platform fabrication workflow. (A) Patterned NS platforms were fabricated starting with track-etched polycarbonate membranes with 150-nm-diameter pores and a pore density of 1 × 108 pores/cm2 (GVS). (B) A 10-nm film of aluminum oxide was coated on all the nanoporous surface of the polycarbonate membranes by Savannah ALD (Cambridge Nanotech) using trimethyl aluminum and water as precursors. A pulse form of a-b-c was used for deposition, where a is the precursor exposure time (0.025 s), b is time for the precursor to remain in the ALD chamber (30 s), and c is the N2 purge time (30 s). (C) A 5-µm-thick positive photoresist film (MEGAPOSIT SPR2203 i-Line photoresist) was spin coated on the top surface of the ALD-coated polycarbonate membrane using a spinning speed of 3,500 rpm for 60 s. The photoresist-coated membrane was baked at 95 °C for 2 min to evaporate resist solvent. (D) The photoresist-coated membrane was exposed to a square pattern of intense UV light for 5 s and then was developed by immersion into the developer (MF-26A) for 60 s. These two steps create the sampling region. (E) After photolithography, the aluminum oxide surface in the microwell was etched away by Plasma Quest RIE with a fast flow composition of 40 sccm BCl3, 30 sccm Cl2, and 5 sccm Ar at 300 W at room temperature, leaving a polycarbonate surface inside the microwell. During the RIE, the unexposed photoresist served as a protection layer. Because the photoresist is much thicker than the alumina layer, the alumina layer inside the microwell can be fully removed without affecting the overall structure of microwells. (F) Oxygen RIE with 30 sccm O2 at 300 W was used to etch the polymer selectively until the desirable NS height was obtained. The etching rate of alumina from oxygen RIE is slower than the etching rate of the polymer; thus the etching of alumina is negligible. On the other hand, the etching rates of the photoresist and polycarbonate are almost the same, so to make NS 1 µm tall, 1 µm of photoresist also will be etched way. The photoresist was thick enough to allow the fabrication of 3-µm nanostraws inside the microwell without completely removing the photoresist during etching. Thus, the photoresist can still act as the blocking layer to cover the pores outside the microwell.
Cells grown on the NS demonstrate normal cell behavior and mRNA expression (23, 29, 33). Previously, we found that NS with a diameter of 100 nm or smaller spontaneously penetrate the cell membrane, allowing the delivery of small molecules into cells. Larger NS (150 nm and above) instead are engulfed by the cell membrane without causing membrane rupture (23). However, access to the cytoplasm can still be gained by applying short electric pulses (10–35 V) to open temporarily small pores on the cell membrane at the NS–cell interface (29). The cell membrane recovers 2–5 min after the pulses (29, 31, 32), and the cells evolve unperturbed. To prevent systemic cytosolic leakage during our experiments, we chose to use 150-nm-diameter NS and to use the electrical pulsing as a “valve” to gate sampling (Methods). We therefore can choose when cells release contents through the NS while maintaining their membrane integrity throughout the remaining culture period.
Real-Time, Longitudinal Sampling from Cell Subpopulations and Single Cells.
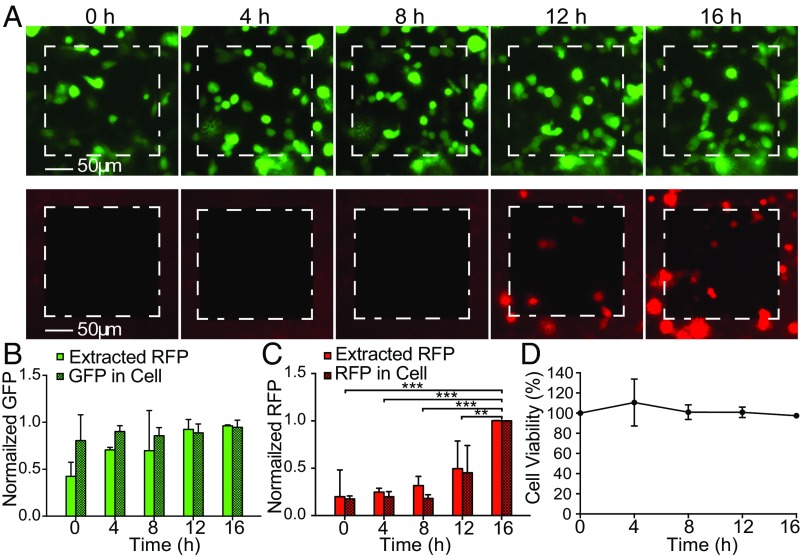
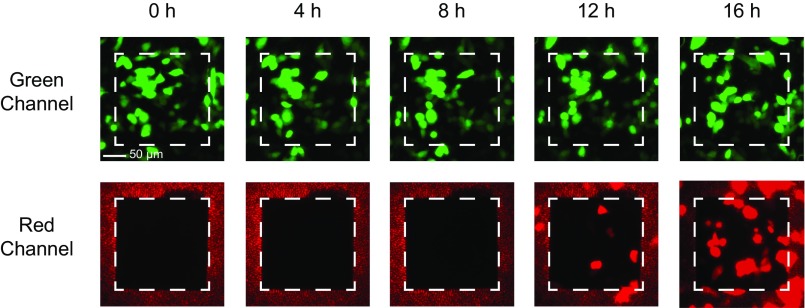
We evaluated the NEX sampling process for quantitative analysis of intracellular protein concentrations within the same set of cells over time. We first chose to compare the NEX-extracted GFP fluorescence with the GFP fluorescence of the sampled cells using microscopy. GFP-expressing CHO cells were cultured on the NS membrane with an active area of 200 × 200 µm mounted on a 2-mm glass cylinder (Methods). Cells were sampled every 4 h for 16 h total (five time points; Fig. 2). Dynamic changes in expression were examined by lipofectamine transfection with a plasmid containing RFP at the 8-h time point, the expression of which became observable at 12 and 16 h. At each sampling point, the NS well was removed from the incubator and was washed with PBS to remove possible contaminants, and the GFP and RFP intensity of the cells on the NS window was measured with fluorescent microscopy (Fig. 2A). A series of short electrical pulses was applied for 20 s, opening small holes in the cell membrane at the NS tips (29), and the cellular proteins were allowed to diffuse through the NS and into the extraction buffer for 10 min. The NS well then was returned to the incubator, and the amount of GFP/RFP in the extraction buffer was analyzed with fluorescence using isotachophoresis (ITP) (34) to concentrate the proteins selectively (Methods) (Fig. S2). Normal cell morphology was observed throughout the experiment, and cell viability was >95% per sampling on average (Fig. 2D), indicating that the cells were healthy during and after the sampling process. Experiments on sister cultures (Fig. S3) did not show qualitative differences.
Fig. 2.
Longitudinal sampling of GFP/RFP from the same subpopulation of CHO cells. (A) Fluorescent microscopy images of GFP (green channel) and RFP (red channel) of a culture of 38 cells on a 200 × 200 µm NS sampling region (white dashed squares). Images were taken every 4 h just before the NEX sampling process was performed. RFP transfection was performed after the 8-h time point. (B) Normalized cellular GFP contents from fluorescence microscopy compared with the NEX-extracted GFP quantities. No statistically significant difference was observed between the relative extracted GFP level and relative intracellular GFP expression level from cells at each time point. Error bars indicate the SD of the underlying signal (n = 2, P > 0.05 for both factors; two-way ANOVA). (C) Normalized RFP fluorescence intensity compared with the extracted quantities. A small background signal was present before RFP transfection, but the clear increase in extracted RFP correlated with the actual increase in RFP expression. Error bars denote SD (n = 2, P < 0.05 between time points; P > 0.05 for extracted to fluorescence; **P < 0.01; ***P < 0.001; post hoc Tukey test). (D) Cell viability as a function of time (sampling points). Cultures showed >95% viability immediately after sampling and >100% over time as the cells divided.
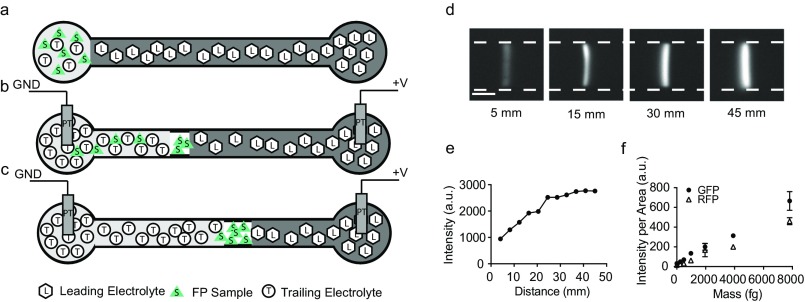
Fig. S2.
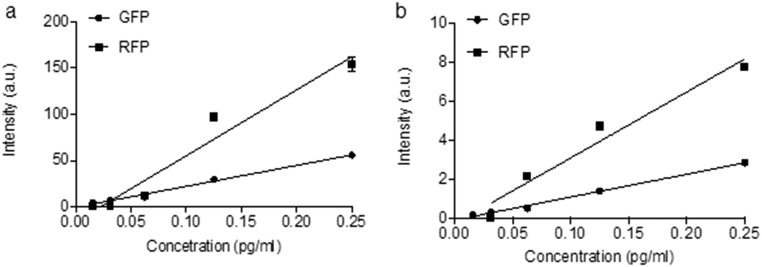
ITP preconcentration schematic and calibration. We used ITP to concentrate GFP and RFP 10,000 times (43, 44). ITP is a form of electrophoresis that focuses the target molecules in a focusing zone between two electrolytes (the LE and TE) with different ionic mobility. After the complete formation of the ITP focusing zone, total FP mass can be quantified by measuring the fluorescent intensity of the ITP zone images obtained with a fluorescence microscope and a CCD camera. (A–C) Assay workflow. (A) Three to ten microliters of LE were injected at the LE reservoir and filled the entire channel (20 μm × 50 μm × 53 mm). One to five microliters of the sample and TE mixture were injected in the TE reservoir. (B) DC (1,100 V) was applied between the two platinum electrodes (Pt) placed at the TE and LE reservoirs. GND, ground. (C) Two minutes after the formation of the ITP zone, the fluorescence intensity of the ITP zone was stabilized, indicating complete focusing of all GFP molecules in the microfluidic chamber. (D) Time-resolved images showing the formation of ITP zone (the white dashed line indicates channel edges) at increasing distances in the microfluidic channel as indicated in the figure. (E) GFP intensity in the ITP zone over time (increasing distance). The fluorescent intensity value at each distance was extracted by averaging the intensity of ITP zone. (F) GFP and RFP standard curves were generated based on the measured intensity (n = 3) from GFP and RFP standards (Abcam) diluted in the TE buffer. a.u., arbitrary units.
Fig. S3.
Longitudinal sampling of GFP/RFP from the same subpopulation of GFP-expressing CHO cells (second dataset). Fluorescent microscopy images of GFP (green channel) and RFP (red channel) of a culture of 48 cells on a 200 × 200 µm NS sampling region (white dashed squares). Images were obtained every 4 h just before the NS sampling process was performed. RFP expression was observed starting at the 12-h time point, 4 h after RFP transfection using lipofectamine.
Fig. 2B shows the quantitative comparison of the cells’ GFP fluorescence by microscopy and the NS-extracted GFP/RFP intensities of the 38 cells in the active NS region. The measurements were normalized to the highest value in each run to account for the different number of cells present and were averaged to provide SDs. The mean GFP expression level in the sampled cells did not show a significant change, as expected for a stably expressing protein. The NEX-extracted GFP followed this trend accurately. The relative NEX-measured GFP levels did not show significant statistical difference with the GFP expression level in cells at any of the five time points (P > 0.05 for both time and extracted vs. fluorescence comparison; two-way ANOVA). However, in most NEX experiments, the extracted GFP signal was significantly lower at the first time point, suggesting that the initial extraction is less efficient and that a pre-electroporation process might be needed to active the NEX system. Thus, although not rising to the level of statistical deviation, the initial data point usually should be discarded; however, we show all samples in this work.
The NEX also can follow temporal dynamics, namely the change in RFP as the cells begin to express RFP fluorescent proteins after transfection (Fig. 2C). Extracted RFP levels were equal to the background fluorescence for the first three time points and then increased quickly at 3 and 4 d, in agreement with microscopy images (P < 0.001; two-way ANOVA). No significant difference was observed between NEX-extracted amounts and the fluorescence imaging (P > 0.05; two-way ANOVA). Thus the sampling process also could measure dynamic changes in cell expression over time.
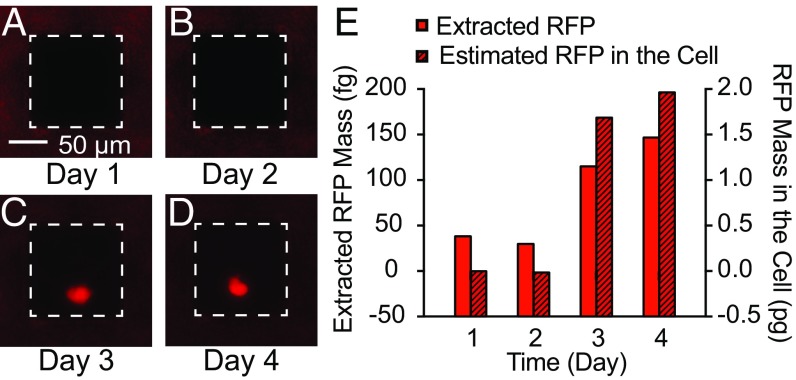
Encouraged by the results on this subpopulation of 38 cells, the active NS area was reduced to 100 × 100 µm to sample a single cell (Fig. 3). The cell was sampled once a day for a 4-d period; RFP contents were analyzed using ITP (Fig. 3 A–D) and were compared with fluorescence microscopy images (Fig. 3E). After sampling at day 2, the cells were transfected with an RFP plasmid using lipofectamine. One cell in the NS active area fluoresced on day 3 and intensified on day 4.
Fig. 3.
Longitudinal sampling of RFP from a single CHO cell. (A–D) Fluorescent microscopy images of a single RFP-expressing cell on a 100 × 100 µm sampling region (white dashed squares). RFP transfection was performed after the day 2 sample point. (E) Calibrated RFP quantities in the cell fluorescence compared with the extracted quantities. The extracted RFP amounts followed the increase in RFP expression within the cell.
The absolute quantity of RFP in both cells and the ITP focus zone was determined by calibrated analyses of the fluorescence intensity. Calibration curves using known concentrations of RFP/GFP in microfluidic channels were measured and showed a linear relationship between concentration and fluorescence intensity (Methods and Fig. S4). Fluorophore quantities then were calculated by accounting for the volume of the channel or cell and the integrated fluorescence intensity per unit area. Fig. 3E shows the calibrated mass of cellular and extracted RFP from a single cell. The extracted RFP expression trend and the actual cell concentration were in good quantitative agreement relative to their initial baselines. The total RFP mass inside the cell was 1.7 pg and 2.0 pg at day 3 and 4, respectively, compared with 120 fg and 150 fg for the extracted RFP at those sampling points. These values correspond to an extraction yield of 7% and 8% of the total cellular RFP at the third and fourth sampling points, respectively.
Fig. S4.
GFP/RFP concentration and intensity calibration curves. GFP and RFP solutions with concentrations of 0.25, 0.13, 0.07, 0.04, and 0.02 pg/mL were injected into a glass microfluidic chamber 10 µm wide and 12 µm deep (PerkinElmer). Microscopic fluorescence images of the GFP/RFP-filled microfluidic chamber were obtained using exposure times of 200 ms (A) and 25 ms (B). The fluorescence intensity of the GFP/RFP solutions was measured by Image J (n = 3; error bars show SD).
Spatial Distribution and Efficiency of Sampling.
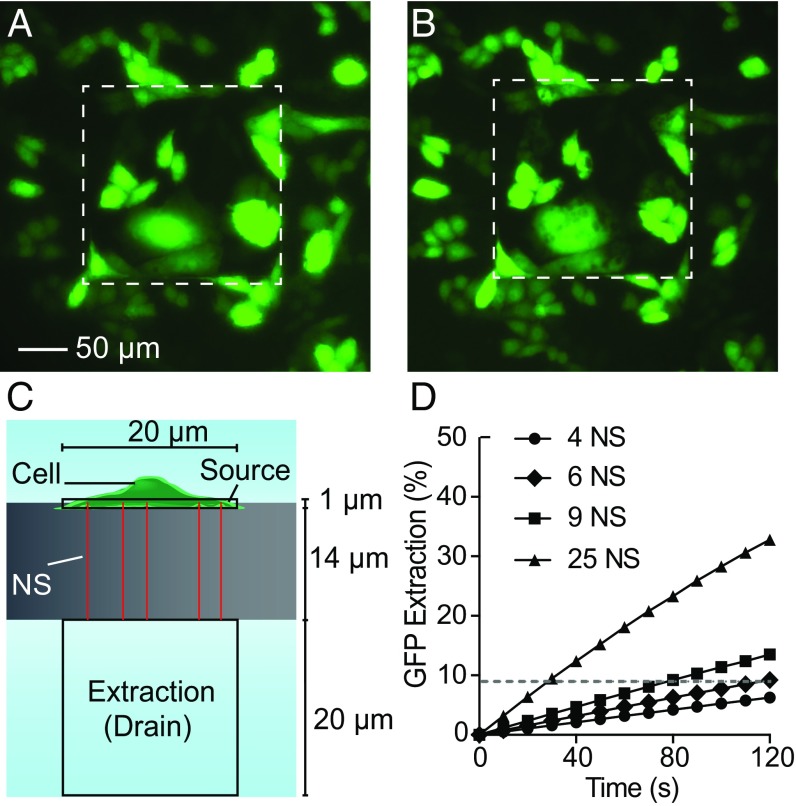
An important question from the longitudinal results is whether the NEX process reflects the contents of the entire cell or samples only a single site. We assessed the spatial distribution of NS extraction from the decrease of GFP intensity within GFP-expressing CHO cells during sampling. CHO cells were cultured overnight on a patterned membrane with a 200 × 200 µm region of approximately 40,000 exposed NS (Fig. 4A). During the 2-min sampling period, GFP diffuses from the cells and through the NS, leaving a region of lower fluorescence intensity (dark spots) in the cell where the membrane was opened (Fig. 4B). Therefore, the location and number of open NS can be visualized by the dark spots in the cells. Twenty-four of twenty-six CHO cells showed spots during sampling, demonstrating that most cells within the sampling region are penetrated and sampled through the NS. The multiple penetrating NS (dark spots) were observed to be distributed throughout the cell bodies, with little difference between the soma and peripheral regions (Fig. S5). No observable change in cell shape was found after the 2-min sampling period, corroborating the notion that the molecular transport through the NS is diffusion driven. NEX thus appears to sample from all regions of the cytoplasm, providing a comprehensive view of the intracellular contents.
Fig. 4.
Sampling spatial distribution. (A and B) Fluorescent microscopy images of GFP of a culture of 26 cells on a 200 × 200 µm NS sampling region (white dashed squares). (Scale bar, 50 µm in A and B.) (A) GFP-expressing CHO cells before sampling. (B) GFP-expressing CHO cells immediately after sampling. Locally diminished GFP intensities (dark spots) were observed in the cells after sampling, corresponding to the locations where GFP was removed from the cells. Brightness was increased to highlight the spots. (C) Diagram of the finite element model of sampling through the NS. The cell was treated as a 20 × 20 × 1 µm source connected to the extraction buffer by varying numbers of NS 14 µm long and 150 nm in diameter. (D) The percentage of the cell’s initial GFP that diffuses into the extraction buffer as a function of time and the number of NS (the dashed line indicates the GFP extraction level after 2 min of diffusion from six NS).
Fig. S5.
Location of dark spots in CHO cells. Fluorescent microscopy image of the GFP of a culture of 26 cells on a 200 × 200 µm NS sampling region (white dashed square). The dark spots as obtained by manual counting were labeled in red to indicate their locations in cells.
The total GFP extracted from the cells during sampling could be measured from the difference in fluorescence intensity before and after sampling. The average GFP in a cell was 0.50 (±0.44) pg before sampling and was 0.47 (±0.38) pg after sampling as calculated from a calibrated volumetric GFP intensity curve (Fig. S4). The GFP extracted from these 24 cells was 680 fg from the change in fluorescence intensity, or 6% of the initial cell concentration. This fraction is also similar to the percentages in single-cell extraction (7% and 8%). Because we know the amount of material the cells lost, we also calculated the collection efficiency. The calibrated amount of GFP measured in the extraction buffer during this same experiment was 230 fg, or ∼30% of the total amount lost from the cell, a reasonable collection efficiency indicating that the material loss during extraction and handling is not limiting. Together, these results show that extractions were made from most of the cells within the sampling region, multiple NS penetrate the cell at one time, molecules were sampled from multiple regions of a cell, and cell contents were extracted and analyzed with reasonable collection efficiency
In theory, the amount of material extracted should be a function of the cellular concentration, the diffusivity of the species, and the NS geometry (23). The extraction was simulated as a purely diffusive transport process using a finite-element model (COMSOL Multiphysics) of a cellular volume 20 μm in diameter and 1 μm tall connected to the 1× PBS extraction buffer through a set of NS 14 μm long and 150 nm in diameter (Fig. 4C). The expected percentage of the total GFP extracted from the cell as a function of time for a GFP diffusivity of 87 µm2/s (35) is shown in Fig. 4D and agrees with our experimental observations. For six penetrating NS, close to the observed number of spots per cell (Fig. 4B), approximately 9% of the total GFP diffuses into the extraction buffer over the 10-min extraction interval; this value corresponds well with the 7% and 8% GFP measured from the single-cell experiments.
Longitudinal Sampling of Proteins from hiPSC-Derived Cardiomyocytes and Astrocytes.
NEX can be used to sample contents not only of cell lines but also of cell types derived in vitro from hiPSCs, an ability that will be essential in future applications related to cell differentiation and disease modeling. We assessed longitudinal extraction and off-platform analyses of nonfluorescent heat-shock protein 27 (HSP27) from hiPSC-derived cardiomyocytes (hiPSC-CMs) measured with ELISA (Fig. 5). Heat-shock protein is up-regulated when exposed to external stressors and is thus suitable for studying transient processes (36).
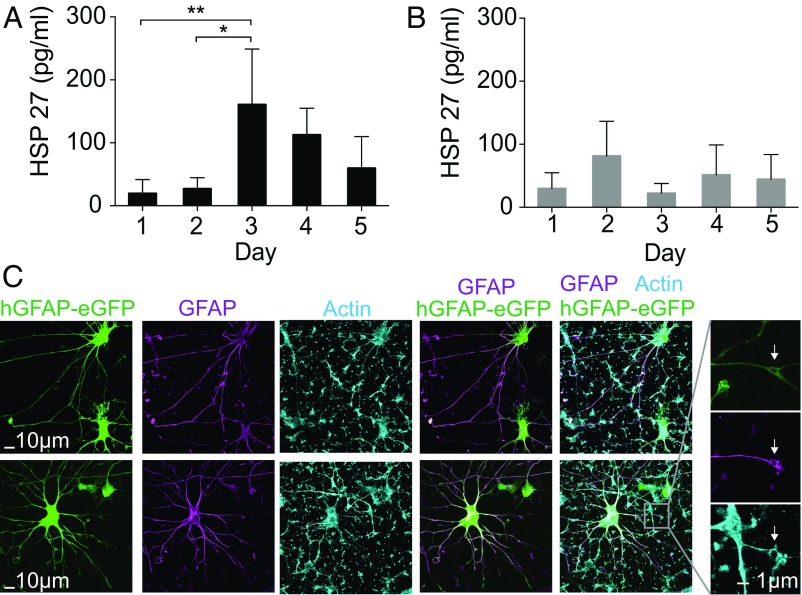
Fig. 5.
Longitudinal sampling from hiPSC-derived cardiomyocytes and astrocytes. (A and B) Longitudinal HSP27 extraction from the same hiPSC-CMs for 4 d. (A) The cardiomyocytes were stimulated by increasing the temperature to 44 °C for 30 min before sampling at day 2. An up-regulation of HSP27 was observed at day 3 (n = 4; *P < 0.05; Tukey’s post hoc test, one-way ANOVA). The HSP27 level started to drop at day 4. (B) Non–heat-shocked hiPSC-CMs were longitudinally sampled for 4 d (n = 4, P > 0.05; one-way ANOVA). (C) Representative images of astrocytes derived from hiPSCs in 3D cultures (hCS) and cultured in monolayer on NS. Astrocytes are labeled fluorescently with a lentiviral reporter (hGFAP::eGFP) and immunostained with an anti-GFAP antibody. The morphology of astrocytes was maintained even after 20 d of culture and repeated sampling on the NS platform. Arrowheads indicate structures that are approximately 1 µm in size, possibly NS that are surrounded by processes.
To obtain a detectable protein signal, we increased the hiPSC-CM plating on our NS platform to 100,000 (± 25,000) cells because of the 10.9 pg/mL detection limit of HSP27 ELISA (Affymetrix), which is not sensitive enough to detect the intracellular extraction from small cell populations. After 7 d in culture on the NS, the hiPSC-CMs began beating. Even under stress from the continuous beating, the NS did not break and were not pulled from the cells, allowing sampling even from this actively moving tissue (Fig. S6). Intracellular extractions were obtained every 24 h for 5 d. At day 2, the cells were stressed by exposure to a heat shock (44 °C for 30 min), which is expected to up-regulate the synthesis of HSP27. A sister culture not exposed to the heat-shock perturbation was sampled at the same time points as a negative control.
Fig. S6.
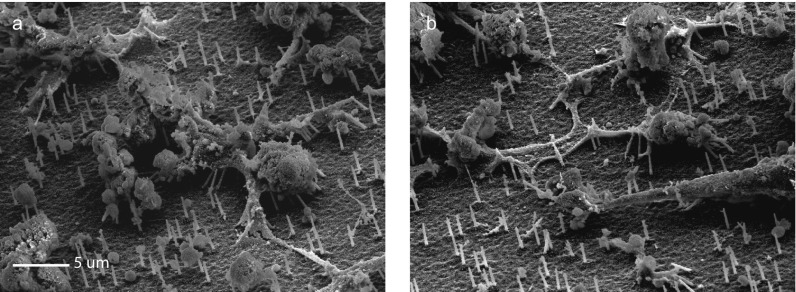
SEM images of cardiac cells grown on NS. (A and B) hiPSC-CMs spread out on the NS platform. The NS are not broken off by the beating of the cardiomyocytes but instead bend to accommodate the stress. Note that the cells are dehydrated during the SEM preparation process so that the images are not indicative of cell morphology in culture. Instead, these images show that NS survive after culture with beating cardiomyocytes. (The scale bar in A is also valid in B.)
The NS platform followed the temporal expression and up-regulation of HSP27 in human CMs. Starting with a relatively low concentration at day 1, there was a small, not statistically significant increase in the HSP27 level 2 h after heat-shock perturbation at day 2, suggesting delayed expression of HSP27. At day 3, the HSP27 level increased to about five times higher than on days 1 and 2 (n = 4, P < 0.05; one-way ANOVA) and then decreased at day 4 and 5. In contrast, the HSP27 level in the control was relatively constant all five days (n = 4; P > 0.05; one-way ANOVA). The first extraction point showed lower extraction levels in both sets of data, similar to the observations in the GFP sampling experiment. The small up-regulation of HSP27 in the control samples indicates a minimal stress response because of sampling, and Calcein AM labeling confirmed >90% cell viability for both the sample and negative control. These results demonstrate the feasibility of using NS to extract and measure nonfluorescent proteins from beating hiPSC-CMs.
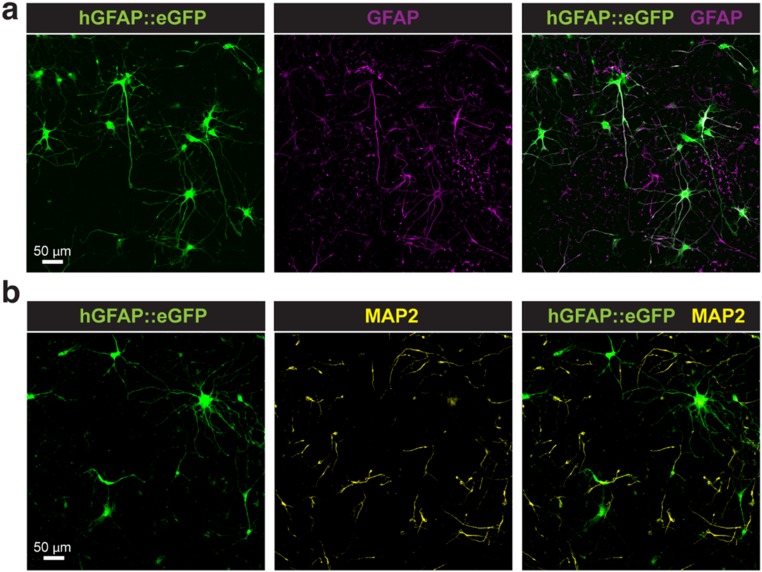
To assess the influence of the long-term culture on the NS platform and repeated sampling process on neural cell types, we examined the viability of astrocytes derived from hiPSC in 3D human cortical spheroids (hCSs) (37). Approximately 50,000 astrocytes and neurons derived from 132-d-old hCSs were plated on the NS platform (Fig. S7) and were electroporated using the protocol used for the cardiomyocytes once per day for 20 d. Astrocytes were fluorescently labeled with a cell-specific reporter (hGFAP::eGFP), as previously described (38). Cell morphology was followed every day during the sampling period, and despite their high overall reactivity to various stimuli and cell injuries, human astrocytes cultured on the NS tolerated the platform and the daily sampling well, with insignificant morphological changes between day 1 and day 20 (37, 38).
Fig. S7.
Human neurons and astrocytes derived in hCS and cultured on NS. Representative images of iPSC-derived astrocytes and neurons generated in 3D hCS and cultured in monolayer on NS. (A) Astrocytes are labeled with the hGFAP::eGFP reporter and coimmunostained with an anti-GFAP antibody. (B) Neurons are immunostained with an anti-MAP2 antibody and do not colocalize with hGFAP::eGFP+ cells. (Scale bars: 50 µm.)
mRNA Expression Levels in hiPSC-CMs.
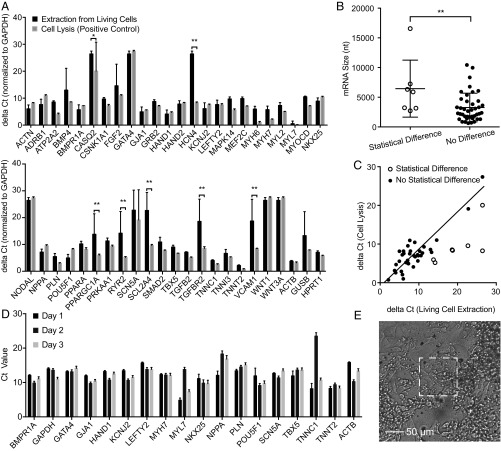
mRNA transcriptomics is a powerful method to detect gene expression, cell phenotype, and cell-to-cell heterogeneity. With the advent of efficient reverse transcription and single-cell sequencing, multiple mRNA sequences can be detected simultaneously and with higher sensitivity than proteins (39). To test whether the mRNA extracted from primary hiPSC-CMs are statistically related to the actual concentrations inside the cell, we first performed a NEX extraction of mRNA for 2 min and compared it with the mRNA expression for a lysed sister cell preparation (Methods). The NEX extract was pipetted from below the well, amplified with RT-PCR using a random primer, and sequenced with a single-cell BioMark system (Fluidigm). The results were compared with the positive control (n = 2) obtained by lysing a sister culture of hiPSC-CMs and a negative control without the electroporation step (n = 4). Both controls were amplified and analyzed in the same manner as the mRNA samples. Among the 48 genes, 25 were cardiac-related genes, including potassium voltage-gated channel subfamily J, member 2 (KCNJ2) and several integral membrane proteins [e.g., phospholamban (FLN) and sodium voltage-gated channel alpha subunit 5 (SCN5A)], 13 were genes related to stem cell differentiation, and 10 were housekeeping genes.
The mRNA from the NEX extracts were in good quantitative agreement with the lysis control samples. All genes with nonzero quantities from lysis were also detected in the NEX extraction, that is, 44 gene detections for each sample. No false positives were observed, because the four mRNA that were not detected in the positive controls were also not detected in the cell extraction. The change in the cycle threshold (ΔCt) of each gene (excluding the statistically unmatched genes) in the extraction was strongly correlated with the positive control (R = 0.89, P < 0.0001) (Fig. 6C). Notably even large genes such as SMAD family member 2 (SMAD2) (10,428 nt) were sampled successfully (Table S1). After sampling, the hiPSC-CMs showed healthy morphology, and the green fluorescence from the calcein AM stain indicated >95% cell viability. Negative controls without an electric field showed no detectable signal (Fig. S8).
Fig. 6.
mRNA expression level in extractions from hiPSC-CMs. (A) mRNA expression level in NS-extractions from living cells (black bars) and from lysed cells (gray bars) in ΔCt (normalized to GAPDH) (42). A higher ΔCt represents lower mRNA expression; the baseline ΔCt was set to 40 cycles to indicate no expression. Of the 48 genes, seven genes are found to be statistically different from the lysis control (n = 4 for living cell extraction; n = 2 for cell lysis; *P < 0.05; **P < 0.01; t test with 95% CI, pooled SD. (B) mRNA sizes of matched and unmatched genes; *P < 0.05, t test, shows a slight difference in the detection of larger and smaller genes. (C) Correlation of ΔCt of the 40 matched genes in living cell extractions with the mRNA expression level in cell lysis (R2 = 0.89, P < 0.0001). Unmatched genes are indicated as unfilled circles. (D) Longitudinal mRNA expression level of approximately 15 hiPSC-CMs on the 100 × 100 µm NS platform over 3 d. Fifteen of eighteen genes showed consistent gene expression, demonstrating the reliability of the sampling process. Error bars indicate the SD of technical duplicates. (E) Microscopy image of approximately 15 hiPSC-CMs after day 3 sampling. These cells were actively beating (Movie S1) but could still be sampled successfully.
Table S1.
Names and number of nucleotides in the 48 genes analyzed in Fig. 6
| Name | Length, nt | Name | Length, nt | Name | Length, nt |
| ACTN1 | 3,306 | LEFTY2 | 1,230 | RYR2 | 16,562 |
| ADRB1 | 2,862 | MAPK14 | 1,083 | SCN5A | 8,343 |
| ATP2A2 | 4,359 | MEF2C | 5,915 | SCL2A4 | 3,159 |
| BMP4 | 1,774 | MYH6 | 351 | SMAD2 | 10,428 |
| BMPR1A | 3,631 | MYH7 | 6,069 | TBX5 | 3,828 |
| CASQ2 | 2,716 | MYL2 | 630 | TGFB2 | 5,882 |
| CSNK1A1 | 2,100 | MYL7 | 830 | TGFBR2 | 4,704 |
| FGF2 | 6,774 | MYOCD | 3,093 | TNNC1 | 705 |
| GAPDH | 1,407 | NKX25 | 1,669 | TNNI3 | 866 |
| GATA4 | 2,662 | NODAL | 2,086 | TNNT2 | 1,307 |
| GJA1 | 3,169 | NPPA | 858 | VCAM1 | 3,220 |
| GRB2 | 3,303 | PLN | 1,742 | WNT1 | 2,284 |
| HAND1 | 1,749 | POU5F1 | 2,075 | WNT3A | 2,988 |
| HAND2 | 675 | PPARA | 1,302 | ACTB | 1,852 |
| HCN4 | 7,245 | PPARGC1A | 6,318 | GUSB | 2,228 |
| KCNJ2 | 1,622 | PRKAA1 | 5,085 | HPRT1 | 1,435 |
Boldface type indicates unmatched genes.
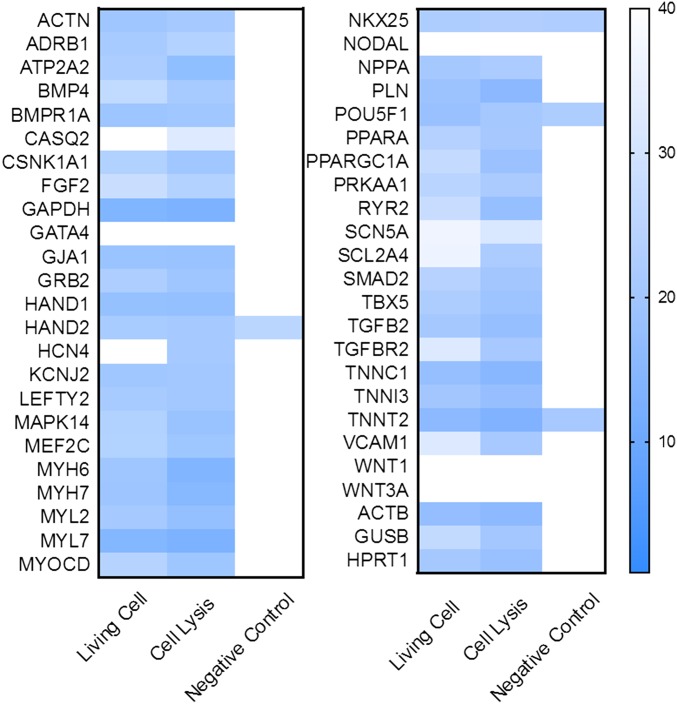
Fig. S8.
Heat map of the ΔCt level of 48 genes from NEX, cell lysis, and negative control (without electrical pulsing). The Ct level of 48 genes from living cells matched well with cell lysis results. Very low or no gene signal was detected in the negative control collected after washing (without electroporation), indicating that the washing step is sufficient to remove the background contamination.
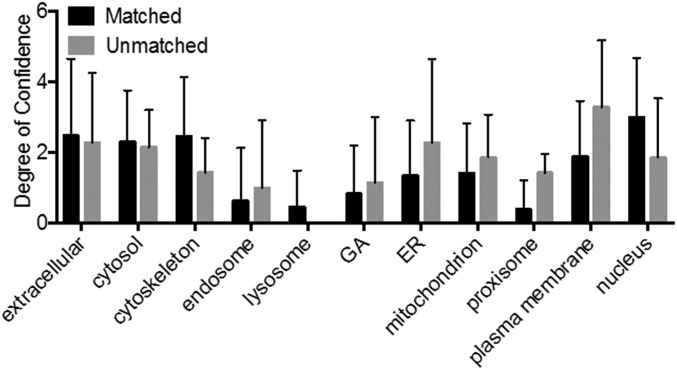
Statistical t test analyses of the 44 detected mRNA sequences revealed that only seven genes were significantly different from the control (P < 0.05, t test). The lower detection efficiency of the seven unsuccessful genes could be caused by several factors, including lower diffusion rates resulting from size or binding to structures within the cell. We intentionally did not include a statistical multiple testing correction, because such a procedure, although maintaining the overall type I error rate, increases the chance of type II errors (the chance that differentially extracted genes are not discovered) (40). Fig. 6B shows a slight statistical difference between the sizes of the unmatched and matched mRNA (P < 0.01; t test), suggesting larger molecules are more difficult to extract during the 10-min extraction period (although the result was heavily influenced by ryanodine receptor 2 (RYR2) at 16,562 nt). Subcellular localization of the mRNA also may affect the sampling efficiency, because mRNAs that code for proteins found in the plasma membrane were unmatched statistically more often (P < 0.05; t test) (Fig. S9).
Fig. S9.
Intracellular localization of matched and unmatched mRNA. The subcellular localization of the mRNA also may affect the sampling efficiency. To examine this issue, we assume that the mRNA is localized to the same regions as the proteins they code for (3). We then compared the protein localization in 12 subcellular locations of the specific unmatched gene to the matched gene. mRNAs that code for proteins found in the plasma membrane were statistically shown to be unmatched more often (P < 0.05, t test), indicating that the specific mRNA sequences are more likely to be immobilized within the cytoplasm, resulting in a low detection rate.
Significantly, NEX sampling could be repeated on the same set of actively behaving primary cells to provide longitudinal mRNA measurements. Fig. 6D shows the mRNA expression levels of 18 different genes from approximately 15 hiPSC-CMs measured once per day for 3 d. Note that these cells were active and beating at the time they were sampled (Movie S1). The presence of additional cell monolayers makes exact cell counting difficult; however, on average 15–20 cells were observed in the 100 × 100 μm sampling region. The measured expression levels were remarkably consistent, demonstrating the precision of the sampling and analysis process. Of the 18 genes, 15 were highly consistent over 3 d, thus indicating that the variations in myosin light chain 7 (MYL7), troponin C1, slow skeletal and cardiac type (TNNC1), and actin beta (ACTB) are likely significant and may reflect fluctuations in gene expression (7–9).
Although the NEX process extracts only a fraction of the total contents of the cell, the extraction quantities in Fig. 6 show these sampling events are consistent and can provide a meaningful representation of the quantities within the cell. It is not yet known whether the quantity extracted for each individual cell is always the same, although initial results from the single-cell measurements (Fig. 3) and fluorescence microscopy of individual cells after sampling (Fig. 4) suggest they may be. More work is necessary to determine the relationship between the cellular and extracted quantities for a particular mRNA sequence to report internal concentrations quantitatively based on the NEX results alone, but it is clear the NEX process has the capacity to measure the change or lack of change in mRNA expression over time in the same set of cells.
Currently the sensitivity of mRNA sequencing systems is not able to measure NEX extracts from single cells, instead requiring approximately 15–20 cells. This requirement agrees well with the approximately 7% extraction efficiency, corresponding to approximately 1.1–1.4 cellular equivalents per sample. With the increasing sensitivity of single-cell mRNA assays, this limitation may soon be overcome, enabling repeated mRNA measurements from a single cell over an extended time period.
Discussion
Measuring dynamic intracellular processes and capturing cellular heterogeneity, especially at the single-cell level, has become an area of active investigation in molecular and cellular biology. Despite rapid technological advancement in the sampling modalities and sensitivity (4, 5, 41), these methods are still limited by the need to lyse the cell for sample extraction. Here, we describe an NS-based sampling platform for longitudinal, nondestructive extraction and quantification of proteins and mRNAs from living cells. The procedure itself is quite straight-forward, locally porating a small area of the cellular membrane near the NS and allowing the cellular contents to diffuse into an underlying extraction buffer. The process requires simple equipment and a common voltage supply and should be feasible in most laboratories.
There was some initial concern that extraction through passive diffusion would bias the results significantly and would sample only a few of the possible species; however such has not been the case. Proteins and mRNA were sampled consistently over a wide range of sizes and with similar percentages of the cellular content as the smaller species. Although there was a slight preference for smaller mRNA (Fig. 6B), this preference was largely the result of the influence of RYR2; without that data point there would have been no statistical difference. Further tests over a much larger range of proteins and genes will be needed to establish quantifiably reliable species. However, the successful analysis of a large fraction (41/48) of the mRNA sequences suggests that a sufficient number of genes can be monitored to make meaningful biological assessments.
Another important aspect is that the NEX process was nondestructive, as evidenced by >95% cell viability after each sampling event. Minimal morphological changes were observed even after daily sampling for 20 d of human astrocytes, which are known to react promptly to perturbations. Cell viability is a critical metric for longitudinal studies. For example, 80% cell viability per sample can take an average of three samples before cell death, whereas 95% viability gives an average of 14 samples. This consideration will be especially important for longitudinal measurements of single cells in which cell apoptosis terminates the experiment.
Several other key features make NEX suitable for longitudinal studies of cell biology. First, the extracted molecules are spatially separated from the cell culture, allowing simple collection using a pipette or microfluidic device. This collection method ensures that future analytical technology improvements can be combined with the NEX process as the sampling platform. Second, the patterned NS sampling region allows scalable numbers of cells to be analyzed while maintaining the normal cell-to-cell connectivity and communication important for cell development and differentiation. Third, the NS platform was compatible with all cell types tested, including CHO cells and hiPSC-derived cells (cardiomyocytes or neural cells derived in 3D cultures).
We believe that this sampling technique will be useful for studying dynamic cellular activity or transformations, for example, for tracking signaling pathways during the differentiation of pluripotent stem cells in vitro and capturing cellular heterogeneity. The throughput could be increased by integrating microfluidics to sample from a number of independent cellular wells at once, similar to 96-chamber single-cell analysis designs. Such systems could increase the understanding of the cellular mechanisms behind cell development, differentiation, and disease pathology from bulk populations down to single cells.
Methods
NS and Patterned NS Fabrication.
The NS membrane is based on 15-µm-thick (±15%) track-etched polycarbonate membranes (GVS) with 1 × 108 pores/cm2, often used for water filtration and cell culture. A 10-nm-thick Al2O3 layer is deposited on the membrane using atomic layer deposition (ALD) at 110 °C, including the insides of the track-etched pores which will become the NS walls. The NS are formed by reactive ion etching (RIE) of the Al2O3 with BCl3 and Cl2 in argon [300 W, 40 standard cm3/min (sccm) BCl3, 30 sccm Cl2, 5 mTorr, 5 min] from the top surface to reveal the polymer, followed by oxygen plasma etching to remove the polymer and expose the inorganic NS. To fabricate the photolithographically defined sampling regions, a 5-µm-thick positive photoresist film (MEGAPOSIT SPR2203 i-Line photoresist; Dow) was spin coated on the surface of the ALD-coated polycarbonate membrane using a spinning speed of 3,500 rpm for 60 s (Fig. S1). Next, the photoresist-coated membrane was baked at 95 °C for 2 min to evaporate resist solvent and then was exposed to a square pattern of intense UV light for 5 s. After exposure, the membrane was developed by immersion in the MF-26A developer (Shipley) for 60 s. The aluminum oxide surface in the sampling region was etched away by RIE, leaving a polycarbonate surface inside the sampling region. Finally, the polymer was etched away by oxygen RIE to form the NS.
Sample Preparation for SEM.
The NS membrane was prepared for SEM imaging by sputter coating with about 10 nm of Au/Pd. Biological samples were prepared by fixing in 2% glutaraldehyde with 4% paraformaldehyde (PFA) in 0.1 M Na Cacodylate buffer (pH 7.3) for at least 4 h, staining with 1%OsO4 for 10 min, and dehydrating in a series of 30, 50, 70, 90, and 100% ethanol with 10 min of incubation at room temperature for each solution. The dehydrated sample was dried by critical point drying in 100% EtOH with liquid CO2 and then was sputter coated with about 10 nm of Au/Pd for SEM. Samples were imaged in an FEI Sirion scanning electron microscope.
General Sampling Protocol.
To perform the sampling process, the cells of interest were first cultured on the NS membrane within a 2- to 5-mm glass cylinder with appropriate cell medium on top (Fig. 1B). This culture tube remained within a 96- (or 48-) well plate in an incubator until a sample was desired. The sampling process was performed in five steps. First, the cells were washed three times with 1× PBS, and the cell-culture medium was changed to PBS to eliminate possible contaminants. Second, the NS cylinder was placed on top of a droplet (1–15 µL) of extraction buffer consisting of PBS (or TE buffer for the ITP assay, as explained below) on an indium tin oxide (ITO) electrode (Fig. 1A). A platinum wire was immersed into the cell-culture buffer to act as the counter electrode. Third, 10–35 V (between anode and cathode) square electric pulses (200-µs pulse duration, 20-Hz repetition rate) were applied between the two electrodes (across the NS membrane) for 20–60 s. The pulses temporarily opened pores in the cell membrane, allowing freely diffusing intracellular proteins and mRNA to diffuse through the NS to the extraction buffer on the underside of the NS membrane. We have observed an increased flux of molecules when the polarity of the electric field is the opposite of the charge of the analytes. When extracting negatively charged molecules (e.g., mRNA), the ITO electrode was kept at positive potential, and it was kept at negative potential when extracting positively charged molecules. Fourth, after the electroporation, the sampling device remained on the extraction buffer for another 10 min to allow diffusion of additional cytosolic content. Because the extraction process depended mainly on molecular diffusion, the total amount of extracted molecules was expected to depend on both diffusivity and concentration. Therefore, a longer diffusion time was expected to be required for the extraction of larger molecules, because of their lower mobility. Finally, the extracted molecules were collected beneath the NS membrane for further analysis by pipetting up the extraction buffer.
ITP Preconcentration.
ITP was conducted in a 50-µm-wide, 20-µm-deep cross-channel design glass microfluidic chip (NS 12A; PerkinElmer). The leading electrolyte (LE) and trailing electrolyte (TE) buffers were 200 mM Tris and 100 mM HCl, and 25 mM Tris and 150 mM of glycine, respectively. One percent polyvinylpyrrolidone (PVP) was added to both the LE and the TE to suppress electro-osmotic flow. All reagents were obtained from Sigma Aldrich. The TE buffer also was used as the extraction buffer in cell sampling. To preconcentrate GFP, first, the microfluidic channel and LE reservoir were filled with 3–10 µL of LE, and then the 1- to 5-µL mixture of TE and GFP sample solution was injected in the TE reservoir. Next, the anode and cathode were placed in the LE and TE reservoirs, respectively. An electric field with 1,100 V (Keithley) was applied between the electrodes. The GFP ITP focusing zone formed and electromigrated toward the anode right after the electric field was applied. The GFP intensity was stabilized 2 min after ITP began.
GFP and RFP Concentration and Intensity Calibration and Quantification of GFP/RFP Total Mass in a Cell.
GFP/RFP standard solutions (Abcam) were injected in a 10-µm-wide, 12-µm-deep glass microfluidic channel (PerkinElmer). The average fluorescence intensity of each standard solution in the channel was recorded and normalized to the channel dimensions to determine the intensity per unit volume. GFP/RFP calibration curves were generated based on the fluorescence intensity versus protein concentration, which were linear (Fig. S4). To measure the GFP/RFP concentration in a cell, an average cell height of 2 µm was taken based on the imaging of a number of different cells. To quantify the total GFP/RFP mass in a cell, the measured GFP/RFP intensity from the cell was first divided by the assumed height to get the intensity per unit of height and then was calibrated by the standard curve to get the GFP/RFP concentration. Finally, the total mass of GFP/RFP in the cell is the product of the concentration of the protein and the measured area and assumed height of the cell (i.e., the cell volume).
HSP27 Extraction from hiPSC-CMs.
To obtain a detectable protein signal, we increased the cell population on our NS platform to 50,000 (±25,000) cells because of the detection limit of HSP27 ELISA (10.9 pg/mL) (Affymetrix), which is not sensitive enough to detect the intracellular extraction from small cell populations. Intracellular extractions were obtained every 24 h for 5 d. Cells were washed in PBS before each sampling to remove loosely adsorbed proteins. At day 2, the cells were stressed by exposing them to a 30-min heat shock at 44 °C, which is expected to up-regulate the synthesis of HSP27. The day 2 extraction was collected 2 h after the heat shock. Cells not exposed to the heat-shock perturbation were also sampled as a negative control.
mRNA Extraction from hiPSC-CMs.
mRNAs were extracted using the NS followed by amplification and analysis using RT-PCR and quantitative PCR (qPCR). The generation and use of iPSCs and their derived cells were approved by the Institutional Review Board at Stanford University School of Medicine. Fibroblasts for reprogramming were collected and de-identified following informed consent. To average out stochastic fluctuations associated with small numbers of cells, we chose to culture 100,000 (± 50,000) cells in an NS well. The cells were rinsed with PBS buffer to remove extraneous or excreted material; then sampling was performed for 2.0 min as described above. Because mRNA degrades rapidly in the presence of RNase, carrier RNA (Sigma Aldrich) and RNase inhibitor (Thermo Fisher Scientific) were added to the extraction buffer to make a mixture with 1 µg/mL carrier RNA and 1 U/µL RNase inhibitor before sampling. The mRNAs in the extraction buffer were reverse transcribed to cDNAs with Oligo(dT)20 (Thermo Fisher Scientific). Next, the cDNAs were preamplified for 15 cycles and purified using DNA Clean & Concentrator-5 (Zymo Research). The preamplified cDNAs were amplified with the 48 gene primers and analyzed by qPCR in an integrated fluidic circuits (IFC) Fluidigm chip following standard protocols (Fluidigm).
mRNA Extraction Analysis.
To preamplify the extracted mRNA, 0.5 µL diethylpyrocarbonate -treated water, 0.5 µL Oligo(dT), and 0.5 µL dNTP reagent were mixed with 5 µL mRNA extraction. The mRNA solution was mixed and briefly centrifuged and then was heated at 65 °C for 5 min and incubated on ice for at least 1 min. Two microliters of 5× SuperScript IV buffer (Thermo Fisher Scientific), 0.5 µL DTT, 0.5 µL RNase inhibitor, and 0.5 µL SuperScript IV Reverse Transcriptase (Thermo Fisher Scientific) were added into the annealed RNA solution. The combined reaction mixture was incubated at 50 °C for 20 min; then the reaction was inactivated by incubating the mixture at 80 °C for 10 min. Ten microliters of pooled assay mix and 20 µL TaqMan PreAmp Master Mix (Applied Biosystems) were added to the combined reaction mixture. The mixture was preamplified at the conditions shown in Table 1.
Table 1.
Preamplification conditions
| Parameters | Hold | 15∼20 cycles | |
| Temperature | 95 °C | 95 °C | 60 °C |
| Time | 10 min | 15 s | 4 min |
Finally, the preamplified cDNA was purified using DNA Clean & Concentrator-5 and was eluted in the provided 6 µL DNA elution buffer. The preamplified cDNA was detected by using Fluidigm Dynamic Array IFC for Gene Expression.
Culturing, GFP Extraction, and Immunocytochemistry of hCS-Derived Astrocytes.
hCSs were derived from iPSCs as previously described (37). The generation and use of iPSCs and their derived cells were approved by the Institutional Review Board at Stanford University School of Medicine. Fibroblasts for reprogramming were collected and deidentified following informed consent. The generation of neurons of deep and superficial cortical layers is followed by astrogenesis in hCS, and after approximately 10 wk in vitro cortical neurons are accompanied by a network of nonreactive astrocytes. For sampling experiments, hCS at day 132 in vitro were enzymatically dissociated and plated on NS at a density of 500,000–750,000 cells per device. The day after plating, plated astrocytes were labeled with a viral reporter (hGFAP::eGFP). For GFP sampling, cells were maintained on the NS for up to 20 d with daily medium changes. Short electrical pluses (45 V, 200-μs pulse width, 20-s duration) were applied to cells every day. For immunocytochemistry, the cells on NS were fixed with 4% PFA for 10 min and were immunostained with an anti-GFAP antibody to label astrocytes, an anti-MAP2 antibody to label neurons (MAP2), and anti-Actin antibody to label filaments (37).
Supplementary Material
Acknowledgments
This work was supported by a National Institute of Standards and Technology Joint Initiative for Metrology in Biology Training Grant (to Y.C.) and the Knut and Alice Wallenberg Foundation (M.H.); NIH Grants R01 HL133272 and NIH R01 HL128170 (to J.C.W.); the Bio-X Interdisciplinary Initiatives Program (Y.C., N.A.M., and J.C.W.); a Progenitor Cell Biology Consortium Jump Start Award (to Y.C. and H.C.); National Institute of Mental Health BRAINS Award R01MH107800, an MQ Fellows Award, and a Donald E. and Delia B. Baxter Foundation Award (to S.P.P.); and Child Health Research Institute Postdoctoral Fellowship UL1-TR001085 (to F.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1615375114/-/DCSupplemental.
References
- 1.Desplats P, et al. Inclusion formation and neuronal cell death through neuron-to-neuron transmission of alpha-synuclein. Proc Natl Acad Sci USA. 2009;106(31):13010–13015. doi: 10.1073/pnas.0903691106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collado M, Blasco MA, Serrano M. Cellular senescence in cancer and aging. Cell. 2007;130(2):223–233. doi: 10.1016/j.cell.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 4.Deng Q, Ramsköld D, Reinius B, Sandberg R. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science. 2014;343(6167):193–196. doi: 10.1126/science.1245316. [DOI] [PubMed] [Google Scholar]
- 5.Gawad C, Koh W, Quake SR. Single-cell genome sequencing: Current state of the science. Nat Rev Genet. 2016;17(3):175–188. doi: 10.1038/nrg.2015.16. [DOI] [PubMed] [Google Scholar]
- 6.Burridge PW, et al. Chemically defined generation of human cardiomyocytes. Nat Methods. 2014;11(8):855–860. doi: 10.1038/nmeth.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297(5584):1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 8.Imayoshi I, et al. Oscillatory control of factors determining multipotency and fate in mouse neural progenitors. Science. 2013;342(6163):1203–1208. doi: 10.1126/science.1242366. [DOI] [PubMed] [Google Scholar]
- 9.Maheshri N, O’Shea EK. Living with noisy genes: How cells function reliably with inherent variability in gene expression. Annu Rev Biophys Biomol Struct. 2007;36:413–434. doi: 10.1146/annurev.biophys.36.040306.132705. [DOI] [PubMed] [Google Scholar]
- 10.Frommer WB, Davidson MW, Campbell RE. Genetically encoded biosensors based on engineered fluorescent proteins. Chem Soc Rev. 2009;38(10):2833–2841. doi: 10.1039/b907749a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Depry C, Mehta S, Zhang J. Multiplexed visualization of dynamic signaling networks using genetically encoded fluorescent protein-based biosensors. Pflugers Arch. 2013;465(3):373–381. doi: 10.1007/s00424-012-1175-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kerppola TK. Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nat Protoc. 2006;1(3):1278–1286. doi: 10.1038/nprot.2006.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kerppola TK. Multicolor bimolecular fluorescence complementation (BiFC) analysis of protein interactions with alternative partners. Cold Spring Harb Protoc. 2013;2013(9):798–803. doi: 10.1101/pdb.top077164. [DOI] [PubMed] [Google Scholar]
- 14.Tantama M, Hung YP, Yellen G. Optogenetic reporters: Fluorescent protein-based genetically encoded indicators of signaling and metabolism in the brain. Prog Brain Res. 2012;196:235–263. doi: 10.1016/B978-0-444-59426-6.00012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michalet X, et al. Quantum dots for live cells, in vivo imaging, and diagnostics. Science. 2005;307(5709):538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaiswal JK, Mattoussi H, Mauro JM, Simon SM. Long-term multiple color imaging of live cells using quantum dot bioconjugates. Nat Biotechnol. 2003;21(1):47–51. doi: 10.1038/nbt767. [DOI] [PubMed] [Google Scholar]
- 17.Delehanty JB, et al. Spatiotemporal multicolor labeling of individual cells using peptide-functionalized quantum dots and mixed delivery techniques. J Am Chem Soc. 2011;133(27):10482–10489. doi: 10.1021/ja200555z. [DOI] [PubMed] [Google Scholar]
- 18.Tyagi S, Kramer FR. Molecular beacons: Probes that fluoresce upon hybridization. Nat Biotechnol. 1996;14(3):303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 19.Tian B, et al. Three-dimensional, flexible nanoscale field-effect transistors as localized bioprobes. Science. 2010;329(5993):830–834. doi: 10.1126/science.1192033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singhal R, et al. Multifunctional carbon-nanotube cellular endoscopes. Nat Nanotechnol. 2011;6(1):57–64. doi: 10.1038/nnano.2010.241. [DOI] [PubMed] [Google Scholar]
- 21.Robinson JT, et al. Vertical nanowire electrode arrays as a scalable platform for intracellular interfacing to neuronal circuits. Nat Nanotechnol. 2012;7(3):180–184. doi: 10.1038/nnano.2011.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan R, et al. Nanowire-based single-cell endoscopy. Nat Nanotechnol. 2011;7(3):191–196. doi: 10.1038/nnano.2011.226. [DOI] [PubMed] [Google Scholar]
- 23.VanDersarl JJ, Xu AM, Melosh NA. Nanostraws for direct fluidic intracellular access. Nano Lett. 2012;12(8):3881–3886. doi: 10.1021/nl204051v. [DOI] [PubMed] [Google Scholar]
- 24.Shalek AK, et al. Vertical silicon nanowires as a universal platform for delivering biomolecules into living cells. Proc Natl Acad Sci USA. 2010;107(5):1870–1875. doi: 10.1073/pnas.0909350107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Na YR, et al. Probing enzymatic activity inside living cells using a nanowire-cell “sandwich” assay. Nano Lett. 2013;13(1):153–158. doi: 10.1021/nl3037068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Actis P, et al. Compartmental genomics in living cells revealed by single-cell nanobiopsy. ACS Nano. 2014;8(1):546–553. doi: 10.1021/nn405097u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guillaume-Gentil O, et al. Tunable single-cell extraction for molecular analyses. Cell. 2016;166(2):506–516. doi: 10.1016/j.cell.2016.06.025. [DOI] [PubMed] [Google Scholar]
- 28.Yang Z, et al. Molecular extraction in single live cells by sneaking in and out magnetic nanomaterials. Proc Natl Acad Sci USA. 2014;111(30):10966–10971. doi: 10.1073/pnas.1411802111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie X, et al. Nanostraw-electroporation system for highly efficient intracellular delivery and transfection. ACS Nano. 2013;7(5):4351–4358. doi: 10.1021/nn400874a. [DOI] [PubMed] [Google Scholar]
- 30.Xu AM, et al. Quantification of nanowire penetration into living cells. Nat Commun. 2014;5:3613. doi: 10.1038/ncomms4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang DC, Reese TS. Changes in membrane structure induced by electroporation as revealed by rapid-freezing electron microscopy. Biophys J. 1990;58(1):1–12. doi: 10.1016/S0006-3495(90)82348-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin ZC, Xie C, Osakada Y, Cui Y, Cui B. Iridium oxide nanotube electrodes for sensitive and prolonged intracellular measurement of action potentials. Nat Commun. 2014;5:3206. doi: 10.1038/ncomms4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chiappini C, et al. Biodegradable silicon nanoneedles delivering nucleic acids intracellularly induce localized in vivo neovascularization. Nat Mater. 2015;14(5):532–539. doi: 10.1038/nmat4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rogacs A, Marshall LA, Santiago JG. Purification of nucleic acids using isotachophoresis. J Chromatogr A. 2014;1335:105–120. doi: 10.1016/j.chroma.2013.12.027. [DOI] [PubMed] [Google Scholar]
- 35.Elowitz MB, Surette MG, Wolf PE, Stock JB, Leibler S. Protein mobility in the cytoplasm of Escherichia coli. J Bacteriol. 1999;181(1):197–203. doi: 10.1128/jb.181.1.197-203.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Venkatakrishnan CD, et al. Heat shock protects cardiac cells from doxorubicin-induced toxicity by activating p38 MAPK and phosphorylation of small heat shock protein 27. Am J Physiol Heart Circ Physiol. 2006;291(6):H2680–H2691. doi: 10.1152/ajpheart.00395.2006. [DOI] [PubMed] [Google Scholar]
- 37.Paşca AM, et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat Methods. 2015;12(7):671–678. doi: 10.1038/nmeth.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deverman BE, et al. Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat Biotechnol. 2016;34(2):204–209. doi: 10.1038/nbt.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nolan T, Hands RE, Bustin SA. Quantification of mRNA using real-time RT-PCR. Nat Protoc. 2006;1(3):1559–1582. doi: 10.1038/nprot.2006.236. [DOI] [PubMed] [Google Scholar]
- 40.Perneger TV. What’s wrong with Bonferroni adjustments. BMJ. 1998;316(7139):1236–1238. doi: 10.1136/bmj.316.7139.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sarkar A, Kolitz S, Lauffenburger DA, Han J. Microfluidic probe for single-cell analysis in adherent tissue culture. Nat Commun. 2014;5:3421. doi: 10.1038/ncomms4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feng M, et al. Myc/miR-378/TOB2/cyclin D1 functional module regulates oncogenic transformation. Oncogene. 2011;30(19):2242–2251. doi: 10.1038/onc.2010.602. [DOI] [PubMed] [Google Scholar]
- 43.Bercovici M, Han CM, Liao JC, Santiago JG. Rapid hybridization of nucleic acids using isotachophoresis. Proc Natl Acad Sci USA. 2012;109(28):11127–11132. doi: 10.1073/pnas.1205004109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bottenus D, et al. 10,000-fold concentration increase of the biomarker cardiac troponin I in a reducing union microfluidic chip using cationic isotachophoresis. Lab Chip. 2011;11(5):890–898. doi: 10.1039/c0lc00490a. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.