Significance
Protein C-mannosylation is restricted to animals and some related species. The C-mannosyltransferases probably evolved from enzymes responsible for N-glycosylation in the endoplasmic reticulum and are therefore expected to provide similar functions. Here we show that mammals have, in contrast to most lower animals, at least two C-mannosyltransferases with different recognition sequences, allowing glycosylation of all three tryptophans in the WxxWxxW motif of thrombospondin repeats, which are small modules found in many different proteins. We demonstrate that one of the C-mannosyltransferases, able to modify two tryptophans, is required and sufficient for the netrin receptor to reach the cell surface.
Keywords: glycosylation, thrombospondin type 1 repeat, C-mannosylation
Abstract
Thrombospondin type 1 repeats (TSRs) occur in diverse proteins involved in adhesion and signaling. The two extracellular TSRs of the netrin receptor UNC5A contain WxxWxxWxxC motifs that can be C-mannosylated on all tryptophans. A single C-mannosyltransferase (dumpy-19, DPY-19), modifying the first two tryptophans, occurs in Caenorhabditis elegans, but four putative enzymes (DPY-19–like 1–4, DPY19L1–4) exist in mammals. Single and triple CRISPR-Cas9 knockouts of the three homologs that are expressed in Chinese hamster ovary cells (DPY19L1, DPY19L3, and DPY19L4) and complementation experiments with mouse homologs showed that DPY19L1 preferentially mannosylates the first two tryptophans and DPY19L3 prefers the third, whereas DPY19L4 has no function in TSR glycosylation. Mannosylation by DPY19L1 but not DPY19L3 is required for transport of UNC5A from the endoplasmic reticulum to the cell surface. In vertebrates, a new C-mannosyltransferase has apparently evolved to increase glycosylation of TSRs, potentially to increase the stability of the structurally essential tryptophan ladder or to provide additional adhesion functions.
Thrombospondin type 1 repeats (TSRs) are small modules found in diverse membrane-bound and secreted proteins in metazoan and some related species. Over 60 distinct human proteins contain from 1 to up to 25 TSRs (1). The roles of TSRs in these proteins have only been addressed to a very limited degree. TSRs are generally considered to be binding modules (2–4). Attachment to sulfated glycans of cell surfaces and extracellular matrix has been observed for mammalian and Plasmodium proteins (5–7). In addition, the three TSRs of thrombospondin-1 have been shown to interact with the cell surface receptor CD36 with resultant antiangiogenic effects (8) and are being explored as an anticancer therapy (9).
The 50–60 amino acids of TSRs have a three-stranded fold with typically three conserved disulfide bridges (10). Most TSRs carry at least one, but often two or three, regularly spaced tryptophans upstream of the cysteine, marking the end of the A strand. In the most extended form, a WxxWxxWxxC sequence is present. The tryptophans play an essential role in the structure of TSRs as they associate with the side chains of conserved arginines of the B strand (10). This structural motif is referred to as a tryptophan–arginine ladder or a tryptophan ladder (11).
TSRs are target sites for two types of glycosylation, protein-O-fucosylation of serine/threonine and α–C-mannosylation of the above-mentioned tryptophans. Both modifications have been shown to be required for secretion of at least some proteins containing TSRs (12–18). Initially, C-mannosylation was described as an endoplasmic reticulum (ER) localized, enzyme-catalyzed process that modifies the first tryptophan in the consensus motif WxxW (19–21). In TSRs, however, mannosylation of more than one tryptophan has also been observed (22–26). The WxxWxxWxxC sequence has even been found to be mannosylated on all three tryptophans (22, 23).
The recently identified C-mannosyltransferase from Caenorhabditis elegans, a multitransmembrane protein related to the enzyme responsible for N-glycosylation, has been shown to mannosylate W1 and W2 of this motif in C. elegans UNC-5 (uncoordinated-5) and MIG-21 (abnormal cell migration-21) (14). In mammals, four homologs of the C. elegans enzyme (dumpy-19, DPY-19) exist, with human protein symbols DPY19L1 (DPY-19–like 1), DPY19L2, DPY19L3, and DPY19L4. DPY19L2 is specifically expressed in sperm and is mutated in the most common form of globozoospermia (27–29). It is only found in mammals and apparently appeared relatively late in evolution by duplication of DPY19L1 (30). All other vertebrates lack DPY19L2, but have DPY19L1, -L3, and -L4, whereas most invertebrate genomes, like that of C. elegans, encode one protein only. Apart from metazoans, the encoding gene is found only in alveolates, including the malaria parasite Plasmodium (14, 31). In the course of evolution, the single ancestor gene has obviously been duplicated several times, before and during vertebrate evolution. Based on sequence homology, the first duplication probably resulted in DPY19L1 separating from DPY19L3/L4, which, before the divergence of fish, deviated further into two distinct proteins. DPY19L1 remained the most similar to C. elegans DPY-19, suggesting functional conservation (30).
Our aim was to determine the activity of the mammalian enzymes of the family. Using CRISPR-Cas9 mutagenesis (32, 33), we generated Chinese hamster ovary (CHO) single knockout cells as well as a triple knockout lacking DPY19L1, -L3, and -L4 and reintroduced the respective mouse enzymes in these cells. This process revealed that DPY19L1 activity is indeed conserved in mammals and C. elegans, whereas DPY19L3 has acquired a new activity—mannosylation of W3 of the WxxWxxWxxC motif in TSRs.
Results
UNC5A Is a Target Protein for C-Mannosylation.
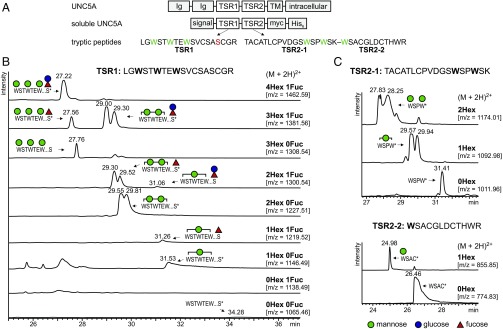
To monitor C-mannosylation, we expressed a soluble mouse netrin receptor UNC5A construct composed of the two TSRs in wild-type CHO cells. A cleavable N-terminal signal sequence allowed the protein fragment to enter the secretory pathway, and a C-terminal myc-His6 tag enabled detection and purification (Fig. 1A). Secreted UNC5A was purified by nickel-affinity chromatography, and tryptic peptides were analyzed by liquid chromatography/mass spectrometry (LC/MS). The analyzed peptide of TSR1 contains an O-fucosylation site downstream of the three putative C-mannosylation sites (Fig. 1A). TSR2 has no O-fucosylation site, but has a trypsin cleavage site between the second and third tryptophans, allowing separate detection of W3 C-mannosylation (Fig. 1A).
Fig. 1.
C-mannosylation pattern of UNC5A in wild-type CHO cells. (A) Soluble UNC5A construct and proteolytic fragments analyzed by mass spectrometry. Membrane-bound UNC5A contains two Ig domains, and two thrombospondin type 1 repeats (TSRs) in the extracellular part of the protein. The two TSRs were used in an expression construct that allowed secretion of the protein fragment. Analyzed trypsin proteolytic fragments with putative C-mannosylation (green) and O-fucosylation (red) sites are depicted. TM, transmembrane domain. (B) Extracted ion chromatograms of observed glycoforms of the UNC5A TSR1 peptide. For signals marked by *, MS/MS spectra confirmed the nature of the peptide and presence of C-mannose (SI Appendix, Fig. S1). Based on MS/MS data and elution time, peaks were attributed to the indicated glycoforms. All graphs are shown with equal intensities (12,000 counts per second). (C) Extracted ion chromatograms of observed glycoform masses of the UNC5A TSR2 peptides. Corresponding MS/MS spectra are shown in SI Appendix, Fig. S2 A and B for the first and second peptides of TSR2, respectively.
Extracted ion chromatograms (EICs) showed that several glycoforms can be observed for both TSR1 and TSR2. Glycosylation of TSR1 ranged from C-mannosylation of a single tryptophan to C-mannosylation of all three tryptophans. In addition, a fucose (Fuc) or a glucose–fucose (Glc–Fuc) disaccharide (Fig. 1B) was found, presumably on the consensus serine residue (24). The EIC profiles suggested that di- and trimannosylated forms were most abundant species and that fucosylation occurred preferably on these glycoforms. It was not possible to distinguish between the two hexoses mannose and glucose by MS alone, but C-mannosylation of a tryptophan resulted in elution of the peptide from the hydrophobic LC column approximately 2 min earlier, most likely due to covering of the hydrophobic surface of tryptophan residues, whereas fucosylation and additional glucosylation only caused slight shifts in elution time (Fig. 1B). This observation made it possible to assign individual peaks even when their masses were identical, for example, the Man3Fuc1 and Man2Fuc1Glc1 peptides from TSR1 (second row of Fig. 1B). In addition, MS/MS analysis (SI Appendix, Fig. S1), during which O-fucose is lost but C-mannose retained, confirmed the peak assignments. Thus, evaluation of extracted ion chromatograms and MS/MS analysis allowed us to distinguish between the different types of glycosylation.
MS/MS of monomannosylated species (SI Appendix, Fig. S1B) indicated the absence of C-mannose on the first and third tryptophans (W1 and W3), suggesting it was mannosylated at W2, but neither the b- nor y-ion series reached the middle tryptophan to confirm this. W1 and W3 mannosylation was detected on dimannosylated species, indicating that there was no problem in detecting mannosylated tryptophans by MS/MS (SI Appendix, Fig. S1 C–E). Dimannosylated peptides obtained from TSR1 are represented by double peaks in EIC graphs. Although the peaks could probably not be completely separated, MS/MS sequencing showed that the fraction that eluted first had almost complete mannosylated W3 and variable mannosylation of W1, indicating that this was a mixture of W1–3 and W2–3 mannosylation. The second fraction contained mainly nonmannosylated W3 and fully mannosylated W1, suggesting that this was a peptide with W1–2 mannosylation (SI Appendix, Fig. S1C). Combined information from EIC and MS/MS profiles thus allowed the C-mannoses to be linked to specific tryptophans.
The tryptic cleavage site in TSR2 of UNC5A enabled detection of the first two and the third tryptophan separately. As in TSR1, C-mannosylation was found on each tryptophan of the motif, with, based on EIC quantification, high rates of mannosylation of W1 and W2 and apparently less of W3 (Fig. 1C). MS/MS peptide sequencing showed that the monomannosylated peptide (containing W1 and W2) was exclusively mannosylated at W2 (SI Appendix, Fig. S2A); i.e., what was suggested above for TSR1 could be directly shown for TSR2. The presence of one mannose on each tryptophan could be confirmed for the dimannosylated peptide (SI Appendix, Fig. S2A). For W3 peptides, the peptide sequence could be confirmed via the y-ion series in MS/MS, but due to the lack of b-ions, the presence of a mannose could not be directly shown (SI Appendix, Fig. S2B).
In conclusion, the TSRs of mouse UNC5A can be fully mannosylated on all three tryptophans by CHO cell enzymes and the distinct tryptic peptides allowed both the rate of mannosylation and the distribution of mannoses on partly mannosylated TSRs to be addressed.
DPY19L1 and DPY19L3 Are C-Mannosyltransferases with Distinct Activities.
The single C. elegans C-mannosyltransferase DPY-19 has been shown to mannosylate the first two tryptophans of a WxxWxxW motif (14), whereas CHO cell enzymes were capable of mannosylating all three. To investigate which of the mammalian DPY-19 homologs were responsible for the separate mannosylation reactions, we individually knocked out DPY19L1, -L3, and -L4 genes in CHO cells using CRISPR-Cas9 technology. Two independent clones, all having frameshift mutations, were analyzed for each knockout. The DPY19L2 homolog was deliberately left out because its expression has been described as being restricted to developing sperm cells (34). As expected, we were not able to detect its expression in CHO cells by RT-PCR, whereas the other three genes were confirmed to be expressed (SI Appendix, Fig. S3).
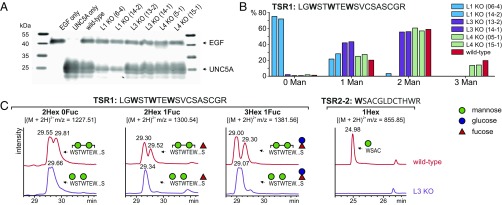
The soluble UNC5A construct was expressed in each knockout clone, and protein secretion as well as the degree of C-mannosylation was analyzed by Western blot and LC/MS as described above for wild-type CHO cells. In cells lacking DPY19L1, the secretion of UNC5A TSRs was markedly reduced, confirming the putative role of C-mannosylation in protein secretion (Fig. 2A). The small amount of secreted UNC5A protein, however, was found to be partly C-mannosylated. Quantification of the EIC profiles suggested that C-mannosylation of the protein was disrupted (Fig. 2B), with the nonmannosylated form producing the strongest signal and trimannosylated forms were undetectable. These results showed that lack of DPY19L1 resulted in reduction of C-mannosylation and that at least one additional enzyme was responsible for residual C-mannosylation in CHO cells.
Fig. 2.
UNC5A secretion and C-mannosylation in single knockouts of DPY19 enzymes. (A) Secreted UNC5A in wild-type and single knockout cells. Purified UNC5A TSRs secreted from equal numbers of WT cells or two independent knockouts of DPY19L1 (L1 KO), DPY19L3 (L3 KO), and DPY19L4 (L4 KO) were analyzed using Western blotting with anti-myc antibody. A construct of myc-tagged EGF repeats 9–14 of mouse Notch1 was used as transfection and secretion control. (B) Quantification of extracted ion chromatograms of DPY19 knockouts. EIC profiles for the different glycoforms of TSR1 were quantified by integration and presented as percentage of the total TSR1 intensity. All peptides, independent of fucosylation (see Fig. 1B), were divided into species without or with one, two, or three mannoses. (C) Comparison of dimannosylated species of TSR1 and W3 mannosylation of TSR2 in wild-type and DPY19L3 (clone 14-1) knockout cells. MS/MS analysis of the dimannosylated TSR1 peptides with m/z = 1381.56 (SI Appendix, Fig. S4) confirmed that the DPY19L3 knockout completely lacks mannosylation of W3. All graphs are shown with equal intensities (13,000 counts per second).
Knocking out DPY19L3 did not affect secretion of UNC5A TSRs (Fig. 2A) but did also result in a reduction of C-mannosylation (Fig. 2B). No trimannosylated TSR1 was observed, and the W3 of TSR2 was detected only without mannose (Fig. 2 B and C). In addition, EIC profiles showed that all TSR1 peptides with two mannoses, appearing as double peaks in wild-type cells as a result of mannosylation at different positions, were reduced to single species in DPY19L3 knockouts (Fig. 2C). MS/MS analysis established that the dimannosylated form in the DPY19L3 knockout was lacking C-mannose on W3 (SI Appendix, Fig. S4).
Deletion of DPY19L4 did not substantially affect C-mannosylation. All C-mannosylated glycoforms detected in wild-type cells were also detected at similar levels in this knockout (Fig. 2B).
These results showed that the lack of either DPY19L1 or DPY19L3 influenced glycosylation of UNC5A, with the DPY19L1 knockout having the most severe effects on both secretion and mannosylation. DPY19L3 was shown to be responsible for C-mannosylation of W3, but to have no influence on secretion. As knocking out DPY19L4 did not affect glycosylation, DPY19L1 and -L3 seem sufficient to fully glycosylate UNC5A.
Restitution of DPY19L1, -L3, and -L4 in the Triple Knockout Confirms Specificity of DPY19L1 and -L3.
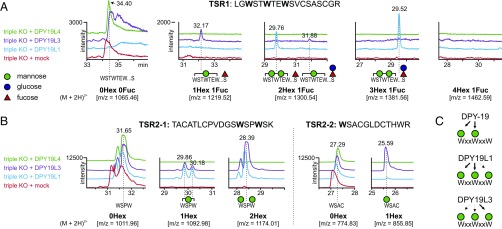
To study the activities of mammalian DPY-19 homologs separately, we generated triple knockouts of DPY19L1, -L3 and -L4 in CHO cells, expressed individual DPY-19 homologs and analyzed the resulting UNC5A C-mannosylation patterns. TSR1 was modified with a single C-mannose only, in cells overexpressing DPY19L3 (Fig. 3A, purple lines). MS/MS analysis showed that the mannose was mainly on the third tryptophan of the WxxWxxW motif (SI Appendix, Fig. S5A). The TSR2 peptide containing the third tryptophan was also C-mannosylated in DPY19L3 expressing cells but not in the triple knockout cells (Fig. 3B). The ratio of this C-mannosylated peptide to the nonmannosylated peptide was higher in cells overexpressing DPY19L3 than in wild-type cells (compare Fig. 3B and Fig. 1B). Overexpression of DPY19L1 resulted in detection of TSR1 peptides with a maximum of two C-mannoses (Fig. 3A, blue lines). In contrast to the EIC of TSR1 expressed in wild-type CHO cells, which displayed a double peak of dimannosylated peptide (Fig. 1B), a single peak assigned to a peptide lacking C-mannose on W3 was observed in the triple knockout cells expressing DPY19L1 (Fig. 3A and SI Appendix, Fig. S5B), exactly as was detected in the DPY19L3 knockout (Fig. 2C).
Fig. 3.
C-mannosylation of UNC5A TSRs in triple knockouts reexpressing single DPY19 enzymes. (A) TSR1 EIC profiles. Triple knockouts (red graph) showed only the unmodified peptide and no C-mannosylation. In cells overexpressing DPY19L1 (blue graph) the most abundant glycoforms had two mannoses and fucosylation (with or without additional glucose), whereas no trimannosylated forms were detected. Cells overexpressing DPY19L3 exclusively showed monomannosylated species. (B) TSR2 EIC profiles. In TSR2, both DPY19L1 and DPY19L3 can mannosylate the first two tryptophans (Left). C-mannosylated W3 (Right) was predominantly detected in triple knockout cells reexpressing DPY19L3. (C) Proposed substrate specificity of C. elegans DPY-19 and mammalian DPY19L1 and -L3 on the WxxWxxW motif of TSRs.
These results confirmed that DPY19L1 and DPY19L3 act on the first two and the third tryptophan, respectively (Fig. 3C). When the enzymes were overexpressed, however, this specificity no longer seemed to be absolute. DPY19L1 overexpression resulted in a residual C-mannosylation of W3 (Fig. 3B, blue line), and DPY19L3 seemed, upon overexpression, to be almost as efficient in modifying W1 and W2 of TSR2 as DPY19L1. Overexpression of DPY19L4 did not result in any change in mannosylation compared with the triple knockout (Fig. 3, green lines). This putative C-mannosyltransferase, therefore, seems not to be able to mannosylate the thrombospondin repeats of UNC5A.
Subcellular Localization of UNC5A Is Dependent on DPY19L1 Activity.
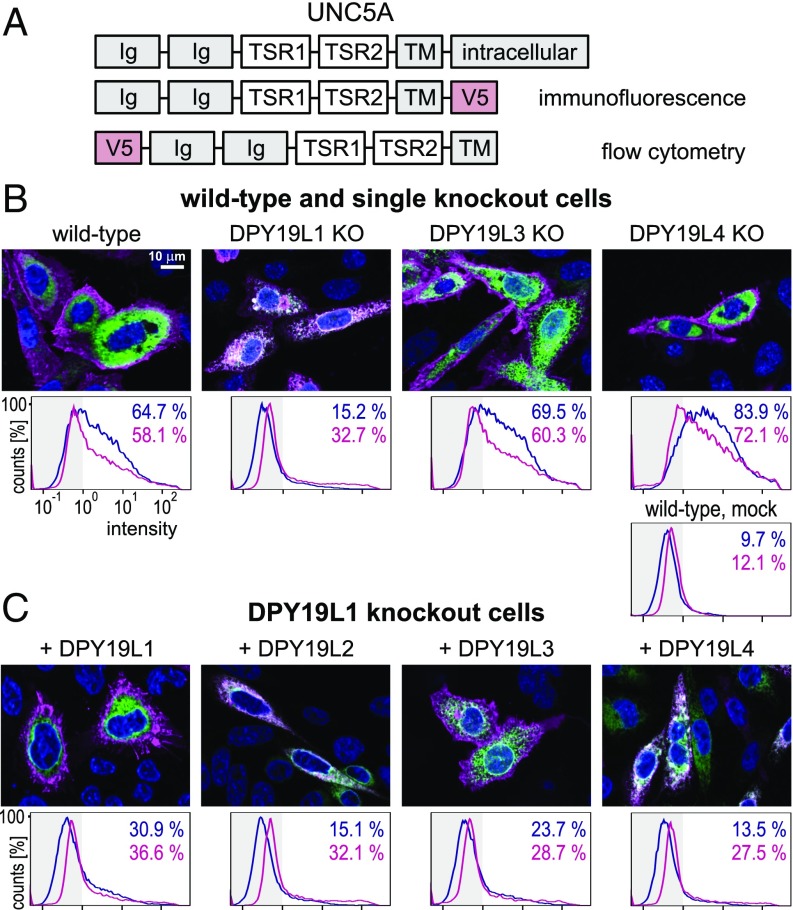
As mentioned above, secretion of UNC5A TSRs was markedly reduced in the DPY19L1 knockout but not in the DPY19L3 knockout. To study the effect on the membrane bound netrin receptor, two constructs with a V5 tag on the internal or external side of the membrane were generated (Fig. 4A). Using immunofluorescence and the internal V5 tag, UNC5A was shown to be localized on the cell surface in wild-type, DPY19L3, and -L4 knockout cells (Fig. 4B). In DPY19L1 knockouts, however, the protein colocalized with an ER marker. By flow cytometry using the external V5 tag, the same was observed in a quantitative manner (Fig. 4B). Cells were either cell surface stained or permeabilized for total cellular staining. Whereas in wild type and other mutants cell surface staining was comparable to total staining, DPY19L1 mutants showed lower surface staining. Reexpression of mouse DPY19L1 in the mutant cells restored the cell surface expression (Fig. 4C). Quantification by flow cytometry confirmed that the majority of staining was found on the cell surface, although, probably due to lower transfection efficiency of the large enzyme plasmids, the total number of positive cells remained lower than in wild-type cells. Surprisingly, overexpression of mouse DPY19L3 in DPY19L1 knockout cells also led to cell surface localization of UNC5A, whereas overexpression of DPY19L2 and DPY19L4 did not compensate for the lack of DPY19L1 (Fig. 4C).
Fig. 4.
Subcellular localization of membrane-bound UNC5A. (A) UNC5A expression constructs. (B) Localization of UNC5A in knockout cells. (Upper) Immunofluorescence of transiently expressed C-terminal V5 tagged UNC5A construct (magenta) in wild-type CHO cells and in single knockouts of DPY19L1, -L3, and -L4. KDEL-tagged GFP (green) was used as an ER marker. (Lower) Flow cytometry of the same cells expressing N-terminally V5-tagged UNC5A. Cells were stained either alive on the cell surface by anti-V5 (blue line) or after fixation and permeabilization (magenta). Percentage of cells with a staining intensity above 100 [less than 1% in unstained cells and around 10% in mock transfected, but antibody-treated cells [mock WT)] are indicated in blue and magenta, respectively. (C) Effect of overexpression of DPY19 proteins in the DPY19L1 knockout. The four mouse DPY19 proteins were transiently expressed in the DPY19L1 knockout and immunostained as in B. (Scale bar, 10 μm.)
These results indicated that the activity of DPY19L1 was crucial for UNC5A to leave the ER, but that, although the natural level of DPY19L3 seems insufficient, overexpression of DPY19L3 could restore this function. This result fully agreed with the observed effects on C-mannosylation described above. Lack of DPY19L1 resulted in the strongest reduction of C-mannosylation, whereas in DPY19L3 knockouts, only mannosylation of W3 was lacking, which does not appear to affect localization. Upon overexpression, DPY19L3 partly mannosylated W1 and W2 of TSR2 (Fig. 3B) and in addition increased mannosylation of W3. This apparently provides enough mannosylation to allow UNC5A to exit the ER.
Discussion
C-mannosylation was initially described as a modification of the first tryptophan in the WxxW consensus sequence (20, 21), but was later also observed on tryptophans that were not part of this motif (35). In addition to WxxW, WxxC is also considered to be a consensus sequence for C-mannosylation (36). We have now shown that the mammalian C-mannosyltransferases DPY19L1 and DPY19L3 do not have a dual WxxW/C recognition motif, but that these paralogous enzymes are active C-mannosyltransferases with distinct substrate specificities. Knockout experiments in CHO cells showed that DPY19L1 can C-mannosylate the first two tryptophans in the WxxWxxWxxC sequence of UNC5A TSRs, and can therefore presumably act on the first tryptophan of a WxxW sequence. DPY19L3, on the other hand, mannosylates the third tryptophan of this sequence and might require a WxxC motif. Although our experiments clearly show that DPY19L1 and DPY19L3 have distinct substrate specificity, the exact requirements of both enzymes, especially the necessity of the cysteine, have to be confirmed by other motifs than WxxWxxWxxC. At endogenous levels, DPY19L1 and DPY19L3 seem highly specific with very little cross-reactivity. However, upon overexpression, DPY19L3 can mannosylate the first two tryptophans and DPY19L1 can modify W3 to a limited extent, suggesting that this specificity is not absolute.
In line with our observations, human DPY19L3 was recently shown to C-mannosylate W2 but not W1 of the WxxWxxC sequence of R-spondin1 (15). The authors used Drosophila S2 cells, as we previously did to demonstrate the activity of C. elegans DPY-19 (14). S2 cells lack C-mannosyltransferase activity but can grow only at temperatures up to 28 °C. Upon overexpression of DPY19L1 together with R-spondin1 in this system, Niwa et al. did not observe any C-mannosylation of R-spondin1 (15). We also observed a limited efficiency of mouse C-mannosyltransferases in S2 cells and therefore generated knockouts in mammalian CHO cells to study the activities of mammalian C-mannosyltransferases. Nevertheless, we were not able to show any C-mannosyltransferase activity for DPY19L2 and -L4 on our target protein UNC5A (Figs. 3 and 4, and SI Appendix, Fig. S6). As DPY19L2 seems to be a sperm-specific protein, it is possible that it mannosylates only specific sperm proteins or that the protein is adapted to the specific secretory pathway of sperm cells and is not appropriately expressed in other cells. An argument for the latter possibility is that DPY19L2 has been attributed to the nuclear membrane (34). The lack of activity of DPY19L4 is more surprising. DPY19L4 is properly ER localized (SI Appendix, Fig. S7), but might have a different target sequence or may not be a C-mannosyltransferase at all.
Our results show that inactivation of DPY19L1 but not DPY19L3 strongly reduced the secretion of UNC5A TSRs and led to the accumulation of the membrane bound protein in the ER. In contrast, using RNAi, Niwa et al. showed that down-regulation of DPY19L3 affected secretion of R-spondin1 (15). As R-spondin1 has a WxxWxxC motif and UNC5A a WxxWxxWxxC motif, it is conceivable that in the case of UNC5A, mannosylation of the first two of three tryptophans is sufficient for secretion, whereas in R-spondin1, mannosylation of the second tryptophan by DPY19L3 is required.
The importance of C-mannosylation for protein secretion might reflect a function in protein folding. Pulse-chase experiments have shown that C-mannosylation occurs early, potentially before or during folding (13) and likely precedes O-fucosylation, which takes place on folded proteins (37, 38). This hypothesis is consistent with our observation that we never see fucose on nonmannosylated peptides. The latter observation suggests that C-mannosylation is a prerequisite for O-fucosylation of UNC5A and potentially also for its folding. In summary, we can conclude that the mammalian proteins of the DPY19 family are not enzymes with redundant activities but have evolved to carry out distinct reactions. The CHO cell knockouts generated here will now allow the study of the effects of C-mannosylation on various proteins. In addition to TSR-containing proteins, another important class of proteins that are C-mannosylated are the type I cytokine receptors (39, 40). Many other unrelated proteins have WxxW or WxxC consensus sequences (36), suggesting a possible more widespread role for C-mannosylation.
Materials and Methods
Generation of CRISPR-Cas9 Knockout Clones.
CRIPSR-Cas9 vector pX330-U6-Chimeric_BB-CBh-hSpCas9 (33) was a gift from Feng Zhang, Broad Institute, Cambridge, MA (Addgene, plasmid 42230). The following target sites were used (protospacer-adjacent motif underlined): ACAGCAACATAAAAGCAAGC-AGG (exon 6, DPY19L1), ATACTACAAGGAGATGCTGC-AGG (exon 3, DPY19L3), and GCACTGCATTGATCGTCTTC-AGG (exon 7, DPY19L4).
To generate knockouts, CHO cells were grown in DMEM/Ham’s F-12 (Biochrom) with 5% (vol/vol) FCS (Biochrom). For transfection, 1 × 105 cells were seeded in 2 mL in a six-well plate. Next day, cells were transfected with 1.5 µg CRISPR-Cas9 vector and 0.5 µg peGFP-C1 (Clontech) with 10 µg polyethyleneimine (PEI MAX 40000, Polysciences) diluted in 200 µL Opti-MEM (Thermo Fisher). After 6 h, medium was exchanged. On day 2, cells were transferred to 96-well plates at a concentration of 0.7 cells per well. After 3–4 d, plates were screened for wells with single green fluorescent colonies. Expanded colonies (2 × 105 cells) were lysed in 20 µL of lysis buffer (10 mM Tris⋅HCl pH 7.6, 50 mM NaCl, 6.25 mM MgCl, 0.045% Nonidet P-40, 0.45% Tween-20) containing 2 mg/mL Proteinase K for 1 h at 56 °C and 15 min at 95 °C (41). A total of 1 µL of lysate was used in two PCR reactions, one using the forward primer of the CRISPR-Cas target site and a primer at a 200- to 300-bp distance. For samples without or with reduced PCR product, a second PCR product covering about 400 bp around the target site was sequenced. Clones with frameshift-causing indels were selected. The obtained clones were subcloned using the same procedure.
To generate triple knockout clones, CHO DPY19L3 knockout cells (clone 14-1) were used to knock out DPY19L1 and subsequently DPY19L4 genes. At least two independent clones were obtained for all knockouts (SI Appendix).
Transient UNC5A Expression in CHO Cells.
TSRs of mouse UNC5A were cloned into HindIII and XbaI sites of the pSecTag B vector (Invitrogen), having an N-terminal signal sequence and a C-terminal myc-His6 tag. Complete mouse DPY19L1, -L2, -L3, and -L4 ORFs were PCR amplified and cloned into pcDNA3.1/myc-His A vector (Invitrogen) via HindIII/XhoI (DPY19L1–3) and HindIII/XbaI (-L4).
For transient transfection, 3 × 106 cells were seeded in 18 mL of medium and on the next day transfected with 10 µg UNC5A and 10 µg DPY19 plasmid or empty vector using 100 µg polyethyleneimine in 2 mL Opti-MEM. Medium was exchanged for 30 mL fresh medium after 6 h and secreted UNC5A was purified from the medium by nickel-affinity chromatography after 3 d. Therefore, medium was supplemented to generate 20 mL 500 mM NaCl, 20 mM Tris⋅HCl pH 8 and 20 mM imidazole. Samples were filtered through a 0.2-µm membrane (Waters) and pumped over a 1-mL HisTrap HP column (GE Healthcare) at 1 mL/min. After a 10-mL washing step in the above buffer, a linear gradient (20–350 mM imidazole) was applied over 7 mL. Eluted UNC5A was precipitated with acetone, resuspended in Laemmli buffer containing 5% (vol/vol) 2-mercaptoethanol and incubated at 95 °C for 10 min. The sample was loaded on a 15% (wt/vol) SDS/PAGE gel and stained with Coomassie. For Western blotting, a control construct containing mouse Notch EGF repeat 9–14 was cotransfected in a 1:2 ratio to UNC5A and 1/10 of purified sample was loaded on gel. Mouse anti-myc 9E10 antibody and IRDye 800 conjugated goat anti-mouse antibody (LI-COR) were used for detection.
Mass Spectrometry.
Protein bands were excised from gels, treated with DTT and iodoacetamide and digested with trypsin at 37 °C in an overnight reaction. The peptides were extracted and analyzed by mass spectrometry as described (14), using a Waters nanoACQUITY UPLC System equipped with an analytical column (Waters, BEH130 C18; 100 µm × 100 µm, 1.7-µm particle size) coupled online to an ESI-Q-TOF Ultima (Waters). Spectra were explored with MassLynx V4.1 software (Waters).
Immunofluorescent Microscopy and Flow Cytometry.
UNC5A constructs were cloned into pcDNA3.1/V5-HisB and pSecTag (Invitrogen) via HindIII and XhoI sites (sequence in SI Appendix). As an ER marker, pAcGFP1 (Clontech), elongated by a KDEL sequence, was used.
The 0.8 × 105 CHO cells were seeded on glass coverslips in a 12-well plate and transfected with 0.5 µg UNC5A vector, 0.25 µg GFP-KDEL vector, and 0.25 µg DPY19 plasmid or empty vector as described above. Cells were incubated for 2 d, fixed with 4% (wt/vol) formaldehyde, permeabilized, and blocked using 0.1% saponin (Sigma) and 0.1% BSA. UNC5A-V5-His was detected using mouse anti-V5 (Acris; 1:500) as primary and Cy3-conjugated sheep anti-mouse (Sigma, 1:1,000) as secondary antibodies. Images were acquired using a Zeiss Axio Observer.Z1 microscope using the ApoTome module.
For flow cytometry, cells were stained using the same anti-V5 antibody and Alexa-488 goat anti-mouse (Molecular Probes). The 5 × 105 cells were either stained alive or fixed in 4% (wt/vol) paraformaldehyde and permeabilized with 0.1% saponin. Samples were measured with a CyFlow ML flow cytometer (Partec). A total of 20,000 events, gated based on forward and side scatter of each sample, were analyzed with FlowJo (Ashland).
Supplementary Material
Acknowledgments
This work was supported by the DFG (Deutsche Forschungsgemeinschaft), Grant BA4091/5-1, and Cluster of Excellence REBIRTH (Regenerative Biology to Reconstructive Therapy, EXC 62/2).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. G.W.H. is a Guest Editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1613165114/-/DCSupplemental.
References
- 1.Tucker RP. The thrombospondin type 1 repeat superfamily. Int J Biochem Cell Biol. 2004;36(6):969–974. doi: 10.1016/j.biocel.2003.12.011. [DOI] [PubMed] [Google Scholar]
- 2.Tzarfaty-Majar V, et al. Plasmin-mediated release of the guidance molecule F-spondin from the extracellular matrix. J Biol Chem. 2001;276(30):28233–28241. doi: 10.1074/jbc.M102585200. [DOI] [PubMed] [Google Scholar]
- 3.Klar A, Baldassare M, Jessell TM. F-spondin: A gene expressed at high levels in the floor plate encodes a secreted protein that promotes neural cell adhesion and neurite extension. Cell. 1992;69(1):95–110. doi: 10.1016/0092-8674(92)90121-r. [DOI] [PubMed] [Google Scholar]
- 4.Prater CA, Plotkin J, Jaye D, Frazier WA. The properdin-like type I repeats of human thrombospondin contain a cell attachment site. J Cell Biol. 1991;112(5):1031–1040. doi: 10.1083/jcb.112.5.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holt GD, Pangburn MK, Ginsburg V. Properdin binds to sulfatide [Gal(3-SO4)beta 1-1 Cer] and has a sequence homology with other proteins that bind sulfated glycoconjugates. J Biol Chem. 1990;265(5):2852–2855. [PubMed] [Google Scholar]
- 6.Tortorella M, et al. The thrombospondin motif of aggrecanase-1 (ADAMTS-4) is critical for aggrecan substrate recognition and cleavage. J Biol Chem. 2000;275(33):25791–25797. doi: 10.1074/jbc.M001065200. [DOI] [PubMed] [Google Scholar]
- 7.Tossavainen H, et al. The layered fold of the TSR domain of P. falciparum TRAP contains a heparin binding site. Protein Sci. 2006;15(7):1760–1768. doi: 10.1110/ps.052068506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klenotic PA, et al. Molecular basis of antiangiogenic thrombospondin-1 type 1 repeat domain interactions with CD36. Arterioscler Thromb Vasc Biol. 2013;33(7):1655–1662. doi: 10.1161/ATVBAHA.113.301523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Russell S, et al. Combined therapy with thrombospondin-1 type I repeats (3TSR) and chemotherapy induces regression and significantly improves survival in a preclinical model of advanced stage epithelial ovarian cancer. FASEB J. 2015;29(2):576–588. doi: 10.1096/fj.14-261636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan K, et al. Crystal structure of the TSP-1 type 1 repeats: A novel layered fold and its biological implication. J Cell Biol. 2002;159(2):373–382. doi: 10.1083/jcb.200206062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olsen JG, Kragelund BB. Who climbs the tryptophan ladder? On the structure and function of the WSXWS motif in cytokine receptors and thrombospondin repeats. Cytokine Growth Factor Rev. 2014;25(3):337–341. doi: 10.1016/j.cytogfr.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 12.Niwa Y, Suzuki T, Dohmae N, Simizu S. O-Fucosylation of CCN1 is required for its secretion. FEBS Lett. 2015;589(21):3287–3293. doi: 10.1016/j.febslet.2015.09.012. [DOI] [PubMed] [Google Scholar]
- 13.Perez-Vilar J, Randell SH, Boucher RC. C-Mannosylation of MUC5AC and MUC5B Cys subdomains. Glycobiology. 2004;14(4):325–337. doi: 10.1093/glycob/cwh041. [DOI] [PubMed] [Google Scholar]
- 14.Buettner FF, Ashikov A, Tiemann B, Lehle L, Bakker H. C. elegans DPY-19 is a C-mannosyltransferase glycosylating thrombospondin repeats. Mol Cell. 2013;50(2):295–302. doi: 10.1016/j.molcel.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 15.Niwa Y, Suzuki T, Dohmae N, Simizu S. Identification of DPY19L3 as the C-mannosyltransferase of R-spondin1 in human cells. Mol Biol Cell. 2016;27(5):744–756. doi: 10.1091/mbc.E15-06-0373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vasudevan D, Takeuchi H, Johar SS, Majerus E, Haltiwanger RS. Peters plus syndrome mutations disrupt a noncanonical ER quality-control mechanism. Curr Biol. 2015;25(3):286–295. doi: 10.1016/j.cub.2014.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ricketts LM, Dlugosz M, Luther KB, Haltiwanger RS, Majerus EM. O-fucosylation is required for ADAMTS13 secretion. J Biol Chem. 2007;282(23):17014–17023. doi: 10.1074/jbc.M700317200. [DOI] [PubMed] [Google Scholar]
- 18.Wang LW, et al. O-fucosylation of thrombospondin type 1 repeats in ADAMTS-like-1/punctin-1 regulates secretion: Implications for the ADAMTS superfamily. J Biol Chem. 2007;282(23):17024–17031. doi: 10.1074/jbc.M701065200. [DOI] [PubMed] [Google Scholar]
- 19.Hofsteenge J, et al. New type of linkage between a carbohydrate and a protein: C-glycosylation of a specific tryptophan residue in human RNase Us. Biochemistry. 1994;33(46):13524–13530. doi: 10.1021/bi00250a003. [DOI] [PubMed] [Google Scholar]
- 20.de Beer T, Vliegenthart JF, Löffler A, Hofsteenge J. The hexopyranosyl residue that is C-glycosidically linked to the side chain of tryptophan-7 in human RNase Us is alpha-mannopyranose. Biochemistry. 1995;34(37):11785–11789. doi: 10.1021/bi00037a016. [DOI] [PubMed] [Google Scholar]
- 21.Doucey MA, Hess D, Blommers MJ, Hofsteenge J. Recombinant human interleukin-12 is the second example of a C-mannosylated protein. Glycobiology. 1999;9(5):435–441. doi: 10.1093/glycob/9.5.435. [DOI] [PubMed] [Google Scholar]
- 22.Hofsteenge J, Blommers M, Hess D, Furmanek A, Miroshnichenko O. The four terminal components of the complement system are C-mannosylated on multiple tryptophan residues. J Biol Chem. 1999;274(46):32786–32794. doi: 10.1074/jbc.274.46.32786. [DOI] [PubMed] [Google Scholar]
- 23.Hartmann S, Hofsteenge J. Properdin, the positive regulator of complement, is highly C-mannosylated. J Biol Chem. 2000;275(37):28569–28574. doi: 10.1074/jbc.M001732200. [DOI] [PubMed] [Google Scholar]
- 24.Hofsteenge J, et al. C-mannosylation and O-fucosylation of the thrombospondin type 1 module. J Biol Chem. 2001;276(9):6485–6498. doi: 10.1074/jbc.M008073200. [DOI] [PubMed] [Google Scholar]
- 25.Wang LW, Leonhard-Melief C, Haltiwanger RS, Apte SS. Post-translational modification of thrombospondin type-1 repeats in ADAMTS-like 1/punctin-1 by C-mannosylation of tryptophan. J Biol Chem. 2009;284(44):30004–30015. doi: 10.1074/jbc.M109.038059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gonzalez de Peredo A, et al. C-mannosylation and o-fucosylation of thrombospondin type 1 repeats. Mol Cell Proteomics. 2002;1(1):11–18. doi: 10.1074/mcp.m100011-mcp200. [DOI] [PubMed] [Google Scholar]
- 27.Harbuz R, et al. A recurrent deletion of DPY19L2 causes infertility in man by blocking sperm head elongation and acrosome formation. Am J Hum Genet. 2011;88(3):351–361. doi: 10.1016/j.ajhg.2011.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koscinski I, et al. DPY19L2 deletion as a major cause of globozoospermia. Am J Hum Genet. 2011;88(3):344–350. doi: 10.1016/j.ajhg.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elinati E, et al. Globozoospermia is mainly due to DPY19L2 deletion via non-allelic homologous recombination involving two recombination hotspots. Hum Mol Genet. 2012;21(16):3695–3702. doi: 10.1093/hmg/dds200. [DOI] [PubMed] [Google Scholar]
- 30.Carson AR, Cheung J, Scherer SW. Duplication and relocation of the functional DPY19L2 gene within low copy repeats. BMC Genomics. 2006;7:45. doi: 10.1186/1471-2164-7-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cova M, Rodrigues JA, Smith TK, Izquierdo L. Sugar activation and glycosylation in Plasmodium. Malar J. 2015;14:427. doi: 10.1186/s12936-015-0949-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pierre V, et al. Absence of Dpy19l2, a new inner nuclear membrane protein, causes globozoospermia in mice by preventing the anchoring of the acrosome to the nucleus. Development. 2012;139(16):2955–2965. doi: 10.1242/dev.077982. [DOI] [PubMed] [Google Scholar]
- 35.Ervin LA, Ball LE, Crouch RK, Schey KL. Phosphorylation and glycosylation of bovine lens MP20. Invest Ophthalmol Vis Sci. 2005;46(2):627–635. doi: 10.1167/iovs.04-0894. [DOI] [PubMed] [Google Scholar]
- 36.Julenius K. NetCGlyc 1.0: Prediction of mammalian C-mannosylation sites. Glycobiology. 2007;17(8):868–876. doi: 10.1093/glycob/cwm050. [DOI] [PubMed] [Google Scholar]
- 37.Luo Y, Koles K, Vorndam W, Haltiwanger RS, Panin VM. Protein O-fucosyltransferase 2 adds O-fucose to thrombospondin type 1 repeats. J Biol Chem. 2006;281(14):9393–9399. doi: 10.1074/jbc.M511975200. [DOI] [PubMed] [Google Scholar]
- 38.Valero-González J, et al. A proactive role of water molecules in acceptor recognition by protein O-fucosyltransferase 2. Nat Chem Biol. 2016;12(4):240–246. doi: 10.1038/nchembio.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hamming OJ, et al. Crystal structure of interleukin-21 receptor (IL-21R) bound to IL-21 reveals that sugar chain interacting with WSXWS motif is integral part of IL-21R. J Biol Chem. 2012;287(12):9454–9460. doi: 10.1074/jbc.M111.311084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Furmanek A, Hess D, Rogniaux H, Hofsteenge J. The WSAWS motif is C-hexosylated in a soluble form of the erythropoietin receptor. Biochemistry. 2003;42(28):8452–8458. doi: 10.1021/bi034112p. [DOI] [PubMed] [Google Scholar]
- 41.van der Burg M, et al. EU-supported EuroChimerism Consortium project QLRT-2001-01485 Standardization of DNA isolation from low cell numbers for chimerism analysis by PCR of short tandem repeats. Leukemia. 2011;25(9):1467–1470. doi: 10.1038/leu.2011.118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.