Significance
Studies of differing female and male demographic histories on the basis of ancient genomes can provide insight into the social structures and cultural interactions during major events in human prehistory. We consider the sex-specific demography of two of the largest migrations in recent European prehistory. Using genome-wide ancient genetic data from multiple Eurasian populations spanning the last 10,000 years, we find no evidence of sex-biased migrations from Anatolia, despite the shift to patrilocality associated with the spread of farming. In contrast, we infer a massive male-biased migration from the steppe during the late Neolithic and Bronze Age. The contrasting patterns of sex-specific migration during these two migrations suggest that different sociocultural processes drove the two events.
Keywords: admixture, migration, Neolithic, sex bias, X chromosome
Abstract
Dramatic events in human prehistory, such as the spread of agriculture to Europe from Anatolia and the late Neolithic/Bronze Age migration from the Pontic-Caspian Steppe, can be investigated using patterns of genetic variation among the people who lived in those times. In particular, studies of differing female and male demographic histories on the basis of ancient genomes can provide information about complexities of social structures and cultural interactions in prehistoric populations. We use a mechanistic admixture model to compare the sex-specifically–inherited X chromosome with the autosomes in 20 early Neolithic and 16 late Neolithic/Bronze Age human remains. Contrary to previous hypotheses suggested by the patrilocality of many agricultural populations, we find no evidence of sex-biased admixture during the migration that spread farming across Europe during the early Neolithic. For later migrations from the Pontic Steppe during the late Neolithic/Bronze Age, however, we estimate a dramatic male bias, with approximately five to 14 migrating males for every migrating female. We find evidence of ongoing, primarily male, migration from the steppe to central Europe over a period of multiple generations, with a level of sex bias that excludes a pulse migration during a single generation. The contrasting patterns of sex-specific migration during these two migrations suggest a view of differing cultural histories in which the Neolithic transition was driven by mass migration of both males and females in roughly equal numbers, perhaps whole families, whereas the later Bronze Age migration and cultural shift were instead driven by male migration, potentially connected to new technology and conquest.
Genetic data suggest that modern European ancestry represents a mosaic of ancestral contributions from multiple waves of prehistoric migration events. Recent studies of genomic variation in prehistoric human remains have demonstrated that two mass migration events are particularly important to understanding European prehistory: the Neolithic spread of agriculture from Anatolia starting ∼9,000 y ago and migration from the Pontic-Caspian Steppe ∼5,000 y ago (1–7). These migrations are coincident with large social, cultural, and linguistic changes, and each has been inferred to have replaced more than half of the contemporaneous gene pool of resident central Europeans.
During such events, males and females often experience different demographic histories owing to cultural factors, such as norms regarding inheritance and the residence locations of families in relation to parental residence, social hierarchy, sex-biased admixture, and inbreeding avoidance (8–12). Empirical evidence suggests that sex-specific differences in migration and admixture have shaped patterns of human genomic variation worldwide, with notable examples occurring in Africa, Austronesia, Central Asia, and the Americas (13–16). These sex-specific behaviors leave signatures in the patterns of variation in genetic material that is differentially inherited between males and females in a population. Therefore, contrasting patterns of genetic variation for differentially inherited genetic material can be informative about past sociocultural and demographic events (8–12, 17).
Analyses of the maternally inherited mitochondrial DNA (mtDNA) and the paternally inherited Y chromosome have lent differential support to the hypothesis that the Neolithic spread of agriculture from Anatolia occurred through a large population migration rather than a spread of technology (18–22). In general, studies of Y-chromosomal data more than mtDNA have supported Anatolian migration. This pattern of results has been interpreted as evidence for male-biased migration of the population that introduced farming (18, 20, 21). The hypothesis of male-biased migration of farming populations is consistent with ethnographic studies showing a higher frequency of patrilocality in farming than in hunter-gatherer (HG) populations, because an inheritance model through the paternal lineage would favor the persistence of farming-associated Y chromosomes as the source population would have greater flexibility in female mates. Isotopic studies from Neolithic European archeological sites suggest more female than male migration on a local scale, supporting the shift to patrilocality in the region (10, 23). However, genetic evidence has been mixed; both Near Eastern-related mitochondrial and Y-chromosome haplotypes have been observed in European populations, which could indicate comparable male and female migration during the Neolithic spread of agriculture. For example, Haak et al. (22) find that mitochondrial haplotype N1a, associated with Near Eastern farmers, occurs at about ∼25% frequency in Neolithic central Europeans. Later migrations from the steppe, which were previously not accounted for, may have obscured signal and interpretation (22).
Based on archeological data, as well as ancient and modern Y chromosome data, the later migration from the Pontic-Caspian Steppe has also been hypothesized to be male-biased (5, 24–29). In particular, multiple large-scale studies of modern Y-chromosome data infer a rapid growth of R1a and R1b haplotypes ∼5,000 y ago (27–29). Similarly, Haak et al. (5) provide evidence that R1a and R1b were rare in central Europe before ∼4,500 y ago, but common soon thereafter. They also observe multiple R1b haplotypes in ancient Yamnaya individuals from the steppe. Populations in the Pontic-Caspian Steppe region, such as the Yamnaya or Pit Grave culture, are thought to have strong male-biased hierarchy, as inferred by overrepresentation of male burials, male deities, and kinship terms (26, 30). The region is a putative origin for the domesticated horse in Europe, and the culture is known for its use of horse-driven wagons, a potential male-biased mechanism of dispersal into central Europe (30).
Recent analytical advances in the understanding of admixture on the autosomes and the sex-specifically–inherited X chromosome and technological advances that have generated genome-wide data from many ancient samples now make it possible to consider the contrasting male and female genetic histories of prehistoric Europe. We test the hypotheses that migrations from Anatolia during the Neolithic transition and from the Pontic Steppe during the late Neolithic/Bronze Age period were male biased.
Results
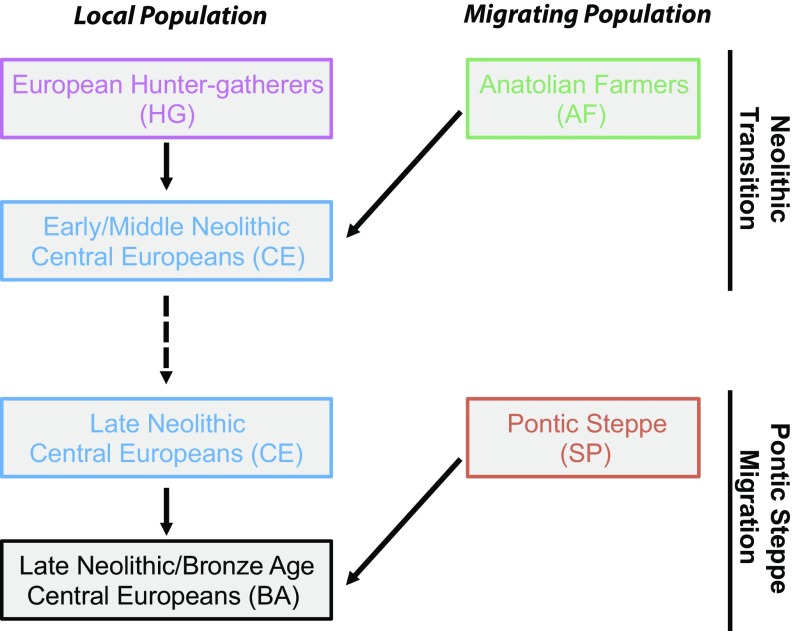
Fig. 1 provides a schematic of the population admixture events that have previously been inferred (1–7). Previous studies have inferred the relationship between the various ancient populations shown in the figure, but they did not consider a population history model. We compare genetic differentiation of the autosomes and the X chromosome between the migrating and admixed populations for each migration event: Anatolian farmers (AF) to early Neolithic central Europeans (CE) and Pontic Steppe pastoralists (SP) to late Neolithic and Bronze Age central Europeans (BA). We compute the statistic Q (31, 32), which is an estimator of the ratio of effective population size of the X chromosome with the ratio of effective population size of the autosomes based on the measure of genetic differentiation (Materials and Methods). Under a simple demographic model with equal male and female effective sizes, Q is expected to be 3/4, because there are three X chromosomes for every four autosomes in the population. Deviations from 3/4 may therefore show sex-biased effective population sizes, which indicate different population histories for males and females. Comparing AF and CE populations for the Neolithic transition, the ratio of X and autosomal differentiation is similar to what is expected for a non–sex-biased process (Q = 0.700; Table 1). In contrast, there is high relative differentiation on the X chromosome between SP and BA populations (Q = 0.237; Table 1), indicating strong male bias during the Pontic Steppe migration.
Fig. 1.
Schematic of the admixture history of central European farmers during the Neolithic and Bronze Age. First, a migration from Anatolia occurred during the Neolithic transition, and, second, a late Neolithic/Bronze Age migration occurred from the Pontic-Caspian Steppe to central Europe. In both cases, the migrating population mixed with the contemporaneous local population upon entering central Europe.
Table 1.
Comparisons of on the X chromosome and autosomes
| Populations | Q | ||
| CE-AF | 0.004 | 0.005 | 0.700 |
| CE-HG | 0.053 | 0.057 | 0.911 |
| BA-SP | 0.008 | 0.032 | 0.237 |
| BA-AF | 0.025 | 0.030 | 0.826 |
| BA-HG | 0.031 | 0.035 | 0.872 |
The quantity Q compares genetic differentiation, calculated from , on the X chromosome and autosomes. For an ideal population with no changes in effective size, Q is expected to be 3/4 (Materials and Methods). Notably, Q is close to 3/4 for the CE-AF comparisons, but it is considerably lower for the BA-SP comparisons.
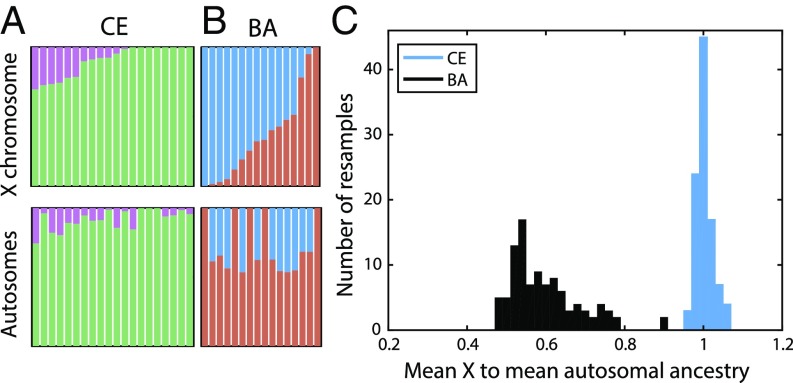
To infer sex-specific admixture rates and compare potential migration models, we estimated ancestry proportions on the X chromosome and autosomes separately, with a model-based clustering algorithm (33), using the ancient genomes as proxies for the ancient source groups in our population model and using supervised clustering (Materials and Methods, Fig. 2, and SI Appendix, Figs. S1 and S2 and Tables S1–S7). For an admixture process with equally many males and females contributing, the ratio of mean X-chromosomal admixture to mean autosomal admixture is expected to be 1. An admixture process with more contributing males leads to a reduction of the migrating population’s ancestry on the X chromosome compared with the autosomes.
Fig. 2.
Comparisons of estimated X and autosomal ancestry on the basis of model-based supervised clustering. (A) Early/middle Neolithic Europeans (CE). (B) Late Neolithic/Bronze Age Europeans (BA). Individuals are ordered by X-chromosomal ancestry, with corresponding autosomal ancestry for the same individual shown below. Clustering results by individual are presented in SI Appendix, Table S1. (C) Histograms of the ratio of the mean across individuals of X-chromosomal ancestry to the mean across individuals of autosomal ancestry for 100 autosomal resampled estimates using random sets of SNPs equal in size to the set of X-chromosomal SNPs for the corresponding population (Materials and Methods). Colors for all panels correspond to ancestry groups given in Fig. 1.
Neolithic Migration.
For the Neolithic transition, we estimated the ratio of mean AF ancestry on the X chromosome (across individuals) to the mean on the autosomes as . The corresponding ratio for European HG ancestry is . Comparing the mean X-chromosomal AF ancestry with the mean autosomal AF ancestry in each of 100 estimates from resampled autosomal SNPs (Materials and Methods), the median ratio of X to autosomal AF ancestries is 1.00 (Fig. 2C). The mean X-chromosomal admixture ± 1 SE estimated by bootstrapping the admixture estimates in 100 resamples of blocks of SNPs largely overlaps with the distribution of mean autosomal ancestry in the population over the 100 estimates (SI Appendix, Fig. S2). The distributions of X and autosomal ancestry within the sampled population are not significantly different (P = 0.493, Wilcoxon signed-rank test; SI Appendix, Fig. S3).
We additionally considered the fraction of individuals in the admixed population with higher X-chromosomal than autosomal ancestry. This measure is indicative of sex bias, with less emphasis on the exact values of the ancestry proportions. Excluding three individuals with 100% ancestry estimated to be from Anatolian-related populations on both the X and autosomes, nine of 17 individuals have higher X than autosomal ancestry (P = 0.500, binomial test).
Notably, the four middle Neolithic individuals (SI Appendix, Table S1) have higher HG ancestry than earlier CE individuals, consistent with a previously described resurgence of HG ancestry during the middle Neolithic (5, 34). The ratio of the mean X to the mean autosomal ancestry for this group of four samples is , supporting no sex bias in farming contributions to CE individuals. Similarly, we see a significant relationship between sample age and ancestry when fit to a linear model (Materials and Methods), although the similarity of X and autosomal ancestry holds over time (SI Appendix, Fig. S1A).
We find no statistical support for differences in X and autosomal ancestry; however, we cannot exclude low levels of sex-specific mating between early farmers and hunter-gatherers. Therefore, we evaluated the magnitude of differences in male and female contributions that would be consistent with observed X-to-autosomal ancestry ratios. We determined this range of sex bias values by simulating ancestry under a mechanistic admixture model, including genetic drift and sampling at specified sample sizes (17, 35, 36) (Materials and Methods and Fig. 3A). Even for a small admixed population, the largest bias consistent with the observed X and autosomal ancestries is less than 1.2 males for every female, with a median over 1,000 simulations of 1.07.
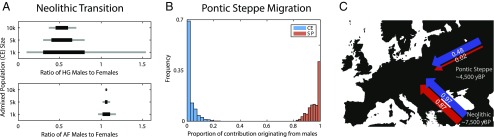
Fig. 3.
Estimated levels of sex bias during the Neolithic transition and Pontic Steppe migration. (A) Neolithic transition. The range of sex bias, measured as the ratio of males to females from a source population, that is consistent with the observed ratio of X and autosomal ancestries (Materials and Methods). Total contributions from the source population, the fraction of admixed individuals with a parent from that source population, are specified based on autosomal ancestry as 0.913 from AF and 0.087 from HG. Lines indicate that the observed ratios of X to autosomal ancestry in our dataset were present in the middle 50% (black) or middle 80% (gray) of 1,000 simulated admixed populations for specified CE population sizes. (B) Pontic Steppe migration. Under a model of constant admixture over time, the fraction of the total contribution of genetic material originating from males for each source population: CE and SP. Contributions are estimated from the migration parameter sets that have the smallest 0.1% Euclidean distance between observed and model-calculated ancestries. (C) Schematic of sex-specific migrations during the early Neolithic and later Neolithic/Bronze Age. Female contributions in are shown in red, and male contributions are shown in blue. Parameters are estimated under a single pulse migration model from Anatolia and under a constant migration model for the Pontic Steppe migration. The total contribution of each population is the average of female and male contributions from that source.
Based on the slightly larger X than autosomal ancestry observed for HG ancestry, under the simulation framework, we estimate a median of 1.91 females for every male from the HG population to the early CE population. The signal of female bias in contributions from HG to CE populations might be caused by a male-biased inheritance structure in the new farming population; that is, it is possible that the migration from Anatolia involved substantial contributions from both men and women, but once in central Europe, a shift to patrilocality might have made absorption of local HG females easier than absorption of HG males. However, the absolute difference between estimated male and female contributions is small (∼0.06). Correspondingly, differences in the numbers of female and male migrants would be small or are potentially a result of sampling.
Considering these analyses together, we find no statistical support for a male-biased migration from Anatolia. Only a small range of possible sex bias is consistent with the data; however, owing to the small total contribution from the HG population, we see female-biased contributions from the HG to CE populations (Fig. 3A).
Pontic-Caspian Steppe Migration.
We next considered female and male migration histories during the late Neolithic/Bronze Age migration from the Pontic-Caspian Steppe (Fig. 1). In contrast to the CE population during the early Neolithic expansion from Anatolia, we find a strikingly lower distribution of SP ancestry on the X chromosome than the autosomes (in accordance with results; Fig. 2 and SI Appendix, Fig. S2), suggesting extreme male-biased migration from SP during the late Neolithic/Bronze Age migration from the Pontic-Caspian Steppe. Using an approach that is similar to the approach used for the early Neolithic migration event, the ratio of mean X-chromosomal SP ancestry to mean autosomal SP ancestry in the BA population is . The ratio of mean X-chromosomal CE ancestry to mean autosomal CE ancestry in the BA population is . Of 16 admixed BA individuals, 12 have more SP ancestry on the autosomes than on the X chromosome (binomial test, P = 0.038). Similarly, the distribution of P values of the Wilcoxon signed-rank test comparing the estimated X-chromosomal ancestries with the autosomal ancestries in each of 100 resamples of autosomal SNPs is highly skewed toward zero, with a median of P = 0.02 (Materials and Methods and SI Appendix, Fig. S3).
To interpret the values of sex-specific admixture that can produce the observed ratio of X to autosomal SP ancestry of about 0.6, we considered four models for the admixture process. The first model is a single admixture event, in which an SP population quickly mixes with central European farmers, with no further migration from either population to the admixed BA population. Under this model, however, the level of sex bias is too high to have been produced by a single admixture event; no solution for the female and male migration rates exists within the possible admixture contribution range from 0 to 1 (Materials and Methods). In other words, in a pulse migration and admixture scenario in a single generation, even a male-only migration event is not extreme enough to generate the observed X-to-autosome bias in the data. Ongoing male migration from the steppe over multiple generations is therefore required to explain observed patterns of X and autosomal ancestry.
We therefore considered a model of constant contributions over time from the SP population and early Neolithic farmers (CE). We follow the method of Goldberg and Rosenberg (17), comparing expected X and autosomal ancestry (equations 5, 17, and 18 of ref. 17 and equation 30 of ref. 35) with observed ancestry in our data over a grid of possible parameter values. We present results from the 0.1% of parameter sets closest to observed data using a Euclidean distance between model-based and observed population mean ancestries on the X and autosomes (Materials and Methods). Other cutoffs (0.5%, 1%, and 5%) produced similar trends.
SI Appendix, Fig. S4 plots the range of sex-specific contributions from the SP and CE populations that produce estimates close to the estimates observed in the BA population. Males from the steppe and central European females show substantial ongoing migration, with continuing admixture rates of almost 1/2; that is, almost half of the male parents in each generation of BA individuals are new migrants from the SP population. Females from the steppe and early Neolithic European males, however, are estimated to have contributed negligibly to the BA population. Fig. 3B plots the proportional contribution of males from each source population, with a median of about 94% of SP ancestry in the BA population coming from male SP migrants and all local CE ancestry originating in CE females. This result corresponds to ∼14 male migrants for every female migrant from the steppe contributing to the ancestry of the BA population. Considering the smallest 0.5%, 1% , and 5% of Euclidean distances instead, this ratio is about 8.5, 7.5, and 5.1, respectively, males per female migrating from the steppe. These estimates are similar to estimates from modern Y-chromosome data, suggesting a reduction in the male effective population size by more than fivefold about 5,000 y ago (28).
The signature of X-chromosomal to autosomal ancestry is driven by the last few generations of admixture. Testing other models of time-dependent admixture, with the contributions from one or both of the source populations increasing or decreasing over time, we find that the data fit model-based estimates approximately equally well when the admixture contributions at the last few generations are similar to the admixture contributions estimated from a constant admixture model (Materials and Methods).
The signal of a large male bias holds when analyzing late Neolithic Corded Ware individuals and later Bronze Age Unetice individuals separately, with mean X-to-autosomal ancestry ratios in the two groups of 0.716 and 0.474, respectively. Ancestry and sex bias do differ between the groups, with a larger male bias and lower SP ancestry for the later Unetice, although the trend is not statistically significant (SI Appendix, Fig. S1B). Individuals from Bell Beaker archeological sites, a culture that overlapped with Corded Ware and Unetice but occurred over a wider geographic scale, show levels of X and autosomal ancestry suggestive of overall ancestry contributions and levels of sex bias that are similar to Corded Ware and Unetice, with mean X and autosomal ancestry of 0.28 and 0.56, respectively (SI Appendix, Table S7).
The signal of male-biased contributions from SP to BA over time is consistent with an admixture scenario in which a massive male-biased migration from the steppe initially looks to local European farmer females for wives, and with a paternal mode of inheritance, the BA population disproportionately absorbs females from local “unadmixed” farmers. Admixture from the steppe population continues over time, although mainly men migrate, perhaps expanding using the male-dominated modes of horses and wagons (24, 30).
Discussion
Overall, the model-based ancestry results show remarkable similarity to our original comparisons of relative genetic drift on the X versus autosomes using a measure of genetic differentiation, (Table 1). Combining observations from both migrations, a picture of sex-specific migrations in central European prehistory emerges (Fig. 3C), with large numbers of males and females migrating with the early Neolithic spread of farming, but almost exclusively male contributions during the later Neolithic and Bronze Age expansion from the Pontic-Caspian Steppe.
Owing to the large ancestry contribution and lack of sex-biased admixture, the massive cultural change that accompanied the shift to agriculture is consistent with a large-scale migration of an entire subset of a population, perhaps families, and a slower rate of spread. Minimal differences in sex-specific migration and the high overall AF ancestry in CE individuals support this scenario. Given the probable patrilocality of the migrating AF population (10, 23), this result suggests that the residence and descent rules were not determining factors in long-distance, sex-specific migration. The lack of sex bias is not, in fact, inconsistent with previous indications of sex bias during the Neolithic based on mtDNA diversity. Earlier work focused on measures of diversity rather than ancestry, which estimate the effective population size rather than admixture. Therefore, earlier single-locus studies are likely seeing the signal of patrilocality rather than the migration process from Anatolia (20).
In contrast, our results, combined with the archeological evidence, suggest that the rapid migration from the Pontic Steppe was strongly male-biased, potentially via newly domesticated horses in multiple waves (24, 25, 30). Such differences in sex-specific migration patterns are suggestive of fundamentally different types of interactions between invading and local populations during the two migration events. Our results demonstrate the power of joint X chromosome and autosome analyses for inferring important processes in human prehistory.
Materials and Methods
Genetic Samples and Populations.
We analyzed published (6) ancient samples that have been genotyped for a set of 1,240,000 SNPs, including 49,711 on the X chromosome. Under notation from a study by Mathieson et al. (6), for the early Neolithic migration from Anatolia, we considered individuals from the CEM population label for “selection label 2”; for the late Neolithic/Bronze Age migrations from the Pontic Steppe, we considered individuals with the “archeological culture” label “central_LNBA.” These subsets of the data geographically restrict analyses to central Europeans, decreasing potential variation from spatial variation within Europe. We further classify individuals within each group using cultural, temporal, and geographic information; archeological labels follow the labels used by Mathieson et al. (6) and Lazaridis et al. (37) (SI Appendix, Table S1).
Additional genomic filtering and analyses were done in PLINK v1.90 (38). We removed the pseudoautosomal region of the X chromosome, and removed one SNP from each pair with a correlation greater than 0.4 using the command “indep-pairwise 200 25 0.4” following previous ancient DNA studies (4, 6). We considered admixed individuals with at least 1,000 SNPs on the X chromosome. SI Appendix, Tables S1 and S2 show the admixed individuals and their population classifications. More information on the samples is available in the study by Mathieson et al. (6).
Sex-Biased Genetic Differentiation.
As a first line of evidence for the sex-specific relationships between the two sets of migrating and admixed populations, AF-CE and SP-BA, we compared genetic differentiation on the X versus autosomes, and (Table 1). We followed the method of Keinan et al. (31) and Waldman et al. (32), computing the statistic Q, which measures relative genetic drift between the X and autosomes, , where and are autosomal and X-chromosomal values. We calculated and in Plink v1.9, using a ratio of averages approach to combine SNPs and the estimator used by Weir and Cockerham (equation 10 of ref. 39).
Values of Q are suggestive, because deviations from 3/4 can also be produced by population histories with population size or migration changes even in the absence of sex bias (40, 41). Additionally, Q lacks a clear framework for quantitative interpretation. Therefore, we used a mechanistic admixture model comparing ancestry on the X chromosome and autosomes to infer sex-specific admixture rates and compare potential migration models.
Estimating Ancestry Components.
Evidence of admixture and migration events in the population history of central Europeans, as well as current best proxy populations for their sources, has been extensively presented in other studies (1–6). Therefore, we assumed these migration events occurred, and used the best samples/populations currently available as representatives of relatives of the admixed populations. A schematic of the migration events is shown in Fig. 1, with estimated ancestry components shown in Fig. 2. Results by individual are presented in SI Appendix, Tables S2, S4, and S5.
We estimated ancestry components of the two admixed populations, CE and BA. For the early Neolithic transition to agriculture, we assumed the number of ancestry components, K, was 2 with AF and HG source populations. For the later migration from the steppe, we assumed three ancestry components (K = 3) with contributions from the SP represented by the Yamnaya Samara population, as well as contributions from AF and HG populations.
Multiple methods exist to infer individual ancestry proportions. We considered two of the most common clustering methods: Admixture (33) v1.3, which is a maximum likelihood method, and Structure (42) v2.3, which is a Bayesian algorithm. Both methods rely on a similar underlying model, with different estimation techniques. In each program, we tested supervised and unsupervised clustering and compared individual ancestry estimates and population-level summary statistic estimates between methods. Results are summarized in SI Appendix, Tables S4 and S5, for the autosomes and X chromosome, respectively. We describe ancestry estimation methods and compare results from both supervised and unsupervised clustering in Admixture and Structure in SI Appendix. Based on these analyses, we use ancestry estimated in Admixture from supervised clustering for main text analyses.
Assessing Ancestry Estimation.
To compare the X chromosome with the autosomes, we estimated autosomal ancestry on 100 sets of SNPs resampled from the autosomes. For each of the two migration events, we resampled autosomal ancestry to match the number of SNPs used in X-chromosomal analyses. For the Neolithic transition, the number of SNPs was 3,763. For the steppe migration, the number of SNPs was 4,605. We also down-sampled X-chromosomal SNPs for BA individuals 10 times to 3,763 to compare estimates of ancestry between the two migration events. Ancestry estimates based on the down-sampled data were within 5% of the original full data.
To test if the ancestry estimates are stable over the choice of individuals in the source populations of Mathieson et al. (6), we tested multiple subsets of source population individuals (SI Appendix, Table S3): (i) all individuals from the study of Mathieson et al. (6) for the respective categories, using the original population descriptions of Anatolians, Western + Scandinavian HG populations, and Yamnaya Samara, and (ii) the subset of individuals whose genetic population assignment matches their known cultural association in an average of 10 independent unsupervised admixture runs for both the X and the autosomes.
For the first event, the Neolithic transition, the estimated ancestry components are roughly constant with varying choice of individuals. For the migration from the steppe, however, we see a range of values for estimated ancestries over different seeds, suggesting variation in the likelihood surface. The qualitative results are consistent through all analyses. The population means for the X chromosome and autosomes range from 0.27 to 0.44 and from 0.54 to 0.73, respectively. The ratio of X to autosomal ancestry for a given seed varies between 0.38 and 0.61. Although the magnitude of ancestry estimates varies, the signal of substantial sex bias based on the ratio of X to autosomal ancestry is seen for all scenarios.
For all analyses, we used ancestry estimated from the mean per individual of X-chromosomal estimates over the 10 seeds. Autosomal ancestry is estimated as the median of 100 estimates from resampled SNP sets, which is in the lower range of autosomal estimates. For the steppe migration, this estimate leads to a ratio of mean X-chromosomal to mean autosomal ancestry of 0.59, which is on the conservative (closer to 1) end of the range of estimates.
We also considered the impact of source population on inference of ancestry by estimating ancestry proportions of alternate Near Eastern farming reference individuals as reported by Lazaridis et al. (37) (SI Appendix, Table S6). We include Natufian and Neolithic Levantine individuals, although these groups are unlikely the source for the Neolithic migration into Europe (7, 43). For admixed CE populations, the mean X ancestry is 0.97 and the mean autosomal ancestry is 0.91, leading to an X-to-autosomal ancestry ratio of 1.06. This ratio of X to autosomal ancestry is slightly elevated compared with the original 0.989, but it leads to the same conclusion of minimal to no sex bias from the Near East into central Europe. Notably, the elevated ratio is driven by higher X-chromosomal ancestry (0.97 vs. original 0.90), which may be an effect of the reduction in the number of X-chromosome SNPs to 1,988.
Ancestry Over Time.
Within each admixed population, samples span thousands of years; therefore, we considered temporal heterogeneity in ancestry by fitting linear models of ancestry over time in MATLAB (fitlm) (SI Appendix, Fig. S1). We fit X and autosomal ancestry separately for each population. Sampling age is defined as the midpoint of the two-sigma calibrated date range given by Mathieson et al. (6). For individual ancestry y and sampling age x, for the Neolithic migration, we have and for the X and autosomes, respectively. Similarly, for the steppe migration, we have and . The relationship between ancestry and sampling age is significant for X and autosomal ancestry for the Neolithic migration (P = 0.02 and P < 0.001), but not for the steppe migration (P = 0.21 and P = 0.13).
Statistical Significance of X and Autosomal Differences.
We tested for statistical significance of the difference between the population means of X and autosomal ancestry within the admixed Neolithic population using the Wilcoxon signed-rank test. We did 100 comparisons of the distribution of ancestry on the X chromosome within the population with the distribution of autosomal ancestry estimated using each resample of M SNPs, where M is the number of X-chromosomal SNPs for the associated population.
The Wilcoxon signed-rank test is a nonparametric paired difference test. For a statistically significant difference in the within-population distribution of X and autosomal ancestry, one would expect an excess of small P values. Rather, for the Neolithic transition, comparing X and autosomal AF-related ancestry, the P value distribution over the 100 calculations is approximately uniform (SI Appendix, Fig. S3). Similarly, comparing X and autosomal ancestry when autosomal ancestry is estimated from all SNPs together (M = 331,515), the Wilcoxon signed-rank test is not significant (P = 0.493). In contrast, for the later migration from the Pontic Steppe, P = 0.002 comparing the distribution of ancestry on the X chromosome with the distribution of ancestry estimated for the autosomes with all SNPs together (M = 375,243), and we see an excess of small P values for the comparisons with 100 resampled autosomal estimates (SI Appendix, Fig. S3).
Simulations to Estimate Range of Sex Bias During Neolithic Transition.
For a constant admixed population of size , with , we simulated the ancestry proportion of individuals in the admixed population recursively for 40 generations (g), or ∼1,000 y, assuming a single admixture event followed by no further migration (Fig. 1). We set the total contributions from each population based on their autosomal ancestry levels (17, 35, 36), with HG as 0.087 and AF as 0.913, and we define the level of sex bias as the ratio of male to female contributions from a given source population, , considering . Adapting equation 1 of ref. 35 to calculate the sex-specific contribution parameters, female and male contribution parameters can then be exactly solved. For a given set of sex-specific contributions, we did 1,000 replicate simulations to test the range of X and autosomal ancestry produced in the population. Individual ancestry is deterministic based on the random sampling of their parents from the previous generation; that is, for autosomal ancestry and X-chromosomal ancestry in females, individuals are the average of their parent’s ancestries, and for X-chromosomal ancestry in males, individuals have the same ancestry as their mother.
At generations, we randomly sampled 20 individuals, and calculated the mean autosomal ancestry and mean X-chromosomal ancestry in the sample. Fig. 3A shows the values of sex bias, , for which the observed X-to-autosomal ancestry ratio is within the middle 50% and 80% of ratios calculated from the 1,000 simulated populations with that level of specified sex bias. Details of the simulation are described in the SI Appendix, Supporting Methods.
Admixture Models for Migration from the Steppe.
We used recursive expressions for X and autosomal ancestry as a function of sex-specific admixture rates to interpret observed ancestry (17, 36). We considered four general models of admixture over time: (i) single admixture event with no further migration, (ii) constant migration over time, (iii) increasing migration from SP over time, and (iv) decreasing migration from SP over time.
First, we considered a single pulse admixture event, analogous to the admixture event used for the early Neolithic migration from Anatolia. For mean autosomal ancestry within the admixed population of 0.618 and mean X-chromosomal ancestry of 0.366, under the model of a single admixture event with no further migration, we used equations 22 and 23 of ref. 17 to write X and autosomal ancestries as a function of sex-specific contribution parameters. We have and However, no solution exists within the bounds on migration contributions of
We next considered constant admixture over time. Assuming , we computed the mean female and male X-chromosomal and autosomal admixture components (equations 5, 17, and 18 of ref. 17) on a grid of possible sex-specific contribution parameter values in 0.02 increments. We fixed initial values to be equal and without sex bias. Mean ancestry levels approach a limit around 15 generations; therefore, initial conditions do not significantly impact final ancestries (17, 35, 36). For time-dependent admixture rates, admixture per generation is calculated as a linear function of the number of generations spanning 0 to the contribution specified by that point on the grid corresponding to the constant admixture scenario. We used the recursive expressions from (equations 5, 17, and 18 of ref. 17) to calculate mean X and autosomal ancestry for each point on the grid.
Because the number of males in both admixed populations is small, mean sample ancestry estimates may not be representative of the population mean. Therefore, we followed equation 5 of ref. 17, calculating a pooled female and male Euclidean distance between model-based ancestry calculations and observed ancestry estimates. Fig. 2 presents results based on the smallest 0.1% of Euclidean distances on the grid, with estimated sex bias values for other cutoffs in the text.
These scenarios, however, are few of many that are possible, and further work is needed to describe the spatiotemporal variation in admixture during these migrations. Although spatial and temporal resolution will refine admixture models, the signals of sex-specific admixture during the prehistory of central Europe will persist. Similarly, other processes may also differentially affect the X chromosome and autosomes, including recombination, mutation, and selection, but these forces are unlikely to have a large impact on the chromosome-wide, ancestry-based summary statistics we base analyses on over the short time scales considered.
Variance in Ancestry.
Our analyses focus on comparisons of mean X-chromosomal and autosomal ancestry. However, the variance can also be informative about the admixture history (35, 36). The variance in ancestry with the admixed Neolithic individuals is quite low (0.013 for the X chromosome and 0.005 for the autosomes), with a higher variance in the admixed BA population (0.102 for the X chromosome and 0.039 for the autosomes). Larger X than autosomal variance is expected owing to the difference in the number of chromosomes inherited per generation. The higher variance in ancestry across individuals associated with the Pontic Steppe migration is consistent with recent or ongoing migration within the past few generations, particularly because sex bias would decrease the variance (36). Additionally, with recent or ongoing male-biased migration, one would expect lower steppe ancestry on X chromosomes in admixed males than in admixed females, because females receive an X chromosome from their fathers. The mean X-chromosomal ancestry of BA males is roughly half the mean X-chromosomal ancestry of BA females, although the difference is not statistically significant with only four individuals. Although consistent with inferences from mean ancestry components, strong conclusions cannot be drawn from the variance or differences in male and female ancestry, given the current sample sizes.
Supplementary Material
Acknowledgments
We thank Jonathan Kang for bioinformatics support with the HapMap data. We acknowledge support from National Science Foundation (NSF) Graduate Research and Achievement Rewards for College Scientists fellowships (to A.G.), a Wenner–Gren Foundation fellowship (to T.G.), NSF Grant BCS 1515127 (to N.A.R.), European Research Council Grant 311413 and Göran Gustafsson Foundation Prize (to M.J.), as well as an NSF-Swedish Research Council Graduate Research Opportunities Worldwide award (to A.G. to visit Uppsala University).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. W.H. is a Guest Editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1616392114/-/DCSupplemental.
References
- 1.Ammerman AJ, Cavalli-Sforza LL. The Neolithic Transition and the Genetics of Populations in Europe. Princeton Univ Press; Princeton: 1984. [Google Scholar]
- 2.Skoglund P, et al. Origins and genetic legacy of Neolithic farmers and hunter-gatherers in Europe. Science. 2012;336(6080):466–469. doi: 10.1126/science.1216304. [DOI] [PubMed] [Google Scholar]
- 3.Lazaridis I, et al. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature. 2014;513(7518):409–413. doi: 10.1038/nature13673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allentoft ME, et al. Population genomics of Bronze Age Eurasia. Nature. 2015;522(7555):167–172. doi: 10.1038/nature14507. [DOI] [PubMed] [Google Scholar]
- 5.Haak W, et al. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature. 2015;522(7555):207–211. doi: 10.1038/nature14317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mathieson I, et al. Genome-wide patterns of selection in 230 ancient Eurasians. Nature. 2015;528(7583):499–503. doi: 10.1038/nature16152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Günther T, Jakobsson M. Genes mirror migrations and cultures in prehistoric Europe-a population genomic perspective. Curr Opin Genet Dev. 2016;41:115–123. doi: 10.1016/j.gde.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 8.Wilkins JF, Marlowe FW. Sex-biased migration in humans: what should we expect from genetic data? BioEssays. 2006;28(3):290–300. doi: 10.1002/bies.20378. [DOI] [PubMed] [Google Scholar]
- 9.Heyer E, Chaix R, Pavard S, Austerlitz F. Sex-specific demographic behaviours that shape human genomic variation. Mol Ecol. 2012;21(3):597–612. doi: 10.1111/j.1365-294X.2011.05406.x. [DOI] [PubMed] [Google Scholar]
- 10.Haak W, et al. Ancient DNA, Strontium isotopes, and osteological analyses shed light on social and kinship organization of the Later Stone Age. Proc Natl Acad Sci USA. 2008;105(47):18226–18231. doi: 10.1073/pnas.0807592105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bustamante CD, Ramachandran S. Evaluating signatures of sex-specific processes in the human genome. Nat Genet. 2009;41(1):8–10. doi: 10.1038/ng0109-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seielstad MT, Minch E, Cavalli-Sforza LL. Genetic evidence for a higher female migration rate in humans. Nat Genet. 1998;20(3):278–280. doi: 10.1038/3088. [DOI] [PubMed] [Google Scholar]
- 13.Tishkoff SA, et al. History of click-speaking populations of Africa inferred from mtDNA and Y chromosome genetic variation. Mol Biol Evol. 2007;24(10):2180–2195. doi: 10.1093/molbev/msm155. [DOI] [PubMed] [Google Scholar]
- 14.Ségurel L, et al. Sex-specific genetic structure and social organization in Central Asia: insights from a multi-locus study. PLoS Genet. 2008;4(9):e1000200. doi: 10.1371/journal.pgen.1000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cox MP, Karafet TM, Lansing JS, Sudoyo H, Hammer MF. Autosomal and X-linked single nucleotide polymorphisms reveal a steep Asian-Melanesian ancestry cline in eastern Indonesia and a sex bias in admixture rates. Proc R Soc Lond B Biol Sci. 2010;277(1687):1589–1596. doi: 10.1098/rspb.2009.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bryc K, Durand EY, Macpherson JM, Reich D, Mountain JL. The genetic ancestry of African Americans, Latinos, and European Americans across the United States. Am J Hum Genet. 2015;96(1):37–53. doi: 10.1016/j.ajhg.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldberg A, Rosenberg NA. Beyond 2/3 and 1/3: The complex signatures of sex-biased admixture on the X chromosome. Genetics. 2015;201(1):263–279. doi: 10.1534/genetics.115.178509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bentley RA, Layton RH, Tehrani J. Kinship, marriage, and the genetics of past human dispersals. Hum Biol. 2009;81(2-3):159–179. doi: 10.3378/027.081.0304. [DOI] [PubMed] [Google Scholar]
- 19.Balaresque P, et al. A predominantly neolithic origin for European paternal lineages. PLoS Biol. 2010;8(1):e1000285. doi: 10.1371/journal.pbio.1000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rasteiro R, Chikhi L. Female and male perspectives on the neolithic transition in Europe: Clues from ancient and modern genetic data. PLoS One. 2013;8(4):e60944. doi: 10.1371/journal.pone.0060944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szécsényi-Nagy A, et al. Tracing the genetic origin of Europe’s first farmers reveals insights into their social organization. Proc R Soc Lond B Biol Sci. 2015;282(1805):20150339. doi: 10.1098/rspb.2015.0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haak W, et al. Ancient DNA from the first European farmers in 7500-year-old Neolithic sites. Science. 2005;310(5750):1016–1018. doi: 10.1126/science.1118725. [DOI] [PubMed] [Google Scholar]
- 23.Bentley RA, et al. Community differentiation and kinship among Europe’s first farmers. Proc Natl Acad Sci USA. 2012;109(24):9326–9330. doi: 10.1073/pnas.1113710109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gimbutas M. The Indo-Europeanization of Europe: The intrusion of steppe pastoralists from south Russia and the transformation of Old Europe. Word. 1993;44(2):205–222. [Google Scholar]
- 25.Anthony DW. Migration in archeology: The baby and the bathwater. Am Anthropol. 1990;92(4):895–914. [Google Scholar]
- 26.Kristiansen K, Larsson TB. The Rise of Bronze Age Society: Travels, Transmissions and Transformations. Cambridge Univ Press; Cambridge, UK: 2005. [Google Scholar]
- 27.Batini C, et al. Large-scale recent expansion of European patrilineages shown by population resequencing. Nature Comm. 2015;6:7152. doi: 10.1038/ncomms8152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karmin M, et al. A recent bottleneck of Y chromosome diversity coincides with a global change in culture. Genome Res. 2015;25(4):459–466. doi: 10.1101/gr.186684.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poznik GD, et al. Punctuated bursts in human male demography inferred from 1,244 worldwide Y-chromosome sequences. Nat Genet. 2016;48(6):593–599. doi: 10.1038/ng.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anthony DW. The Horse, the Wheel, and Language: How Bronze-Age Riders from the Eurasian Steppes Shaped the Modern World. Princeton Univ Press; Princeton: 2007. [Google Scholar]
- 31.Keinan A, Mullikin JC, Patterson N, Reich D. Accelerated genetic drift on chromosome X during the human dispersal out of Africa. Nat Genet. 2009;41(1):66–70. doi: 10.1038/ng.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Waldman YY, et al. The genetics of Bene Israel from India reveals both substantial Jewish and Indian ancestry. PLoS One. 2016;11(3):e0152056. doi: 10.1371/journal.pone.0152056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19(9):1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bollongino R, et al. 2000 years of parallel societies in Stone Age Central Europe. Science. 2013;342(6157):479–481. doi: 10.1126/science.1245049. [DOI] [PubMed] [Google Scholar]
- 35.Verdu P, Rosenberg NA. A general mechanistic model for admixture histories of hybrid populations. Genetics. 2011;189(4):1413–1426. doi: 10.1534/genetics.111.132787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goldberg A, Verdu P, Rosenberg NA. Autosomal admixture levels are informative about sex bias in admixed populations. Genetics. 2014;198(3):1209–1229. doi: 10.1534/genetics.114.166793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lazaridis I, et al. Genomic insights into the origin of farming in the ancient Near East. Nature. 2016;536(7617):419–424. doi: 10.1038/nature19310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang CC, et al. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience. 2015;4(7):7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38(6):1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 40.Ramachandran S, Rosenberg NA, Feldman MW, Wakeley J. Population differentiation and migration: Coalescence times in a two-sex island model for autosomal and X-linked loci. Theor Popul Biol. 2008;74(4):291–301. doi: 10.1016/j.tpb.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emery LS, Felsenstein J, Akey JM. Estimators of the human effective sex ratio detect sex biases on different timescales. Am J Hum Genet. 2010;87(6):848–856. doi: 10.1016/j.ajhg.2010.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kılınç GM, et al. The demographic development of the first farmers in Anatolia. Curr Biol. 2016;26(19):2659–2666. doi: 10.1016/j.cub.2016.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.