Significance
This season marks the 50th anniversary of the discovery of polynucleotide ligases, the sine qua non enzymes of nucleic acid repair and the enabling reagents of molecular biology, reported in a series of seminal papers in PNAS. Ligases react with ATP or NAD+ to form a covalent enzyme–adenylate intermediate in which AMP is linked via a P–N bond to a lysine. This work reports crystal structures of the Michaelis complexes of an exemplary ATP-dependent RNA ligase (bacteriophage T4 Rnl1) and an NAD+-dependent DNA ligase (Escherichia coli LigA) that illuminate the chemical and structural basis for lysine adenylylation, via distinctive two-metal (ATP) and one-metal (NAD+) mechanisms.
Keywords: metal catalysis, covalent nucleotidyltransferase, lysyl-AMP
Abstract
Polynucleotide ligases comprise a ubiquitous superfamily of nucleic acid repair enzymes that join 3′-OH and 5′-PO4 DNA or RNA ends. Ligases react with ATP or NAD+ and a divalent cation cofactor to form a covalent enzyme-(lysine-Nζ)–adenylate intermediate. Here, we report crystal structures of the founding members of the ATP-dependent RNA ligase family (T4 RNA ligase 1; Rnl1) and the NAD+-dependent DNA ligase family (Escherichia coli LigA), captured as their respective Michaelis complexes, which illuminate distinctive catalytic mechanisms of the lysine adenylylation reaction. The 2.2-Å Rnl1•ATP•(Mg2+)2 structure highlights a two-metal mechanism, whereby: a ligase-bound “catalytic” Mg2+(H2O)5 coordination complex lowers the pKa of the lysine nucleophile and stabilizes the transition state of the ATP α phosphate; a second octahedral Mg2+ coordination complex bridges the β and γ phosphates; and protein elements unique to Rnl1 engage the γ phosphate and associated metal complex and orient the pyrophosphate leaving group for in-line catalysis. By contrast, the 1.55-Å LigA•NAD+•Mg2+ structure reveals a one-metal mechanism in which a ligase-bound Mg2+(H2O)5 complex lowers the lysine pKa and engages the NAD+ α phosphate, but the β phosphate and the nicotinamide nucleoside of the nicotinamide mononucleotide (NMN) leaving group are oriented solely via atomic interactions with protein elements that are unique to the LigA clade. The two-metal versus one-metal dichotomy demarcates a branchpoint in ligase evolution and favors LigA as an antibacterial drug target.
Polynucleotide ligases join 3′-OH and 5′-PO4 RNA termini via a series of three nucleotidyl transfer steps. In step 1, a ligase reacts with ATP or NAD+ to form a covalent ligase-(lysyl-Nζ)–AMP intermediate and release pyrophosphate (PPi) or nicotinamide mononucleotide (NMN). In step 2, AMP is transferred from ligase-adenylate to the 5′-PO4 DNA or RNA end to form a DNA–adenylate or RNA–adenylate intermediate (AppDNA or AppRNA). In step 3, ligase catalyzes attack by a DNA or RNA 3′-OH on the polynucleotide–adenylate to seal the two ends via a phosphodiester bond and release AMP. All steps in the ligase pathway require a divalent cation cofactor.
The autoadenylylation reaction of polynucleotide ligases is performed by a nucleotidyltransferase (NTase) domain that is conserved in ATP-dependent DNA and RNA ligases and NAD+-dependent DNA ligases (1, 2). The NTase domain includes defining peptide motifs that form the nucleotide-binding pocket. Motif I (KxDG) contains the lysine that becomes covalently attached to the AMP. As Robert Lehman pointed out in 1974 (3), it is unclear how lysine (with a predicted pKa value of ∼10.5) loses its proton at physiological pH to attain the unprotonated state required for attack on the α phosphorus of ATP or NAD+. In principle, a ligase might use a general base to deprotonate the lysine. Alternatively, the pKa could be driven down by positive charge potential of protein amino acids surrounding lysine-Nζ. Several crystal structures of ligases absent metals provided scant support for either explanation. In these structures, the motif I lysine nucleophile is located next to a motif IV glutamate or aspartate side chain (2, 4–6). The lysine and the motif IV carboxylate form an ion pair, the anticipated effect of which is to increase the pKa of lysine by virtue of surrounding negative charge. It is unlikely that a glutamate or aspartate anion could serve as a general base to abstract a proton from the lysine cation. A potential solution to the problem would be if a divalent cation abuts the lysine-Nζ and drives down its pKa.
A metal-driven mechanism was revealed by the recent crystal structure of Naegleria gruberi RNA ligase (NgrRnl) as a step 1 Michaelis complex with ATP and manganese (its preferred metal cofactor) (7). The key to capturing the Michaelis-like complex was the replacement of the lysine nucleophile by an isosteric methionine. The 1.9-Å structure contained ATP and two manganese ions in the active site. The “catalytic” metal was coordinated with octahedral geometry to five waters that were, in turn, coordinated by the carboxylate side chains of conserved residues in motifs I, III, and IV. The sixth ligand site in the catalytic metal complex was occupied by an ATP α phosphate oxygen, indicative of a role for the metal in stabilizing the transition state of the autoadenylylation reaction. A key insight, fortified by superposition of the Michaelis complex on the structure of the covalent NgrRnl-(Lys-Nζ)–AMP intermediate, concerned the role of the catalytic metal complex in stabilizing the unprotonated state of the lysine nucleophile before catalysis, via local positive charge and atomic contact of Lys-Nζ to one of the metal-bound waters (7).
The NgrRnl Michaelis complex revealed a second metal, coordinated octahedrally to four waters and to ATP β and γ phosphate oxygens. The metal complex and the ATP γ phosphate were engaged by an ensemble of amino acid side chains (unique to NgrRnl) that collectively orient the PPi leaving group apical to the lysine nucleophile. Consistent with a single-step in-line mechanism, the α phosphate was stereochemically inverted during the transition from NgrRnl•ATP Michaelis complex to lysyl–AMP intermediate (7).
NgrRnl exemplifies one of five distinct families of ATP-dependent RNA ligases that arose independently by fusion of an ancestral ATP-using NTase domain to structurally unique flanking domains (7). An important issue for ligase evolution is whether the two-metal mechanism and lysine adenylylation strategy exploited by NgrRnl—an Rnl5 family ligase that seals nicks in duplex RNAs (8)—applies to other ATP-dependent polynucleotide ligases. Here, we address the issue by capturing a structure of the Michaelis complex of bacteriophage T4 RNA ligase 1 (Rnl1). T4 Rnl1 was the first RNA ligase discovered (9, 10), and it spearheads a phage-encoded pathway of tRNA break repair that thwarts a tRNA-damaging host antiviral response (11). We find that T4 Rnl1 adheres to a two-metal mechanism of lysine adenylylation, driven by: (i) a catalytic metal–water complex that engages the lysine nucleophile and the ATP α phosphate and (ii) a second metal that orients the PPi leaving group, albeit in a ligase-assisted manner distinct from that seen for NgrRnl.
DNA ligases are thought to have evolved separately from RNA ligases, initially by fusion of an ancestral ATP-using NTase domain to a C-terminal OB domain (to comprise the minimal catalytic core of a DNA ligase) and, subsequently, via the fusion of additional structural modules to the NTase-OB core (7). NAD+-dependent DNA ligases (LigA enzymes), which are ubiquitous in bacteria and essential for bacterial viability in all cases tested, acquired their specificity for NAD+ via fusion of an NMN-binding Ia domain module to the N terminus of the NTase domain (4, 12, 13). Escherichia coli DNA ligase (EcoLigA) was the first cellular DNA ligase discovered and characterized (14–20), and it remains the premier model for structural and functional studies of the NAD+-dependent DNA ligase family (21–27). Interest in LigA mechanism is propelled by the promise of targeting LigA (via its signature NAD+ substrate specificity and unique structural features vis à vis human DNA ligases) for antibacterial drug discovery (24, 28). Here, we report the structure of a Michaelis complex of EcoLigA with NAD+ and magnesium, which reveals a one-metal mechanism in which a catalytic metal–water complex engages the lysine nucleophile and the NAD+ α phosphate, whereas the NMN leaving group is positioned by enzymic contacts without agency of a second metal.
Results
A Michaelis Complex of T4 Rnl1 with ATP and Magnesium.
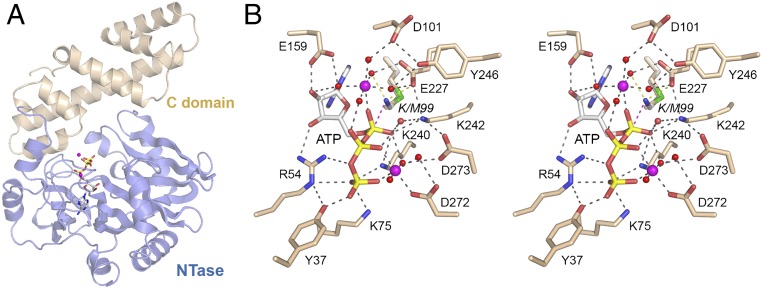
The initial structure of T4 Rnl1 with the unreactive analog AMPCPP had a calcium ion in the putative catalytic metal site, coordinated to six waters and one AMPCPP α phosphate oxygen (29). Our aim here was to capture a mimetic of the Michaelis complex of T4 Rnl1 with ATP and its physiological metal cofactor magnesium. To accomplish this goal, we exploited a mutated version, K99M, in which the Lys99 nucleophile was replaced with methionine (effectively isosteric to lysine, minus the ε-amino group). Rnl1-K99M was preincubated with 1 mM ATP and 2 mM MgCl2 before crystallization. The refined 2.2-Å structure (R/Rfree = 0.189/0.261) (Table S1) comprised the entire T4 Rnl1 polypeptide, composed of an N-terminal NTase domain (amino acids 1–254) and a C-terminal domain (amino acids 255–374) unique to Rnl1 (Fig. 1A). The NTase domain per se is capable of autoadenylylation and of “nonspecific” joining of RNA 3′-OH and 5′-PO4 ends; the distinctive C domain of T4 Rnl1 confers specificity for the repair of tRNAs with breaks in the anticodon loop (30). Electron density for ATP and two magnesium ions was evident in the active site (Figs. S1A and S2A). Fig. 1B shows a stereoview of the active site of the ATP•(Mg2+)2 complex, highlighting atomic interactions relevant to catalysis. For comparison, we superimposed the Lys99 side chain from the T4 Rnl1•AMPCPP structure (29) on Met99 of the Michaelis complex. The modeled Lys99-Nζ is situated 3.1 Å from the ATP α phosphorus, in an apical orientation to the pyrophosphate leaving group (Nζ–Pα–O3α angle = 169°).
Table S1.
Crystallographic data and refinement statistics
| T4 Rnl-K99M•ATP•Mg2+ | EcoLigA-K115M•NAD+•Mg2+ | |
| Data collection | ||
| Space group | C2 | P212121 |
| Cell dimensions | ||
| a, b, c, Å | 107.34, 39.97, 108.29 | 40.91, 94.31, 150.93 |
| α, β, γ (°) | 90.0, 108.27, 90.0 | 90.0, 90.0, 90.0 |
| Resolution, Å | 50–2.2 (2.25–2.2) | 50–1.55 (1.58–1.55) |
| Wavelength, Å | 0.97920 | 0.97920 |
| Rpim | 0.048 (0.274) | 0.038 (0.323) |
| CC(1/2) | 0.997(0.840) | 0.996 (0.758) |
| <I>/<σI> | 15.3 (2.0) | 22.1 (2.2) |
| Completeness, % | 90.1 (69.3) | 98.0 (82.7) |
| Redundancy | 3.1 (3.1) | 4.2 (3.7) |
| Unique reflections | 19235 | 83987 |
| Phasing | ||
| MR search model | 2C5U | 2OWO |
| Refinement | ||
| Rwork/Rfree | 0.189/0.261 (0.229/0.314) | 0.177/0.205 (0.289/0.313) |
| B factors (Å2) Average/Wilson | 50.4/35.3 | 22.93/17.4 |
| RMS deviations | ||
| Bond lengths, Å | 0.009 | 0.007 |
| Bond angles, ° | 1.05 | 0.924 |
| Ramachandran plot | ||
| % favored | 95.2 | 98.3 |
| % allowed | 4.5 | 1.7 |
| % outliers | 0.3 | 0 |
| Model contents | ||
| Protomers/Asymmetric unit | 1 | 1 |
| Protein residues | 375 | 581 |
| Water | 93 | 840 |
| Ligands/Ions | 1 ATP, 2 Mg2+ | 1 NAD+, 1 Mg2+, 1 Zn2+ |
| PDB ID code | 5TT6 | 5TT5 |
Values in parentheses refer to the highest resolution shell. Rfree set consists of 10% of data chosen randomly against which structures was not refined.
Fig. 1.
Structure of a Michaelis complex of T4 Rnl1 with ATP and magnesium. (A) Tertiary structure of T4 Rnl1-K99M, which consists of an N-terminal nucleotidyltransferase (NTase) domain (blue) and a unique C-terminal domain (beige). The ATP in the active site is rendered as a stick model. Mg2+ ions are depicted as magenta spheres. (B) Stereoview of the active site of Rnl1-(K99M)•ATP•(Mg2+)2 highlighting the catalytic Mg2+(H2O)5 coordination complex at the α phosphate, a second Mg2+ bridging the β and γ phosphates, and enzymic contacts to the ATP and metal complexes. Amino acids and ATP are shown as stick models with beige and gray carbons, respectively. Mg2+ ions and associated waters are depicted as magenta and red spheres, respectively. Atomic contacts are indicated by black dashed lines. The superimposed Lys99 side chain from the structure of wild-type Rnl1 (29) is shown with its proximity of Nζ to the ATP α phosphorus indicated by a magenta dashed line and to a metal-bound water and Glu227-Oε indicated by yellow dashed lines.
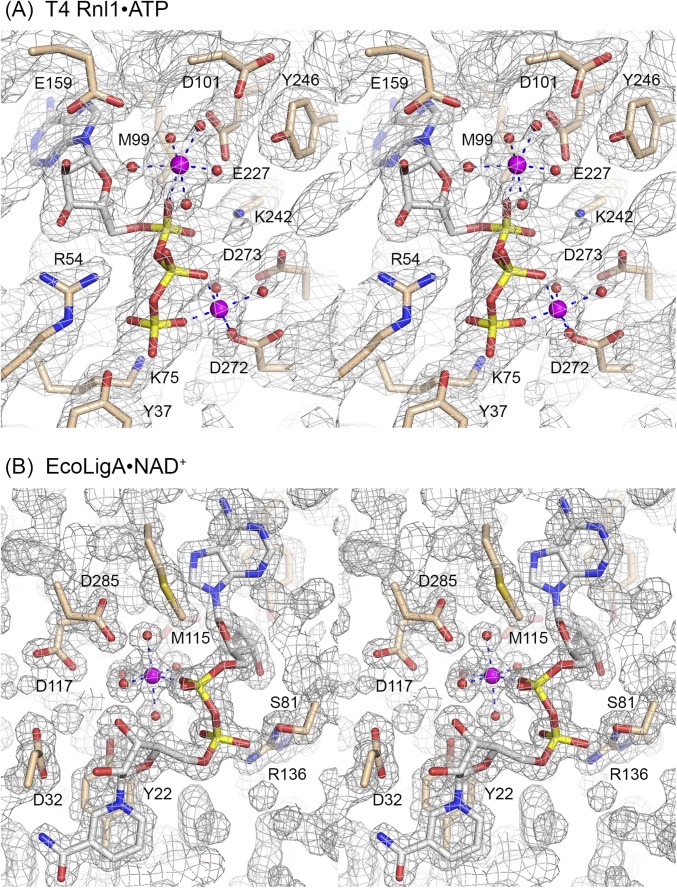
Fig. S1.
Stereoviews of active site electron density. The 2Fo–Fc maps of the T4 Rnl1•ATP (A, contoured at 1σ) and EcoLigA•NAD+ (B, contoured at 1.5σ) complexes are shown in gray mesh. The ATP and NAD+ nucleotides, metal ions, and waters were omitted during refinement and phase calculation. Ligase amino acids are depicted as stick models with beige carbons. ATP and NAD+ are depicted as stick models with gray carbons. Metal ions and coordinated waters are rendered as magenta spheres and red spheres, respectively. The atomic contacts of the octahedrally coordinated magnesium ions are indicated by dashed lines.
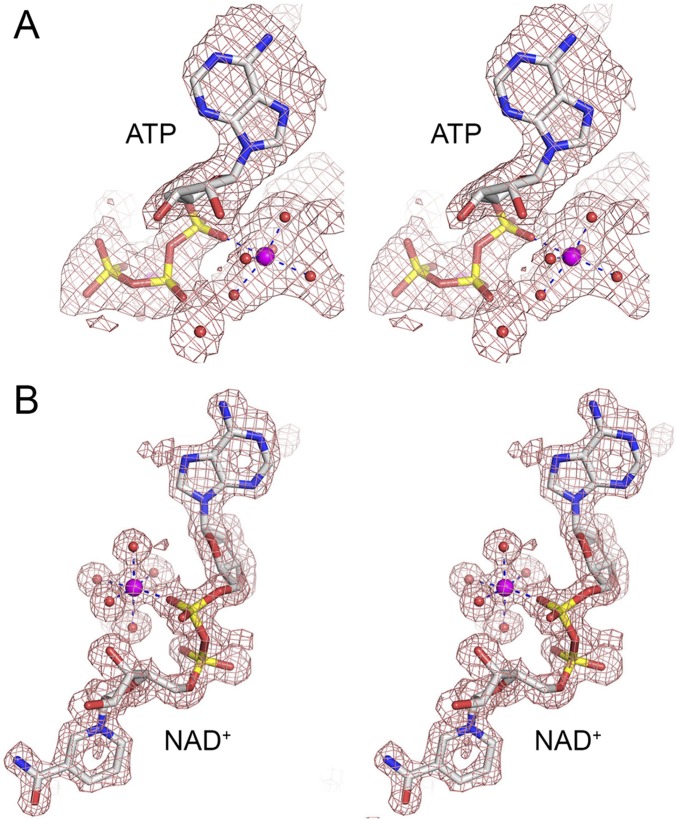
Fig. S2.
Stereoviews of simulated annealing omit maps (Fo-Fc, shown in red mesh) of the nucleotide-metal complexes of T4 Rnl1 (A) with ATP contoured at 1σ and EcoLigA (B) with NAD+ contoured at 2σ, oriented to highlight the catalytic metal (magenta sphere).
Two-Metal Mechanism of T4 Rnl1 Lysine Adenylylation.
The adenosine nucleoside of ATP is in the syn conformation. The ribose 3′-OH and 2′-OH make hydrogen bonds to Arg54 and Glu159, respectively. The ATP α phosphate is engaged by a catalytic magnesium. Waters occupy five of the ligand sites in the octahedral Mg2+ complex. Rnl1 binds the Mg2+(H2O)5 complex via water-mediated contacts to Asp101 (motif I), Glu159 (motif III), Glu227 (motif IV), and Tyr246. The sixth Mg2+ ligand site is occupied by one of the nonbridging α phosphate oxygens (Fig. 1B). The other nonbridging α phosphate oxygen is contacted by Lys240 and Lys242 (motif V). The structure suggests that Lys240, Lys242, and Mg2+ stabilize a pentavalent transition state of the ATP α phosphate during the lysine adenylylation reaction. The structure also reveals how the catalytic Mg2+(H2O)5 complex stabilizes the unprotonated state of the Lys99 nucleophile before catalysis, via local positive charge and an atomic contact of Lys99-Nζ to one of the metal-bound waters (indicated by a yellow dashed line in Fig. 1B). The modeled Lys99-Nζ is also proximal to Glu227-Oε, as seen in the T4 Rnl1•AMPCPP structure (29).
There are no direct enzymic contacts to the ATP β phosphate, only water-bridged interactions of one β phosphate oxygen with the catalytic Mg2+ and of the other β phosphate oxygen with Lys242. A second Mg2+ is coordinated octahedrally to three waters, to ATP β and γ phosphate oxygens, and to the Asp272 carboxylate (Fig. 1B). One of the second Mg2+-bound waters makes a bifurcated bridge to the ATP α phosphate and the Asp273 carboxylate. Direct contacts to the ATP γ phosphate are made by Tyr37, Arg54, Lys75, and Lys240. The Rnl1 residues that binds the γ phosphate and the Mg2+ complex with the β and γ phosphates together achieve a conformation of the ATP triphosphate moiety conducive to expulsion of the PPi•Mg2+ leaving group. The structure of the Michaelis complex accords with extensive mutational studies showing that Arg54, Lys75, Glu159, Glu227, Lys240, Lys242, and Tyr246 are essential for RNA ligase activity (31, 32).
When we align the previous Rnl1•AMPCPP structure (29) to that of our Michaelis complex with ATP (Fig. S3), we see that the AMP moieties and the amino acids (Asp101, Glu159, Glu227, Tyr246) that coordinate the waters in the catalytic metal complex superimpose nicely, but there is a difference in the trajectory of the β and γ phosphates of AMPCPP and ATP, whereby the β phosphorus atoms are offset by 1.8 Å and the γ phosphorus atoms are separated by 2.4 Å. This difference results in a 2.6-Å movement of the Tyr37-OH that contacts the γ phosphate and a shift in the contact made by Lys75 from the γ phosphate of ATP to the β phosphate of AMPCPP (Fig. S3). We surmise that the bridging α–β methylene carbon in the ATP analog perturbs the geometry of the triphosphate moiety of the nucleotide. There are noteworthy differences in the atomic interactions of the PPi leaving group and the noncatalytic Mg2+ in the AMPCPP structure versus the ATP structure. For example, in the AMPCPP structure, the noncatalytic Mg2+ makes direct contact with the β phosphate, but not with γ phosphate; rather, two of the waters coordinated by the second metal bridge to the γ phosphate (Fig. S3). Insofar as classic Mg2+ binding to an NTP directly engages the β and γ phosphate oxygens, we regard the ATP structure as reflective of the disposition of the second metal during the lysine adenylylation reaction.
Fig. S3.
Comparison of the active sites of Rnl1•AMPCPP and Rnl1•ATP complexes. The Rnl1 structures were aligned in Pymol, and the superimposed active site components are shown in a stereo image. For the Rnl1•AMPCPP structure, the amino acids are rendered as stick models with gray carbons; AMPCPP is depicted as a stick model with gray carbons and green phosphorus atoms; the calcium and magnesium ions and associated waters are depicted as green spheres. Atomic contacts are indicated by green dashed lines. For the Rnl1•ATP structure, amino acids have beige carbons; ATP has beige carbons and yellow phosphorus atoms; magnesium ions and waters are depicted as magenta spheres; waters are red spheres. Atomic contacts are indicated by black dashed lines.
Michaelis Complex of E. coli LigA with NAD+ and Magnesium.
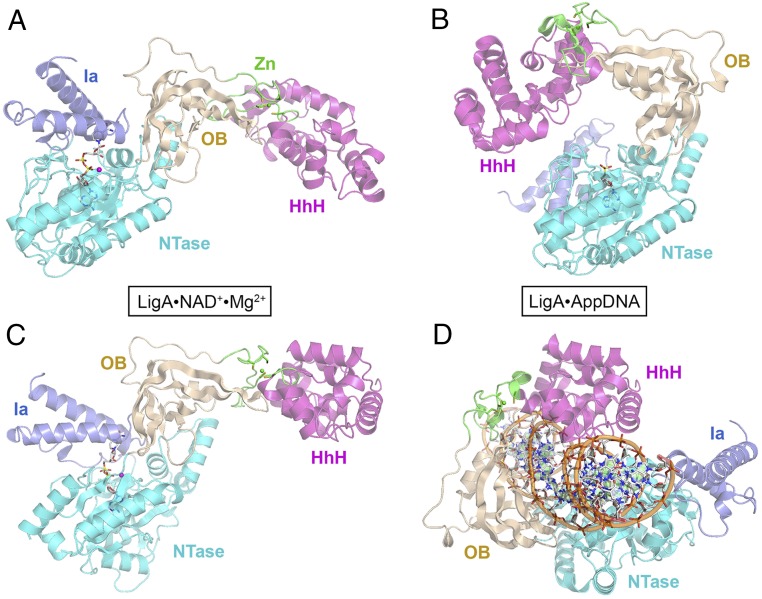
The 671-aa EcoLigA protein consists of a catalytic core composed of an NTase domain (amino acids 70–316) and an OB domain (amino acids 317–404). These two modules are common to NAD+- and ATP-dependent DNA ligases. The LigA core is flanked by an N-terminal “Ia” domain (amino acids 1–69) that confers NAD+ specificity and by three C-terminal modules: tetracysteine Zn-finger (amino acids 405–432), helix-hairpin-helix (HhH) (amino acids 433–586), and BRCT (amino acids 587–671). Each step of the ligation pathway depends on a different subset of the LigA domains, with only the NTase domain being required for all steps. The 2.3-Å crystal structure of EcoLigA bound to a nicked DNA-adenylate intermediate (AppDNA) showed that the ligase encircles the DNA helix as a C-shaped protein clamp (24) (Fig. 2 B and D). The LigA-DNA interface entails extensive DNA contacts by the NTase, OB, and HhH domains over a 19-bp segment of duplex DNA centered about the nick, many of which are essential for DNA ligase activity (25, 26).
Fig. 2.
Structure of a LigA•NAD+ Michaelis complex and comparison with DNA-bound LigA. (A and B) The tertiary structures of EcoLigA-(K115M)•NAD+•Mg2+ (A, with ATP shown as a stick model) and EcoLigA•AppDNA (B, with AMP shown as a stick model, absent the nicked DNA) were aligned with respect to their NTase domains and then offset horizontally. (C and D) The structures of EcoLigA-(K115M)•NAD+•Mg2+ (C) and EcoLigA•AppDNA (D, with AppDNA) were aligned with respect to their HhH domains and then offset horizontally. The Ia (blue), NTase (cyan), OB (beige), Zn (green, with the Zn atom shown as a green sphere), and HhH (magenta) domains are color-coded.
The Ia and NTase domains suffice for LigA enzymes to react with NAD+ and magnesium to form the covalent ligase–AMP intermediate (13, 33, 34). The 2.7-Å crystal structure of the Ia-NTase segment of Enterococcus faecalis LigA in a complex with NAD+ (generated in crystallo from AMP and NMN) provided the first insights to a bipartite mode of NAD+ recognition, whereby the AMP moiety of NAD+ is engaged by the NTase domain, whereas the NMP moiety is bound by the Ia domain (4). However, the EfaLigA Ia-NTase structure, devoid of metals, and with the lysine nucleophile making a bifurcated salt bridge to an aspartate (thus enforcing the protonated state of lysine), left many issues unsettled anent catalysis of the lysine adenylylation reaction of LigA enzymes.
Here, we aimed to capture a Michaelis complex of EcoLigA with NAD+ and magnesium, using a mutated version in which the Lys115 nucleophile was replaced by methionine. LigA-K115M was preincubated with 0.3 mM NAD+ and 5 mM MgCl2 before crystallization. The refined 1.55-Å structure (R/Rfree = 0.177/0.205) (Table S1) spanned from Met1 to Pro586 (embracing the Ia, NTase, OB, Zn, and HhH domains; Fig. 2A) with a 5-aa gap between Leu102 and Val108 in a surface loop of the NTase domain for which there was no interpretable electron density. Electron density for NAD+ and one magnesium ion was evident in the active site (Figs. S1B and S2B). No electron density was observed for the C-terminal 85-aa BRCT domain. The BRCT domain was also disordered in the crystal structures of EcoLigA bound to nicked DNA-adenylate (24) and of Thermus filiformis LigA–AMP (ref. 35, see corrected PDB ID code 1V9P).
The arrangement of the component domains of the EcoLigA Michaelis complex is shown in Fig. 2A. The N-terminal Ia module packs against the NTase domain to form a binding pocket for the NAD+ substrate akin to that seen in the EfaLigA Ia-NTase•NAD+ complex. The C-terminal OB, Zn, and HhH domains of the EcoLigA Michaelis complex adopt an extended conformation distinct from any previous LigA structure. A superposition of the NTase domains of the EcoLigA•NAD+•Mg2+ Michaelis complex (Fig. 2A) and the EcoLigA•DNA-adenylate complex (Fig. 2B; minus the nicked DNA) highlights the large movements of each of the flanking domains in the transition between these two functional states along the nick sealing pathway. For example: Met1 in the Ia domain moves 62 Å; Arg333 in the OB domain and the Zn2+ atom in the Zn-finger each move by 57 Å; and Pro586 at the end of the HhH domain moves 102 Å. Superposition of the HhH domains of the Michaelis complex (Fig. 2C) and the DNA–adenylate complex (Fig. 2D; with the nicked DNA) provides a different view of the massive conformation rearrangement between functional states of EcoLigA, entailing a 106-Å movement of the Met1 residue of the Ia domain.
One-Metal Mechanism of LigA Lysine Adenylylation.
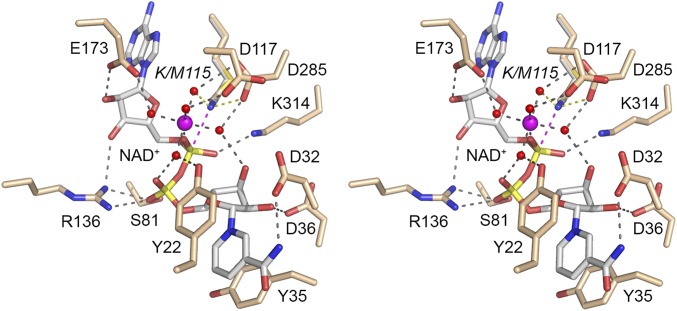
Fig. 3 shows a stereoview of the active site of the EcoLigA Michaelis complex, featuring atomic interactions relevant to NAD+ recognition and catalysis. For comparison, we superimposed on Met115 of the Michaelis complex the Lys115 side chain from the LigA•AppDNA structure. The Lys115-Nζ is poised 2.9 Å from the NAD+ α phosphorus, in an apical orientation to the NMN leaving group (Nζ–Pα–O3α angle = 175°). The adenosine nucleoside of NAD+ is in the syn conformation. The ribose 3′-OH and 2′-OH make hydrogen bonds to Arg136 and Glu173, respectively. The NAD+ α phosphate is engaged by a catalytic Mg2+(H2O)5 complex. LigA binds the Mg2+(H2O)5 complex via water-mediated contacts to Asp117 (motif I), Glu173 (motif III), and Asp285 (motif IV). The sixth Mg2+ ligand site is filled by one of the nonbridging α phosphate oxygens (Fig. 1B). The other nonbridging α phosphate oxygen is contacted by Lys314 (motif V). The structure suggests that Lys314 and Mg2+ stabilize the pentavalent transition state of the NAD+ α phosphate during the lysine adenylylation reaction. The catalytic Mg2+(H2O)5 complex also stabilizes the unprotonated state of the Lys115 nucleophile before catalysis, via local positive charge and atomic contact of Lys115-Nζ to one of the metal-bound waters (indicated by the yellow dashed line in Fig. 3). The modeled Lys115-Nζ is also proximal to Asp285-Oδ, as seen in the LigA•AppDNA structure (24).
Fig. 3.
Active site of the EcoLigA Michaelis complex. Stereoview of the active site of LigA-(K115M)•NAD+•Mg2+ featuring the catalytic Mg2+(H2O)5 coordination complex at the NAD+ α phosphate and enzymic contacts to the NAD+ and metal complexes. Amino acids and NAD+ are shown as stick models with beige and gray carbons, respectively. Mg2+ and associated waters are depicted as magenta and red spheres, respectively. Atomic contacts are indicated by black dashed lines. The superimposed Lys115 side chain adduct from the structure of wild-type LigA (24) is shown with its proximity of Nζ to the NAD+ α phosphorus indicated by a magenta dashed line and to a metal-bound water and Asp285-Oδ indicated by yellow dashed lines.
The NAD+ β phosphate (i.e., the phosphate of the NMN leaving group) is coordinated directly on the same nonbridging oxygen by the Ser81 and Arg136 side chains of the EcoLigA NTase domain (Fig. 3). The other nonbridging β phosphate oxygen coordinates one of the Mg2+-bound waters.
The nicotinamide nucleoside of NAD+ is in the anticonformation and is held in place solely by enzymic contacts to the Ia domain. Tyr22 and Tyr35 make a π sandwich above and below the nicotinamide ring (Fig. 3). The amide nitrogen of nicotinamide donates a hydrogen bond to Asp32. Asp36 accepts a hydrogen bond from the ribose 2′-OH of NMN, whereas the ribose 3′-OH coordinates one of the Mg2+-bound waters (Fig. 3).
The atomic interactions of EcoLigA with NAD+ and Mg2+ in the Michaelis complex accord with mutational studies showing that domain Ia residues Tyr22, Asp32, Tyr35, and Asp36 and NTase domains residues Asp117, Arg136, Glu173, Asp285, and Lys314 are essential for DNA ligase activity (12, 22, 23, 26). Of the NTase residues, Arg136, which interacts with the β phosphate and adenosine ribose of NAD+, stands out as essential for the reaction of EcoLigA with NAD+ to form the covalent LigA-AMP intermediate (step 1), but dispensable for phosphodiester formation at a preadenylylated nick (step 3) (26).
Discussion
Metal Catalysis of Lysine Adenylylation.
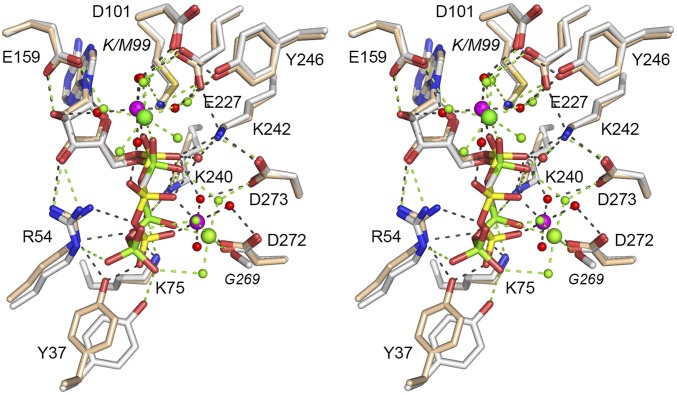
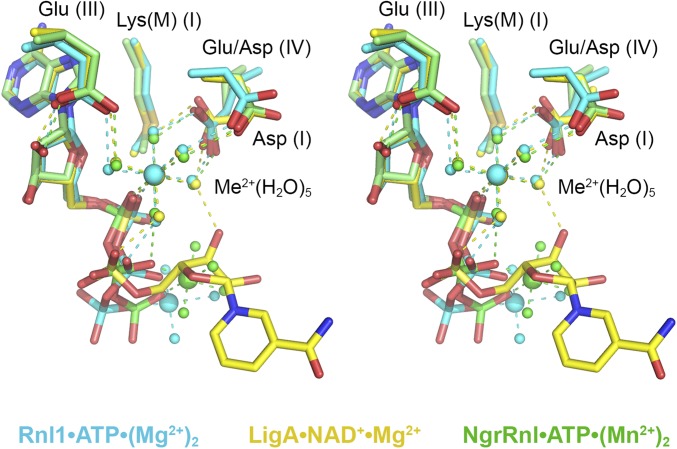
The present study illuminates a conserved mechanism of lysine adenylylation by ATP-dependent RNA ligases and NAD+-dependent DNA ligases via a catalytic metal–(H2O)5 complex that favors the unprotonated form of the lysine nucleophile and stabilizes the transition state of the ATP or NAD+ α phosphate. The shared structural basis for catalysis was gleaned from crystal structures of the Michaelis-like complexes of T4 Rnl1 (the exemplary Rnl1-family ligase), NgrRnl (Rnl5-family), and EcoLigA (the prototypal NAD+-dependent DNA ligase), each of which was captured by replacing the motif I lysine nucleophile by an isosteric methionine. Modeling the lysine in Rnl1 and LigA (based on structure of the wild-type enzymes) confirmed mimesis of the Michaelis complex of the adenylylation reaction in which the Lys-Nζ nucleophile and the PPi or NMN leaving groups are oriented apically for in-line catalysis. Superposition of the available Michaelis complexes shows that the positions and enzymic interactions of catalytic Me2+(H2O)5 complexes that engage the lysine nucleophile and α phosphate are virtually identical in the active sites of Rnl1, NgrRnl, and LigA (Fig. 4). Three carboxylate side chains of the signature nucleotidyltransferase motifs I, III, and IV bind the pentahydrated metal cofactor. A motif IV (EGYVA in Rnl1; DGVVI in LigA; EGLVF in NgrRnl) acidic side chain coordinates two of the metal-bound waters, including the water that interacts with the lysine nucleophile of motif I (KEDG in Rnl1; KLDG in LigA; KLDG in NgrRnl). A motif I aspartate coordinates a third metal-bound water. A motif III glutamate makes bidentate hydrogen bonds to a fourth metal-bound water and the ribose 2′-OH of the adenosine nucleoside (Fig. 4). The fifth metal-bound water bridges to the β phosphate in all three structures.
Fig. 4.
Comparison of the Michaelis complexes of ATP-dependent and NAD+-dependent polynucleotide ligases. Stereoview of the superimposed active sites of T4 Rnl1-(K99M)•ATP•(Mg2+)2 (with carbon atoms, waters, and Mg2+ colored cyan), EcoLigA-(K115M)•NAD+•Mg2+ (yellow), and the NgrRnl-(K170M)•ATP•(Mn2+)2 (green). Atomic contacts to the nucleotides and catalytic Me2+(H2O)5 complexes are shown as color-coded dashed lines.
Although we do not have structures of T4 Rnl1 or EcoLigA as the covalent ligase–AMP products of the step 1 adenylylation reaction, the equivalent structure of the covalent NgrRnl–AMP•Mn2+ covalent intermediate revealed that the position and enzymic contacts of the catalytic metal-(H2O)5 complex are unchanged vis à vis the Michaelis complex (7). In this vein, it is instructive to note that a recent 2.2-Å crystal structure of Methanobacterium thermoautotrophicum RNA ligase (MthRnl; a prototypal Rnl3-family enzyme), captured as the MthRnl–AMP•Mg2+ intermediate, disclosed a virtually identical catalytic Mg2+(H2O)5 complex, contacting the motif I lysine and the AMP phosphate and engaged to the enzyme via water bridges to motif IV and III glutamates and a motif I asparagine (36). Similarly, the structure of Clostridium thermocellum RNA ligase (the founder of the Rnl4 family) as the ligase–AMP•Mg2+ covalent intermediate also featured a catalytic Mg2+(H2O)5 complex at the AMP phosphate (39). Thus, a pentahydrated catalytic metal is likely to be an ancestral feature shared by all modern polynucleotide ligases that use lysyl–AMP intermediates.
Distinct Strategies for Leaving Group Orientation.
Our Michaelis complex structures are not consistent with the catalytic metal being involved in expulsion of the PPi or NMN leaving groups, with which the catalytic metal makes no direct contacts. In the case of ATP-dependent RNA ligases, a second metal bridges the β and γ phosphates to orient the PPi leaving group apical to the lysine nucleophile, aided by manifold enzymic interactions with the ATP γ phosphate (Fig. 1B). Whereas the ADP portion of the ATP substrate is virtually identical in the T4 Rnl1 and NgrRnl Michaelis complexes, the γ phosphates and the second metals are displaced slightly (by ∼2.3 Å with respect to the second metal; Fig. 4), consistent with the complete divergence of the structural components of the respective RNA ligases that bind the ATP γ phosphate. In T4 Rnl1, the γ phosphate and second metal are held in place by constituents that are unique to the Rnl1 clade (Fig. 1B). In NgrRnl, the γ phosphate and second metal are coordinated by Rnl5-specific elements derived from the signature N-terminal domain and the NTase domain (7). We envision that other families of ATP-dependent polynucleotide ligases (RNA and DNA) also exploit a two-metal mechanism of lysine adenylylation, with the second metal bound to the PPi leaving group, but with each family of ATP-dependent ligase evolving its own set of enzymic contacts to the PPi•Me2+ complex. Although there are no structures available for a step 1 Michaelis complex of an ATP-dependent DNA ligase, functional studies of Chlorella virus DNA ligase suggest that conserved elements of the C-terminal OB domain are likely candidates to orient the PPi leaving group and coordinate a second metal (38, 39).
Mechanistic Divergence in NAD+-Dependent DNA Ligase.
The salient insights from the structure of the EcoLigA Michaelis complex are that whereas the catalytic metal is shared with ATP-dependent RNA ligases, the NAD+-dependent DNA ligases eschew a second metal for leaving group orientation. Indeed, the nicotinamide nucleoside of NAD+ clashes sterically with the second metal site of ATP-dependent RNA ligases (Fig. 4). The metal-independent leaving group strategy of LigA is completely diverged from the ATP-dependent enzymes in two key respects. First, LigA makes essential contacts to the NAD+ β phosphate via an arginine of the NTase domain (Fig. 3), in contrast to T4 Rnl1 and NgrRnl, which make no enzymic contacts to the ATP β phosphate (Fig. 1B) (7). Second, LigA has acquired a defining Ia domain, the only known function of which is to bind and orient the NMN leaving group apical to the motif I lysine nucleophile. Thus, replacement of an PPi•Me2+ leaving group in the (putatively ancestral) ATP-dependent ligase lineage (7) with an NMN leaving group in NAD+ was presumably coupled during ligase evolution in bacteria to the abandonment of a second metal and the gain of a novel NMN-binding module.
NAD+-dependent DNA ligases are promising targets for antibacterial drug discovery efforts, most of which have identified LigA inhibitors that occupy the adenosine site of the NTase domain and the nearby hydrophobic “C2 drug tunnel” unique to the LigA family (24, 28, 40–45). The structure of the LigA Michaelis complex, together with previous LigA structures (4, 24), makes a strong case for targeting small molecule inhibitors to the NMN site (composed of domain Ia and residues in the NTase domain). Such compounds might be especially effective when chemically linked to molecules that occupy the adenosine site. In addition, there would be value in identifying molecules that interfere with the various LigA domain movements that are coupled to progression through the LigA adenylylation and DNA binding steps of the ligation pathway.
Methods
Crystallization of T4 Rnl1.
Mutant Rnl1-(K99M) was produced in E. coli BL21(DE3) as His10•Rnl1 fusion and isolated from a soluble bacterial extract by Ni-affinity chromatography as described (31). Rnl1-(K99M) was purified further by gel filtration through a column of Superdex-200 equilibrated in 20 mM Tris⋅HCl (pH 8.0), 100 mM NaCl. The peak fractions were pooled, concentrated by centrifugal ultrafiltration, and stored at −80 °C. A solution of 6.5 mg/mL Rnl1-(K99M), 1 mM ATP, and 2 mM MgCl2 was preincubated for 30 min on ice before mixture with an equal volume of reservoir solution containing 0.2 M MgCl2, 16% (vol/vol) PEG 3350. Crystals were grown at 22 °C by hanging drop vapor diffusion. Crystals appeared after 1–2 d. Single crystals were harvested, cryoprotected with 0.2 M MgCl2, 20% PEG 3350, 15% PEG 400, and then flash-frozen in liquid nitrogen.
Crystallization of EcoLigA.
Mutant LigA-(K115M) was produced in E. coli BL21(DE3) as His10•EcoLigA fusion and isolated from a soluble bacterial extract by Ni-affinity chromatography as described (22). LigA-(K115M) was purified further by Superdex-200 gel filtration. The peak fractions were pooled, concentrated by centrifugal ultrafiltration, and stored at −80 °C. A solution of 20 mg/mL LigA-(K115M), 0.3 mM NAD+, and 5 mM MgCl2 was preincubated for 30 min on ice before mixture with an equal volume of reservoir solution containing 0.2 M Na acetate, 20% PEG 3350. Crystals were grown at 22 °C by sitting drop vapor diffusion. Crystals appeared after 10 d. Single crystals were harvested, cryoprotected with 0.2 M Na acetate, 25% PEG 3350, 15% PEG 400, and then flash-frozen in liquid nitrogen.
Diffraction Data Collection and Structure Determination.
Diffraction data were collected continuously from single crystals at Argonne National Laboratory Advanced Photon Source beamlines 24ID-E for Rnl1-(K99M) and 24ID-C for LigA-K115M) at 100 K and processed with HKL-2000 (46). The structures were solved by molecular replacement. The search probe for Rnl1-(K99M) was 2C5U, and phasing was performed with Phenix (47). For LigA-(K115M), the search was done successively with separate domains of E. coli LigA (2OWO). NTase domain (residues 72–315), HhH domain (residues 429–586), and OB together with Zn-finger (residues 316–428) were found by using MOLREP (48). The N-terminal Ia domain was built into difference electron density maps. Interactive model building was done by using O (49). Both structures were refined with Phenix (47). Data collection and refinement statistics are presented in Table S1.
Acknowledgments
This research was supported by NIH Grant GM63611 (to S.S.). The Memorial Sloan–Kettering Cancer Center structural biology core laboratory is supported by National Cancer Institute Grant P30-CA008748. X-ray diffraction data were collected at synchrotron facilities supported funded by National Institute of General Medical Sciences Grants P41GM103403, P41GM103473, and P41GM111244; NIH Office of Research Infrastructure Programs Grant HEI S10RR029205; and Department of Energy Grants DE-AC02-06CH11357, DE-AC02-98CH10886, and E-SC0012704).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 5TT6 and 5TT5).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1619220114/-/DCSupplemental.
References
- 1.Shuman S, Lima CD. The polynucleotide ligase and RNA capping enzyme superfamily of covalent nucleotidyltransferases. Curr Opin Struct Biol. 2004;14(6):757–764. doi: 10.1016/j.sbi.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 2.Ho CK, Wang LK, Lima CD, Shuman S. Structure and mechanism of RNA ligase. Structure. 2004;12(2):327–339. doi: 10.1016/j.str.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 3.Lehman IR. DNA ligase: Structure, mechanism, and function. Science. 1974;186(4166):790–797. doi: 10.1126/science.186.4166.790. [DOI] [PubMed] [Google Scholar]
- 4.Gajiwala KS, Pinko C. Structural rearrangement accompanying NAD+ synthesis within a bacterial DNA ligase crystal. Structure. 2004;12(8):1449–1459. doi: 10.1016/j.str.2004.05.017. [DOI] [PubMed] [Google Scholar]
- 5.Brooks MA, et al. The structure of an archaeal homodimeric ligase which has RNA circularization activity. Protein Sci. 2008;17(8):1336–1345. doi: 10.1110/ps.035493.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith P, Wang LK, Nair PA, Shuman S. The adenylyltransferase domain of bacterial Pnkp defines a unique RNA ligase family. Proc Natl Acad Sci USA. 2012;109(7):2296–2301. doi: 10.1073/pnas.1116827109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Unciuleac MC, Goldgur Y, Shuman S. Structure and two-metal mechanism of a eukaryal nick-sealing RNA ligase. Proc Natl Acad Sci USA. 2015;112(45):13868–13873. doi: 10.1073/pnas.1516536112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Unciuleac MC, Shuman S. Characterization of a novel eukaryal nick-sealing RNA ligase from Naegleria gruberi. RNA. 2015;21(5):824–832. doi: 10.1261/rna.049197.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silber R, Malathi VG, Hurwitz J. Purification and properties of bacteriophage T4-induced RNA ligase. Proc Natl Acad Sci USA. 1972;69(10):3009–3013. doi: 10.1073/pnas.69.10.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cranston JW, Silber R, Malathi VG, Hurwitz J. Studies on ribonucleic acid ligase. Characterization of an adenosine triphosphate-inorganic pyrophosphate exchange reaction and demonstration of an enzyme-adenylate complex with T4 bacteriophage-induced enzyme. J Biol Chem. 1974;249(23):7447–7456. [PubMed] [Google Scholar]
- 11.Amitsur M, Levitz R, Kaufmann G. Bacteriophage T4 anticodon nuclease, polynucleotide kinase and RNA ligase reprocess the host lysine tRNA. EMBO J. 1987;6(8):2499–2503. doi: 10.1002/j.1460-2075.1987.tb02532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sriskanda V, Shuman S. Conserved residues in domain Ia are required for the reaction of Escherichia coli DNA ligase with NAD+ J Biol Chem. 2002;277(12):9695–9700. doi: 10.1074/jbc.M111164200. [DOI] [PubMed] [Google Scholar]
- 13.Sriskanda V, Moyer RW, Shuman S. NAD+-dependent DNA ligase encoded by a eukaryotic virus. J Biol Chem. 2001;276(39):36100–36109. doi: 10.1074/jbc.M105643200. [DOI] [PubMed] [Google Scholar]
- 14.Olivera BM, Lehman IR. Linkage of polynucleotides through phosphodiester bonds by an enzyme from Escherichia coli. Proc Natl Acad Sci USA. 1967;57(5):1426–1433. doi: 10.1073/pnas.57.5.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olivera BM, Lehman IR. Diphosphopyridine nucleotide: A cofactor for the polynucleotide-joining enzyme from Escherichia coli. Proc Natl Acad Sci USA. 1967;57(6):1700–1704. doi: 10.1073/pnas.57.6.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olivera BM, Hall ZW, Lehman IR. Enzymatic joining of polynucleotides, V. A DNA-adenylate intermediate in the polynucleotide-joining reaction. Proc Natl Acad Sci USA. 1968;61(1):237–244. doi: 10.1073/pnas.61.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gellert M. Formation of covalent circles of lambda DNA by E. coli extracts. Proc Natl Acad Sci USA. 1967;57(1):148–155. doi: 10.1073/pnas.57.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zimmerman SB, Little JW, Oshinsky CK, Gellert M. Enzymatic joining of DNA strands: A novel reaction of diphosphopyridine nucleotide. Proc Natl Acad Sci USA. 1967;57(6):1841–1848. doi: 10.1073/pnas.57.6.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Little JW, Zimmerman SB, Oshinsky CK, Gellert M. Enzymatic joining of DNA strands, II. An enzyme-adenylate intermediate in the dpn-dependent DNA ligase reaction. Proc Natl Acad Sci USA. 1967;58(5):2004–2011. doi: 10.1073/pnas.58.5.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gumport RI, Lehman IR. Structure of the DNA ligase-adenylate intermediate: Lysine (ε-amino)-linked adenosine monophosphoramidate. Proc Natl Acad Sci USA. 1971;68(10):2559–2563. doi: 10.1073/pnas.68.10.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Modrich P, Lehman IR. Deoxyribonucleic acid ligase. A steady state kinetic analysis of the reaction catalyzed by the enzyme from Escherichia coli. J Biol Chem. 1973;248(21):7502–7511. [PubMed] [Google Scholar]
- 22.Sriskanda V, Schwer B, Ho CK, Shuman S. Mutational analysis of Escherichia coli DNA ligase identifies amino acids required for nick-ligation in vitro and for in vivo complementation of the growth of yeast cells deleted for CDC9 and LIG4. Nucleic Acids Res. 1999;27(20):3953–3963. doi: 10.1093/nar/27.20.3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu H, Shuman S. Structure-guided mutational analysis of the nucleotidyltransferase domain of Escherichia coli NAD+-dependent DNA ligase (LigA) J Biol Chem. 2005;280(13):12137–12144. doi: 10.1074/jbc.M413685200. [DOI] [PubMed] [Google Scholar]
- 24.Nandakumar J, Nair PA, Shuman S. Last stop on the road to repair: Structure of E. coli DNA ligase bound to nicked DNA-adenylate. Mol Cell. 2007;26(2):257–271. doi: 10.1016/j.molcel.2007.02.026. [DOI] [PubMed] [Google Scholar]
- 25.Wang LK, Nair PA, Shuman S. Structure-guided mutational analysis of the OB, HhH, and BRCT domains of Escherichia coli DNA ligase. J Biol Chem. 2008;283(34):23343–23352. doi: 10.1074/jbc.M802945200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang LK, Zhu H, Shuman S. Structure-guided mutational analysis of the nucleotidyltransferase domain of Escherichia coli DNA Ligase (LigA) J Biol Chem. 2009;284(13):8486–8494. doi: 10.1074/jbc.M808476200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chauleau M, Shuman S. Kinetic mechanism and fidelity of nick sealing by Escherichia coli NAD+-dependent DNA ligase (LigA) Nucleic Acids Res. 2016;44(5):2298–2309. doi: 10.1093/nar/gkw049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shuman S. DNA ligases: Progress and prospects. J Biol Chem. 2009;284(26):17365–17369. doi: 10.1074/jbc.R900017200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.El Omari K, et al. Molecular architecture and ligand recognition determinants for T4 RNA ligase. J Biol Chem. 2006;281(3):1573–1579. doi: 10.1074/jbc.M509658200. [DOI] [PubMed] [Google Scholar]
- 30.Wang LK, Nandakumar J, Schwer B, Shuman S. The C-terminal domain of T4 RNA ligase 1 confers specificity for tRNA repair. RNA. 2007;13(8):1235–1244. doi: 10.1261/rna.591807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang LK, Ho CK, Pei Y, Shuman S. Mutational analysis of bacteriophage T4 RNA ligase 1. Different functional groups are required for the nucleotidyl transfer and phosphodiester bond formation steps of the ligation reaction. J Biol Chem. 2003;278(32):29454–29462. doi: 10.1074/jbc.M304320200. [DOI] [PubMed] [Google Scholar]
- 32.Wang LK, Schwer B, Shuman S. Structure-guided mutational analysis of T4 RNA ligase 1. RNA. 2006;12(12):2126–2134. doi: 10.1261/rna.271706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Timson DJ, Wigley DB. Functional domains of an NAD+-dependent DNA ligase. J Mol Biol. 1999;285(1):73–83. doi: 10.1006/jmbi.1998.2302. [DOI] [PubMed] [Google Scholar]
- 34.Kaczmarek FS, et al. Cloning and functional characterization of an NAD(+)-dependent DNA ligase from Staphylococcus aureus. J Bacteriol. 2001;183(10):3016–3024. doi: 10.1128/JB.183.10.3016-3024.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee JY, et al. Crystal structure of NAD(+)-dependent DNA ligase: Modular architecture and functional implications. EMBO J. 2000;19(5):1119–1129. doi: 10.1093/emboj/19.5.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gu H, et al. Structural and mutational analysis of archaeal ATP-dependent RNA ligase identifies amino acids required for RNA binding and catalysis. Nucleic Acids Res. 2016;44(5):2337–2347. doi: 10.1093/nar/gkw094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang P, et al. Molecular basis of bacterial protein Hen1 activating the ligase activity of bacterial protein Pnkp for RNA repair. Proc Natl Acad Sci USA. 2012;109(33):13248–13253. doi: 10.1073/pnas.1209805109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sriskanda V, Shuman S. Role of nucleotidyl transferase motif V in strand joining by Chlorella virus DNA ligase. J Biol Chem. 2002;277(12):9661–9667. doi: 10.1074/jbc.M110613200. [DOI] [PubMed] [Google Scholar]
- 39.Samai P, Shuman S. Kinetic analysis of DNA strand joining by Chlorella virus DNA ligase and the role of nucleotidyltransferase motif VI in ligase adenylylation. J Biol Chem. 2012;287(34):28609–28618. doi: 10.1074/jbc.M112.380428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mills SD, et al. Novel bacterial NAD+-dependent DNA ligase inhibitors with broad-spectrum activity and antibacterial efficacy in vivo. Antimicrob Agents Chemother. 2011;55(3):1088–1096. doi: 10.1128/AAC.01181-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stokes SS, et al. Discovery of bacterial NAD+-dependent DNA ligase inhibitors: Optimization of antibacterial activity. Bioorg Med Chem Lett. 2011;21(15):4556–4560. doi: 10.1016/j.bmcl.2011.05.128. [DOI] [PubMed] [Google Scholar]
- 42.Gu W, et al. Design, synthesis and biological evaluation of potent NAD+-dependent DNA ligase inhibitors as potential antibacterial agents. Part I: Aminoalkoxypyrimidine carboxamides. Bioorg Med Chem Lett. 2012;22(11):3693–3698. doi: 10.1016/j.bmcl.2012.04.037. [DOI] [PubMed] [Google Scholar]
- 43.Wang T, et al. Design, synthesis and biological evaluation of potent NAD+-dependent DNA ligase inhibitors as potential antibacterial agents. Part 2: 4-amino-pyrido[2,3-d]pyrimidin-5(8H)-ones. Bioorg Med Chem Lett. 2012;22(11):3699–3703. doi: 10.1016/j.bmcl.2012.04.038. [DOI] [PubMed] [Google Scholar]
- 44.Surivet JP, et al. Structure-guided design, synthesis and biological evaluation of novel DNA ligase inhibitors with in vitro and in vivo anti-staphylococcal activity. Bioorg Med Chem Lett. 2012;22(21):6705–6711. doi: 10.1016/j.bmcl.2012.08.094. [DOI] [PubMed] [Google Scholar]
- 45.Murphy-Benenato KE, et al. Negishi cross-coupling enabled synthesis of novel NAD(+)-dependent DNA ligase inhibitors and SAR development. Bioorg Med Chem Lett. 2015;25(22):5172–5177. doi: 10.1016/j.bmcl.2015.09.075. [DOI] [PubMed] [Google Scholar]
- 46.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 47.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vagin A, Teplyakov A. MOLREP: An automated program for molecular replacement. J Appl Cryst. 1997;30:1022–1025. [Google Scholar]
- 49.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]